Abstract
EVER1 and 2 confer resistance to cutaneous oncogenic human papillomavirus infections by downregulating the activating protein 1 (AP-1) signaling pathway. Defects in their expression are associated with susceptibility to epidermodysplasia verruciformis, which is characterized by persistent β-HPV infection, tumor necrosis factor alpha (TNF-α) overproduction in keratinocytes and the development of skin cancers. TNF-α-induced apoptosis is a key defense strategy, preventing the persistence of the virus within cells, but the role of EVER proteins in this cell death mechanism triggered by extrinsic stimuli is unknown. We show here that EVER2 induces TNF-α- and TRAIL-dependant apoptosis. It interacts with the N-terminal domain of TRADD, impairs the recruitment of TRAF2 and RIPK1 and promotes apoptosis. The skin cancer-associated EVER2 I306 allele results in an impaired TRADD–EVER2 interaction, with lower levels of cell death following treatment with TNF-α. These data highlight a new, critical function of EVER2 in controlling cell survival in response to death stimuli.
Keywords: EVER2, TRADD, apoptosis, polymorphism
Epidermodysplasia verruciformis (EV) is rare autosomal recessive dermatosis (OMIM 226400) leading to an abnormal susceptibility to β-human papillomavirus (HPV) infections. EV patients have disseminated, persistent, flat wart-like or macular skin lesions in early childhood and frequently develop squamous cell carcinoma induced by HPV5 and 8 early in adulthood.1 They display defective cell-mediated immunity, resulting in a predisposition to persistent lesions and high viral loads of infecting β-HPVs.1, 2 Homozygous mutations in EVER1 (TMC6) or EVER2 (TMC8) gene are associated with EV.3 The single nucleotide polymorphism (SNP) rs7208422 in exon 8 of the EVER2 gene (c.917 A>T), leading to an amino-acid substitution (p.N306I), may have a role in EV.4, 5 It has also been associated with seropositivity for β-HPV5 and -8, and squamous cell carcinoma.6
EVER2 regulates zinc balance and has also been reported to downregulate the activity of activating protein 1 (AP-1), a transcription factor having a key role in the life cycle of HPV.7 EVER2 is expressed by T and B lymphocytes, natural killer cells, endothelial cells, bone marrow myeloid cells and dendritic cells,8 consistent with a role in immunity to HPV.2 Patients with EV overproduce tumor necrosis factor alpha (TNF-α) in lesions and have a defect in AP-1 signaling in keratinocytes.7, 9 Both these mechanisms are controlled by TNF receptor 1 (TNFR-1).
TNFR-1 is a death receptor from the TNF receptor family. It is activated by its ligand, TNF-α, a cytokine involved principally in host defense to bacterial, parasitic and viral infections.10 Physiologically, TNF-α is important for the normal response to infection, due to particularly its role in triggering cell death of infected cells. Inappropriate responses to TNF-α can be harmful and may contribute to carcinogenesis by promoting tumor cell proliferation, invasion and metastasis.11 The TNF-α signal transduction pathway is complex, being able to trigger both pro-survival and pro-apoptotic pathways, and it remains incompletely understood. However, it has become clear that TNFR-associated death domain protein (TRADD) has a key role in pro-survival complex I formation.12 TRADD recruits TNF receptor-associated factor 2 (TRAF2) protein, which in turn interacts with the E3 ligases' cellular inhibitor of apoptosis protein (cIAP)-1 and cIAP-2, leading to receptor-interacting serine/threonine-protein kinase 1 (RIPK1) ubiquitination and nuclear factor kappa B (NF-κB) activation platform formation. However, any RIPK1 that is not ubiquitinated is released from complex I, and associates with Fas-associated protein with death domain (FADD) and caspase-8 to form the pro-apoptotic complex II.13 Therefore, an ongoing question is to determine if the pro-survival or the pro-apoptotic pathways will predominate in response to TNF-α.
We investigated the role of EVER2 in death receptor-mediated apoptosis in EVER2-expressing human embryonic kidney 293 T (HEK-293T) cells and in EVER2-silenced Jurkat cells, focusing on the TNF-α-induced survival/death decision. We also analyzed the effect of EVER2 p.N306I polymorphism on these mechanisms.
We found that EVER2 triggered TNF-α- and TRAIL-induced apoptosis, but had no effect on FasL stimulation. It interacted with TRADD, impairing the recruitment of TRAF2 and RIPK1 to complex I, and promoting pro-apoptotic complex II formation. The protein encoded by the EVER2 306I allele interacted less strongly with TRADD, with consequences for TNF-α-induced apoptosis and NF-κB activation.
Results and discussion
EVER2 sensitizes cells to both TNF-α- and TRAIL-induced cell death
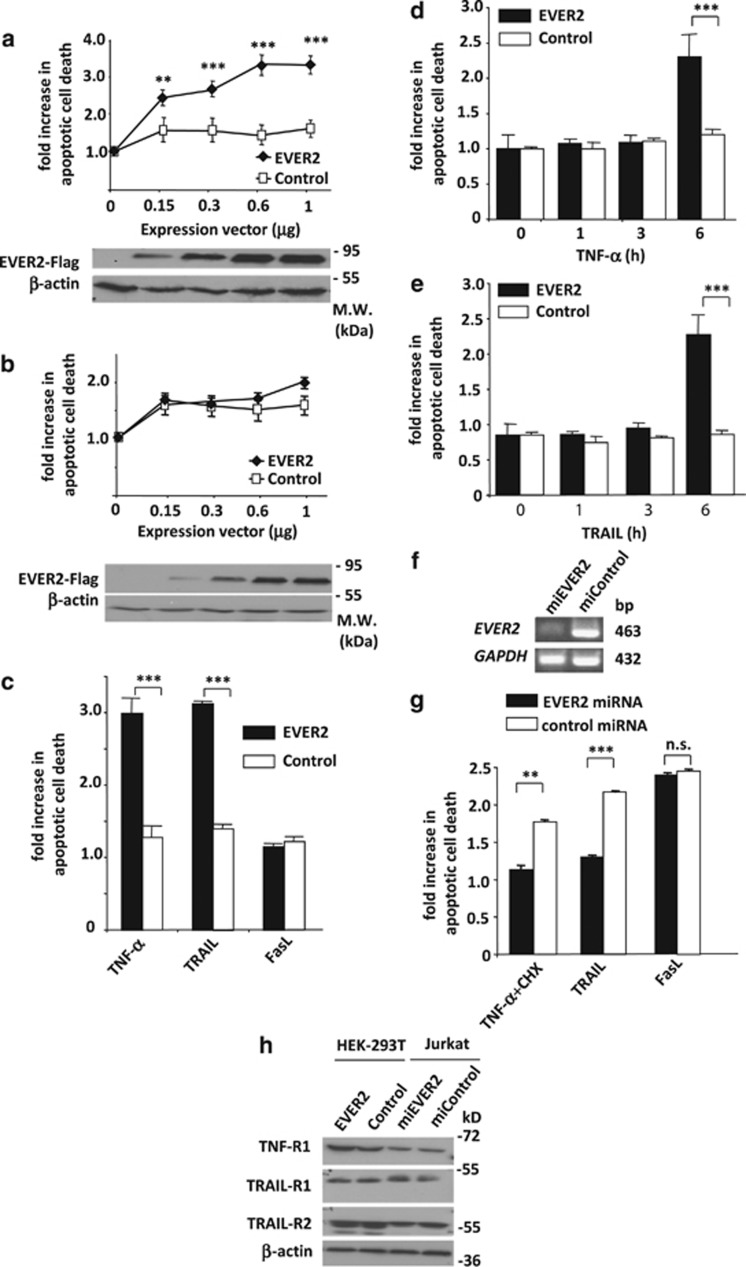
While studying EVER2 properties in cells, we observed a high mortality rate in EVER2-overexpressing HEK-293T cells. We first quantified the effect of EVER2 expression in HEK-293T cells grown in complete medium supplemented with 10% fetal calf serum (FCS), and confirmed that this protein had a dose-dependent effect on cell death rates (Figure 1a). Then, cells were starved from FCS in order to stop the signaling pathways potentially induced by extrinsic cytokines and growth factors brought by the serum. In these conditions, we assessed the role of extrinsic stimuli on EVER2 pro-apoptotic effects. Tsutsumi et al.14 used a similar procedure for assessing the pro-apoptotic effect of TGF-β in serum. In conditions of FCS deprivation (0.2% FCS), we almost totally abrogated EVER2 effects on cell death even at high concentrations, suggesting that EVER2-induced cell death was mainly triggered by extrinsic factors activating cell surface receptors (Figure 1b). In addition, western blot analysis was performed to monitor EVER2 expression in cells grown in complete or serum-deprived medium. No significant difference in EVER2 expression was observed between these two cell culture conditions when compared to β-actin (Figures 1a and b).
