Abstract
The study of genetics is providing new and exciting insights into the pathogenesis, diagnosis, and treatment of disease. Both normal sleep and several types of sleep disturbances have been found to have significant genetic influences, as have traits of normal sleep, such as those evident in EEG patterns and the circadian sleep-wake cycle. The circadian sleep-wake cycle is based on a complex feedback loop of genetic transcription over a 24-h cycle. Restless legs syndrome (RLS) and periodic limb movements in sleep (PLMS) have familial aggregation, and several genes have a strong association with them. Recent genome-wide association studies have identified single nucleotide polymorphisms linked to RLS/PLMS, although none has a definite functional correlation. Narcolepsy/cataplexy are associated with HLA DQB1*0602 and a T-cell receptor α locus, although functional correlations have not been evident. Obstructive sleep apnea is a complex disorder involving multiple traits, such as anatomy of the oropharynx, ventilatory control, and traits associated with obesity. Although there is clear evidence of familial aggregation in the obstructive sleep apnea syndrome, no specific gene or locus has been identified for it. Angiotensin-converting enzyme has been proposed as a risk variant, but evidence is weak. Fatal familial insomnia and advanced sleep phase syndrome are sleep disorders with a definite genetic basis.
One of the most exciting and interesting frontiers in medical research is the study of the genetics of disease. Understanding the genetic basis of sleep disorders is important because it leads to insights about their pathogenesis; it also confirms the biologic basis of these disorders, leads to new tests for their diagnosis, and, more importantly, leads to novel therapies for, and individualized treatment of, patients with sleep disorders. The beginning of the modern scientific study of sleep is usually marked by Nathaniel Kleitman’s studies on sleep and the effects of sleep deprivation in the 1920s.1 This was followed in 1937 by descriptions by Blake and Gerard2 of EEG patterns associated with wake, light sleep, and deep sleep. Subsequent years have been marked by major advances in the observation, description, and clinical features of sleep and its disorders. The next major horizon in sleep medicine will be the discovery and description of the genetic basis of normal and abnormal sleep. The genetics of sleep disorders represents an important and exciting area of research that attempts to define mechanisms of disease at the level of DNA. Several excellent, comprehensive reviews on the genetics of sleep and sleep disorders have been published recently.3-5
Research into the genetic influence of any trait or disease, including sleep, begins with careful observation and recording of familial aggregation of a given trait or disorder. Additionally, studies of traits or disorders occurring in monozygotic and dizygotic twins provide valuable information about genetic transmission. Heritability, defined as the fraction of variance in a phenotype, trait, or disease explainable by genetic influence, can be estimated in families by using more advanced techniques such as segregation analysis and genome-wide linkage studies. Recently, a powerful tool called population-based, case-control, genome-wide association studies (GWASs) has provided insight into previously unknown genetic loci, including the genetic influences in sleep disorders.6 Different gene alleles (risk variants) can be characterized by their frequency of occurrence and by how large an effect they have on a phenotype, trait, or disease. Risk variants range from rare (allele frequency <1%) to common (allele frequency >5%) and may be associated with a range of effects from small (increased risk by a factor of 0.1) to large (increased risk by a factor of >100) on a given phenotype, trait, or disease.7-9
Genetic Influence on Normal Sleep Traits
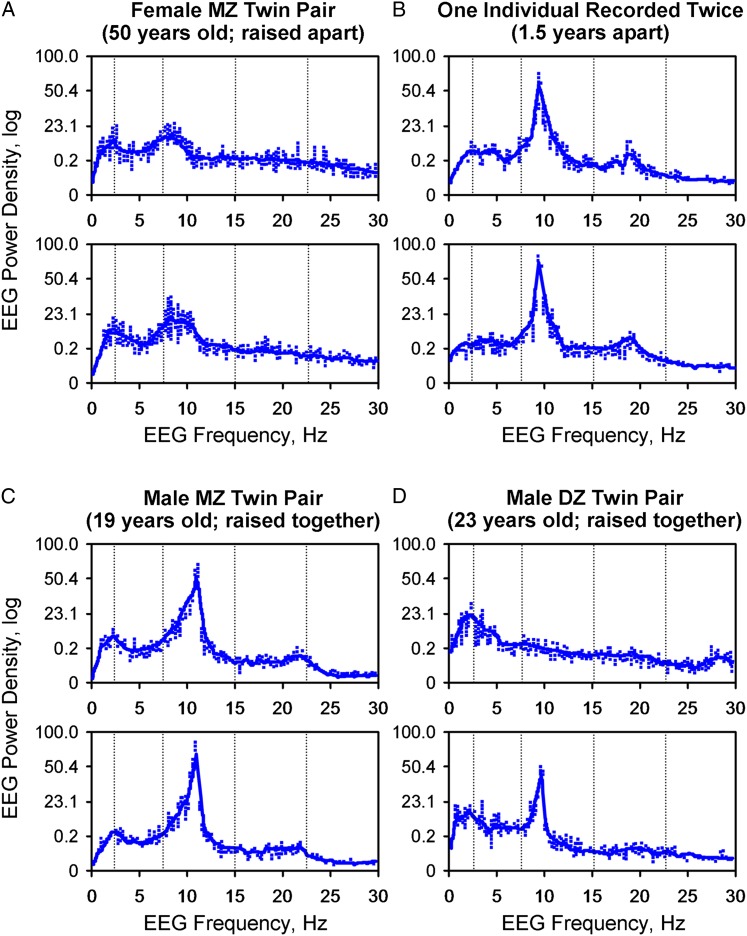
There is no single sleep gene. Sleep is a complex phenotype involving a recurrent behavioral state, characteristic EEG changes, timing during the 24-h clock, and responses to deprivation. As such, sleep may be controlled or influenced by many genes—many still not yet defined. Initial investigations have attempted to focus on traits easily measured, such as EEG patterns during sleep, and have compared monozygotic twins, dizygotic twins, and unrelated control subjects. For example, preliminary investigations of genetic influences examined EEG patterns and the power spectrum of monozygotic and dizygotic twins (Fig 1).5,10,11 If no genetic linkage existed, then EEG frequencies in monozygotic twins should be no more similar than those in dizygotic twins or unrelated individuals. Several studies have demonstrated that EEG frequencies are much more similar in monozygotic twins than in dizygotic twins or in unrelated control subjects, which indicates significant genetic determination.12
Figure 1.
