Abstract
Germline testing for familial cases of myeloid leukemia in adults is becoming more common with the recognition of multiple genetic syndromes predisposing people to bone marrow disease. Currently, Clinical Laboratory Improvement Amendments approved testing exists for several myeloid leukemia predisposition syndromes: familial platelet disorder with propensity to acute myeloid leukemia (FPD/AML), caused by mutations in RUNX1; familial AML with mutated CEBPA; familial myelodysplastic syndrome and acute leukemia with mutated GATA2; and the inherited bone marrow failure syndromes, including dyskeratosis congenita, a disease of abnormal telomere maintenance. With the recognition of additional families with a genetic component to their leukemia, new predisposition alleles will likely be identified. We highlight how to recognize and manage these cases as well as outline the characteristics of the major known syndromes. We look forward to future research increasing our understanding of the scope of inherited myeloid leukemia syndromes.
Keywords: familial leukemia predisposition, RUNX1, CEBPA, GATA2, dyskeratosis congenita, Fanconi anemia
Introduction
Familial cases of adult myelodysplastic syndrome and acute myeloid leukemia (MDS/AML) are considered rare, but are likely to be more common than currently appreciated. Understanding the recognized syndromes is critical for clinicians to have a high index of suspicion. Familial MDS/AML can be divided into three groups: those for which Clinical Laboratory Improvement Amendments (CLIA) approved testing exists, those emerging from basic research and requiring validation in additional families or development of clinical testing, and those without an identified genetic basis. Examples of familial MDS/AML syndromes for which clinical testing is available include: familial platelet disorder (FPD) with propensity to AML, caused by mutations in RUNX1; familial AML with mutated CEBPA; familial MDS and acute leukemia (AL) with mutated GATA2; and the inherited bone marrow failure (IBMF) syndromes, including dyskeratosis congenita (DC), a disease of abnormal telomere maintenance, and Fanconi anemia (FA) (Figures 1 and 2). Clinical care of adult patients with a suspected inherited predisposition to MDS/AML is complicated by a paucity of clinical guidelines for both testing and clinical management of patients found to have a predisposing mutation. Recently, our group collaborated with others in the field to begin formulating such recommendations for adult patients [Churpek et al. 2013]. For recommendations regarding pediatric patients, please refer to Seif [Seif, 2011]. In this review, we discuss the current understanding of the genetic basis and clinical presentation of the known adult familial MDS/AML syndromes, and propose guidelines to determine when genetic testing is indicated.
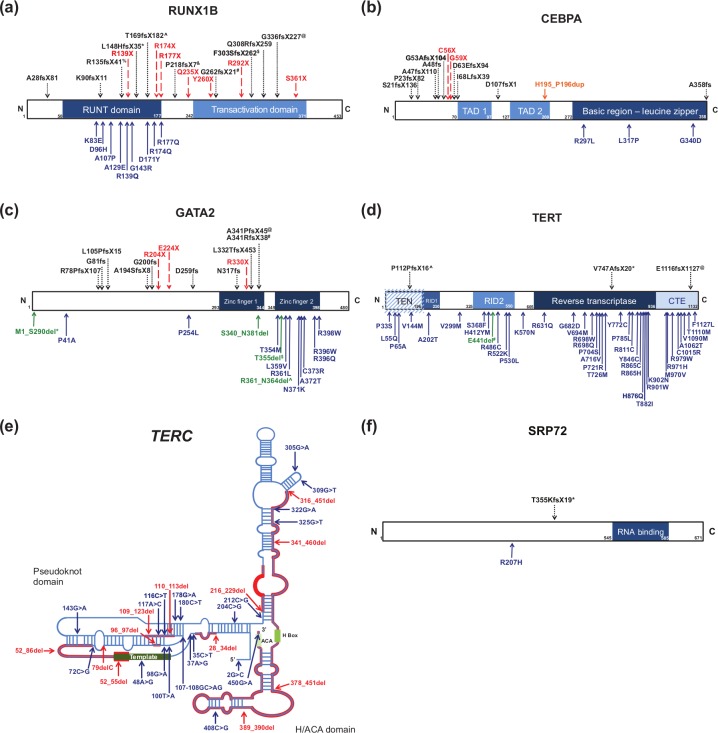
Figure 1.
Missense, nonsense, frameshift, duplication, and deletion mutations that confer familial predisposition to inherited leukemias. Missense mutations are shown in blue; nonsense mutations in red; frameshift mutations in black; duplications in orange; and deletions in green. Superscripts used for the deletion mutations delineate those shown in greater detail in Figure 2. (a) Protein schematic of RUNX1 isoform B (NP_001001890.1) (adapted from Owen et al. [2008b]). (b) Protein schematic of CEBPA (NP_004355.2) (adapted from Ho et al. [2009]). (c) Protein schematic of GATA2 (NP_116027.2) (adapted from Greif et al. [2012]). (d) Protein schematic of TERT (NP_937983.2) (adapted from Wyatt et al. [2009]). (e) Predicted RNA secondary structure of TERC RNA (NR_001566.1) (adapted from Vulliamy and Dokal [2008]).(f) Protein schematic of SRP72 protein (NP_008878.3) (adapted from Iakhiaeva et al. [2010]). CTE, C-terminal extension; RID, RNA interaction domain; TAD, transactivation domain; TEN, TERT essential N-terminal domain.
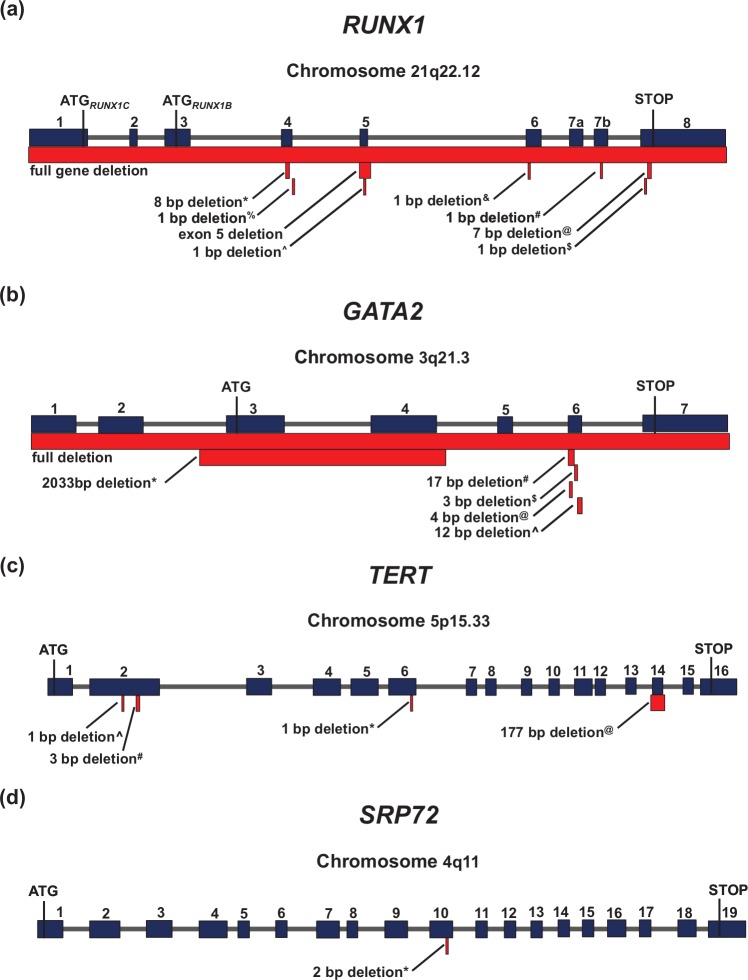
Figure 2.