Figure 1.
EVER2 sensitizes cells to both TNF-α- and TRAIL-induced cell death. (a) EVER2 sensitizes cells to cell death in a dose-dependent manner. Cells were transiently transfected with various amounts of a construct encoding EVER2 or with an empty vector as a control, and allowed to grow in complete medium supplemented with 10% FCS. Cells were lysed and cell death evaluated. The fold-increase is the OD ratio of empty vector- or EVER2-transfected cells rapported to untransfected cells. EVER2–Flag expression in transfected HEK-293T was monitored using western blot analysis. (b) EVER2-expressing cells cultured in the presence of 0.2% FCS were no more sensitive to cell death than control cells. (c) Cells expressing EVER2 were sensitized to TNF-α- and TRAIL- but not to FasL-induced cell death. Cells were transfected with the EVER2 construct or with empty vector. Cells were stimulated with TNF-α (10 ng/ml), TRAIL (10 ng/ml) and FasL (100 ng/ml). Cell death was evaluated after stimulation with TNF-α (d), or TRAIL (e). In Figures 1c, d and e, cells were plated in 24-well plates and transfected with 0.6 μg per well EVER2 plasmid. The fold-increase in apoptotic cell death represents the OD ratio of treated cells rapported to unstimulated cells. (f) Monitoring of EVER2 silencing in Jurkat cells by semi-quantitative PCR. (g) EVER2 silencing protects Jurkat cells from cell death induced by TNF-α+CHX and TRAIL, but not by FasL. Stable Jurkat cell lines producing control or EVER2 miRNA were generated and treated with TNF-α (25 ng/ml)+CHX (0.2 μg/ml), TRAIL (50 ng/ml) or FasL (50 ng/ml), and cell death was assessed. (h) Monitoring of TNFR-1, TRAIL-R1 and TRAIL-R2 expression by western blotting in HEK-293T cells transfected with empty or EVER2-expressing vector, and in Jurkat cells transfected with control or EVER2 miRNA-expressing vector. Values are means±S.E. (n=3 separated experiments). Student's t-test: ***P<0.001; **P<0.005
Four of the six death receptors (DR1–6) – TNFR-1 (DR1), the Fas ligand-activated Fas (CD95/DR2) and the two TNF-related apoptosis-inducing ligand (TRAIL)-activated DR4 and DR515, 16 – have been studied extensively. We stimulated EVER2-expressing HEK-293T cells with TNF-α, TRAIL and Fas ligand (FasL) in the absence of ‘sensitizer' adjuvants (e.g. cycloheximide, CHX), to determine which stimuli induced cell death in our model. TNF-α and TRAIL robustly triggered cell death at low concentrations, whereas FasL did not (Figure 1c). By contrast, none of the three cytokines triggered cell death in control cells. Experimentations also showed that cell death occured within 6 h of TNF-α stimulation (Figures 1d and e).
In addition, we assessed the extent of cell death achieved in the various culture conditions by performing an annexin V-FITC/propidium iodide (PI) assay and evaluated the percentage of apoptotic dead cells (annexin V-positive, PI-positive) by flow cytometry. The viability of cells was determined following transfection with EVER2 plasmid construct or empty vector as control. Data from a representative experiment are shown in Supplementary Figure S1. About 13% of EVER2-transfected cells were dead by apoptosis after 48 h of incubation in complete medium, whereas only 1.2% entered cell death in control condition. Upon serum deprivation, only 4.5% of EVER2-transfected cells were still undergoing cell death, whereas 17.5% of these cells died following TNF-α stimulation. Interestingly, the addition of TRAIL induced similar results. In contrast, FasL did not induce significant cell death following EVER2 transfection. This is consistent with previous data obtained with an ELISA analysis of DNA fragmentation (Figure 1c), providing the confirmation that EVER2 sensitizes HEK-293T cells to TNF-α and TRAIL-induced apoptosis, but has no effect on FasL stimulation. Nevertheless, in 0.2% FCS, we observed a small percentage of dead cells (4.5%) and apoptotic cells (annexin V-positive, PI-negative) (7.1%) following EVER2 transfection, as compared with control (0.4 and 1%, respectively). This may be due to the remaining FCS in culture medium or may indicate that EVER2 could also slightly induce an apoptotic cell death through an intrinsic process.
For confirmation of the role of EVER2 in these mechanisms, we downregulated EVER2 expression with microRNA (miRNA) in a Jurkat cell line (Figure 1f) known to be naturally sensitive to FasL- and TRAIL-induced cell death.17 As expected, a stimulation with TRAIL, FasL or a combination of TNF-α with CHX induced cell death in Jurkat control cells. Conversely, in EVER2-silenced Jurkat cells, this effect was almost entirely abolished for TNF-α and TRAIL, but not for FasL (Figure 1g). We cannot exclude the possibility that EVER2 has a role in the regulation of death receptor expression. Thereby, we monitored the expression of the two TRAIL receptors (TRAIL-R1 and -R2) and the TNFR-1 in extracts from wild-type HEK-293T and EVER2-transfected cells, as well as cells transfected with miRNA-targeting EVER2. No variation was observed in the expression of the death receptors ruling out a possible regulation of their expression by EVER2 or the lack of receptor expression in HEK-293T. In addition, several studies demonstrated that HEK-293T expressed Fas.18, 19
We conclude that EVER2 sensitizes HEK-293T cells to TNF-α and TRAIL-induced apoptosis, but has no effect on FasL stimulation.