Power spectral analysis demonstrating the genetic influence on sleep EEG frequency power density. A, A female MZ twin pair with virtually identical EEG frequency power density. B, A single individual recorded on two different occasions. C, EEG frequency of a male MZ twin pair with virtually identical EEG power density. D, A male DZ twin pair with different EEG power spectrums. DZ = dizygous; MZ = monozygous. (Adapted with permission from Andretic et al.5)
Genetic Determination of Circadian Rhythm
Circadian rhythm is one of the first sleep traits found both in animal models and in humans to be controlled by genetic factors and to have its molecular mechanism elucidated. In the Drosophila melanogaster fruit fly, genetic mutations can be induced that result in a longer or shorter circadian sleep-wake cycle than the mean of the fly population.13 Gene mutations have been identified that correlate with changes in circadian sleep-wake cycle length, and a model of its molecular mechanism has been developed.14 In a series of remarkable findings, researchers have determined that the Drosophila genes are quite similar to those that influence the circadian sleep-wake cycles of animals, such as mice, and of humans (Table 1).15,16
Table 1.
—Mammalian Genes That Influence Circadian Rhythm and Their Close Counterparts in the Drosophila melanogaster Fruit Fly
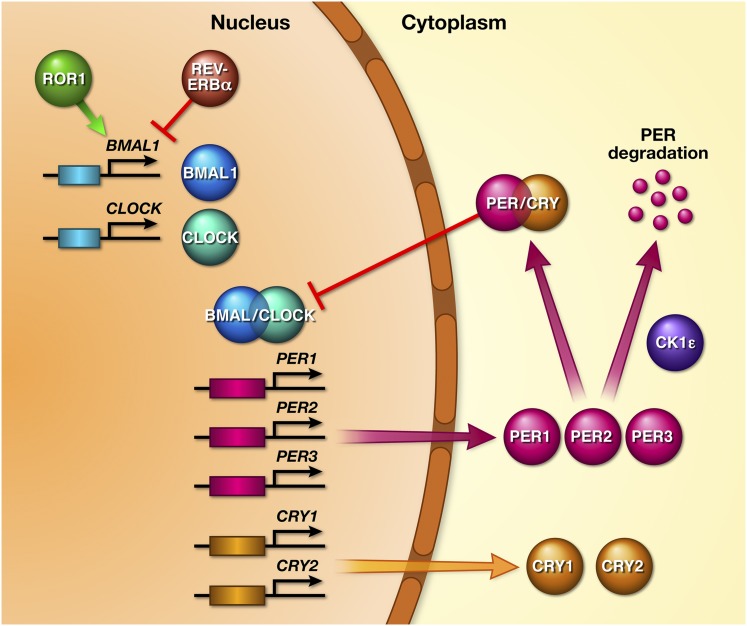
The circadian rhythm is controlled by light input transmitted from the retina to the suprachiasmatic nucleus in the hypothalamus. Cells within the suprachiasmatic nucleus contain a molecular feedback loop with a cycle of approximately 24 h. The first gene discovered that involved the circadian rhythm was called CLOCK; it was found on chromosome 5, which encodes for the CLOCK protein.17 Additional genes involved in the sleep-wake cycle are BMAL1 (or ARNTL), PERIOD (PER1, PER2, and PER3), and cryptochrome 1 and 2 (CRY1 and CRY2). The molecular basis of the circadian clock has a feedback loop involving the CLOCK/BMAL1, PER, and CRY proteins produced by these genes. Within the nucleus of the cell in the suprachiasmatic nucleus, the CLOCK protein forms a heterodimer with the BMAL1 protein, the product of the BMAL1 gene.18 CLOCK/BMAL1 increases transcription of the PER1, PER2, and PER3 genes, encoding the PER1, PER2, and PER3 proteins, and the CRY gene encodes the CRY protein. The PER and CRY proteins are transported from the nucleus into the cytoplasm and combine to form a PER/CRY dimer. Over a 24-h period, the PER/CRY dimer gradually increases to a critical concentration in the cytoplasm, and then moves back into the nucleus where it provides negative feedback to the CLOCK/BMAL1 dimer, thus inhibiting its own transcription19 (Fig 2). Additional feedback loops exist to regulate the sleep-wake cycle. Casein kinase 1, ε (CSNK1E or CK1ε) genes produce protein products that regulate the PERIOD protein. BMAL1 appears to be modulated by unique proteins known as REV-ERBα and ROR1, which are transcribed in the nucleus of the suprachiasmatic nucleus and are products of the genes NR1D1 (previously termed REV-ERBα) and ROR1. CRY also has proteins that modulate its expression.20
Figure 2.
Diagram of the molecular basis of the circadian sleep-wake cycle. The BMAL1 and CLOCK genes are transcribed to proteins BMAL1 and CLOCK, which combine to create a transcription factor BMAL1/CLOCK, which facilitates transcription of the PER and CRY proteins from their respective genes. PER and CRY form a dimer in the cytoplasm that gradually increases in concentration in the cytoplasm and reenters the nucleus, providing negative feedback. CK1ε CSNK1E facilitates degradation of PER. ROR1 and REV-ERBα regulate BMAL1. (Adapted with permission from Lowrey et al.83)
Genetic Influence on Clinical Disorders of Sleep
Restless Legs Syndrome and Periodic Limb Movements in Sleep
Restless legs syndrome (RLS) and periodic limb movements in sleep (PLMS) have significant evidence of a genetic basis. RLS/PLMS represents the sensory and motor manifestations of a common, yet poorly understood, sleep disorder. Diagnostic criteria consist of four main symptoms: (1) a persistent urge to move or stretch the legs, (2) symptoms that occur with rest or inactivity, (3) relief by stretching or walking, and (4) occurrence in the evening or night. RLS may be either primary or secondary, that is, related to a medical condition such as iron deficiency, pregnancy, or renal failure.
