Abstract
Reactive oxygen species, the by-products of oxidative energy metabolism, are considered a main proximate cause of aging. Accordingly, overexpression of the enzyme Cu–Zn superoxide dismutase 1 (SOD1) can lengthen lifespan of Drosophila melanogaster in the laboratory. However, the role of SOD1 as a main determinant of lifespan has been challenged on the grounds that overexpression might be effective only in compromised genetic backgrounds. Moreover, interspecific comparisons show lower levels of antioxidant activities in longer-lived species, suggesting that life-span extension may evolve through less reactive oxygen species generation from the mitochondria rather than higher expression of SOD1. The tremendous variation in lifespan between ant castes, ranging over 2 orders of magnitude, coupled with the fact that all individuals share the same genome, provides a system to investigate the role of SOD1 in the wild. We used the ant Lasius niger as a model system, because queens can reach the extreme age of 28 years, whereas workers and males live only 1–2 years and a few weeks, respectively. We cloned SOD1 and found that long-lived queens have a lower level of expression than workers and males. Specific enzyme-activity assays also showed higher SOD1 activity levels in males and workers compared with queens, which had SOD1 activity levels similar to that of D. melanogaster. Altogether, these data show that increased expression of SOD1 is not required for the evolution of extreme lifespan, even in a system in which differential gene expression is the only way to express phenotypes with great lifespan differences.
To date, most of the genes implicated in aging have been identified in laboratory populations of short-lived model organisms such as the fruit fly Drosophila melanogaster, the nematode Caenorhabditis elegans, and the yeast Saccharomyces cerevisiae (1–4). However, it is not known whether these genes are functionally important in underlying lifespan differences in the wild or whether they may also influence lifespan of longer-lived organisms.
One of the more prominent antioxidant enzymes thought to be involved in lifespan extension is encoded by the gene superoxide dismutase 1 (SOD1). SOD1 overexpression was originally found to extend lifespan in D. melanogaster (5–10), and some D. melanogaster lines selected for extended longevity also show increased levels of expression of SOD1 (11, 12). However, additional experiments showed that the effect of SOD1 overexpression depends on the genetic background of the particular lines used (13–16). Most recently, it has been shown that SOD1 overexpression positively effects lifespan in some but not all natural genetic backgrounds (17).
Experiments in other organisms have also provided contrasting results. In yeast, increased lifespan has been obtained by overexpression of SOD1 and the mitochondrial form of SOD (18), but in C. elegans, contradictory results were obtained for chemical mimetics of SOD dependent on laboratory conditions (19, 20). In mice, manipulation of SOD1 levels reduces lifespan, although probably by mimicking one of the effects of Down's syndrome (21). Finally, comparative studies between species differing in lifespan showed that the level of SOD1 is not correlated with lifespan (22, 23). Importantly, however, these comparative studies of distant species suggest that increased lifespan is associated with reduced levels of reactive oxygen species production or leakage from the electron transport chain (24). The emerging picture is that SOD1 levels might influence lifespan under some conditions in some organisms, but it is yet unclear whether increased levels of SOD1 have evolved as a mechanism to increase lifespan in the wild.
The tremendous naturally evolved variation in lifespan between ant castes within one colony, ranging over 2 orders of magnitude, provides a unique system to determine whether the evolution of increased lifespan requires an increase in the level of SOD1 expression. In ants, the same genome, through differential gene expression, can result in phenotypes with distinct morphological and lifespan differences between social castes. Moreover, the level of gene expression is likely to be optimally adjusted, because it is the product of 100 millions of years of natural selection. Indeed, the power and strength of natural selection is demonstrated by the fact that queens can live 100 times longer than their solitary ancestor (25). Thus, the ant caste system provides a uniquely valuable but complementary model to study the role of specific genes on aging processes.
We used the ant Lasius niger as a model system, because queens can reach the extreme age of >28 years, whereas workers and males live only 1–2 years and a few weeks, respectively (26, §). By using this system, we tested whether the level of expression of SOD1 is higher in queens than workers and males. Activity assays were done also to test whether the patterns would be similar to that observed in interspecific comparative studies.
Methods
Collection and Colony Rearing. Queens and workers were collected from colonies kept 1–2 years under standard laboratory conditions. Colonies were started by a single queen collected in summer just after a mating flight on the campus of the University of Lausanne. Queens and workers were sampled simultaneously from each colony. Because males cannot be produced under laboratory conditions, they were collected from their natal colony on the campus of the University of Lausanne ≈1 week before mating flights. We also collected workers directly from the field and found similar levels of expression of SOD1 with workers collected from the laboratory (unpublished data).