Endogenous EVER2 directly interacts with TRADD
TRADD plays a key role in TNFR-1 signaling20 and is essential for resistance to TRAIL-induced cell death. Hence, TRADD binding and subsequent RIPK1 recruitment to the receptor-associated complex, are determinant in the life and death balance decision of TRAIL signaling.21, 22 On the other hand, it is now well established that Fas activation triggers the formation of a complex formed of the receptor Fas, FADD, procaspases -8 and -10 and cFLIP23, 24 and is totally independent from TRADD and RIPK1 binding.18 This led us to hypothesize that EVER2 might act by interacting with TRADD.
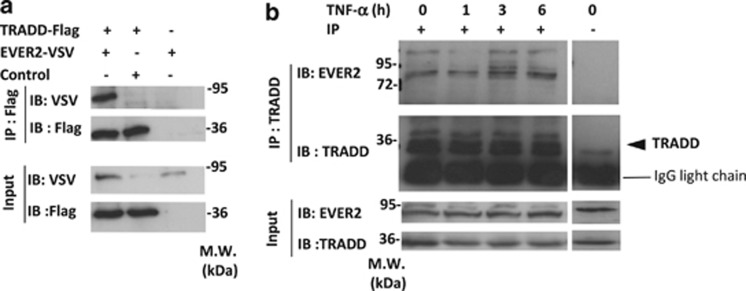
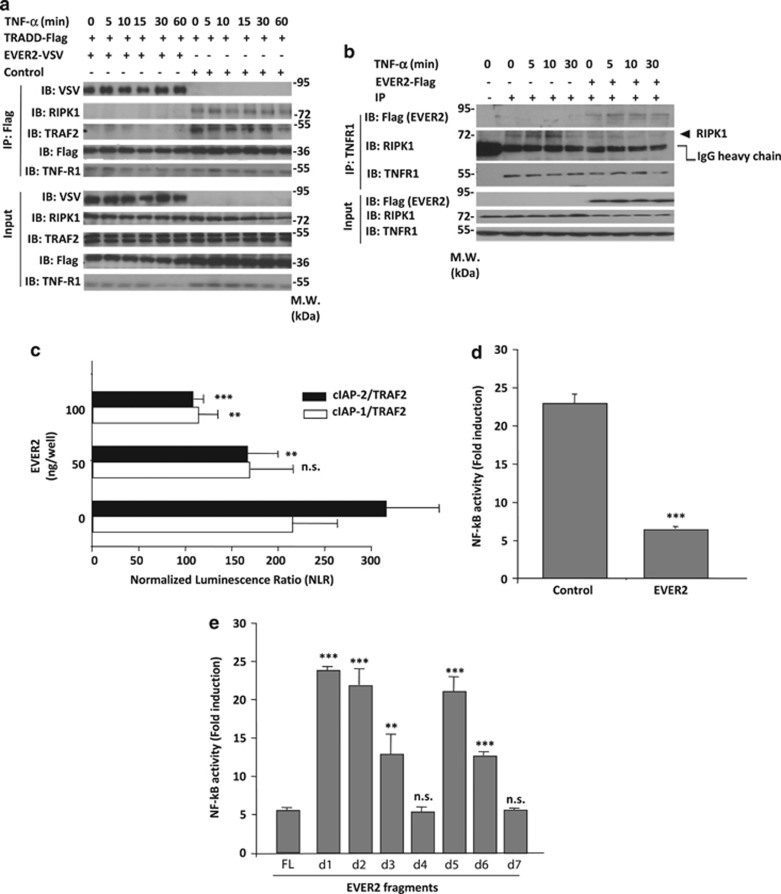
Figure 2a shows that EVER2-Flag and TRADD-VSV co-immunoprecipitate when over-expressed in HEK-293T cells. We then investigated whether endogenous EVER2 can interact with TRADD and if the interaction is constitutive or inducible. We therefore immunoprecipitated endogenous TRADD protein in FCS starved HEK-293T, untreated or treated with TNF-α for various times. Figure 2b shows that EVER2 and TRADD interact in untreated HEK-293T demonstrating that this interaction is constitutive. Furthermore, this interaction was unaffected by TNF-α treatment, ruling out the possibility of an inducible event.
Figure 2.
EVER2 directly interacts with TRADD. (a) EVER2 interacts with TRADD. HEK-293T cells were cotransfected with EVER2–VSV and TRADD–Flag constructs. Cells were lysed and subjected to co-immunoprecipitation (IP) with an anti-Flag antibody. Immunoblotting (IB) was performed with specific antibodies directed against VSV and Flag. (b) Interaction of endogenous EVER2 with endogenous TRADD is constitutive and independent of TNF-α stimulation. HEK-293T cells were cultured for 2 h in 0.2% FCS-containing medium then stimulated with TNF-α (10 ng/ml) for various time periods and subjected to IP with an anti-TRADD antibody or non-specific isotype IgG (shown as −). IB was performed with specific antibodies directed against EVER2 and TRADD
TNF-α-induced cell death is mediated by interaction of EVER2 TMC region with TRADD
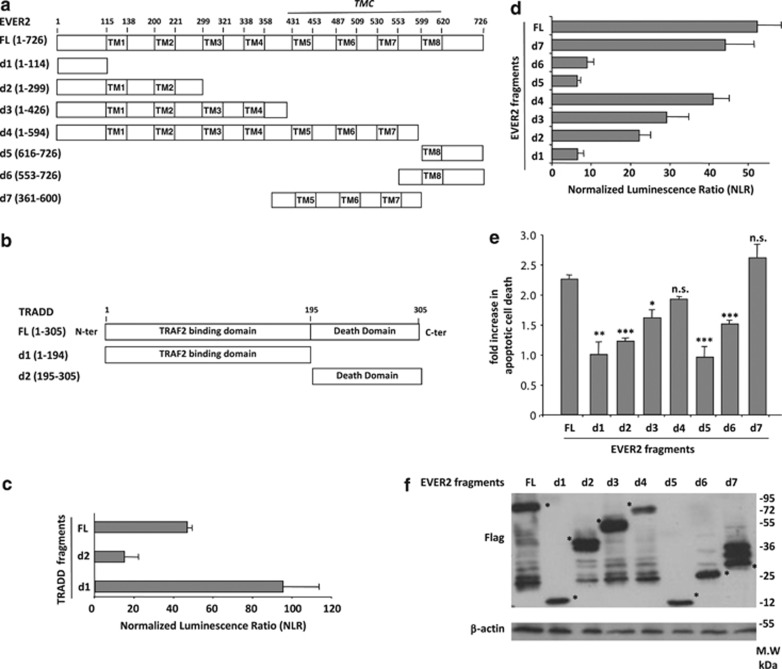
For identification of the domains of the two proteins involved in this interaction, we generated vectors encoding seven fragments of EVER2 (Figure 3a) and two fragments of TRADD (Figure 3b). We then mapped the interaction between the two proteins by the high-throughput Gaussia protein complementation assay (HT-GPCA) technique.25 EVER2 interacted principally with the N-terminal domain of TRADD, which contains the TRAF2 binding site (d1) (Figure 3c). By contrast, TRADD interacted only weakly with the N-terminal (d1) and C-terminal (d5 and d6) fragments of EVER2, moderately with the first two and four putative transmembrane domains (d2 and d3) and strongly with the d4 and d7 fragments (Figure 3d). These last two regions contain the TMC domain, which is highly conserved among transmembrane channel-like (TMC) family proteins.26 We then investigated the relationship between the interaction of EVER2 fragments with TRADD and TNF-α-induced cell death. High mortality rates were recorded for cells expressing the TMC-containing fragments (d4 and d7), whereas no cell death was detected in cells expressing the d1 and d5 fragments (Figure 3e). These results were not explained by the difference between expression of EVER2 fragments, as shown by d4 fragment that induced the strongest cell death despite lower expression (Figure 3f). These experiments highlight the importance of EVER2 binding to TRADD for its pro-apoptotic effect.