Familial forms of RLS and PLMS have been identified. Most patients (40%-60%) have a family history of a relative with RLS. Persons with a positive family history tend to have symptom onset at an earlier age compared with those without a family history.20 Genetic linkage studies in these families have identified several associated chromosomal loci: RLS1 on chromosome 12q, discovered in a French Canadian family21; RLS2 on 14q, in an Italian family22; RLS3 on chromosome 9p, in 15 extended American families; RLS4 on chromosome 2q; and RLS5 on chromosome 20p.23 In these families, transmission appears to be autosomal dominant with incomplete penetrance. A high concordance has been found in monozygotic twins. Since dopaminergic medications are used to treat RLS, candidate genes that code for receptors or enzymes related to dopamine transmission have been studied but have not been found to be significant.
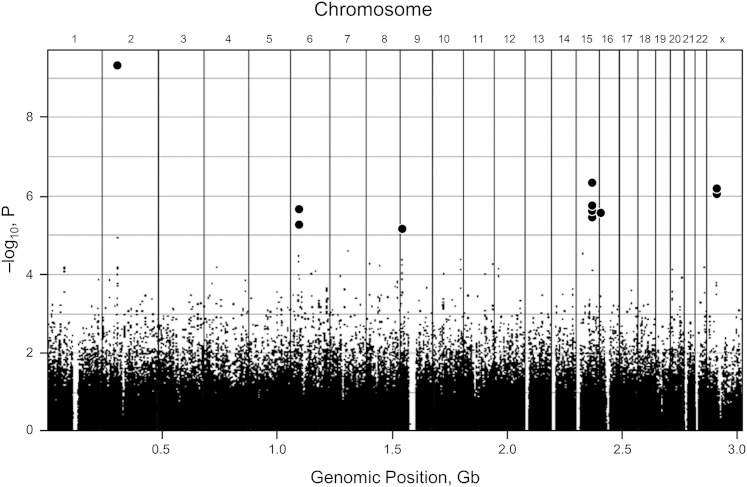
GWAS has been used successfully to identify candidate genes associated with RLS. In one large study, PLMS was strongly associated with four genes: MEIS1, BTBD9, MAP2K5, and SKOR1 (previously called LBXCOR1) (Fig 3).24 Each genetic variant of these alleles carries an increased risk for RLS of approximately 50%. BTBD9 was associated with PLMS but not with RLS, which suggests that this variant is associated with motor manifestations of the syndrome but not necessarily with the sensory part.25 Each at-risk allele of BTBD9 is also associated with a 13% decrease in ferritin, a known risk factor for RLS.25 Also of interest is that the MEIS1 gene is involved with a regulatory network important in motor neuron development.26 SKOR1 is involved with regulation of the development of dorsal horn sensory pathways. Additional risk variants have been identified recently, consisting of two single nucleotide polymorphisms (SNPs) expressed in the protein tyrosine phosphatase, receptor type, D (PTPRD) gene, a gene involved with neuronal development that may also provide a possible functional association.27
Figure 3.
Genome-wide association study (GWAS) of restless legs syndrome (RLS). A Manhattan plot diagram of a GWAS demonstrating a common method of displaying data from a GWAS. The x axis shows various chromosome locations. The y axis displays the negative logarithm of the association P value. Each dot in the display represents different single nucleotide polymorphisms (SNPs), possibly linked to RLS. The stronger the association, the smaller the P value and the higher the negative logarithm. The higher the dot on the display, the more likely it is that the SNP is statistically different among those with the disease and those without it. In this display, 13 SNPs are significant and highlighted in bold (three SNPs on chromosome 15 are similar and appear as a single dot.) Gb = gigabases. (Adapted with permission from Winkelmann et al.24)
The importance of these studies is that they point toward a molecular mechanism for this condition. Little is known at this time about the exact function of most of these genes and how they play a role in the development of RLS/PLMS. The next challenge in research is to develop relevant animal models of these variants and attempt to identify how they contribute to RLS/PLMS.9 Further studies should fill in the gaps in the exact pathophysiology of this sleep disorder, leading to objective diagnostic testing, and to the development of new treatments.
Genetic Basis of Narcolepsy
Narcolepsy is a relatively uncommon disorder characterized by hypersomnolence, cataplexy, hypnagogic hallucinations, and sleep paralysis. Its prevalence is approximately 25 to 50 per 100,000 people in the general population.28,29 Familial clustering has been observed: First-degree relatives have 10- to 40-fold higher risk (0.9%-2.3%) than that observed in the general population.30 Monozygotic twins show low concordance (about 32%).28 In a canine model, a genetic basis of narcolepsy has been observed; in humans a definite genetic basis has not yet been determined. However, recent investigations provide significant evidence of a genetic predisposition associated with an autoimmune pathogenesis.