Detailed schematic diagrams of the deletion mutations that confer familial predisposition to inherited leukemias. The genomic location of each gene is listed under the gene names. Superscripts for each deletion mutation corresponds to that used in Figure 1. (a) Genomic structure of RUNX1 demonstrating locations of disease-causing deletions. ATG1, the first start codon, corresponds to full-length RUNX1 isoform C (NP_001116079.1), and ATG2, the second start codon, corresponds to RUNX1 isoform B (NP_ 001001890.1). (b) Genomic structure of GATA2 demonstrating locations of disease-causing deletions. (c) Genomic structure of TERT demonstrating locations of disease-causing deletions. (d) Genomic structure of SRP72 demonstrating locations of disease-causing deletions. bp, base pair.
How to recognize and manage patients with a familial myeloid leukemia syndrome
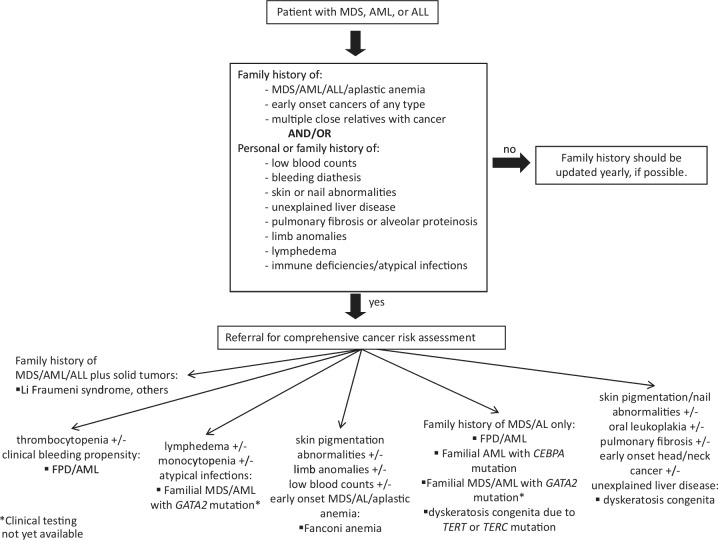
To recognize which patients may have an inherited predisposition to myeloid leukemia, clinicians must take a complete family history and be familiar with the characteristics of familial MDS/AML syndromes. Certain features of the history or laboratory values may increase the likelihood of a particular syndrome over others in individual cases. In the next section, we review the clinical pathological characteristics of each syndrome. Figure 3 shows a recently published model for detection of leukemia predisposition syndromes [Churpek et al. 2013]. Consultation with a certified genetic counselor and the documentation of a complete family history are integral to this assessment.
Figure 3.
Guidelines for the clinical detection of familial myeloid leukemia predisposition syndromes. This figure is reprinted with permission from Churpek et al. [2013]. AL, acute leukemia; ALL, acute lymphoblastic leukemia; AML; acute myeloid leukemia; FPD, familial platelet disorder; MDS, myelodysplastic syndrome.
The assembly of a family pedigree aids in developing a differential diagnosis to determine the most likely genetic syndrome in the family, allowing construction of a genetic testing plan appropriate for the patient’s clinical situation. Patients under consideration for allogeneic stem-cell transplantation with a human leukocyte antigen (HLA)-matched relative warrant expedited genetic testing to rule out a familial MDS/AL predisposition syndrome and avoid use of stem cells from a relative carrying a mutation in the same predisposition gene. For these cases, testing of multiple genes at once, rather than sequential testing, is appropriate. During pretesting genetic counseling, patients and their families should receive education on the principles of human genetics and details about what is known about the specific hereditary syndromes being considered in their case. This information forms the basis of the informed consent process for genetic testing, ensuring that patients understand the risks and benefits of genetic testing, the possible testing outcomes, alternatives to genetic testing (such as screening tests without testing), and the potential impact of test results on patients and their families.
When genetic testing is sent for patients with MDS/AML, it is important to remember that blood is an affected tissue, and many of the genes mutated in the germline are also found mutated somatically, confounding the interpretation of genetic testing on blood or bone marrow samples. For this reason, we advocate genetic testing using skin fibroblasts for germline analysis. In practice, a skin biopsy is easily performed with minimal local anesthetic and a punch biopsy or a small shaving of skin collected at the site of the skin incision during a bone marrow biopsy. Skin fibroblasts are then grown in vitro in a CLIA-approved laboratory. In our experience, growing skin fibroblasts in vitro takes a minimum of 2 weeks, but more often up to 4 weeks, and in some laboratories even longer, all depending on culture conditions and biopsy handling. This is a significant length of time that is added to the turnaround time for germline testing, and in certain clinical settings may not be acceptable when cases require expedited results for patient management decisions. In this situation, buccal swab or saliva samples, which can be obtained immediately, may be preferred. However, both buccal swab and saliva samples contain up to 50% contaminating lymphocytes, possibly confounding results.
All individuals who undergo genetic testing should have a follow-up plan for disclosure of genetic test results when they become available. A disclosure visit for either negative or positive results should ideally occur in a face-to-face setting and include post-testing genetic counseling focused on educating patients about the meaning of their test results, any additional testing if needed, the implications of the results for the patient’s health, and any screening or management recommendations based on the results or the family history.
In the event of a positive genetic test result, it is important to discuss the implications of these results for other at-risk family members and to inform patients of their duty to share this information with at-risk family members. Individual genetic counseling would also be recommended for these family members. Additional health implications should be discussed, such as the risk of nonhematologic malignancies associated with certain germline disorders, including telomere biology disorders (TBDs) or FA, as well as the implications for treatment sensitivity and the possibility of alternative treatment protocols for these individuals. All mutation carriers should be counseled to avoid exposure to known bone marrow toxins, such as smoking and heavy alcohol use. Other genetic issues, including recurrence risk and preimplantation genetic testing for potential parents, should be addressed [Churpek et al. 2013].
We recommend that all mutation carriers undergo a baseline bone marrow biopsy to assess for occult malignancy, as well as twice annual physical examinations and complete blood count (CBC) with differential testing. If there is a significant change from baseline, the CBC should be repeated 1–2 weeks later. If the change persists, a bone marrow evaluation should be repeated. If a mutation carrier develops a bone marrow malignancy, we recommend against using stem cells from any donor who also carries the familial MDS/AML predisposition mutation, since poor outcomes including poor engraftment, graft failure, and donor-derived leukemia have been reported [Buijs et al. 2001; Fogarty et al. 2003; Owen et al. 2008b]. For families with a clinical history consistent with a predisposition to MDS/AML, but who do not carry a mutation in one of the defined familial MDS/AML predisposition genes, the treating physician should consider the strength of the confirmed family history as well as the availability of unrelated donors and the urgency of the transplant when deciding whether or not to use an HLA-matched related donor.
In addition to genetic testing, phenotypic assays exist for the detection of some of the familial MDS/AML predisposition syndromes and are sometimes the preferred manner to detect underlying germline disorders. Telomere length testing detects TBDs [Alter et al. 2012], including DC, for which genetic testing detects abnormalities in about 60% of patients [Walne et al. 2013; Young, 2012]. The diagnosis of FA is made through the diepoxybutane (DEB) breakage assay [Auerbach, 2003; Kee and D’Andrea, 2012]. When indicated, these phenotypic assays can be followed by additional testing to determine the precise genetic mutation present in most cases.
