Abstract
While the majority of leukemia cases occur in the absence of any known predisposing factor, there are germline mutations that significantly increase the risk of developing hematopoietic malignancies in childhood. In this review article, we describe a number of these mutations and their clinical features. These predispositions can be broadly classified as those leading to bone marrow failure, those involving tumor suppressor genes, DNA repair defects, immunodeficiencies or other congenital syndromes associated with transient myeloid disorders. While leukemia can develop as a secondary event in the aforementioned syndromes, there are also several syndromes that specifically lead to the development of leukemia as their primary phenotype. Many of the genes discussed in this review can also be somatically mutated in other cancers, highlighting the importance of understanding shared alterations and mechanisms underpinning syndromic and sporadic leukemia.
Keywords: genetic predisposition, germline, hematologic malignancy, leukemia, oncogene, cancer syndrome, tumor suppressor
Introduction
The risk of developing childhood leukemia is significantly increased in the setting of germline genetic mutations, which can either be inherited or occur de novo in the patient’s germline. This review focuses on describing the molecular basis of multiple germline syndromes that confer an increased risk of leukemia, and will specifically highlight several genes that appear to increase this risk in the absence of other significant clinical manifestations (Table 1). These discoveries have led to improved surveillance guidelines for affected children and their families, and have also facilitated a deeper understanding of the normal function of these mutated genes. A number of genes discussed in this paper are also somatically mutated in other human cancers and thus further understanding of the biochemical consequences of these lesions can shed light on disease pathogenesis and the identification of novel therapeutic targets in syndromic and nonsyndromic cancer alike.
Table 1.
Inherited predispositions to childhood hematologic malignancy.
| Gene | Locus | Inheritance | Type of leukemia | Incidence of leukemia | |
|---|---|---|---|---|---|
| Pure familial leukemia | |||||
| CEBPA | CEBPA | 19q13.1 | AD | MDS/AML | Unknown |
| Familial platelet disorder/AML | RUNX1 | 21q22.12 | AD | MDS/AML | 35% |
| MonoMAC syndrome | GATA2 | 3q21.3 | AD | MDS/AML | Unknown |
| Monosomy 7 | Unknown | 7p/q | AD | ALL, MDS/AML | Unknown |
| Bone marrow failure syndromes | |||||
| Diamond Blackfan anemia | RPS19, RPL5, RPL11 | 19q13.2, 1p22.1, 1p35 | Sporadic, AD, AR | MDS/AML, ALL | 2% |
| Shwachman–Diamond syndrome | SBDS | 7q11.21 | AR | MDS/AML, ALL | 19–36% |
| Amegakaryocytic thrombocytopenia | c-MPL | 1p34.2 | AR | MDS/AML | 2% |
| Thrombocytopenia and absent radii syndrome | RBM8A | 1q21.1 | AR, Sporadic | AML, ALL | 1% |
| Dyskeratosis congenita | DKC1, TIN2, TERC, TERT, NOP10 | Xq28, 14q11, 3q26.2, 5p15, 15q14 | X-linked, AD, AR | MDS/AML | 2% |
| Severe congenital neutropenia | ELA2, HAX1, G6PC3, WASP | 19p13, 1q21.3 17q21, Xp11 | AD, AR, X-linked | MDS/AML | 10% |
| Tumor suppressor gene syndromes | |||||
| Li-Fraumeni syndrome | TP53 | 17p13.1 | AD | ALL | 2–4% |
| Neurofibromatosis type 1 | NF1 | 17q11.2 | AD | JMML, MDS/AML | Unknown |
| Noonan syndrome | PTPN11 | 12q24.13 | AD | TMD, JMML, CMML | <1% |
| CBL syndrome | CBL | 11q23.3 | AD | JMML | <1% |
| DNA repair gene syndromes | |||||
| Mismatch repair cancer syndrome | PMS2, MSH6, MLH1, MSH2 | 7p22.2, 2p16, 3p22.2, 2p21 | AR | ALL | Unknown |