The neuropeptide hypocretin (orexin) has been the subject of considerable interest in the syndrome of narcolepsy/cataplexy, and evidence that depletion of hypocretin (orexin) is important in narcolepsy has accumulated in the past several years. A gene mutation in a canine model resulting in orexin-2 receptor dysfunction leads to narcolepsy that may be autosomal recessive.31 A narcolepsylike phenotype has been seen in hypocretin/orexin knockout mice.32 Patients with narcolepsy/cataplexy syndrome lack hypocretin in cerebrospinal fluid.33 Autopsy studies of the brains of persons with narcolepsy have found a marked reduction of hypocretin neurons in the lateral hypothalamus.34 Hypocretin is important in maintaining sleep and wake homeostasis, and its deficiency leads to hypersomnolence and dissociation of rapid eye movement (REM) sleep and wakefulness. The hypothesis that destruction of hypocretin cells is caused by an autoimmune event has gained wide appeal. Recent observations have found that onset of narcolepsy is associated with upper airway infections due to Streptococcus infections, H1N1 influenza infections, and even H1N1 vaccinations.35
Narcolepsy has been associated with the human leukocyte antigen (HLA) system, a 4-megabase region of chromosome 6, which is divided into three subregions: class I, class II, and class III. Class II HLA antigens have been strongly associated with autoimmune diseases. The class II antigen, DQB1*0602, has been associated with narcolepsy.30,36 Most (90%-100%) patients with narcolepsy/cataplexy across all ethnic groups have this allele. In addition, 35% to 56% of patients who have narcolepsy without cataplexy also have this allele. However, 20% of the normal population also has the allele, reducing its usefulness in diagnostic testing. Homozygotes, carrying two alleles, have a risk of narcolepsy two to four times greater than heterozygotes and may have more severe symptoms.37,38 Several other alleles have been identified that also predispose to narcolepsy (DQB1*0301, DQA1*06, DRB1*04, DRB1*08, DRB1*11, and DRB1*12), whereas several alleles seem to be protective (DQB1*0601, DQB1*0501, and DQA1*01).39
Recent investigations have found strong evidence pointing to an autoimmune pathogenesis of narcolepsy. Recent GWASs have found an association between narcolepsy and a specific SNP on the T-cell receptor α locus.40 The T-cell receptor α locus has been identified as the unique protein on T lymphocytes that interacts with HLA classes I and II, including the HLA-DQB1*0602 allele.41 The exact function is indeterminate but supports a strong genetic influence in narcolepsy and again suggests the possibility of an autoimmune disease. T-cell receptor is important in the immune response because it interacts with protein-bound HLA antigens, which likely have a role in an autoimmune reaction resulting in the destruction of hypocretin cells, in the context of some type of antigenic trigger. Autoantibodies directed against a protein expressed in hypocretin cells, Tribbles homolog 2 (Trib 2), have been discovered in patients with newly diagnosed narcolepsy. There is not direct evidence that these antibodies actually kill hypocretin cells, but it may be a consequence of cell injury or may be incidental.42,43 GWAS technology with replication and fine mapping methodologies have shown an SNP associated with the purinergic receptor (P2RY11) to be significantly associated with narcolepsy.44 The allele associated with narcolepsy is important in the function of CD8+ T lymphocytes and natural killer cells, which may be implicated in cell death. This finding suggests that immune mechanisms have a role in the pathogenesis of narcolepsy.44
Genetics of Obstructive Sleep Apnea
Obstructive sleep apnea (OSA) is a complex disorder with multiple predisposing factors (eg, obesity, age, male sex, and craniofacial abnormalities) and with physiologic variables such as apnea-hypopnea index, oxyhemoglobin saturation, and other clinical variables such as sleepiness, obesity, and hypertension. In such a complex syndrome, it would be unlikely for there to be a single genetic predisposition. However, several traits most likely influence the predisposition to OSA, such as anatomy of the oropharynx, ventilatory control, or traits associated with obesity. The most common anatomic abnormality found in patients with OSA is mild retrognathism and inferior position of the hyoid bone, which are likely determined genetically. However, distinct genetic loci for this trait have not yet been identified.
Many studies have been undertaken on the genetic influences on OSA since the documentation of numerous familial forms of this sleep disorder.45 Inherited factors likely account for approximately 40% of the risk of OSA.46 Redline and Tishler46 noted that the prevalence of OSA in first-degree relatives of patients with OSA has been reported to range from 22% to 84%. The OR of a first-degree relative having OSA ranges from 2 to 46.46
Several authors have noted familial aggregation of cases of OSA.47-50 Among these, the Cleveland Family Study provided the strongest data in support of familial aggregation, and it was the first to observe a major difference in heritability between whites and blacks.49 Obesity, which has both environmental and genetic influences, is a significant risk factor for OSA. Genetic factors are important in the determination of BMI (weight in kg/height in m2), and they may explain as much as 50% to 90% of the variance in the BMIs of siblings in twin studies. Furthermore, family studies provide a heritability estimate of 20% to 80% for BMI.51,52 In the Cleveland Family Study, first-degree relatives of patients with OSA had a higher relative risk of OSA even when obesity was controlled for as measured by BMI. Another study of a population in Scotland also demonstrated significant familial aggregation in OSA, with the authors observing that maxillofacial structure was a more important factor than obesity.53 The first-degree relatives of patients with OSA have smaller mandibular and maxillary bone structure than do control subjects, which suggests that maxillofacial anatomy is an important inherited trait that contributes to the development of OSA.
Many candidate gene association studies have been done on OSA using different populations. Among the most promising genetic associations are genes with different alleles for apolipoprotein E4 (ApoE4), tumor necrosis factor (TNF), and angiotensin-converting enzyme (ACE). On the basis of two reports, ApoE4 has been proposed as a candidate gene for development of OSA.54,55 However, a recent systematic review found only 1 TNF polymorphism, TNFA rs1800629, to have a statistically significant association with OSA. There was not sufficient data in this review to support an association with ACE or ApoE4. Many studies were found to be underpowered and thus were unable to detect significant ORs.56 Furthermore, a lack of association of ApoE4 with OSA was found in another meta-analysis of eight studies (N =6,508).57 In another study, certain polymorphisms of TNF-α were found to occur more frequently in patients with OSA than in control subjects,58 and to correlate with the presence of sleepiness in those patients.59
ACE polymorphisms have been associated with the development of hypertension, which has provoked interest in a hypothesis that certain polymorphisms in patients with OSA might identify those patients in whom hypertension would develop. There have been mixed results in studying this hypothesis. For example, in the Wisconsin Sleep Cohort, the D allele of the insertion/deletion polymorphism was correlated with the presence of hypertension in patients who had mild to moderate OSA,60 whereas in the Cleveland Family Study, the D allele seemed to reduce the risk for hypertension.61
Congenital central hypoventilation syndrome is a rare, lifelong condition that typically presents in infancy, and is characterized by absence of spontaneous breathing or by shallow, inadequate breathing. Hypoventilation, confirmed by a Paco2 level >45 mm Hg during wakefulness, is due to abnormal central respiratory drive. Other abnormalities of the autonomic nervous system are often present as well. Some method of ventilatory support is usually necessary during wakefulness and sleep. Congenital central hypoventilation syndrome is associated with defects in the PHOX2B gene, which is important in neuronal development, specifically in the neurons determining central ventilatory control.62
Fatal Familial Insomnia
Two sleep disorders have been identified as having a definite genetic basis (Table 2). The first is fatal familial insomnia, which has been described as a rare autosomal neurodegenerative disease initially reported in 1986.63 The clinical manifestations consist of progressive cognitive impairment, hallucinations, severe insomnia, and, eventually, death. The age of onset is 30 to 60 years of age, and the natural history is approximately three years. Autopsy has shown selective degeneration of thalamic nuclei associated with abnormal accumulation of the prion protein. In these persons, a single mutation has been identified on the prion protein gene (PRNP), at position 178, in combination with another mutation on position 129.64
Table 2.