Known causes of familial myeloid leukemia with available clinical testing
Familial platelet disorder with propensity to acute myelogenous leukemia (germline RUNX1 mutations; OMIM 601399; RUNX1 is composed of nine exons on chromosome 21q22.12)
Case 1
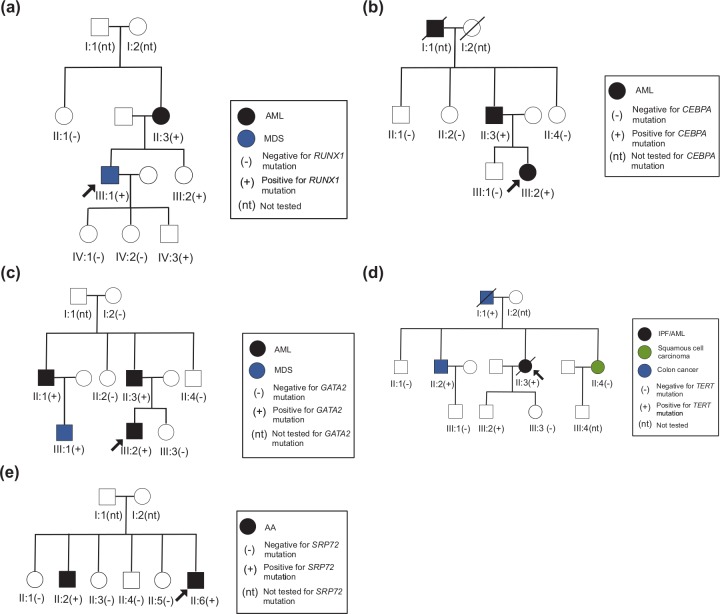
The proband was initially diagnosed with 5q– syndrome at the age of 37, with a history of thrombocytopenia since age 4 (Figure 4(a)). A complete family history revealed that the proband’s mother had developed a normal karyotype AML at the age of 49. The proband’s sister was noted to have a history of extensive bleeding following minor trauma. Germline RUNX1 testing revealed a nonsense mutation in the proband, the proband’s mother, sister, and daughter, who at age 15, demonstrated no clinical signs of bleeding disorders.
Figure 4.
Pedigrees for case studies of families with germline mutations in (a) RUNX1, (b) CEBPA, (c) GATA2, (d) TERT, and (e) SRP72. AA, aplastic anemia; AML; acute myeloid leukemia; IPF, idiopathic pulmonary fibrosis; MDS, myelodysplastic syndrome.
Background
FPD/AML is a rare Mendelian disorder characterized by clinical bleeding due to platelet dysfunction, mild to moderate thrombocytopenia, and a propensity to develop myeloid malignancies. Among the more than 30 families described with FPD/AML (Figures 1 and 2), most carriers have a hemizygous mutation in the RUNX1 gene, which encodes one subunit of a heterodimeric transcription factor that controls genes essential for hematopoiesis. Within individual FPD/AML families, carriers of the same RUNX1 mutation display heterogeneity in their degree of platelet dysfunction, with some family members having moderate thrombocytopenia and bleeding, whereas others have neither. In addition, some family members develop myeloid malignancies, although not always the same type, and other mutation carriers do not succumb to leukemia. Furthermore, different FPD/AML families have varying risks of progressing to myeloid malignancy, which likely reflects the fact that each family carries unique mutations that disrupt various domains within the protein.
As more families have been examined, a spectrum of RUNX1 mutations has been identified (Figure 1(a)), and as noted above, the molecular characteristics of different mutations may account for some of the varying risk of leukemia. Currently, the reported RUNX1 mutations resulting in FPD/AML include gene deletions or duplications, frameshift and nonsense mutations throughout the gene, and missense point mutations within the highly conserved RUNT domain at residues important for DNA binding or heterodimerization. Truncating mutations within the RUNT domain result in hypomorphic alleles, whereas those that truncate the protein at the extreme C terminus activate the baseline transcriptional activity of the protein. Although biochemically these mutations have opposing effects, interestingly they result in the same clinical syndrome [Churpek et al. 2010]. The mechanisms that underlie progression to leukemia in RUNX1 mutation carriers remain to be discovered, but proposed mechanisms include haploinsufficiency for tumor suppression, dominant-negative effects on normal RUNX1 function, acquisition of a de novo mutation in the nonmutated germline allele, and acquisition of cooperating mutations. Germline testing on individuals with suspected RUNX1 mutations should include tests sensitive to deletions, duplications, and rearrangements, which may go undetected by standard sequencing techniques [Jongmans et al. 2010; Katzaki et al. 2010; Shinawi et al. 2008]. These alterations may be detected through genomic arrays or multiplex ligation-probe dependent amplification analysis. Microdeletions of chromosome 21q22, a region containing RUNX1, are associated with growth and developmental delay, dysmorphic features, congenital heart defects, and platelet abnormalities with predisposition to MDS/AML [Izumi et al. 2012; Katzaki et al. 2010; Melis et al. 2011].
Presentation
As described above, FPD/AML families may be identified by the presence of qualitative and quantitative platelet defects in a family with multiple individuals who have developed myeloid leukemias. Of note, T-cell acute lymphoblastic leukemia and one case of lymphoma have also been described within these families. The variability seen within and among families makes this syndrome particularly difficult to diagnose. Not all families or family members demonstrate a low platelet count, and a clinical bleeding history can be difficult to elicit. For this reason, we recommend testing individuals for germline RUNX1 mutations if more than two individuals in the family have been diagnosed with a myeloid malignancy, in particular when at least one individual is thrombocytopenic [Churpek et al. 2013; Owen et al. 2008b].
Management
The management of a patient with a germline RUNX1 mutation and a bone marrow derived malignancy is dictated by the nature of the leukemia. However, the diagnosis of an underlying germline mutation raises the issue of using an allogeneic stem-cell transplant for consolidation therapy, since transplant is the only way to rid the bone marrow of the underlying predisposing allele. In this case, as we describe above, great care must be given to performing RUNX1 mutation testing in HLA-matched relatives in a timely manner, and if none exists, to the use of an unrelated donor.
It is much more difficult to manage RUNX1 mutation carriers who have not developed myeloid malignancies, since clinical guidelines for following these patients have only been discussed recently [Churpek et al. 2013]. Because FPD/AML displays strong anticipation (the phenomenon in which members of younger generations present with disease at earlier ages than those of prior generations), close clinical follow up for members of the youngest generations in the family is particularly critical. Generally, we recommend performing a baseline CBC with differential and a bone marrow biopsy. It is also prudent to screen all first-degree relatives to identify those who are HLA identical before malignancy is diagnosed, since this information will expedite the clinical management of the affected individual. We recommend performing a CBC yearly, and repeating a bone marrow biopsy if there is any significant change in the peripheral blood counts.
Familial acute myelogenous leukemia with mutated CEBPA (OMIM 116897; CEBPA is composed of one exon on chromosome 19q13.11)
Case 2
The proband was initially diagnosed with AML at age 22 (Figure 4(b)). The proband’s father had been diagnosed with AML at age 52, and her grandfather had died of AML in his 70s. There was no history of unusual bleeding in any family member, and no one remembered having been told of any abnormal blood counts prior to their diagnoses of AML. Genetic testing revealed a germline CEBPA mutation.