Figure 3.
TNF-α-induced cell death in EVER2-expressing cells is mediated by interaction of EVER2 TMC domain with TRADD. (a) Schematic representation of the putative EVER2 domains and fragments used. TM: transmembrane domain; TMC: transmembrane conserved domain; FL: full-length; d1–d7: EVER2 fragments. (b) Schematic representation of the TRADD fragments used. FL: full-length; d1–d2: TRADD fragments. (c) Mapping of the interaction of EVER2 with TRADD fragments by HT–GPCA. (d) Mapping of the interaction of TRADD with EVER2 fragments by HT–GPCA. (e) Impact of EVER2 fragments on TNF-α-induced apoptosis. Cells were transfected with constructs encoding the various EVER2 fragments and TNF-α-induced apoptosis was assayed as described above. (f) Detection of EVER2 fragments expression in transfected HEK-293T using western blot analysis. Asterisks indicate the band corresponding to the expected molecular weight of each EVER2 fragment. Means±S.E.'s (n=3 separate experiments) are shown. The effect of the full-length protein was taken as the reference in panels D and E. Student's t-test comparing effect of EVER2 FL with that of other EVER2 fragments: ***P<0.001; **P<0.005; *P<0.01
Thus, EVER2 exerts its sensitizing effect principally through interactions of the TMC containing region with the N-terminal, TRAF2-binding domain of TRADD.
EVER2 sensitizes cells to apoptosis by facilitating pro-apoptotic complex II formation
Apoptosis and necroptosis are two types of cell death induced by TNF-α but executed through different pathways.27 Apoptosis is mediated by the intermediate and death domains of RIPK1, leading to caspase-8, -3 and -7 activation, whereas necroptosis signaling requires the kinase activity of RIPK1 for the phosphorylation of RIPK3 and seems to involve the accumulation of reactive oxygen species and the depolarization of mitochondria membrane.27, 28
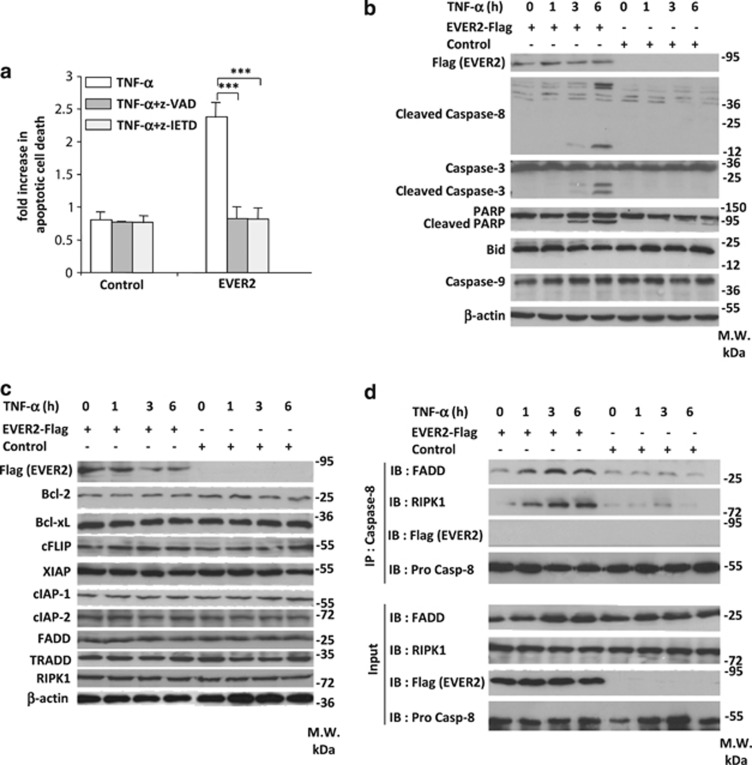
For identification of the type of TNF-α-induced cell death to which cells are sensitized by EVER2, we stimulated EVER2-expressing and control cells with TNF-α in the presence or absence of the caspase inhibitors z-VAD and z-IETD. Both inhibitors abolished completely TNF-α-induced cell death in EVER2-expressing cells (Figure 4a). These results were confirmed by western blotting, which showed the cleavage of caspase-8 and -3 and poly (ADP-ribose) polymerase (PARP) (Figure 4b). Thus, EVER2-expressing cells display all the features of TNF-α-induced apoptosis rather than necroptosis, as no cell death was observed after caspase inhibition (Figure 4a).28 However, no caspase-9 or Bid cleavage was observed on western blots (Figure 4b). These findings point to a type II apoptosis model, in which caspase-8 activation is sufficient to activate caspase-3 and no mitochondrial amplification loop involving Bid and caspase-9 is required, as previously reported.29, 30, 31
Figure 4.