—Summary of Sleep Disorders With Alleles or Loci That Have Been Associated on the Basis of Candidate Gene, Linkage, or Genome-Wide Association Studies
| Sleep Disorder | Associated Gene, Loci, SNP | Comments |
| Narcolepsy/cataplexy | DQB1 and DQA1; primary allele DQB1*0602 | HLA class II allele; effect observed across several ethnic groups; high prevalence (90%) in narcolepsy with cataplexy |
| Narcolepsy/cataplexy | T-cell receptor α | Based on GWAS; identified across several ethnic groups independently; T-cell receptor on lymphocytes interacts with HLA class I and II antigens, including DQB1*0602 allele |
| RLS/PLMS | RLS1 | Identified in familial linkage studies across several nationalities; complex transmission, but often autosomal dominant, with incomplete penetrance; no association with dopamine system or iron metabolism documented |
| RLS2 | ||
| RLS3 | ||
| RLS4 | ||
| RLS5 | ||
| RLS/PLMS | MEIS1 | Based on GWAS; replicated in several studies; MEIS1 associated with motor neuron development |
| RLS/PLMS | BTBD9 | Based on GWAS; associated with PLMS without RLS; allele also associated with reduced ferritin levels |
| RLS/PLMS | MAP2K5/SKOR1 (LBXCOR1) | Based on GWAS; gene associated with development of dorsal-horn sensory pathways |
| RLS/PLMS | PTPRD | Based on GWAS; two SNPs involved with neuronal development |
| Familial advanced sleep phase syndrome | HPER2 | Autosomal dominant transmission; identified through candidate gene sequencing |
| Fatal familial insomnia | Single point mutation in prion gene | Point mutation at position 178 of prion protein gene combined with point mutation at codon 129 |
GWAS = genome-wide association study; HLA = human leukocyte antigen; PLMS = periodic limb movements in sleep; RLS = restless legs syndrome; SNP = single nucleotide polymorphism.
Familial Advanced Sleep Phase Syndrome
The second sleep disorder with a well-defined genetic basis is familial advanced sleep phase syndrome, which is an autosomal dominant disorder. Persons with this disorder have a regular sleep cycle that occurs 4 h earlier than that of unaffected family members. A mutation has been discovered on the human PER2 gene on chromosome 2q, which was referred to previously in the discussion of the circadian clock. In a mouse model, this mutation results in alterations in the phosphorylation of the PER2 protein by casein kinase-1, α1 (CSNK1A1).65 Of great interest is that the mouse model accurately replicates the human phenotype.66
Sleep and the Immune System
The biochemical and molecular mechanisms of sleep have been the subject of intense investigation in the past 2 decades, and the understanding of the function of regulatory mechanisms has increased significantly during that time. Regulation of sleep is extraordinarily complex and multiple systems are involved. Many cytokines intrinsic to the function of the immune response to infection have been found to influence and regulate sleep.67 Additionally, recent studies have found that sleep duration is important in the immune system. Short sleep duration appears to affect the immune system and is associated with alterations in the system and may increase risk of clinical infections. This section reviews some of the most interesting work investigating the relation between sleep and the immune system.
Multiple cytokines and other molecules are involved in both positive and negative feedback loops affecting nonrapid eye movement (NREM) sleep. Most of the evidence revolves around two cytokines: IL-1β and TNF-α. Both IL-1β and TNF-α are found in the brain and cerebrospinal fluid and have diurnal rhythms in the brain; the highest levels of IL-1β and TNF-α are found during periods of sleep. Both IL-1β and TNF-α promote sleep, specifically NREM sleep, if injected either centrally or systemically into rabbits or other animal models, including mice, rats, and cats. Total sleep time is increased, and the normal cycle of REM, NREM, and wake is physiologic.68,69 Substances that block or inhibit the action of IL-1β or TNF-α decrease NREM sleep. Examples include anti-IL-1β or anti-TNF, soluble receptors to the cytokines, and a substance known as small interfering RNA targeting IL-1β and TNF-α. Animals lacking receptors for the cytokines demonstrate less sleep.69 Compounds known to inhibit NREM sleep, such as corticotropin-releasing hormone, glucocorticoids, and prostaglandin E2, also suppress the production of IL-1 or TNF.70 Other substances known to inhibit either IL-1β or TNF-α include IL-4, IL-10, IL-13, and transforming factor growth factor-β, and they also inhibit NREM sleep. Levels of IL-1β mRNA increase in the brain following sleep deprivation. The expected rebound in sleep after sleep deprivation can be blunted by blocking TNF.70
The mechanism of action of IL-1β and TNF-α in enhancing NREM sleep is not fully understood, but there is evidence that these two cytokines activate nuclear factor-κB (NF-κB), a protein involved with DNA transcription. Substances that result in inhibition of NF-κB activity also inhibit sleep. For example, IL-4, IL-10, and the glucocorticoids are known to inhibit NF-κB activity and also to inhibit NREM sleep.68 Furthermore, evidence exists that clinical symptoms associated with sleep deprivation, such as sleepiness, fatigue, cognitive impairment, and enhanced pain sensitivity, can be elicited by exogenous injection of IL-1β or TNF-α.68,71 Clinical syndromes such as sleep apnea, chronic fatigue, rheumatoid arthritis, and chronic fatigue seen with chronic inflammation are associated with increased brain and circulating levels of IL-1β and TNF-α.72 Several intriguing case series report that the fatigue of rheumatoid arthritis and the sleepiness of sleep apnea can be attenuated by clinically available inhibitors of TNF, such as etanercept.68,73