Background
This syndrome is distinguished by a germline mutation in CEBPA (Figures 1 and 2), a gene also found mutated sporadically in AML. The familial form is associated with bi-allelic CEBPA mutation, generally with the first mutation present in the germline within the 5’ end of the gene, accompanied by acquisition of a second 3’ mutation within the leukemia. Germline 3’ CEBPA mutations have also been identified [Taskesen et al. 2011; Udani et al. 2012]. Since CEBPA mutations confer a relatively favorable prognosis, CEBPA mutation testing is becoming routine in leukemia diagnosis, and patients found to have biallelic CEBPA mutations within their leukemic cells should be tested for germline mutations [Pabst and Mueller, 2009; Renneville et al. 2009].
Familial AML with mutated CEBPA is inherited in an autosomal dominant fashion and displays complete or near-complete penetrance for development of AML [Owen et al. 2008a; Pabst et al. 2009; Renneville et al. 2008], meaning almost all patients inheriting the mutated gene go on to develop AML.
Presentation
No specific genotype–phenotype correlations have been described with germline CEBPA mutations, and most patients’ leukemias have a normal karyotype. AML with a germline CEBPA mutation and a normal karyotype confers a favorable prognosis, with overall survival in the 50–65% range compared with 25–40% in normal-karyotype AML without a germline CEBPA mutation [Bienz et al. 2005; Fröhling et al. 2004; Marcucci et al. 2008; Preudhomme et al. 2002]. Approximately 9% of patients with AML and 15–18% of patients with AML with normal karyotype have either a germline, or more commonly, somatic CEBPA mutation [Renneville et al. 2008]. In one series, 18 of 187 consecutive patients presenting with AML were found to have CEBPA mutations, two of which were germline. Both of the patients with germline CEBPA mutations also had a family history of AML [Pabst et al. 2008]. The familial and sporadic forms of the disease share similar pathologic features, including normal cytogenetic analysis, a preponderance of FAB subtypes M1 and M2, numerous Auer rods seen in peripheral blood or bone marrow blasts, and aberrant CD7 expression on blasts demonstrated by flow cytometry. Genetic testing of the single exon comprising CEBPA is clinically available, with greater than 99% sensitivity for mutations within the coding region. No germline mutations causing familial AML with mutated CEBPA have been reported outside the coding region.
Management
Management of cases of familial AML with mutated CEBPA may include the use of allogeneic stem-cell transplant, being the only modality capable of replacing the mutated allele within the bone marrow [Stelljes et al. 2011]. However, given the relative favorable prognosis of familial CEBPA AML, the risks of allogeneic stem cell-transplant must be considered on an individual basis. Of note, patients with familial CEBPA-mutated AML may be at increased risk of developing additional malignant clones after cure of their initial leukemia, often with acquired CEBPA mutations distinctive from those found in the original leukemia [Pabst et al. 2008]. Therefore, both longer and more frequent post-cure surveillance may be appropriate in patients with the familial form of disease.
Genetic counseling is a critical component of management since penetrance is nearly complete, as discussed above. Although thus far all reported patients also had an affected parent, de novo mutations in a proband and early death of a parent from other causes prior to development of AML could potentially confound diagnosis. In a proband with an apparent de novo germline mutation, confirmatory evaluation of parents should be done, including CBC, hematologic indices, peripheral blood smear and testing for the germline CEBPA mutation identified in the proband. Siblings of a proband with a germline CEBPA mutation are at 50% risk of carrying the mutation. Even when parents are clinically unaffected, siblings of a proband are still at increased risk due to the possibility of incomplete penetrance of a germline mutation in a parent or germline mosaicism in a parent (causing fewer than 50% of the germ cells to carry the deleterious mutation). Children of a proband each have a 50% risk of inheriting the mutation. Genetic counseling should be offered to individuals with known familial AML with mutated CEBPA ideally before pregnancy to determine genetic risk in their offspring [Churpek et al. 2013].
Familial myelodysplastic syndrome/acute myelogenous leukemia with mutated GATA2 (OMIM 137295; GATA2 is comprised of seven exons on chromosome 3q21.3)
Case 3
The proband was initially diagnosed with AML containing del(7q) at age 27 (Figure 4(c)). A complete family history identified AML in the proband’s father and paternal uncle. The father’s AML was diagnosed at the same hospital and had also contained a deletion of chromosome 7. The paternal uncle’s AML had been diagnosed elsewhere, and details regarding the cytogenetic analysis of this AML were not available. While genetic testing of the proband was underway, the son of the paternal uncle was diagnosed with MDS. Germline testing of the proband identified a GATA2 mutation. Our genetic counselor advised genetic testing of other family members who had developed hematopoietic malignancies or were at risk of inheritance of the deleterious mutation.
Background
GATA2 encodes a zinc finger transcription factor critical for normal hematopoiesis [Rodrigues et al. 2005; Tsai et al. 1994] and lymphatic vascular development [Kazenwadel et al. 2012]. Germline mutations in GATA2 have been described in association with familial MDS/AML, as well as with several heterogeneous clinical syndromes, including Emberger syndrome and the MonoMAC syndrome (Figures 1 and 2). Interestingly, GATA2 mutation has been described in association with at least one case that combines phenotypic features of the Emberger and MonoMAC syndromes [Ishida et al. 2012]. These syndromes lead to an overall increased risk of developing MDS/AML.
Presentation
Several GATA2 mutation-carrying pedigrees have been described that display no distinguishing phenotypic abnormalities other than early-onset familial MDS or AML [Hahn et al. 2011]. These families displayed highly penetrant autosomal dominant inheritance of early-onset MDS or AML, resulting in poor outcomes unless successfully transplanted. The clinical characteristics of MDS/AML in these pedigrees is variable, with different FAB subtypes as well as variable cytogenetic abnormalities, including monosomy 7, trisomy 8, and trisomy 21.
Emberger syndrome is defined by primary lymphedema, confined to the lower extremities and genitals, with myelodysplasia progressing to AML. It may also present with a low CD4/CD8 T-cell ratio, cutaneous warts, and sensorineural deafness. Emberger syndrome occurs both sporadically and heritably in an autosomal dominant fashion with incomplete penetrance. One series identified eight independent GATA2 variants in 14 individuals with Emberger syndrome [Ostergaard et al. 2011]. The cytogenetic abnormalities seen in these individuals also include monosomy 7, and their MDS often rapidly transforms to AML.
Recently described, the MonoMAC syndrome comprises severe monocytopenia and severe infections with nontuberculous Mycobacteria, typically M. avium complex (MAC). The phenotype may also include natural killer cell and B-cell lymphocytopenia, disseminated viral and opportunistic fungal infections, pulmonary alveolar proteinosis, and severely decreased circulating and tissue dendritic cells, as well as a predisposition to MDS/AML or chronic myelomonocytic leukemia [Vinh et al. 2010]. This syndrome has been associated with mutations in GATA2 in at least 20 patients so far. In these patients, the infectious and pulmonary features of MonoMAC syndrome tend to predate the development of overt MDS by many years and are thought to arise from tissue macrophage dysfunction mediated by GATA2 deficiency.
Compared with patients with de novo MDS, patients with MonoMAC display a significantly younger average age of onset (33 years versus 70–80 years) as well as distinctive bone marrow features, such as hypocellularity, significant fibrosis, and multilineage dysplasia [Hsu et al. 2011]. Common cytogenetic abnormalities include monosomy 7, trisomy 8, and trisomy 1q.
Diagnosis
Genetic sequencing of the entire coding region of GATA2 is clinically available. Given the existence of pure familial MDS/AML with mutated GATA2, this test should be considered in all patients being evaluated for a familial MDS/AML syndrome, regardless of the presence or absence of any of the phenotypic features noted above.