| Fanconi anemia | FANC A-E, BRCA | 16q24.3, 9q22 | AR | ALL, AML | 10% |
| Ataxia telangiectasia | ATM | 11q22.3 | AR | ALL | Unknown |
| Nijmegen breakage syndrome | NBS1 | 8q21.3 | AR | ALL | 50% |
| Bloom syndrome | BLM | 15q26.1 | AR | ALL | 12% |
| Werner syndrome | WRN (RECQL2) | 8p12 | AR | MDS/AML | Unknown |
| Rothmund–Thomson | RECQL4 | 8q24.3 | AR | AML | Unknown |
| Immunodeficiency syndromes | |||||
| Wiskott–Aldrich syndrome | WASP | Xp11.23 | X-linked | ALL | 13% |
| Bruton’s agammaglobulinemia | BTK | Xq22.1 | X-linked | ALL | Unknown |
| Down syndrome | |||||
| Trisomy 21 | Unknown | 21q | Sporadic | TMD, ALL, AML | 10% for TMD, 1–2% for ALL/AML |
The most frequent mutations for each syndrome are listed here. When multiple items are reported in one field, they are listed in order of decreasing frequency.
AD, autosomal dominant; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; AR, autosomal recessive; CMML, chronic myelomonocytic leukemia; JMML, juvenile myelomonocytic leukemia; MDS, myelodysplastic syndrome; TMD, transient myeloproliferative disorder.
Familial leukemia
While it is well known that patients with bone marrow failure disorders, DNA repair defects, and constitutional chromosomal anomalies are predisposed to developing hematologic malignancies, a more recent discovery reveals that there are inherited mutations that innately increase the risk of developing leukemia in the absence of extramedullary phenotypes [Owen et al. 2007]. Some have coined these inherited malignancies as ‘pure familial leukemia’ to differentiate them from leukemias that arise in the context of other comorbidities [Horwitz, 1997]. However, it should be noted that other cytopenias such as neutropenia and thrombocytopenia can be seen without concomitant leukemia in these disorders, thereby displaying other hematologic manifestations. While there have been many case reports detailing family pedigrees with an increased incidence of leukemia in the absence of a known inherited mutation [Goldgar et al. 1994; Gunz et al. 2009], to date there have been three genes identified that can be inherited in an autosomal dominant fashion and specifically predispose people to the development of leukemia: CEPBA, RUNX1, and GATA2. In this section we also discuss familial inheritance of monosomy 7 and its unique association with predisposing individuals to leukemia.
CEBPA
CEBPA resides at 19q13.1 and encodes the granulocytic differentiation factor C/EBPα, which belongs to the bZIP family of proteins and plays a role in myeloid differentiation [Pabst et al. 2001]. While somatic mutations and rearrangements in this gene occur in acute myeloid leukemia (AML) and confer a favorable outcome [Preudhomme, 2002], germline mutations of CEBPA were identified in 2004 [Smith et al. 2004]. After developing AML with a latency period ranging from 10 to 30 years after birth, a father and two children were found to be heterozygous germline carriers of a CEBPA-mutant allele resulting in a frame shift deletion. Interestingly, only one of the three affected family members developed a mutation in the residual wild-type allele of their leukemic cells. A more recent analysis has shown that additional but distinct somatic mutations in CEBPA occasionally occur in patients who inherit germline mutations of the same gene and likely contribute to the development of AML [Pabst et al. 2008]. Since the first report in 2004, there have been several additional reported pedigrees that document that germline transmission of CEBPA mutations leads to familial AML [Sellick et al. 2005; Renneville et al. 2008].