TNF-α induces caspase-8-dependent apoptosis in EVER2-expressing cells, by facilitating FADD/RIPK1/caspase-8 complex II formation. (a) TNF-α-induced apoptosis in EVER2-expressing cells is impaired by the inhibition of the activity of caspases . HEK-293T cells were transfected with EVER2–Flag or empty vector. Cells were stimulated with TNF-α, together with z-VAD or z-IETD. Values are means±S.E.'s (n=3). Student's t-test: ***P<0.001. (b) The cleavage of caspase-8, caspase-3 and PARP was observed after 3 h of TNF-α stimulation in EVER2-expressing cells. Cells were transfected with the EVER2–Flag construct or empty vector. Cells were stimulated with TNF-α for various time periods. Immunoblotting was performed on whole-cell lysates with the indicated antibodies. (c) Profile of apoptosis-regulator protein levels during TNF-α stimulation in EVER2-expressing cells. Cells were treated as previously described and immunoblotting was performed on whole-cell lysates with the indicated antibodies. (d) FADD/RIPK1/caspase-8 complex II formation in EVER2-expressing cells. Cells were transfected with the EVER2–Flag construct or empty vector. Cells were stimulated with TNF-α (10 ng/ml) for various time periods, lysed and subjected to IP with a specific antibody against caspase-8. IB was performed with the indicated antibodies
We then investigated the mechanism of HEK-293T cell sensitization to apoptosis by EVER2. We first investigated the levels of key proteins involved in DR apoptosis: B-cell lymphoma 2 (Bcl-2), B-cell lymphoma-extra large (Bcl-xl), cellular FLICE-inhibitory protein (cFLIP), X-linked inhibitor of apoptosis protein (XIAP), cIAP-1, cIAP-2, FADD, TRADD and RIPK1. No changes in the levels of these proteins were observed (Figure 4c), ruling out a modulation of transcriptional activity.
We then investigated whether EVER2 could trigger cell death by enhancing pro-apoptotic complex formation. Following the co-precipitation of endogenous caspase-8, we observed the formation of the RIPK1/FADD/caspase-8 complex II after 1–6 h of TNF-α stimulation in EVER2-expressing cells, but not in control cells (Figure 4d). EVER2-Flag was not detected in this complex, suggesting an indirect mode of action.
We conclude that EVER2 sensitizes HEK-293T cells to TNF-α-induced apoptosis through formation of the pro-apoptotic complex II, consisting of the caspase-8, FADD and RIPK1 proteins.
EVER2 impairs the association of RIPK1 with TRADD and NF-κB-inducing complex I formation
The TRADD/RIPK1/TRAF2/cIAPs complex I forms rapidly in response to TNF-α stimulation. RIPK1 and TRAF2 interact with the death domain and the N-terminal domain of TRADD, respectively. TRAF2 then recruits cIAP-1/-2 for RIPK1 ubiquitination and initiates the NF-κB activation cascade.32 This last step is essential for cell survival, as the depletion or inhibition of cIAPs leads to the rapid formation of a specific death-inducing complex IIb composed of RIPK1, FADD and caspase-8, in response to TNFR-1 stimulation.33, 34
As EVER2 interacts directly with TRADD, we investigated TRADD/TRAF2/cIAPs complex I formation in the presence of EVER2. Co-immunoprecipitation experiments showed EVER2 to be present in complex I (Figures 5a and b). TRAF2 and RIPK1 were not found in complex I at any stage of TNF-α stimulation in EVER2-expressing cells, whereas they bound TRADD in control cells (Figure 5a). We performed endogenous TNFR-1 co-immunoprecipitation experiments following transfection with empty or EVER2 expression vector (Figure 5b). RIPK1 was found to be less abundant in complex I from EVER2-transfected cells than in the corresponding control cells at any stage of TNF-α stimulation. As EVER2 interacts strongly with the N-terminal/TRAF2-binding domain of TRADD and seems to disrupt TRAF2 interaction with TRADD, we assessed whether EVER2 could influence TRAF2/cIAPs interplay. Indeed, EVER2 disrupts the TRAF2/cIAPs interaction in a dose-dependent manner (Figure 5c). Importantly, this interaction is known to be indispensable for RIPK1 ubiquitination, an essential biochemical event for maintenance of RIPK1 in complex I.35 These observations are in accordance with our data showing a decreased recruitment of RIPK1 in complex I in the EVER2-expressing cells, whereas no RIPK1 degradation was observed, as shown by western blots of whole-cell lysates (Figures 4c and 5a).
Figure 5.
EVER2 interaction with TRADD impairs TRADD/RIPK1/TRAF2/cIAPs complex I formation and TNF-α-induced NF-κB activation. (a) TRADD/RIPK1/TRAF2 complex I formation is impaired in EVER2-expressing cells. Cells were cotransfected with EVER2–VSV or empty vector and TRADD–Flag constructs. Cells were stimulated with TNF-α (10 ng/ml) for various time periods, lysed and subjected to IP with anti-Flag-specific antibody. IB was performed with indicated antibodies. (b) EVER2 overexpression impairs RIPK1 recruitment to TNFR-1 complex I. Cells were transfected with empty or EVER2 expression vector and stimulated with TNF-α (10 ng/ml) for various time periods. Cells were lysed and subjected to IP with anti-TNFR-1-specific antibody or non specific isotype IgG (shown as −). IB was performed with indicated antibodies. (c) TRAF2/cIAP-1 and TRAF2/cIAP-2 interaction in the presence or absence of EVER2, as assessed by HT–GPCA. Cells were transfected with TRAF2–Gluc1 and cIAP-1–Gluc2 or cIAP-2–Gluc2 constructs, in the absence or presence of various amounts of EVER2–Flag. Student's t-test comparing TRAF2–cIAPs interaction in the presence (50–100 ng) and absence of EVER2: ***P<0.001; **P<0.005. (d) Measurement of TNF-α-induced NF-κB activation in the NF-κB plasmid reporter assay. Cells were cotransfected with NF-κB plasmid reporter and EVER2–Flag or empty vector as a control. Cells were stimulated with TNF-α (10 ng/ml) for 16 h. (e) Impact of EVER2 fragments on TNF-α-induced NF-κB activation. Cells were transfected with the various EVER2 fragments and TNF-α-induced NF-κB activation was assayed as described above. Means±S.E.'s (n=3 separate experiments) are shown. Student's t-test comparing the effect of EVER2–FL with that of other EVER2 fragments: ***P<0.001; **P<0.005; *P<0.05
As EVER2 impaired complex I formation, we assessed the impact of EVER2 expression on TNF-α-induced NF-κB activation. EVER2 expression resulted in much lower levels of NF-κB activity upon TNF-α stimulation than that observed in control conditions (Figure 5d). These data were substantiated by differential analysis of TNF-α-induced NF-κB activation following transfection with various EVER2 fragments (Figure 5e). We showed that the impact of EVER2 fragments on TNF-α-induced NF-κB activation depended directly on their ability to trigger an apoptotic cell death (Figure 3e) and to interact with TRADD (Figure 5e).
Thus, EVER2 interacts with TRADD, impairing the recruitment of TRAF2 and RIPK1 to complex I, and destabilizing TRAF2/cIAP-1/cIAP-2 oligomerization.
These results suggest that EVER2 competes with TRAF2 for TRADD binding, disrupting TRADD/TRAF2/cIAPs interaction and, therefore, RIPK1 ubiquitination, resulting in the release of RIPK1 into the cytosol as shown by Darding and Meier.13 The release of RIPK1 from complex I in EVER2-expressing cells has two major effects: a large decrease in TNF-α-induced NF-κB activation and the formation of a specific pro-apoptotic complex IIb, consisting of RIPK1, FADD and caspase-8.