Management
As described above for familial CEBPA mutations, management of an individual’s AML is typically similar to that for AML presenting in the sporadic setting, although special consideration for the use of allogeneic stem-cell transplant should be entertained, given the germline predisposition.
Inherited bone marrow failure syndromes and telomere biology disorders
Case 4
The proband initially presented at the medical center with idiopathic pulmonary fibrosis (IPF) and 2 years later was found to be pancytopenic (Figure 4(d)). She developed acute promyelocytic leukemia 6 months later. The proband’s family history was significant for cancer, including squamous cell nasopharyngeal cancer diagnosed in the proband’s sister at age 45. Age-adjusted telomere length testing of the proband’s peripheral blood cells revealed telomere lengths below the first percentile in all cell types tested. Germline DNA from the proband showed a mutation in TERT. Further germline testing of the father, brother, and sister with malignancies demonstrated the same TERT mutation and age-adjusted telomere lengths below the first percentile.
Background
Within the spectrum of IBMF syndromes, TBDs are associated with abnormal telomere maintenance and predisposition to MDS/AML. DC is the prototypical example of a TBD. Classic features of DC include the mucocutaneous triad of nail dystrophy, abnormal reticular skin pigmentation, and oral leukoplakia, as well as a predisposition to malignancy [Dokal, 2011]. Patients with DC have a high risk of MDS, with an observed expected ratio of 2663 (95% confidence interval 858–6215) and a mean age of onset of 35 years [Alter et al. 2009]. Not all patients with a TBD demonstrate the classic features of DC, since both the genetic causes and clinical presentations of TBDs are heterogeneous. Patients may present initially with bone marrow failure, MDS, or pulmonary fibrosis, without demonstrating mucocutaneous features of the disease [Armanios et al. 2007; Yamaguchi et al. 2003]. Bone marrow failure is the leading cause of death in affected patients [Dokal, 2011].
TBDs result from mutations in at least nine genes in three inheritance patterns. X-linked recessive DC (XLR-DC, OMIM #305000) is associated with mutations in DKC1 [Heiss et al. 1998]. Autosomal recessive DC (AR-DC, OMIM #224230) results from mutations in NOP10, TERT, NPH2, TCAB1 (also known as WRAP53), C16orf57, and RTEL1 [Dokal, 2011; Walne et al. 2013]. Autosomal dominant DC (AD-DC, OMIM #127550) results from mutations in TERT (Figure 1(d)), TERC (Figure 1(e)), TINF2, and RTEL1 [Ballew et al. 2013; Savage et al. 2008; Vulliamy et al. 2001]. Heterozygous mutations in the telomerase reverse transcriptase TERT and RNA-component-encoding gene TERC may present as familial MDS/AL predisposition syndromes [Kirwan et al. 2009]. Of note, biallelic mutations in TERT resulting in AR-DC are generally more severe than mono-allelic TERT mutations leading to AD-DC, with the presence of mucocutaneous features of the disease and greatly reduced telomeric repeat amplification protocol activity, which measures telomerase activity [Marrone et al. 2007]. Mutations in TINF2, which encodes the shelterin complex protein TIN2, lead to extremely short telomeres. TINF2 mutations, which are generally de novo, typically lead to severe DC or other genetic syndromes, with high penetrance and an early age of onset [Savage et al. 2008; Walne et al. 2008]. Recently, mutations in RTEL1, a gene encoding a telomere-associated elongation helicase, were identified in two families with Hoyeraal Hreidarsson syndrome, a clinically severe form of DC associated with cerebellar hypoplasia, immunodeficiency, enteropathy, and intrauterine growth retardation [Ballew et al. 2013].
Presentation
TBDs affect regenerating tissues in which telomere maintenance is important. System-wide clinical features seen in TBDs include IPF, seen in 20.3% of patients; extensive dental caries, 16.9%; esophageal stricture, 16.9%; premature hair loss or graying, 16.1%; and liver disease, 7.1%, including predisposition to cirrhosis [Calado et al. 2009; Dokal, 2011]. In addition, telomere instability predisposes people to malignancies: MDS/AML and a variety of solid tumors, including head and neck squamous cell carcinoma (SCC), skin SCC, and anorectal, stomach, lung, esophageal, and colon cancer [Alter et al. 2009, 2010].
Although XLR-DC, AR-DC, and TINF2 mutations often present with severe phenotypes at young ages, AD-DC caused by TERT and TERC mutations often present later in life without classic mucocutaneous symptoms. Instead, bone marrow failure is a common presenting symptom. TERT and TERC mutations show variable penetrance and variable disease onset and progression. Carriers of the same TERT or TERC mutation may demonstrate few symptoms, with only slight erythrocyte macrocytosis or thrombocytopenia as the only blood abnormalities prior to the onset of aplastic anemia (AA) [Young, 2012]. Both TERT and TERC mutations are associated with anticipation, with progressively shorter telomeres passed down through generations [Vulliamy et al. 2004]. Members of older generations often demonstrate mild disease, whereas those of younger generations experience more severe disease manifestations, such as AA or MDS/AML [Armanios, 2012; Parry et al. 2011; Vulliamy et al. 2005]. The combination of AA and IPF in patients with the absence of classic mucocutaneous features is highly specific for a TBD [Parry et al. 2011].
Diagnosis
TBDs are best diagnosed through telomere length testing, which correlates with disease severity [Alter et al. 2012]. Telomere length testing can be performed based on flow cytometry separation of specific leukocyte subsets, followed by telomere fluorescence in situ hybridization [Martens et al. 2000], and age-adjusted telomere lengths below the first percentile are diagnostic for a TBD [Alter et al. 2012]. The clinical diagnosis of DC is based on the presence of the major features of the disease, which include the mucocutaneous triad and bone marrow failure, as well as the presence of multisystem features of the disease: epiphora, learning difficulties/developmental delay/mental retardation, pulmonary disease, short stature, extensive dental caries, esophageal stricture, premature hair graying or loss, hyperhiderosis, or malignancy. The clinical diagnosis of DC requires at least two of four major features and at least two multisystem features [Dokal, 2011], though some centers conduct telomere length testing when just one major feature is present to account for the heterogeneity of clinical presentations. DC mutation testing is available clinically, but only about 50% of patients with demonstrably very short telomeres will test positive for a mutation in one of the nine known predisposition genes.
Management
Hematopoietic stem-cell transplantation (HSCT) is the only definitive cure for patients with a TBD. Oxymetholone treatment may improve hematopoietic function in some patients through the upregulation of telomerase [de la Fuente and Dokal, 2007]. Diagnosis of a TBD as the cause of bone marrow failure or malignancy is highly important prior to HSCT, since patients with a TBD are at risk of HSCT complications after use of conventional myeloablative conditioning regimens [Dietz et al. 2011], including increased risk of pulmonary complications and venoocclusive disease [de la Fuente and Dokal, 2007; Dokal, 2011].
Low-intensity myeloablation has been recommended, but has also resulted in post-transplant complications [Brazzola et al. 2005]. Although long-term data are lacking, outcomes after fludarabine-based nonmyeloablative conditioning regimens demonstrated reduced pulmonary and vascular complications and increased survival in allogeneic transplants [Dietz et al. 2011]. In addition, due to anticipation, younger generations may present with more severe disease at earlier ages and should be screened appropriately for signs of disease.