RUNX1 familial platelet disorder/acute myeloid leukemia
The underlying genetic defects in the autosomal dominant familial platelet disorder with a propensity to myeloid malignancy (FPD/AML) are mutations in the RUNX1 gene located at 21q22.12 [Song et al. 1999]. RUNX1 is a transcription factor broadly involved in hematopoiesis, and mutations and/or translocations involving the gene have been reported in acute lymphoblastic leukemia (ALL), AML and myelodysplastic syndrome (MDS) [McLean et al. 1996; Gaidzik et al. 2011]. The most common somatic abnormality involving the RUNX1 gene in sporadic AML or ALL is usually a translocation. However, in MDS or FPD/AML associated with RUNX1 alterations, the most common type of mutation is a point mutation resulting in haploinsufficiency [Song et al. 1999; Osato, 2004]. Recently, deletions due to nonallelic homologous recombination have been reported, highlighting the importance of using screening methodologies to detect sequence variation and copy number alterations [Jongmans et al. 2009]. The incidence of progression to MDS/AML is approximately 35% [Owen et al. 2008]. There have been 19 families to date with documented germline mutations in RUNX1, although the low prevalence may be related to the lack of awareness regarding this inherited syndrome. In addition, variable expressivity leading to a range of thrombocytopenia and varied latency for the development of MDS/AML (7–68 years of age at diagnosis) make this condition difficult to identify. However, since treatment of childhood MDS and AML frequently involves hematopoietic stem cell transplant, careful attention should be paid to the family history, as screening potential donor siblings for the same mutation is necessary. Indeed, the development of AML in the donor cells of previously asymptomatic siblings after transplant has been documented [Owen et al. 2008].
GATA2 MonoMAC syndrome
MonoMAC syndrome was first identified in 2010 as an immunodeficiency characterized by nontuberculosis mycobacterial infection, opportunistic fungal infections, human papilloma virus (HPV) infections, and monocytopenia with an increased incidence of MDS/AML [Vinh et al. 2010]. Simultaneously, Hahn and colleagues reported four families displaying an autosomal dominant inheritance pattern with predisposition to MDS/AML that resulted from mutations in the GATA2 gene found on 3q21.3 [Hahn et al. 2011]. Based on the overlapping features in these two reports, Hsu and colleagues screened 13 kindreds with MonoMAC syndrome and identified a germline GATA2 mutation in 10 pedigrees [Hsu et al. 2011]. The majority of mutations in GATA2 affect the zinc finger region and are either heterozygous missense mutations or insertion/deletions, implying that a haploinsufficiency model is sufficient for pathogenesis [Hsu et al. 2011].
GATA2 is both a transcription factor involved in hematopoietic stem cell integrity as well as a regulator of phagocytosis by macrophages [Tsai and Orkin, 1997]. The most common types of opportunistic infections listed to date include mycobacterial disease, fungal infections, and HPV-induced warts. Endogenous defects in macrophages are reported to cause the development of pulmonary alveolar proteinosis, even in the absence of infection.
Patients with MonoMAC syndrome frequently develop MDS but not before several years or even decades of opportunistic infections without any apparent dysplasia in their bone marrow. AML and chronic myelomonocytic leukemia appear to be the most common forms of leukemia in MonoMAC syndrome, and bone marrow transplantation appears to be an effective form of treatment [Cuellar-Rodriguez et al. 2011].
Mutations in GATA2 have also been recognized to underlie specific forms of congenital neutropenia that evolve into MDS or AML [Pasquet et al. 2013]. In the mild chronic form of congenital neutropenia caused by GATA2 mutations, the rate of transformation to MDS or AML at 20 years was 54%. Of 14 patients with GATA2 mutations in the French national registry of chronic neutropenia, less than half displayed the characteristic features of the aforementioned MonoMAC syndrome before evolving into MDS and AML [Pasquet et al. 2013]. Identical GATA2 mutations in different patients exhibit various outcomes, raising the hypothesis that alternative mechanisms (e.g. cooperative mutations or epigenetic modifications) may play a role in determining phenotype as well [Pasquet et al. 2013].