EVER2 alleles have different effects on TNF-α-induced apoptosis and NF-κB activation
Host gene polymorphism is an important risk factor for HPV infection and cancer progression,33, 36 and several EVER2 polymorphisms have recently been implicated in HPV persistence and/or cancer progression.6, 33 One of these polymorphisms, rs7208422, which leads to the replacement of an asparagine with an isoleucine residue, (p.N306I), has been associated with β-HPV persistence, squamous cell carcinoma and EV.4, 5, 6 We therefore investigated the effect of the EVER2 p.I306N polymorphism on TNF-α-induced apoptosis and NF-κB activation.
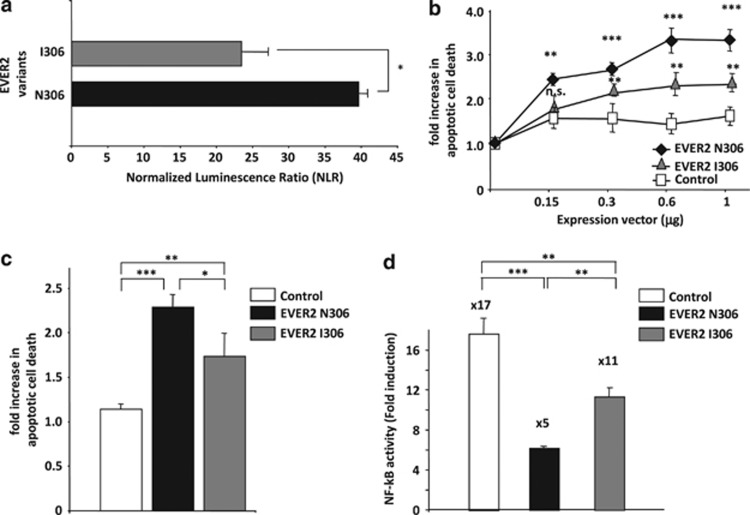
We first used HT-GPCA to compare the ability of the EVER2 N306 and I306 proteins to bind TRADD. The EVER2 I306 protein interacted less strongly with TRADD than did EVER2 N306 (Figure 6a). We then assessed the effect of the two alleles on apoptosis in HEK-293T cells cultured in complete medium supplemented with 10% FCS. Both the proteins encoded by these alleles sensitized cells to apoptosis in a dose-dependent manner, but EVER2 I306 was significantly less effective than EVER2 N306 (Figure 6b). Similarly, EVER2 N306 triggered TNF-α-induced apoptosis more strongly (Figure 6c). We then assessed the effect of the two alleles on TNF-α-induced NF-κB activation. The EVER2 I306 protein decreased NF-κB activation significantly less effectively than EVER2 N306 (Figure 6d). Thus, the p.N306I polymorphism was sufficient to modulate the effect of EVER2 on TNF-α-induced apoptosis and NF-κB activation significantly, probably through slight changes to the conformation of EVER2.
Figure 6.
EVER2 alleles have different effects on TNF-α-induced NF-κB activation and apoptosis. (a) Assessment of the interaction between EVER2 N306 or EVER2 I306 with TRADD by HT–GPCA. (b) Impact of EVER2 alleles on apoptosis. Cells were transiently transfected with various amounts of EVER2 N306, EVER2 I306 or empty vector and allowed to grow in complete medium supplemented with 10% FCS. Cells were lysed and cell death was assessed as previously described. Student's t-test comparing the effect of EVER2 alleles with the control: ***P<0.001; **P<0.005. (c) Impact of EVER2 alleles on TNF-α-induced apoptosis. Cells were transfected with EVER2 N306, EVER2 I306 or empty vector and stimulated with TNF-α (10 ng/ml). Cells were lysed and cell death was assessed as previously described. (d) Impact of EVER2 alleles on TNF-α-induced NF-κB activation. Cells were cotransfected with NF-κB reporter plasmid and EVER2 N306, EVER2 I306 or empty vector and stimulated with TNF-α (10 ng/ml) for 16 h. Values are means±S.E.'s (n=3 separate experiments). Student's t-test: *P<0.05; **P<0.01;***P<0.001
This study provides new insight into the mode of action of EVER2. This protein sensitizes cells to TNF-α-induced apoptosis by interacting with TRADD through the TMC-containing region. This impairs TRAF2 and RIPK1 binding and pro-survival complex I formation. Once released, RIPK1 associates with caspase-8 and FADD to form the pro-apoptotic complex II, triggering the caspase activation cascade and apoptosis. EVER2 may therefore have a determinant role in TNF-α-induced cell death/survival decisions.
TNF-α is a major immune system cytokine. It has a key role in defense against viral infection in general, and HPV infection in particular.37 By triggering cell death, it is involved in the clearance of virus-infected cells by the innate and adaptive immune systems. EVER2 is a strong sensitizer to TNF-α-induced apoptosis. Its inhibition, or the presence of a less effective allele, may therefore be responsible for the persistence of lesions and their progression to tumors, as observed in EV patients.
Materials and Methods
Cell lines, culture conditions and stable cell line production
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FCS and penicillin/streptomycin. The Jurkat cell line, clone E6-1, was obtained from ATCC (Rockville, MD, USA) and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FCS and penicillin/streptomycin.
For EVER2 silencing in Jurkat cells, a miRNA expression vector specific for EVER2 (miEVER2) was generated with the BLOCK-iT Pol II miR RNAi Expression Vector Kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. Two single-stranded DNA oligonucleotides encoding the pre-miRNA were designed with the Invitrogen RNAi Designer, according to the manufacturer's instructions; they target the following sequence of the EVER2 gene (encoding part of the N-terminal region): 5′-CAGGAATTCGGTCCTACTTCA-3′. Cells were transfected with miEVER2 or the control (miCTRL) expression vector containing an EmGFP-coding sequence, by electroporation. Briefly, 10–20 × 106 Jurkat cells were washed and resuspended in 0.4 ml of culture medium. The miRNA construct (5–20 μg) was added to the mixture, which was then incubated for 15 min at room temperature, transferred in a 0.4-cm path-length electroporation cuvette and electroporated at 250 V, 960 μF (time constant 25) in a GenePulser Xcell device (Bio-Rad, Marnes-la-Coquette, France). Cells were allowed to rest for 15 min at room temperature and were then transferred to 15 ml of prewarmed culture medium. Cells producing GFP were then selected by incubation for 2 weeks in medium supplemented with10 μg/ml blasticidin.