Fanconi anemia
FA is an autosomal or X-linked recessive IBMF syndrome associated with growth retardation, organ malformation, and a predisposition to malignancy, in particular AML but also other solid tumors. The clinical presentation of FA is heterogeneous. Common congenital abnormalities include short stature, abnormal skin pigmentation, radial ray defects, and abnormalities of various organs, including arms, head, eyes, ears, and kidneys [Kee and D’Andrea, 2012; Knies et al. 2012]. Additionally, 25–40% of patients lack physical abnormalities associated with the disease [D’Andrea, 2010]. Although the median age of diagnosis is 6.5 years for boys and 8 years for girls, the disease is diagnosed over a wide range of ages, from birth to 48 years old [D’Andrea, 2010]. The median age of onset of bone marrow failure is 7 years [Butturini et al. 1994]. Median survival for patients with FA is 24 years, with a cumulative incidence of 90% of bone marrow failure by age 40 [Kutler et al. 2003]. At least 20% of patients with FA develop malignancy, with AML the most common diagnosis [Kutler et al. 2003].
Currently, 15 FA genes are associated with the disease [Kee and D’Andrea, 2012], with FANCA demonstrating the highest prevalence [Wang, 2007]. FA genes function in repairing DNA crosslinks associated with the FA/BRCA pathway. Subtyping FA complementation groups is useful in the management of patients with FA, since certain groups, such as FANCA, demonstrate milder disease with later onset of bone marrow failure, whereas FANCG generally results in more severe hematologic disease [Faivre et al. 2000]. FANCD1 is identical to BRCA2 [Wang, 2007], with homozygous mutations resulting in FA and heterozygous mutations leading to an increased susceptibility to breast, ovarian, and pancreatic cancer [Howlett et al. 2002].
Although the clinical presentation and genetic causes are highly variable, in general, FA results in hypersensitivity to DNA crosslinking agents. DEB, a DNA crosslinking agent, is used in the DEB-induced chromosome breakage assay to diagnose FA. Mitomycin C may also be used in place of DEB in the assay. Peripheral blood from a patient with suspected FA is cultured and treated with DEB, which induces irreparable DNA crosslinking and chromosome breakage in FA cells visible in Giemsa-stained metaphase cells but is innocuous to wild type cells [Auerbach, 2003]. HSCT offers the only cure for hematopoietic abnormalities associated with FA [Gluckman et al. 1995].
Additional genes implicated in familial leukemia for which clinical gene mutation testing is not yet available
Familial aplastic anemia/myelodysplastic syndrome with SRP72 mutation (OMIM 602122; SRP72 is composed of 19 exons on chromosome 4q11)
Case 5
The proband presented with pancytopenia at age 20 and a bone marrow biopsy confirmed a diagnosis of AA (Figure 4(e)). Of note, the proband’s brother had also been diagnosed with AA at age 16. Based on the occurrence of two family members with AA, a workup for germline mutations leading to familial bone marrow failures syndromes was undertaken, including telomere length testing (which was normal) and SRP72 mutation testing, which was positive for a mutation when performed on a research basis.
Background
SRP72 encodes one of six protein subunits of the signal recognition particle (SRP), part of the cellular apparatus responsible for nascent protein processing and trafficking. By means of whole-exome sequencing, a mutation in SRP72 was identified in a family with apparent autosomal dominant inheritance of bone marrow failure and congenital neural deafness [Kirwan et al. 2012]. Three siblings in this family had AA and deafness, and an additional sibling with normal hearing had normal blood counts. The mother had MDS. All had normal karyotypes. Subsequent screening of 96 additional patients with bone marrow failure identified one more family in which both the index case and her mother had MDS and an SRP72 mutation. In this family, however, neither affected individual had hearing loss, although the index case presented with possible labyrinthitis [Kirwan et al. 2012].
To date, only these two pedigrees have been identified with SRP72 mutations and AA/MDS (Figures 1 and 2). Thus, it is difficult to describe a particular phenotype associated with this mutation. However, as more patients are screened for the mutation, an association with hearing loss or other audiovestibular abnormalities may become evident, as in the case of GATA2 mutation. As with other familial bone marrow failure syndromes, attention to possible inheritance of seemingly unrelated abnormalities will be important to elucidate the phenotype.
Presentation
Since only a small number of families with SRP72 mutations have been identified to date, it is difficult to describe a consistent presentation, but bone marrow failure may precede the development of a myeloid bone marrow malignancy.
Management
As with the other syndromes, no special initial management of the myeloid malignancy is undertaken. However, subsequently, if a germline SRP72 mutation is found, consideration should be given to the use of allogeneic stem-cell transplant using a nonmutated donor.
Thrombocytopenia 2 (ANKRD26; OMIM 610855; ANKRD26 is composed of 34 exons on chromosome 10p12.1)
Background
Thrombocytopenia 2 (THC2) is an inherited thrombocytopenia with mild thrombocytopenia and bleeding tendency with normal in vitro platelet aggregation [Noris et al. 2011]. In 78 people from 21 families known to have THC2, 18 mutations were identified within a 19 base-pair region of the ANKRD26 5’ untranslated region (Figure 1) [Noris et al. 2011; Pippucci et al. 2011]. Expression studies showed that these mutations resulted in increased gene expression, suggesting a gain-of-function mechanism of action. Among 105 people with confirmed or suspected ANKRD26 mutations, 10 developed hematologic malignancies, including seven with acute leukemias, five of which were myeloid and two of which were undefined [Noris et al. 2011]. The overall incidence of development of hematologic malignancies was 240 out of 100,000, and of acute leukemia was 167 out of 100,000, both elevated over expected levels.
Presentation
To date, patients with ANKRD26 mutations are recognized by their familial inheritance of thrombocytopenia. Although there is a suspicion that these mutations may lead to an increased risk of hematologic disease, additional families need to be studied to be confident about this relationship.
Management
The management of hematologic malignancies in these patients is according to the diagnosis. No additional treatment measures need to be instituted. As discussed above, if a firm causative relationship is established between ANKRD26 mutations and the development of hematopoietic diseases, consideration should be given to the use of allogeneic stem-cell transplant.
Conclusion
In summary, we advocate that all physicians caring for patients with MDS/AML become familiar with the inherited predisposition syndromes underlying the development of the malignancy in certain families. Recognition of these syndromes is crucial to proper clinical management of patients with an inherited susceptibility and for genetic screening of additional family members. Although currently CLIA-approved testing exists only for inherited mutations in RUNX1, CEBPA, GATA2, and the IBMF syndromes including DC, we look forward to the development of additional clinical tests as more inherited syndromes are identified.
We recognize that the anticipated rapid incorporation of next-generation sequencing into clinical practice may change our diagnostic approach, if full exome, transcriptome, and genome sequencing become standard practice [Biesecker et al. 2012; Shyr and Liu, 2013]. For the moment, if physicians exhaust CLIA-approved genetic testing for a patient with a high suspicion for an inherited leukemia syndrome, further testing occurs in the research setting. However, many experimental protocols are now written with exchange of medically relevant information back to participating individuals, allowing patients access to the results of research-based testing. We encourage all physicians who care for these patients to refer patients for appropriate genetic counseling, testing, and if appropriate, research studies that could lead to the identification of additional predisposition alleles.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The authors declare no conflicts of interest in preparing this article.
Contributor Information
Eric M. Nickels, Section of Hematology/Oncology, Department of Medicine, University of Chicago, Chicago, IL, USA
Jesse Soodalter, Section of Hematology/Oncology, Department of Medicine, University of Chicago, Chicago, IL, USA.