RNA Isolation. Individuals were sorted on ice and, if necessary, dissected into body sections. They then were frozen with liquid nitrogen and stored at -80°C. Samples were ground with liquid nitrogen and processed as described in the Totally RNA Kit (Ambion) with the optional LiCl precipitation. RNA integrity was always checked on a native agarose gel.
Cloning of SOD1. 3′ extension from the initial fragment was accomplished by using the FirstChoice RNA ligase-mediated rapid amplification of cDNA ends (RACE) kit (Ambion, Austin, TX) with a gene-specific primer (5′-ggacttgctaagggtctgcatggattcca-3′) based on alignment of D. melanogaster, Drosophila willistoni, Ceratitis capitata, and C. elegans SOD1 cDNA sequence downloaded from GenBank (accession nos. M24421, L13281, M76975, and X77020, respectively). The product was subcloned into the vector pCR2.1-TOPO (TopoA, Invitrogen) and sequenced at Microsynth (Balgach, Switzerland). Based on this sequence, new primers were designed to generate 5′ RACE products covering the entire transcript. The transcript was verified by 5′ and 3′ RACE in pools of whole bodies of males, workers, and queens to rule out caste-specific variants.
Genomic Sequencing. Inverse PCR was performed as described by Ping and Knecht (27) but with the restriction enzymes BamHI, EcoRI, and HindIII (GIBCO/BRL) and subcloned with the PCR XL Topo kit (Invitrogen). The sequence was assembled by using the free programs bioedit (www.mbio.ncsu.edu/BioEdit/bioedit.html) and se-al (http://evolve.zoo.ox.ac.uk). All sequence was covered at least twice, and cDNA sequence was checked against the genomic-derived sequence. Polymorphisms were included when verified in at least two sequencing runs consisting of independent clones. The entire annotated sequence is deposited in GenBank under accession number AY309973.
Northern Blots. Northern blots were performed with the NorthernMax kit (Ambion). For whole-body blots, we used 1 queen, 5 males, and 25 workers from each of four colonies. Colonies were treated as the independent replicates in the statistical analyses. For body sections, we used three independent samples, each consisting of 4 queen heads, 4 queen thoraces, 4 queen abdomens, 70 male heads, 20 male thoraces, 10 male abdomens, 50 worker heads, 50 worker thoraces, and 50 worker abdomens. In each of these three samples, queens came from four colonies and workers and males came from one colony. Because the three samples never contained individuals originating from the same colony, there was no pseudoreplication. Four micrograms of RNA was loaded per lane, with transfer verified by methylene-blue staining. The 645-bp probe fragment was amplified by using the primers 5′-cataatccgagctatcgcgcag-3′ and 5′-tggctaatacatcaaagccgaacg-3′, which comprised the entire coding region of SOD1. The fragment was purified (QIAquick, Qiagen, Valencia, CA), and 25 ng was labeled by random priming (Stratagene) in the presence of [32P]dCTP. Hybridization and washes were performed at 48°C. Blots were exposed to a phosphorescent plate and imaged by using a Bio-Rad GS-250 molecular imager. The images were checked for saturation and quantitated by using imagej (http://rsb.info.nih.gov/ij) with normalization to methylene-blue staining of the 28S ribosomal RNA band.
Density values were converted to the percentage signal of each caste (values thus sum to 100% for all comparisons between the three castes), and homogeneity of variances was tested with Bartlett's test (28). Accordingly, means were compared with t tests (SPlus 2000) or Welch's modified t test when variances failed to test for equality. The results were corrected for multiple comparisons by using the Bonferroni correction (28).
SOD1 Assays. Cu–Zn SOD activity was measured by using 2% SDS pretreatment for 30 min to remove Mn SOD activity as described (29). Three independent measurements were made for each of the nests or samples used in the expression analysis. Statistical tests were done as for the expression comparisons.