Other cytokines have been identified that also influence sleep. For example, increased levels of IL-6 are associated with sleep.74 IL-6 demonstrates a biphasic pattern, with nadirs at about 8:00 am and 9:00 pm, and it peaks at about 7:00 pm and 5:00 am. Sleep deprivation delays this circadian pattern, such that the increase in IL-6 occurs during the day.74,75
Sleep Deprivation and the Immune System
The effect of total or partial sleep deprivation on the immune system has created considerable interest. These studies have been recently reviewed.76 The physiologic response to either complete or partial sleep deprivation is to activate components of the inflammatory system. Most studies have found a nonspecific activation of the immune reaction and such as increases in leukocyte and monocyte counts, increases in cytokines such as IL-6 and TNF. Increases in IL-1β and IL-17 have also been documented in response to moderate sleep deprivation.76 Sleep deprivation is associated with reductions in immune response,77 including impairment of T-cell cytokine production.78 These studies of molecular changes in immune function have been supplemented by clinical studies supporting an association of short sleep with risk of infection. On the basis of data from almost 57,000 individuals, both short sleep and long sleep are associated with an increased risk of pneumonia compared with 8-h sleepers.79 Normal sleep enhances, whereas sleep deprivation for one night impairs, the response to hepatitis A vaccine.80 Shorter sleep duration measured by actigraphy in healthy adults was associated with an impaired IgG response to hepatitis B surface antibody and inability to achieve the threshold for clinical protection to hepatitis B vaccine despite correction for other factors.81 In addition, response to influenza vaccine can be impaired by sleep deprivation.82
Conclusion
There have been very exciting and interesting developments in the areas of genetics and immunology related to normal sleep and sleep disorders. These findings will have a significant effect on the clinical practice of sleep medicine in the near future. A clear, genetic, molecular mechanism regulates the circadian rhythm. New information about the genetic basis of RLS/PLMS, narcolepsy, circadian rhythm disorders, and OSA will soon lead to improved diagnostic tests and new individualized treatments. Rapidly advancing research on the relation of the immune system and sleep will lead to more advanced understanding of each area.
Acknowledgments
Financial/nonfinancial disclosures: The author has reported to CHEST that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Other contributions: Editing, proofreading, and reference verification were provided by the Section of Scientific Publications, Mayo Clinic.
Abbreviations
- ACE
angiotensin-converting enzyme
- GWAS
genome-wide association study
- HLA
human leukocyte antigen
- NREM
nonrapid eye movement
- OSA
obstructive sleep apnea
- PLMS
periodic limb movements in sleep
- REM
rapid eye movement
- RLS
restless legs syndrome
- SNP
single nucleotide polymorphism
- TNF
tumor necrosis factor
Footnotes
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians. See online for more details.
References
- 1.Kleitman N. Sleep and Wakefulness as Alternating Phases in the Cycle of Existence. Chicago, IL: University of Chicago Press; 1939. [Google Scholar]
- 2.Blake H, Gerard RW. Brain potential during sleep. Am J Physiol. 1937;119(4):692-703. [Google Scholar]
- 3.Winkelmann J, Kimura M. Genetics of sleep disorders. Handb Clin Neurol. 2011;99:681-693. [DOI] [PubMed] [Google Scholar]
- 4.Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146(2):194-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andretic R, Franken P, Tafti M. Genetics of sleep. Annu Rev Genet. 2008;42:361-388. [DOI] [PubMed] [Google Scholar]
- 6.Raizen DM, Wu MN. Genome-wide association studies of sleep disorders. Chest. 2011;139(2):446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228-237. [DOI] [PubMed] [Google Scholar]
- 8.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19(3):212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pack AI, Pien GW. Update on sleep and its disorders. Annu Rev Med. 2011;62:447-460. [DOI] [PubMed] [Google Scholar]
- 10.Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H. Genetic and environmental determination of human sleep. Sleep. 1983;6(3):179-185. [DOI] [PubMed] [Google Scholar]
- 11.De Gennaro L, Marzano C, Fratello F, et al. The electroencephalographic fingerprint of sleep is genetically determined: a twin study. Ann Neurol. 2008;64(4):455-460. [DOI] [PubMed] [Google Scholar]
- 12.Ambrosius U, Lietzenmaier S, Wehrle R, et al. Heritability of sleep electroencephalogram. Biol Psychiatry. 2008;64(4):344-348. [DOI] [PubMed] [Google Scholar]
- 13.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirelli C, Bushey D. Sleep and wakefulness in Drosophila melanogaster. Ann N Y Acad Sci. 2008;1129:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178(3):1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi JS. Finding new clock components: past and future. J Biol Rhythms. 2004;19(5):339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitaterna MH, King DP, Chang AM, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564-1569. [DOI] [PubMed] [Google Scholar]
- 19.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 (Spec No 2):R271-R277. [DOI] [PubMed] [Google Scholar]
- 20.Schormair B, Winkelmann J. Genetics of restless legs syndrome: Mendelian, complex, and everything in between. Sleep Med Clin. 2011;6(2):203-215. [Google Scholar]