Jane E. Churpek, Section of Hematology/Oncology, Department of Medicine, University of Chicago, Center for Clinical Cancer Genetics, University of Chicago, and University of Chicago Comprehensive Cancer Center, University of Chicago, Chicago, IL, USA
Lucy A. Godley, University of Chicago, Chicago, 5841 South Maryland Avenue, MC 2115, Chicago, IL 60637, USA
References
- Alter B., Giri N., Savage S., Peters J., Loud J., Leathwood L., et al. (2010) Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol 150: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter B., Giri N., Savage S., Rosenberg P. (2009) Cancer in dyskeratosis congenita. Blood 113: 6549–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter B., Rosenberg P., Giri N., Baerlocher G., Lansdorp P., Savage S. (2012) Telomere length is associated with disease severity and declines with age in dyskeratosis congenita. Haematologica 97: 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M. (2012) Telomerase and idiopathic pulmonary fibrosis. Mutat Res 730: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M., Chen J., Cogan J., Alder J., Ingersoll R., Markin C., et al. (2007) Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med 356: 1317–1326 [DOI] [PubMed] [Google Scholar]
- Auerbach A. (2003) Diagnosis of Fanconi anemia by diepoxybutane analysis. Curr Protoc Hum Genet Chapter 8: Unit 8 7. [DOI] [PubMed] [Google Scholar]
- Ballew B., Yeager M., Jacobs K., Giri N., Boland J., Burdett L., et al. (2013) Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in dyskeratosis congenita. Hum Genet 132: 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M., Ludwig M., Leibundgut E., Mueller B., Ratschiller D., Solenthaler M., et al. (2005) Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res 11: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Biesecker L., Burke W., Kohane I., Plon S., Zimmern R. (2012) Next-generation sequencing in the clinic: are we ready? Nat Rev Genet 13: 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazzola P., Duval M., Fournet J., Gauvin F., Dalle J., Champagne J., et al. (2005) Fatal diffuse capillaritis after hematopoietic stem-cell transplantation for dyskeratosis congenita despite low-intensity conditioning regimen. Bone Marrow Transplant 36: 1103–1105; author reply 1105. [DOI] [PubMed] [Google Scholar]
- Buijs A., Poddighe P., van Wijk R., van Solinge W., Borst E., Verdonck L., et al. (2001) A novel CBFA2 single-nucleotide mutation in familial platelet disorder with propensity to develop myeloid malignancies. Blood 98: 2856–2858 [DOI] [PubMed] [Google Scholar]
- Butturini A., Gale R., Verlander P., Adler-Brecher B., Gillio A., Auerbach A. (1994) Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood 84: 1650–1655 [PubMed] [Google Scholar]
- Calado R., Regal J., Kleiner D., Schrump D., Peterson N., Pons V., et al. (2009) A spectrum of severe familial liver disorders associate with telomerase mutations. PLoS One 4: e7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churpek J., Garcia J., Madzo J., Jackson S., Onel K., Godley L. (2010) Identification and molecular characterization of a novel 3′ mutation in RUNX1 in a family with familial platelet disorder. Leuk Lymphoma 51: 1931–1935 [DOI] [PubMed] [Google Scholar]
- Churpek J., Lorenz R., Nedumgottil S., Onel K., Olopade O., Sorrell A., et al. (2013) Proposal for the clinical detection and management of patients and their family members with familial myelodysplastic syndrome/acute leukemia predisposition syndromes. Leuk Lymphoma 54: 28–35 [DOI] [PubMed] [Google Scholar]
- D’Andrea A. (2010) Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med 362: 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J., Dokal I. (2007) Dyskeratosis congenita: advances in the understanding of the telomerase defect and the role of stem cell transplantation. Pediatr Transplant 11: 584–594 [DOI] [PubMed] [Google Scholar]
- Dietz A., Orchard P., Baker K., Giller R., Savage S., Alter B., et al. (2011) Disease-specific hematopoietic cell transplantation: nonmyeloablative conditioning regimen for dyskeratosis congenita. Bone Marrow Transplant 46: 98–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I. (2011) Dyskeratosis congenita. Hematology Am Soc Hematol Educ Program 2011: 480–486 [DOI] [PubMed] [Google Scholar]
- Faivre L., Guardiola P., Lewis C., Dokal I., Ebell W., Zatterale A., et al. (2000) Association of complementation group and mutation type with clinical outcome in Fanconi anemia. European Fanconi Anemia Research Group Blood 96: 4064–4070 [PubMed] [Google Scholar]
- Fogarty P., Yamaguchi H., Wiestner A., Baerlocher G., Sloand E., Zeng W., et al. (2003) Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet 362: 1628–1630 [DOI] [PubMed] [Google Scholar]
- Fröhling S., Schlenk R., Stolze I., Bihlmayr J., Benner A., Kreitmeier S., et al. (2004) CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol 22: 624–633 [DOI] [PubMed] [Google Scholar]
- Gluckman E., Auerbach A., Horowitz M., Sobocinski K., Ash R., Bortin M., et al. (1995) Bone marrow transplantation for Fanconi anemia. Blood 86: 2856–2862 [PubMed] [Google Scholar]
- Greif P., Dufour A., Konstandin N., Ksienzyk B., Zellmeier E., Tizazu B., et al. (2012) GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood 120: 395–403 [DOI] [PubMed] [Google Scholar]
- Hahn C., Chong C., Carmichael C., Wilkins E., Brautigan P., Li X., et al. (2011) Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 43: 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss N., Knight S., Vulliamy T., Klauck S., Wiemann S., Mason P., et al. (1998) X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Ho P., Alonzo T., Gerbing R., Pollard J., Stirewalt D., Hurwitz C., et al. (2009) Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood 113: 6558–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett N., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., et al. (2002) Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609 [DOI] [PubMed] [Google Scholar]
- Hsu A., Sampaio E., Khan J., Calvo K., Lemieux J., Patel S., et al. (2011) Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118: 2653–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakhiaeva E., Iakhiaev A., Zwieb C. (2010) Identification of amino acid residues in protein SRP72 required for binding to a kinked 5e motif of the human signal recognition particle RNA. BMC Mol Biol 11: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Imai K., Honma K., Tamura S., Imamura T., Ito M., et al. (2012) GATA-2 anomaly and clinical phenotype of a sporadic case of lymphedema, dendritic cell, monocyte, B- and NK-cell (DCML) deficiency, and myelodysplasia. Eur J Pediatr 171: 1273–1276 [DOI] [PubMed] [Google Scholar]
- Izumi K., Brooks S., Feret H., Zackai E. (2012) 1.9 Mb microdeletion of 21q22.11 within Braddock-Carey contiguous gene deletion syndrome region: dissecting the phenotype. Am J Med Genet A 158A: 1535–1541 [DOI] [PubMed] [Google Scholar]
- Jongmans M., Kuiper R., Carmichael C., Wilkins E., Dors N., Carmagnac A., et al. (2010) Novel RUNX1 mutations in familial platelet disorder with enhanced risk for acute myeloid leukemia: clues for improved identification of the FPD/AML syndrome. Leukemia 24: 242–246 [DOI] [PubMed] [Google Scholar]
- Katzaki E., Morin G., Pollazzon M., Papa F., Buoni S., Hayek J., et al. (2010) Syndromic mental retardation with thrombocytopenia due to 21q22.11q22.12 deletion: Report of three patients. Am J Med Genet A 152A: 1711–1717 [DOI] [PubMed] [Google Scholar]
- Kazenwadel J., Secker G., Liu Y., Rosenfeld J., Wildin R., Cuellar-Rodriguez J., et al. (2012) Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 119: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., D’Andrea A. (2012) Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest 122: 3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan M., Vulliamy T., Marrone A., Walne A., Beswick R., Hillmen P., et al. (2009) Defining the pathogenic role of telomerase mutations in myelodysplastic syndrome and acute myeloid leukemia. Hum Mutat 30: 1567–1573 [DOI] [PubMed] [Google Scholar]