Recently, Emberger syndrome, an autosomal dominant disorder characterized by primary lymphedema with a predisposition to AML, was also discovered to be associated with mutations in GATA2 [Ostergaard et al. 2011]. This is not entirely surprising considering GATA2 has been shown to play an important role in lymphatic development [Kazenwadel et al. 2012]. Interestingly, the majority of GATA2 mutations in Emberger syndrome are large deletions or nonsense mutations predicted to cause complete loss of function in one allele as opposed to the missense mutations and in-frame insertions seen in the majority of other patients with MonoMAC syndrome without lymphedema. This has led to the hypothesis that the complete deletions seen in Emberger syndrome are more disruptive to lymphatic development compared with the missense mutations seen in MonoMAC syndrome and congenital neutropenia [Kazenwadel et al. 2012].
Of note, clonal cytogenetic abnormalities are a common finding in MonoMAC syndrome, affecting more than 50% of patients. The most frequent abnormality involves monosomy 7, the significance of which is yet to be elucidated [Calvo et al. 2011].
Monosomy 7
Partial or complete loss of chromosome 7 in the leukemic cells of MDS and AML is well established as a poor prognostic factor [Baranger et al. 1990]. In rare cases there have been reports of families in which multiple family members have constitutional loss of chromosome 7 leading to a syndrome with hematologic and neurologic sequelae [Chitambar et al. 1983]. To date, there have been at least 14 pedigrees reported in which multiple family members have had chromosome 7 abnormalities in their bone marrows [Gaitonde et al. 2010]. The pattern of inheritance is typically autosomal dominant and there is a wide variability in terms of disease expression. This heterogeneity underscores our incomplete understanding of the pathogenesis in familial monosomy 7. Several papers have hypothesized that the loss of a tumor suppressor gene on chromosome 7 leads to the development of cytopenias, MDS, and AML, which is frequently seen in these patients at a relatively young age [Shannon et al. 1992; Kratz et al. 2001]. Others hypothesize that a mutator effect resulting in karyotypic instability underlies monosomy 7 [Minelli et al. 2001]. As is true with de novo monosomy 7 associated leukemia, the outcome in congenital monosomy 7 associated leukemias is poor and aggressive therapy including swift transplant is often indicated [Kardos et al. 2003].
Bone marrow failure syndromes
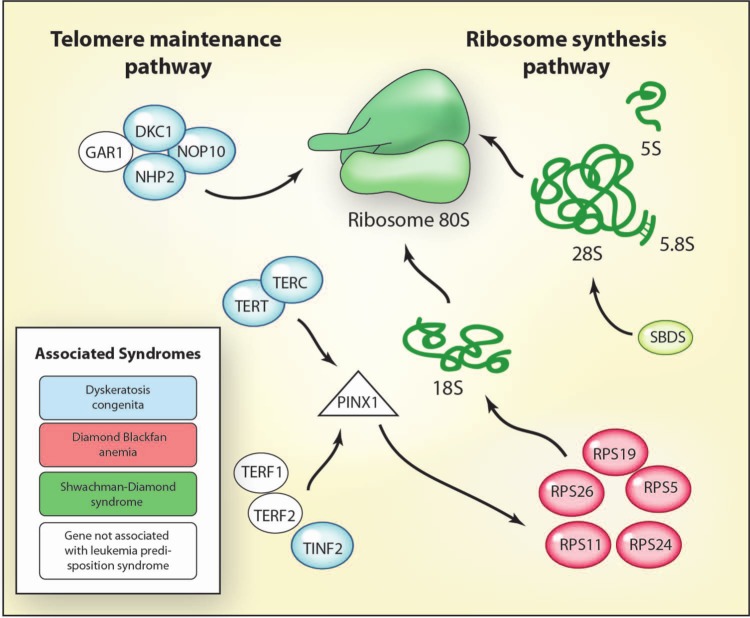
Inherited bone marrow failure syndromes (IBMFS) are a heterogeneous group of disorders that lead to aplastic anemia and have an increased incidence of malignancy [Alter, 2007]. Over 80 different genes have been identified as causing one of the IBMFS in several different pathways [Ahmed and Dokal, 2009; Narla and Ebert, 2010]. The genes are broadly involved in gatekeeping functions that regulate cell cycle. In general, these cell cycle perturbations lead to p53 activation and cell cycle arrest or senescence [Parikh and Bessler, 2012]. Several of the genes are also involved in ribosome synthesis and telomere function (Figure 1).