Antibodies and reagents
Antibodies against TRADD, TRAF2, RIPK1, FADD, Bcl-2, Bcl-xL, XIAP, cIAP1, cIAP2, caspase-3, cleaved caspase-8 (Asp391), caspase-8 (1C12), caspase-9 and PARP were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against cFLIP were obtained from Upstate Biotechnology (Lake Placid, NY, USA). Anti-β-actin and anti-Flag M2 antibodies were obtained from Sigma-Aldrich (St Louis, MO, USA). Antibody against TRADD (A-5) for endogenous co-immunoprecipitation was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibody against EVER2 was purchased from Interchim (Montluçon, France). Recombinant human TNF-α and TRAIL were purchased from Miltenyi Biotec (Paris, France). Recombinant human FasL was purchased from R&D Systems (Minneapolis, MN, USA).
Transient transfection and reporter gene assays
Cells were plated in 24-well plates, grown to 50–80% confluence, and transfected for 5 h by the PEI (polyethylenimine) method, as previously reported.7 For NF-κB luciferase reporter assays, we used 0.3 μg per well NF-κB-firefly luciferase reporter plasmid (Stratagene, San Diego, CA, USA) and 0.3 μg EVER2-Flag plasmid. The Renilla luciferase pRLTK vector (0.03 μg) was used as an internal control for transfection efficiency. Luciferase activity was measured with the Dual-Luciferase Reporter Assay kit (Promega, Madison, CA, USA) and a Centro XS3 LB 960 luminometer (Berthold Technologies, Bad Wildbad, Germany). Data were normalized with respect to Renilla luciferase activity and protein concentration, as determined in the Bio–Rad protein assay (Bio-Rad).
Semi-quantitative PCR
We isolated mRNA with the μmacs mRNA isolation kit (Miltenyi Biotec, Paris, France) and generated cDNA by reverse transcription with the Superscript II First-Strand system (Invitrogen). The oligonucleotide sequences used for semi-quantitative real-time PCR (RT-PCR) were, for EVER2: 5′-ATGGTGAAGGGGCTGCCGCAG-3′ and 5′-CACCACGCACCAGATCAGAGTGAG-3′ and for GAPDH: 5′-GACCACAGTCCATGCCATCACT-3′ and 5′-TCCACCACCCTGTTGCTGTAG-3′.
Co-immunoprecipitation and western blotting
For co-immunoprecipitation, cells (1–2 × 107) were lysed in 700 μl of immunoprecipitation buffer (20 mM Tris, pH 7.2, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1 × complete protease inhibitor cocktail, 0.1 mM Na3VO4 and 10 mM NaF). Cell lysates (600 μl) were incubated overnight with 2–4 μg of antibody at 4 °C. Complexes were precipitated with protein G/protein A-agarose (Calbiochem, Merck Chemicals Ltd, Nottingham, UK), washed three times in immunoprecipitation buffer and suspended in 50 μl of sodium dodecyl sulfate (SDS) sample buffer. Immunoprecipitates were subjected to SDS-PAGE and western blotting.
Whole-cell extracts were prepared by incubating cells with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM ethylene-diaminetetraacetic acid, 1% Nonidet P-40, 1 × complete protease inhibitor cocktail, 0.1 mM Na3VO4 and 10 mM NaF) for 45 min on ice. Nitrocellulose membranes were probed with the appropriate antibodies and developed with the SuperSignal West Pico or West Femto Chemiluminescent Substrate System (Pierce Biotechnology, Rockford, IL, USA).
Apoptosis assays
Apoptosis was quantified first by directly assessing nucleosomal DNA fragmentation with the Cell Death Detection ELISAPLUS kit (Roche Diagnostics, Mannheim, Germany). Cells were plated in 24-well plates at an initial concentration of 3 × 105 cells per well. The next day, cells were transfected with EVER2–Flag and empty plasmids in the morning, and treated with TNF-α(10 or 25 ng/ml), TRAIL (10 or 50 ng/ml) or FasL (50 or 100 ng/ml) for various time periods, or 16 h when not indicated. Cells were then processed according to the manufacturer's instructions. The mono- and oligonucleosome contents of cell lysates were determined with the antihistone–biotin antibody. The concentration of mono- and oligonucleosomes was determined photometrically with an antihistone–biotin antibody, followed by incubation with 2,2′-azino-di(3-ethylbenzthiazolin-sulfonate) (ABTS) as a substrate for the antibody. OD was read on a microplate reader (Bio-Rad) at a wavelength of 405 nm, and the OD ratio of stimulated to non stimulated cells was calculated to determine the fold-increase in apoptosis.
Apoptotic cell death was also determined by flow cytometry with an Annexin-V/PI viability assay. Floating and adherent cells were collected and stained with Annexin V-FITC kit (Miltenyi Biotec) according to the manufacturer's instructions. Cell death was detected using a CyAn flow cytometer (Beckman Coulter, Villepinte, France), and the data were analyzed with FlowJo software (Tree Star, Ashland, OR, USA).
High-throughput Gaussia protein complementation assay (HT-GPCA)
ORF sequences corresponding to proteins of interest were inserted into the SPICA-N1 and SPICA-N2 vectors for expression in fusion with fragments 1–93 (Gluc1) and 94–169 (Gluc2) of Gaussia princeps luciferase, as previously described.25 HEK-293T cells were used to seed 96-well plates at a concentration of 3.5 × 104 cells per well. After 24 h, cells were transfected with 120 ng of SPICA-N1 and SPICA-N2 constructs. The firefly luciferase pFLTK vector (0.03 μg) was used as an internal control for transfection efficiency. Cells were harvested 24 h after infection by incubation with 30 μl of Renilla Lysis Buffer (Promega, E2820) for 30 min, and Renilla luciferase activity was measured with a Berthold Centro XS LB960 luminometer. Results are expressed as a fold-change normalized with respect to the sum for controls, as previously described.25 Data were normalized with respect to firefly luciferase activity and the final result is the mean normalized luminescence ratio (NLR) for triplicate experiments.
Statistics
The statistical significance of differences was determined with Student's t-test. For all analyses, P<0.05 was considered significant.
Acknowledgments
This work was supported by grants from the ANR (contract no. 026 01), ARC (contract no. A09/1/5031) and the Ligue Nationale contre le Cancer (contract no. RS07/75-75) and by a donation from ODYSSE RE Holdings Corp. G GAUD was supported by a fellowship from the ANR (contract no. 026 01). We also thank Alex Edelman and Associates for correcting the English version of the manuscript.