- Kirwan M., Walne A., Plagnol V., Velangi M., Ho A., Hossain U., et al. (2012) Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am J Hum Genet 90: 888–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies K., Schuster B., Ameziane N., Rooimans M., Bettecken T., de Winter J., et al. (2012) Genotyping of Fanconi anemia patients by whole exome sequencing: advantages and challenges. PLoS One 7: e52648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutler D., Singh B., Satagopan J., Batish S., Berwick M., Giampietro P., et al. (2003) A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101: 1249–1256 [DOI] [PubMed] [Google Scholar]
- Marcucci G., Maharry K., Radmacher M., Mrózek K., Vukosavljevic T., Paschka P., et al. (2008) Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol 26: 5078–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A., Walne A., Tamary H., Masunari Y., Kirwan M., Beswick R., et al. (2007) Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood 110: 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens U., Brass V., Engelhardt M., Glaser S., Waller C., Lange W., et al. (2000) Measurement of telomere length in haematopoietic cells using in situ hybridization techniques. Biochem Soc Trans 28: 245–250 [DOI] [PubMed] [Google Scholar]
- Melis D., Genesio R., Cappuccio G., MariaGinocchio V., Casa R., Menna G., et al. (2011) Mental retardation, congenital heart malformation, and myelodysplasia in a patient with a complex chromosomal rearrangement involving the critical region 21q22. Am J Med Genet A 155A: 1697–1705 [DOI] [PubMed] [Google Scholar]
- Noris P., Perrotta S., Seri M., Pecci A., Gnan C., Loffredo G., et al. (2011) Mutations in ANKRD26 are responsible for a frequent form of inherited thrombocytopenia: analysis of 78 patients from 21 families. Blood 117: 6673–6680 [DOI] [PubMed] [Google Scholar]
- Ostergaard P., Simpson M., Connell F., Steward C., Brice G., Woollard W., et al. (2011) Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 43: 929–931 [DOI] [PubMed] [Google Scholar]
- Owen C., Barnett M., Fitzgibbon J. (2008a) Familial myelodysplasia and acute myeloid leukaemia – a review. Br J Haematol 140: 123–132 [DOI] [PubMed] [Google Scholar]
- Owen C., Toze C., Koochin A., Forrest D., Smith C., Stevens J., et al. (2008b) Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 112: 4639–4645 [DOI] [PubMed] [Google Scholar]
- Pabst T., Eyholzer M., Fos J., Mueller B. (2009) Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer 100: 1343–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst T., Eyholzer M., Haefliger S., Schardt J., Mueller B. (2008) Somatic CEBPA mutations are a frequent second event in families with germline CEBPA mutations and familial acute myeloid leukemia. J Clin Oncol 26: 5088–5093 [DOI] [PubMed] [Google Scholar]
- Pabst T., Mueller B. (2009) Complexity of CEBPA dysregulation in human acute myeloid leukemia. Clin Cancer Res 15: 5303–5307 [DOI] [PubMed] [Google Scholar]
- Parry E., Alder J., Qi X., Chen J., Armanios M. (2011) Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood 117: 5607–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippucci T., Savoia A., Perrotta S., Pujol-Moix N., Noris P., Castegnaro G., et al. (2011) Mutations in the 5’ UTR of ANKRD26, the ankirin repeat domain 26 gene, cause an autosomal-dominant form of inherited thrombocytopenia, THC2. Am J Hum Genet 88: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preudhomme C., Sagot C., Boissel N., Cayuela J., Tigaud I., de Botton S., et al. (2002) Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 100: 2717–2723 [DOI] [PubMed] [Google Scholar]
- Renneville A., Mialou V., Philippe N., Kagialis-Girard S., Biggio V., Zabot M.T., et al. (2009) Another pedigree with familial acute myeloid leukemia and germline CEBPA mutation. Leukemia 23(4): 804–806, http://www.ncbi.nlm.nih.gov/pubmed/18946494 [DOI] [PubMed] [Google Scholar]
- Renneville A., Roumier C., Biggio V., Nibourel O., Boissel N., Fenaux P., et al. (2008) Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia 22: 915–931 [DOI] [PubMed] [Google Scholar]
- Rodrigues N., Janzen V., Forkert R., Dombkowski D., Boyd A., Orkin S., et al. (2005) Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106: 477–484 [DOI] [PubMed] [Google Scholar]
- Savage S., Giri N., Baerlocher G., Orr N., Lansdorp P., Alter B. (2008) TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet 82: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif A. (2011) Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis. Cancer Genet 204: 227–244 [DOI] [PubMed] [Google Scholar]
- Shinawi M., Erez A., Shardy D., Lee B., Naeem R., Weissenberger G., et al. (2008) Syndromic thrombocytopenia and predisposition to acute myelogenous leukemia caused by constitutional microdeletions on chromosome 21q. Blood 112: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr D., Liu Q. (2013) Next generation sequencing in cancer research and clinical application. Biol Proced Online 15: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelljes M., Corbacioglu A., Schlenk R., Dohner K., Fruhwald M., Rossig C., et al. (2011) Allogeneic stem cell transplant to eliminate germline mutations in the gene for CCAAT-enhancer-binding protein alpha from hematopoietic cells in a family with AML. Leukemia 25: 1209–1210 [DOI] [PubMed] [Google Scholar]
- Taskesen E., Bullinger L., Corbacioglu A., Sanders M., Erpelinck C., Wouters B., et al. (2011) Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood 117: 2469–2475 [DOI] [PubMed] [Google Scholar]
- Tsai F., Keller G., Kuo F., Weiss M., Chen J., Rosenblatt M., et al. (1994) An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371: 221–226 [DOI] [PubMed] [Google Scholar]
- Udani R., Parlow M., Yin L., Belilissimo D. (2012) Novel germline CEBPA sequence variations in familial AML and cytogenetically normal AML: American Society of Hematology Conference 2012 [Google Scholar]
- Vinh D., Patel S., Uzel G., Anderson V., Freeman A., Olivier K., et al. (2010) Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood 115: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T., Dokal I. (2008) Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex. Biochimie 90: 122–130 [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone A., Goldman F., Dearlove A., Bessler M., Mason P., et al. (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435 [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone A., Szydlo R., Walne A., Mason P., Dokal I. (2004) Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet 36: 447–449 [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Walne A., Baskaradas A., Mason P., Marrone A., Dokal I. (2005) Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol Dis 34: 257–263 [DOI] [PubMed] [Google Scholar]
- Walne A., Vulliamy T., Beswick R., Kirwan M., Dokal I. (2008) TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 112: 3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne A., Vulliamy T., Kirwan M., Plagnol V., Dokal I. (2013) Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet 92: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. (2007) Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8: 735–748 [DOI] [PubMed] [Google Scholar]
- Wyatt H., Tsang A., Lobb D., Beattie T. (2009) Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One 4: e7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Baerlocher G., Lansdorp P., Chanock S., Nunez O., Sloand E., et al. (2003) Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood 102: 916–918 [DOI] [PubMed] [Google Scholar]
- Young N. (2012) Bone marrow failure and the new telomere diseases: practice and research. Hematology 17(Suppl. 1): S18–S21 [DOI] [PubMed] [Google Scholar]