Results and Discussion
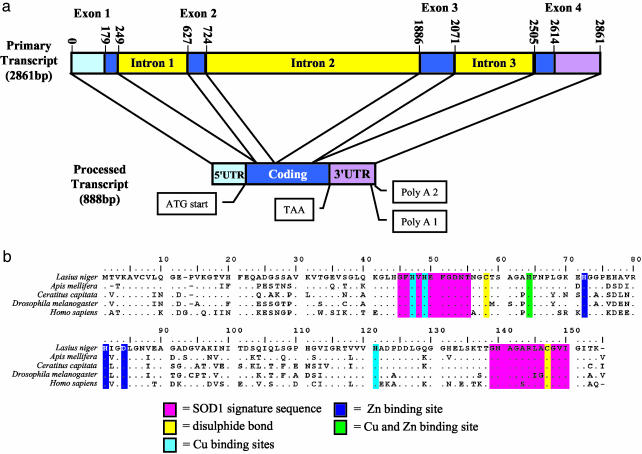
We cloned SOD1 from L. niger by RACE and used inverse PCR to obtain flanking genomic sequence. SOD1 is composed of four exons and three introns, with intron 1 being in the same position as it is in many other organisms (Fig. 1a). Analysis of the 5′ and 3′ RACE products from all three castes allowed us to determine that SOD1 is coded by an 888-base transcript after processing. Two transcript lengths were isolated from the workers: the 888-bp transcript and a slightly smaller transcript that was ≈45 bases shorter at the 3′ end corresponding to an alternative polyadenylation signal site (Fig. 1a). The transcript length was confirmed by Northern blot, although we could not resolve these two length variants in the workers. The only other variation is allelic, with one silent substitution and one 3-base insertion in the 5′ UTR. These allelic variants obtained by RACE were verified by comparison with genomic sequences obtained by inverse PCR. All the functionally important sequences of SOD1 are present in the conceptual translation (Fig. 1b). The greatest homology was with honey bee SOD1 (Apis mellifera, 88% amino acid identity) followed by Mediterranean fruit fly (C. capitata, 72% amino acid identity).
Fig. 1.
(a) Inferred structure of the SOD1 gene primary transcript and the transcript processed by RACE in the ant L. niger.(b) Comparison of the inferred amino acid SOD1 sequence of L. niger with several previously sequenced SOD1 genes in public databases. The important conserved sites are shaded as indicated. The honey bee (A. mellifera) sequence is a conceptual translation of contig 1487 from the Honey Bee Genome EST Project (http://titan.biotec.uiuc.edu/bee/honeybee_project.htm). The others are from GenBank: Mediterranean fruit fly (C. capitata, accession no. AAA57249), fruit fly (D. melanogaster, accession no. CAA68443), and human (Homo sapiens, accession no. DSHUCZ).
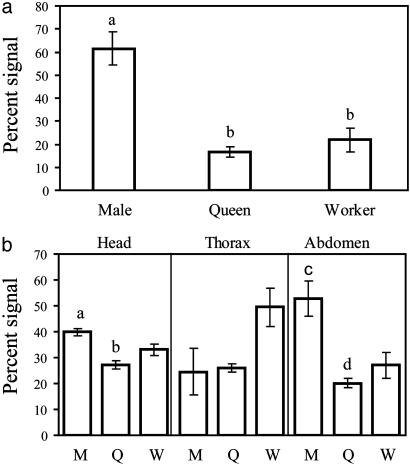
Northern blots of whole individuals using a probe made from this sequence show that males have significantly higher levels of SOD1 expression than queens and workers (Fig. 2a). The difference between queens and workers is not significant, but if anything, there is a tendency for queens to have lower expression than workers. Northern blots of body sections show that the higher level of expression of SOD1 in males is mostly because of a higher level of expression in the abdomen (gaster in ant nomenclature) (Fig. 2b). The heads of males also have significantly higher expression of SOD1 than the heads of queens. There is a trend of SOD1 expression being higher in worker head, thorax, and abdomen than in similar body parts of the queens, although none of these comparisons are significant after correcting for multiple comparisons.
Fig. 2.
SOD1 gene expression as measured by Northern blots. M, males; Q, mature queens; W, workers. Bars in lowercase letters differed significantly after the Bonferroni correction with P < 0.05. (a) SOD1 expression is highest in males in whole-body samples (for male/queen: t = 7.70, df = 6, P < 0.001; for male/worker: t = 4.940, df = 6, P = 0.002). (b) Male expression is higher than queen in both heads (t = 7.61, df = 2.027, P = 0.01) and abdomens (t = 3.94, df = 4, P = 0.01).