- 21.Desautels A, Turecki G, Montplaisir J, Sequeira A, Verner A, Rouleau GA. Identification of a major susceptibility locus for restless legs syndrome on chromosome 12q. Am J Hum Genet. 2001;69(6):1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonati MT, Ferini-Strambi L, Aridon P, Oldani A, Zucconi M, Casari G. Autosomal dominant restless legs syndrome maps on chromosome 14q. Brain. 2003;126(pt 6):1485-1492. [DOI] [PubMed] [Google Scholar]
- 23.Winkelmann J, Lichtner P, Schormair B, et al. Variants in the neuronal nitric oxide synthase (nNOS, NOS1) gene are associated with restless legs syndrome. Mov Disord. 2008;23(3):350-358. [DOI] [PubMed] [Google Scholar]
- 24.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39(8):1000-1006. [DOI] [PubMed] [Google Scholar]
- 25.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357(7):639-647. [DOI] [PubMed] [Google Scholar]
- 26.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123(3):477-491. [DOI] [PubMed] [Google Scholar]
- 27.Schormair B, Kemlink D, Roeske D, et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet. 2008;40(8):946-948. [DOI] [PubMed] [Google Scholar]
- 28.Mignot E. Genetic and familial aspects of narcolepsy. Neurology. 1998;50(2 Suppl 1):S16-S22. [DOI] [PubMed] [Google Scholar]
- 29.Longstreth WT, Jr, Koepsell TD, Ton TG, Hendrickson AF, van Belle G. The epidemiology of narcolepsy. Sleep. 2007;30(1):13-26. [DOI] [PubMed] [Google Scholar]
- 30.Chabas D, Taheri S, Renier C, Mignot E. The genetics of narcolepsy. Annu Rev Genomics Hum Genet. 2003;4:459-483. [DOI] [PubMed] [Google Scholar]
- 31.Mignot E. Sleep, sleep disorders and hypocretin (orexin). Sleep Med. 2004;5(Suppl 1):S2-S8. [DOI] [PubMed] [Google Scholar]
- 32.Chemelli RM, Willie JT, Sinton CM, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98(4):437-451. [DOI] [PubMed] [Google Scholar]
- 33.Mignot E, Lammers GJ, Ripley B, et al. The role of cerebrospinal fluid hypocretin measurement in the diagnosis of narcolepsy and other hypersomnias. Arch Neurol. 2002;59(10):1553-1562. [DOI] [PubMed] [Google Scholar]
- 34.Thannickal TC, Moore RY, Nienhuis R, et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27(3):469-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornum BR, Faraco J, Mignot E. Narcolepsy with hypocretin/orexin deficiency, infections and autoimmunity of the brain. Curr Opin Neurobiol. 2011;21(6):897-903. [DOI] [PubMed] [Google Scholar]
- 36.Mignot E, Hayduk R, Black J, Grumet FC, Guilleminault C. HLA DQB1*0602 is associated with cataplexy in 509 narcoleptic patients. Sleep. 1997;20(11):1012-1020. [PubMed] [Google Scholar]
- 37.Pelin Z, Guilleminault C, Risch N, Grumet FC, Mignot E. HLA-DQB1*0602 homozygosity increases relative risk for narcolepsy but not disease severity in two ethnic groups. US Modafinil in Narcolepsy Multicenter Study Group. Tissue Antigens. 1998;51(1):96-100. [DOI] [PubMed] [Google Scholar]
- 38.Hong SC, Lin L, Lo B, et al. DQB1*0301 and DQB1*0601 modulate narcolepsy susceptibility in Koreans. Hum Immunol. 2007;68(1):59-68. [DOI] [PubMed] [Google Scholar]
- 39.Mignot E, Lin L, Rogers W, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68(3):686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallmayer J, Faraco J, Lin L, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41(6):708-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padhan K, Varma R. Immunological synapse: a multi-protein signalling cellular apparatus for controlling gene expression. Immunology. 2010;129(3):322-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawashima M, Lin L, Tanaka S, et al. Anti-Tribbles homolog 2 (TRIB2) autoantibodies in narcolepsy are associated with recent onset of cataplexy. Sleep. 2010;33(7):869-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim AS, Scammell TE. The trouble with Tribbles: do antibodies against TRIB2 cause narcolepsy? Sleep. 2010;33(7):857-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornum BR, Kawashima M, Faraco J, et al. Common variants in P2RY11 are associated with narcolepsy. Nat Genet. 2011;43(1):66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pack AI. Genetics of sleep apnea. Sleep Med Clin. 2011;6(2):237-245. [Google Scholar]
- 46.Redline S, Tishler PV. The genetics of sleep apnea. Sleep Med Rev. 2000;4(6):583-602. [DOI] [PubMed] [Google Scholar]
- 47.Redline S, Tosteson T, Tishler PV, Carskadon MA, Millman RP. Studies in the genetics of obstructive sleep apnea. Familial aggregation of symptoms associated with sleep-related breathing disturbances. Am Rev Respir Dis. 1992;145(2 pt 1):440-444. [DOI] [PubMed] [Google Scholar]
- 48.Strohl KP, Saunders NA, Feldman NT, Hallett M. Obstructive sleep apnea in family members. N Engl J Med. 1978;299(18):969-973. [DOI] [PubMed] [Google Scholar]
- 49.Redline S, Tishler PV, Tosteson TD, et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med. 1995;151(3 Pt 1):682-687. [DOI] [PubMed] [Google Scholar]
- 50.Guilleminault C, Partinen M, Hollman K, Powell N, Stoohs R. Familial aggregates in obstructive sleep apnea syndrome. Chest. 1995;107(6):1545-1551. [DOI] [PubMed] [Google Scholar]
- 51.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325-351. [DOI] [PubMed] [Google Scholar]
- 52.Silventoinen K, Kaprio J. Genetics of tracking of body mass index from birth to late middle age: evidence from twin and family studies. Obes Facts. 2009;2(3):196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathur R, Douglas NJ. Family studies in patients with the sleep apnea-hypopnea syndrome. Ann Intern Med. 1995;122(3):174-178. [DOI] [PubMed] [Google Scholar]
- 54.Kadotani H, Kadotani T, Young T, et al. Association between apolipoprotein E epsilon4 and sleep-disordered breathing in adults. JAMA. 2001;285(22):2888-2890. [DOI] [PubMed] [Google Scholar]