Figure 1.
Germline syndromes associated with the telomere maintenance and ribosome synthesis pathways.
Schematic diagram detailing the interaction between the telomere maintenance and the ribosome biogenesis pathways. The telomerase complex includes a telomerase reverse transcriptase (TERT) domain as well as a telomerase RNA component (TERC) which enables it to add a six-nucleotide sequence, 5’-TTAGGG, to the 3’ strand of chromosomes [Walne and Dokal, 2009]. Ribosome synthesis involves synthesis of ribosomal proteins in the nucleus, processing of ribosomal RNA, and assembly of ribosomal proteins with subsequent transport into the cytoplasm [Ellis and Gleizes, 2011]. Color coding indicates a specific association between a mutated gene and a syndrome that predisposes people to childhood leukemia. Syndromes include dyskeratosis congenita (OMIM 127550, 305000, 224230); Diamond Blackfan anemia (OMIM 105650); and Shwachman–Diamond syndrome (OMIM 260400). Illustration courtesy of Alessandro Baliani. Copyright © 2013. Adapted with permissions from Ahmed and Dokal [2009].
All IBMFSs are preleukemic conditions with varying degrees of malignant transformation depending on the underlying disorder. However, the exact mechanism for the increased risk of leukemia development has yet to be elucidated.
Diamond Blackfan anemia
Diamond Blackfan anemia (DBA) is characterized by a macrocytic anemia that arises in infancy and manifests distinct physical anomalies [Diamond and Blackfan, 1938; Ball, 2011]. Craniofacial and urogenital abnormalities are most common, followed by thumb, heart and spinal defects [Lipton et al. 2006]. Failure to thrive is a common finding [Vlachos et al. 2008]. Laboratory features include elevated mean corpuscular volume, hemoglobin F, and erythrocyte adenosine deaminase activity. The majority of cases are sporadic in nature, although there are forms of both autosomal dominant and recessive inheritance [Vlachos et al. 2008]. The first identified mutation in DBA was found on chromosome 19q13 [Gustavsson et al. 1997] in the gene RPS19 [Draptchinskaia et al. 1999] which encodes a ribosomal protein. Since then, mutations in several other ribosomal protein genes have been recognized with over 200 individual mutations, accounting for nearly half of all DBA cases [Boria et al. 2010]. Recently, exome sequencing analysis of two siblings with clinical DBA were found to have splice site mutations in GATA1 [Sankaran et al. 2012], a gene encoding for a transcription factor involved in hematopoiesis [Tsai et al. 1989]. These splice site mutations in GATA1 resulted in impaired production of the full-length form of the protein. This is the first demonstration that mutations outside of genes encoding for ribosomal proteins can be involved in DBA pathogenesis.
Recent work shows that the erythroid compartment has a low threshold for p53 activation and that induction of that pathway leads to selective cycle arrest in erythroid progenitor cells after silencing of the RPS19 gene [Dutt et al. 2011]. A proposed haploinsufficiency model stipulates that one functional copy of a ribosomal protein is insufficient at producing enough erythropoietic elements to maintain normal red blood cell counts due to inefficient translation [Dutt et al. 2011]. In addition, erythroid cells in DBA have been seen to express alternative spliced variants of FLVCR1, which are known to play a role in iron metabolism [Rey et al. 2008]. Free heme toxicity may be partially responsible for the erythroid failure seen in DBA secondary to damage by radical oxygen species. At this point no one single proposed mechanism accounts for all facets of DBA and it is likely that the molecular pathogenesis is multifactorial [Ball, 2011].