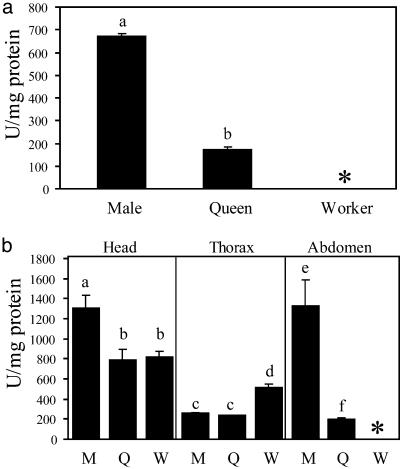
The finding that the greater lifespan of queens is not associated with higher levels of expression of SOD1 is bolstered further by Cu–Zn SOD-specific enzyme-activity assays (Fig. 3) (29). Males again have significantly higher activity than queens for whole-body assays. The high formic acid content of workers interferes with the assay such that whole workers and worker abdomens were excluded. The measured activity level of the queen (171 units/mg of protein ± 41.2) matches that of whole D. melanogaster (159.8 units/mg of protein ± 27.4, from data provided in ref. 14; t = -0.616, df = 17, P = 0.546, Student's t test), whereas male levels are significantly higher (665 units/mg of protein ± 36.8; t = -28.7, P < 0.001, Student's t test). The similar levels of SOD1 among reproducing females of these two species are consistent with another study finding SOD1 levels similar in primates of various longevities (22, 30). When considering body parts, the difference between males and queens is significant for heads and abdomens. In these assays, male heads also have significantly more activity than worker heads. There is no difference for the thorax between males and queens, and both are significantly less than workers. Again, analysis of body parts reveals a slightly higher activity in workers than queens, the difference being significant for thoraces but not heads, paralleling the differences in expression.
Fig. 3.
SOD1 activity assay levels in activity units per milligram. M, males; Q, mature queens; W, workers; *, not done because of interference by formic acid. Bars in lowercase letters differed significantly after the Bonferroni correction with P < 0.05. (a) In whole bodies, males are higher than queens (t = 22.57, df = 16, P < 0.0001). (b) Male heads have higher SOD1 activity than queen (t = 2.75, df = 15.86, P = 0.01) and worker (t = 3.23, df = 11.24, P = 0.007) heads. Males also show higher activity than queens in the abdomens (t = 4.78, df = 8.13, P = 0.001). For thoraces, male and queen activities are the same but significantly lower than worker activity (male/worker: t = -5.94, df = 10.06, P = 0.001; queen/worker: t = 6.79, df = 8.48, P = 0.0001).
This study demonstrates that the dramatic difference in lifespan between queens, workers, and males is not associated with higher levels of SOD1 expression and activity in the longer-lived castes. Rather, there is a tendency for a negative relationship with shorter-lived males having the greater expression and activity and the longer-lived queens having the lowest (or tied for lowest) level of expression and activity across all comparisons. This finding parallels interspecific comparisons of antioxidant activity levels, suggesting that longer-lived organisms produce lower rates of reactive oxygen species but do not necessarily have a higher level of expression of SOD1 than shorter-lived organisms (22, 23). Taken together, these results support the suggestion that the observed effect of overexpression of SOD1 on lifespan is limited to particular genetic strains under specific conditions in the laboratory (13–15). More generally, this study shows that studies in nonmodel organisms with long lifespans such as ants are necessary to test the role of candidate aging genes identified with short-lived organisms in the laboratory.
Acknowledgments
We thank L. Braendli and J. Cosendai for help in maintaining ant colonies and R. Mockett for prepublication access to fly SOD1 activity levels and manuscript comments. We also thank M. Chapuisat, T. Farmer, K. Hughes, S. Jemielity, H. Niculita, P. Reymond, G. Robinson and his research group, and three anonymous reviewers for helpful comments on the manuscript. This work was supported by the AETAS Foundation for Research in Aging (Geneva), the A. R. & J. Leenards Foundation (Lausanne, Switzerland), and the Swiss National Science Foundation.
Abbreviation: SOD, superoxide dismutase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY309973).
Footnotes
Kutter, H. & Stumper, R., Sixth International Congress of the International Union for the Study of Social Insects, September 15–20, 1969, Bern, Switzerland.