- 55.Gottlieb DJ, DeStefano AL, Foley DJ, et al. APOE epsilon4 is associated with obstructive sleep apnea/hypopnea: the Sleep Heart Health Study. Neurology. 2004;63(4):664-668. [DOI] [PubMed] [Google Scholar]
- 56.Varvarigou V, Dahabreh IJ, Malhotra A, Kales SN. A review of genetic association studies of obstructive sleep apnea: field synopsis and meta-analysis. Sleep. 2011;34(11):1461-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thakre TP, Mamtani MR, Kulkarni H. Lack of association of the APOE epsilon 4 allele with the risk of obstructive sleep apnea: meta-analysis and meta-regression. Sleep. 2009;32(11):1507-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riha RL, Brander P, Vennelle M, et al. Tumour necrosis factor-alpha (-308) gene polymorphism in obstructive sleep apnoea-hypopnoea syndrome. Eur Respir J. 2005;26(4):673-678. [DOI] [PubMed] [Google Scholar]
- 59.Khalyfa A, Serpero LD, Kheirandish-Gozal L, Capdevila OS, Gozal D. TNF-α gene polymorphisms and excessive daytime sleepiness in pediatric obstructive sleep apnea. J Pediatr. 2011;158(1):77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin L, Finn L, Zhang J, Young T, Mignot E. Angiotensin-converting enzyme, sleep-disordered breathing, and hypertension. Am J Respir Crit Care Med. 2004;170(12):1349-1353. [DOI] [PubMed] [Google Scholar]
- 61.Patel SR, Larkin EK, Mignot E, Lin L, Redline S. The association of angiotensin converting enzyme (ACE) polymorphisms with sleep apnea and hypertension. Sleep. 2007;30(4):531-533. [DOI] [PubMed] [Google Scholar]
- 62.Healy F, Marcus CL. Congenital central hypoventilation syndrome in children. Paediatr Respir Rev. 2011;12(4):253-263. Epub Apr 16, 2011. [DOI] [PubMed] [Google Scholar]
- 63.Lugaresi E, Medori R, Montagna P, et al. Fatal familial insomnia and dysautonomia with selective degeneration of thalamic nuclei. N Engl J Med. 1986;315(16):997-1003. [DOI] [PubMed] [Google Scholar]
- 64.Cortelli P, Gambetti P, Montagna P, Lugaresi E. Fatal familial insomnia: clinical features and molecular genetics. J Sleep Res. 1999;8(Suppl 1):23-29. [DOI] [PubMed] [Google Scholar]
- 65.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291(5506):1040-1043. [DOI] [PubMed] [Google Scholar]
- 66.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptácek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128(1):59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10(3):199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krueger JM. The role of cytokines in sleep regulation. Curr Pharm Des. 2008;14(32):3408-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clinton JM, Davis CJ, Zielinski MR, Jewett KA, Krueger JM. Biochemical regulation of sleep and sleep biomarkers. J Clin Sleep Med. 2011;7(5)(Suppl):S38-S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Obal F, Jr, Krueger JM. Biochemical regulation of non-rapid-eye-movement sleep. Front Biosci. 2003;8:d520-d550. [DOI] [PubMed] [Google Scholar]
- 71.Krueger JM, Rector DM, Churchill L. Sleep and cytokines. Sleep Med Clin. 2007;2(2):161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Majde JA, Krueger JM. Links between the innate immune system and sleep. J Allergy Clin Immunol. 2005;116(6):1188-1198. [DOI] [PubMed] [Google Scholar]
- 73.Vgontzas AN, Zoumakis E, Lin HM, Bixler EO, Trakada G, Chrousos GP. Marked decrease in sleepiness in patients with sleep apnea by etanercept, a tumor necrosis factor-alpha antagonist. J Clin Endocrinol Metab. 2004;89(9):4409-4413. [DOI] [PubMed] [Google Scholar]
- 74.Vgontzas AN, Bixler EO, Lin HM, Prolo P, Trakada G, Chrousos GP. IL-6 and its circadian secretion in humans. Neuroimmunomodulation. 2005;12(3):131-140. [DOI] [PubMed] [Google Scholar]
- 75.Gudewill S, Pollmächer T, Vedder H, Schreiber W, Fassbender K, Holsboer F. Nocturnal plasma levels of cytokines in healthy men. Eur Arch Psychiatry Clin Neurosci. 1992;242(1):53-56. [DOI] [PubMed] [Google Scholar]
- 76.Faraut B, Boudjeltia KZ, Vanhamme L, Kerkhofs M. Immune, inflammatory and cardiovascular consequences of sleep restriction and recovery. Sleep Med Rev. 2012;16(2):137-149. [DOI] [PubMed] [Google Scholar]
- 77.Lange T, Dimitrov S, Born J. Effects of sleep and circadian rhythm on the human immune system. Ann N Y Acad Sci. 2010;1193:48-59. [DOI] [PubMed] [Google Scholar]
- 78.Irwin M, McClintick J, Costlow C, Fortner M, White J, Gillin JC. Partial night sleep deprivation reduces natural killer and cellular immune responses in humans. FASEB J. 1996;10(5):643-653. [DOI] [PubMed] [Google Scholar]
- 79.Patel SR, Malhotra A, Gao X, Hu FB, Neuman MI, Fawzi WW. A prospective study of sleep duration and pneumonia risk in women. Sleep. 2012;35(1):97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lange T, Perras B, Fehm HL, Born J. Sleep enhances the human antibody response to hepatitis A vaccination. Psychosom Med. 2003;65(5):831-835. [DOI] [PubMed] [Google Scholar]
- 81.Prather AA, Hall M, Fury JM, et al. Sleep and antibody response to hepatitis B vaccination. Sleep. 2012;35(8):1063-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spiegel K, Sheridan JF, Van Cauter E. Effect of sleep deprivation on response to immunization. JAMA. 2002;288(12):1471-1472. [DOI] [PubMed] [Google Scholar]
- 83.Lowrey PL, Shimomura K, Antoch MP, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288(5465):483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.HUGO International Gene Nomenclature Committee database. Human Genome Organisation website. http://www.genenames.org/. Accessed May 2, 2012.
- 85.Allada R. Genetics of sleep in a simple model organism: Drosophila. In: Kryger MH, Roth T, Dement WC, ed. Principles and Practice of Sleep Medicine. 5th ed. St. Louis, MO: Elsevier Saunders; 2011:151-160. [Google Scholar]