Glossary
- AP-1
activating protein 1
- Bcl-2
B-cell lymphoma 2
- Bcl-xl
B-cell lymphoma-extra large
- cIAP
cellular inhibitor of apoptosis protein
- cFLIP
cellular FLICE-inhibitory protein
- CHX
cycloheximide
- DR
death receptor
- ELISA
enzyme-linked immunosorbent assay
- EV
epidermodysplasia verruciformis
- FADD
Fas-associated protein with death domain
- FasL
Fas ligand
- FCS
fetal calf serum
- FITC
fluorescein-5-isothiocyanate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescence protein
- HEK-293T
human embryonic kidney 293T
- HPV
human papillomavirus
- HT-GPCA
high-throughput Gaussia protein complementation assay
- miRNA
microRNA
- NF-κB
nuclear factor kappa B
- OD
optical density
- PARP
poly (ADP-ribose) polymerase
- PI
propidium iodide
- RIPK
receptor-interacting serine/threonine-protein kinase
- SNP
single nucleotide polymorphism
- TGF
transforming growth factor
- TMC
transmembrane channel-like
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- TRADD
TNFR-associated death domain protein
- TRAF2
TNFR-associated factor 2
- TRAIL
TNF-related apoptosis-inducing ligand
- TRAIL-R
TRAIL receptor
- XIAP
X-linked inhibitor of apoptosis protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by P Salomoni.
Supplementary Material
References
- Jablonska S, Majewski S, Obalek S, Orth G. Cutaneous warts. Clin Dermatol. 1997;15:309–319. doi: 10.1016/s0738-081x(96)00170-8. [DOI] [PubMed] [Google Scholar]
- Orth G. Genetics of epidermodysplasia verruciformis: insights into host defense against papillomaviruses. Semin Immunol. 2006;18:362–374. doi: 10.1016/j.smim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Ramoz N, Rueda LA, Bouadjar B, Montoya LS, Orth G, Favre M. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat Genet. 2002;32:579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- Arnold AW, Burger B, Kump E, Rufle A, Tyring SK, Kempf W, et al. Homozygosity for the c.917A-->T (p.N306l) polymorphism in the EVER2/TMC8 gene of two sisters with epidermodysplasia verruciformis Lewandowsky-Lutz originally described by Wilhelm Lutz. Dermatology. 2011;222:81–86. doi: 10.1159/000322536. [DOI] [PubMed] [Google Scholar]
- Hohenstein E, Rady PL, Hergersberg M, Huber AR, Tyring SK, Bregenzer T, et al. Epidermodysplasia verruciformis in a HIV-positive patient homozygous for the c917A-->T polymorphism in the TMC8/EVER2 gene. Dermatology. 2009;218:114–118. doi: 10.1159/000174084. [DOI] [PubMed] [Google Scholar]
- Patel T, Morrison LK, Rady P, Tyring S. Epidermodysplasia verruciformis and susceptibility to HPV. Dis Markers. 2010;29:199–206. doi: 10.3233/DMA-2010-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarczyk M, Pons C, Mendoza JA, Cassonnet P, Jacob Y, Favre M. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J Exp Med. 2008;205:35–42. doi: 10.1084/jem.20071311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski S, Hunzelmann N, Nischt R, Eckes B, Rudnicka L, Orth G, et al. TGF beta-1 and TNF alpha expression in the epidermis of patients with epidermodysplasia verruciformis. J Invest Dermatol. 1991;97:862–867. doi: 10.1111/1523-1747.ep12491539. [DOI] [PubMed] [Google Scholar]
- Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- Moore RJ, Owens DM, Stamp G, Arnott C, Burke F, East N, et al. Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis. Nat Med. 1999;5:828–831. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- Darding M, Meier P. IAPs: guardians of RIPK1. Cell Death Differ. 2012;19:58–66. doi: 10.1038/cdd.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S, Tomisato W, Hoshino T, Tsuchiya T, Mizushima T. Transforming growth factor-beta1 is responsible for maturation-dependent spontaneous apoptosis of cultured gastric pit cells. Exp Biol Med (Maywood) 2002;227:402–411. doi: 10.1177/153537020222700606. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010;29:4752–4765. doi: 10.1038/onc.2010.221. [DOI] [PubMed] [Google Scholar]
- Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- Schnepple DJ, Shepard B, Bren GD, Cummins NW, Natesampillai S, Trushin S, et al. Isolation of a TRAIL antagonist from the serum of HIV-infected patients. J Biol Chem. 2011;286:35742–35754. doi: 10.1074/jbc.M111.274639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura R, Konaka K, Matsumoto N, Hasegawa M, Fukui M, Mukaida N, et al. Fas ligand induces cell-autonomous NF-kappaB activation and interleukin-8 production by a mechanism distinct from that of tumor necrosis factor-alpha. J Biol Chem. 2004;279:46415–46423. doi: 10.1074/jbc.M403226200. [DOI] [PubMed] [Google Scholar]
- Lai YJ, Lin VT, Zheng Y, Benveniste EN, Lin FT. The adaptor protein TRIP6 antagonizes Fas-induced apoptosis but promotes its effect on cell migration. Mol Cell Biol. 2010;30:5582–5596. doi: 10.1128/MCB.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9:1037–1046. doi: 10.1038/ni.1638. [DOI] [PubMed] [Google Scholar]
- Kim JY, Lee JY, Kim DG, Koo GB, Yu JW, Kim YS. TRADD is critical for resistance to TRAIL-induced cell death through NF-kappaB activation. FEBS Lett. 2011;585:2144–2150. doi: 10.1016/j.febslet.2011.05.034. [DOI] [PubMed] [Google Scholar]
- Cao X, Pobezinskaya YL, Morgan MJ, Liu ZG. The role of TRADD in TRAIL-induced apoptosis and signaling. FASEB J. 2011;25:1353–1358. doi: 10.1096/fj.10-170480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH, et al. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol. 2010;6:352. doi: 10.1038/msb.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat Rev Immunol. 2007;7:532–542. doi: 10.1038/nri2115. [DOI] [PubMed] [Google Scholar]
- Cassonnet P, Rolloy C, Neveu G, Vidalain PO, Chantier T, Pellet J, et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat Methods. 2011;8:990–992. doi: 10.1038/nmeth.1773. [DOI] [PubMed] [Google Scholar]
- Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82:300–308. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev. 2010;24:2592–2602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kroemer G. Necroptosis turns TNF lethal. Immunity. 2011;35:849–851. doi: 10.1016/j.immuni.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Van Herreweghe F, Festjens N, Declercq W, Vandenabeele P. Tumor necrosis factor-mediated cell death: to break or to burst, that's the question. Cell Mol Life Sci. 2010;67:1567–1579. doi: 10.1007/s00018-010-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Brink R. Regulation of TNFRSF and innate immune signalling complexes by TRAFs and cIAPs. Cell Death Differ. 2010;17:35–45. doi: 10.1038/cdd.2009.114. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (tnf) to efficiently activate nf-(kappa)b and to prevent tnf-induced apoptosis. J Biol Chem. 2009;284:35906–35915. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson PK, Lichtenstein P, Gyllensten UB. Heritability of cervical tumours. Int J Cancer. 2000;88:698–701. doi: 10.1002/1097-0215(20001201)88:5<698::aid-ijc3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Scott M, Nakagawa M, Moscicki AB. Cell-mediated immune response to human papillomavirus infection. Clin Diagn Lab Immunol. 2001;8:209–220. doi: 10.1128/CDLI.8.2.209-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.