References
- 1.Finkel, T. & Holbrook, N. J. (2000) Nature 408, 239-247. [DOI] [PubMed] [Google Scholar]
- 2.Guarente, L. & Kenyon, C. (2000) Nature 408, 255-262. [DOI] [PubMed] [Google Scholar]
- 3.Kirkwood, T. B. L. & Austad, S. N. (2000) Nature 408, 233-238. [DOI] [PubMed] [Google Scholar]
- 4.Hekimi, S. & Guarente, L. (2003) Science 299, 1351-1354. [DOI] [PubMed] [Google Scholar]
- 5.Klichko, V. I., Radyuk, S. N. & Orr, W. C. (1999) Neurobiol. Aging 20, 537-543. [DOI] [PubMed] [Google Scholar]
- 6.Orr, W. C. & Sohal, R. S. (1994) Science 263, 1128-1130. [DOI] [PubMed] [Google Scholar]
- 7.Parkes, T. L., Elia, A. J., Dickinson, D., Hilliker, A. J., Phillips, J. P. & Boulianne, G. L. (1998) Nat. Genet. 19, 171-174. [DOI] [PubMed] [Google Scholar]
- 8.Sohal, R. S., Agarwal, A., Agarwal, S. & Orr, W. C. (1995) J. Biol. Chem. 270, 15671-15674. [DOI] [PubMed] [Google Scholar]
- 9.Sun, J. & Tower, J. (1999) Mol. Cell. Biol. 19, 216-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tower, J. (2000) Mech. Ageing Dev. 118, 1-14. [DOI] [PubMed] [Google Scholar]
- 11.Dudas, S. P. & Arking, R. (1995) J. Gerontol. Biol. Sci. B1117-B1127. [DOI] [PubMed]
- 12.Arking, R., Buck, S., Novoseltev, V. N., Hwangbo, D. & Lane, M. (2002) Ageing Res. Rev. 1, 209-228. [DOI] [PubMed] [Google Scholar]
- 13.Sohal, R. S., Mockett, R. J. & Orr, W. C. (2002) Free Radical Biol. Med. 33, 575-586. [DOI] [PubMed] [Google Scholar]
- 14.Orr, W. C., Mockett, R. J., Benes, J. J. & Sohal, R. S. (2003) J. Biol. Chem. 278, 26418-26422. [DOI] [PubMed] [Google Scholar]
- 15.Orr, W. C. & Sohal, R. S. (2003) Exp. Gerontol. 38, 227-230. [DOI] [PubMed] [Google Scholar]
- 16.Miller, R. A., Harper, J. M., Dysko, R. C., Durkee, S. J. & Austad, S. M. (2002) Exp. Biol. Med. 227, 500-508. [DOI] [PubMed] [Google Scholar]
- 17.Spencer, C. C., Howell, C. E., Wright, A. R. & Promislow, D. E. L. (2003) Aging Cell 2, 123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabrizio, P., Liou, L., Moy, V. N., Diaspro, A., Valentine, J. S., Gralla, E. B. & Longo, V. D. (2003) Genetics 163, 35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melov, S., Ravenscroft, J., Malik, S., Gill, M. S., Walker, D. W., Clayton, P. E., Wallace, D. C., Malfroy, B., Doctrow, S. R. & Lithgow, G. J. (2000) Science 289, 1567-1569. [DOI] [PubMed] [Google Scholar]
- 20.Keaney, M. & Gems, D. (2003) Free Radical Biol. Med. 34, 277-282. [DOI] [PubMed] [Google Scholar]
- 21.Huang, T. T., Carlson, E. J., Gillespie, A. M., Shi, Y. P. & Epstein, C. J. (2000) J. Gerontol. A 55, B5-B9. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Campo, R., López-Torres, M., Cadenas, E., Rojas, C. & Barja, G. (1998) J. Comp. Physiol. B 168, 149-158. [DOI] [PubMed] [Google Scholar]
- 23.Barja, G. (2002) Free Radical Biol. Med. 33, 1167-1172. [DOI] [PubMed] [Google Scholar]
- 24.Barja, G. (2002) Ageing Res. Rev. 1, 397-411. [DOI] [PubMed] [Google Scholar]
- 25.Keller, L. & Genoud, M. (1997) Nature 389, 958-960. [Google Scholar]
- 26.Hölldobler, B. & Wilson, E. O. (1990) The Ants (Springer, Berlin).
- 27.Ping, K. M. & Knecht, D. A. (1997) Biotechniques 22, 1046-1048. [DOI] [PubMed] [Google Scholar]
- 28.Sokal, R. R. & Rohlf, F. J. (1995) Biometry: The Principles and Practice of Statistics in Biological Research (Freeman, New York).
- 29.Mockett, R. J., Bayne, A. C. V., Sohal, B. H. & Sohal, R. S. (2002) Methods Enzymol. 349, 287-292. [DOI] [PubMed] [Google Scholar]
- 30.Tolmasoff, J. M., Ono, T. & Cutler, R. G. (1980) Proc. Natl. Acad. Sci. USA 77, 2777-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]