Despite the lack of clarity underlying this disorder, there is a clear predisposition to MDS and AML in patients with DBA with an observed-to-expected ratio of 287 for MDS and 28 for AML [Vlachos et al. 2012]. The most common form of leukemia is AML but rare cases of ALL and lymphoma have been reported [Willig et al. 2000]. Patients are also at increased risk of developing osteogenic sarcoma, which is the most common malignancy reported in the North American DBA registry [Lipton et al. 2001].
Shwachman–Diamond syndrome
Shwachman–Diamond syndrome (SDS) is an autosomal recessive disorder which manifests with anemia in infancy, pancreatic insufficiency, short stature and eventually bone marrow failure [Bodian et al. 1964]. Biallelic mutations in 90% of patients with SDS have been identified to occur in the SBDS gene on 7q11.21 [Boocock et al. 2003]. The SBDS protein is involved in several functions, including cell proliferation, mitosis, and maintaining the stromal microenvironment [Myers et al. 2013]. While it has also has been shown that the SBDS protein is involved in ribosomal subunit joining, it is still unclear how this protein is implicated in disease pathogenesis [Narla and Ebert, 2010; Burwick et al. 2012].
The role that SBDS proteins play in mitotic spindle stability and regulating chromosomal segregation may also lead to the varied cytogenetic abnormalities seen in patients with SDS [Austin et al. 2008]. This chromosomal instability may play a role in the known predisposition of patients with SDS to develop hematologic malignancies. The estimated risk of developing MDS or AML is 19% at 20 years and 36% at 30 years [Donadieu et al. 2005].
Amegakaryocytic thrombocytopenia
Congenital amegakaryocytic thrombocytopenia (CAMT) is an autosomal recessive disorder characterized by isolated thrombocytopenia and progression to pancytopenia. Typically, there are no associated physical exam features, which differentiates CAMT from nearly all other bone marrow failure syndromes and often results in delayed diagnosis [Geddis, 2011]. Mutations in c-MPL, the receptor for thrombopoietin, are responsible for the disorder and lead to high levels of dysfunctional thrombopoietin with low or absent numbers of megakaryocytes in the bone marrow [Ihara et al. 1999]. Two classes of mutations exist, with group CAMT I nonsense mutations leading to persistently low platelet counts and rapid progression to aplastic anemia. Group CAMT II missense mutations are associated with less severe thrombocytopenia and a longer latency to aplasia [Germeshausen et al. 2006]. In a review of 96 patients with CAMT, the rate of malignant transformation was 2% [Ballmaier and Germeshausen, 2011].
Thrombocytopenia with absent radii syndrome
Thrombocytopenia with absent radii syndrome (TAR) is distinguished on a clinical basis from CAMT by physical exam features, including absent radii but present thumbs. In 2007, it was shown that of 30 patients with TAR examined, all had a deletion at 1q21.1 [Klopocki et al. 2007]. Most recently, TAR became the first recognized human disease caused by mutations in the exon-junction complex (EJC), which is involved in essential RNA processing tasks. In a study of 55 patients with TAR, 53 were caused by compound inheritance of a null allele and one of two low-frequency single nucleotide polymorphisms in the regulatory regions of RBM8A that encodes the Y14 subunit of EJC [Albers et al. 2012]. In 75% of cases the mode of inheritance is autosomal recessive in which the 200-kb deleted region is passed on from an unaffected parent, with the remainder of cases occurring as de novo mutations. There have been four cases of leukemia (three AML, one ALL) in 300 patients with TAR in the literature [Alter, 2007].
Dyskeratosis congenita
Dyskeratosis congenita (DC) is a bone marrow failure syndrome with a triad of exam findings, including lacey reticulated pigmentation, dysplastic nails, and oral leukoplakia, although patients frequently present early with only one or two findings [Dokal, 2011]. Several other manifestations, including pulmonary fibrosis, liver disease, developmental delay and aplastic anemia, occur with varying incidence [Savage and Bertuch, 2010]. There have been mutations reported in several genes including but not limited to DKC1, TERC, TERT, NOP10, NHP2, TINF2, C16orf57, RTEL1, and TCAB1 which are all involved in telomerase function or the shelterin complex (reviewed by Walne and Dokal) [Walne and Dokal, 2009; Dokal, 2011]. However, roughly 50% of all patients with DC do not have a detectable mutation in any of the aforementioned mutations. The mode of inheritance differs with each mutation type and can be autosomal recessive, dominant or X linked. Anticipation is often seen in the autosomal dominant form [Savage and Bertuch, 2010]. However, identical mutations and telomere length within families have led to different disease phenotypes, suggesting that there are other factors involved in pathogenesis.
Diagnosis now involves Clinical Laboratory Improvement Amendments (CLIA) approved measurement of telomere length via different techniques, including polymerase chain reaction or a flow/fluorescence in situ hybridization based approach. Each are technically challenging, the merits of which have been presented [Aubert et al. 2012].
A cohort study from the National Cancer Institute in 2010 reported that 7 out of 50 patients with DC developed leukemia or MDS [Alter et al. 2010]. However, head and neck squamous cell carcinoma remained the most common solid malignancy. Of note, progressive bone marrow failure will occur in up to 80% of patients and is major contributor to premature mortality [Dokal, 2011].
Severe congenital neutropenia
Severe congenital neutropenia (SCN) encompasses a diverse range of disorders, including Kostmann syndrome which is generally manifest in infants with recurrent infections [Kostmann, 1956]. The most common form of the disease is autosomal dominant and is related to ELA2, which encodes for neutrophil elastase, a serine proteinase involved in neutrophilic function [Dale et al. 2000]. Recently, several other mutations in genes including HAX1, G6PC3, GFI1, GATA2, and WASP have all been implicated in SCN (reviewed by Klein) [Klein, 2011]. The latest data on the long-term risk of developing a myeloid malignancy in this population is 2.3% per year after the first decade [Rosenberg et al. 2010].
Tumor suppressor gene syndromes
Tumor suppressor genes encode for proteins that have repressive effects on regulation of the cell cycle and promote apoptosis. In the classic ‘two hit hypothesis’, tumor suppressor genes require inactivation in each of the two functioning alleles for malignant transformation.
Li-Fraumeni syndrome
TP53 is a widely expressed tumor suppressor gene at 17p13.1 that encodes for a protein involved in numerous functions, including activating DNA repair, inducing cell cycle arrest, and initiating apoptosis. Point, missense and nonsense mutations, the majority of which occur in exons 5–8 of TP53, are found in diverse cancers [Nigro et al. 1989]. In 1969, Li reported a familial cancer predisposition syndrome with a significantly increased incidence of soft tissue sarcomas, breast cancer, and bone tumors [Li, 1969]. In 1990 it was subsequently reported that families with ‘Li-Fraumeni syndrome’ (LFS) harbored germline point mutations in TP53 that, when compounded with somatic TP53 mutations in target tissues, led to the development of malignancy [Malkin et al. 1990].
In an analysis of 91 families with constitutional TP53 mutations, 4.2% of patients had developed ALL or lymphoma with an average age of onset at 25.4 years [Kleihues et al. 1997]. In 2013, it was reported that somatic TP53 alterations are present in 91% of children with low hypodiploid ALL characterized by the presence of 32–39 chromosomes in lymphoblasts [Holmfeldt et al. 2013]. Surprisingly, 43% of these children also harbored germlineTP53 mutations, indicating either an inherited or spontaneous mutation. Several of the identified TP53 mutations were previously reported in LFS, supporting that hypodiploid leukemia of childhood is a more common malignancy in LFS than previously documented. Similarly, in 2013, whole exome sequencing was performed on a family with five cases of leukemia marked by aneuploidy. Affected family members were found to harbor a germline nonsense mutation in TP53 [Powell et al. 2012]. Taken together, these recent reports support that the occurrence of hypodiploid ALL in children should prompt testing for LFS.
Neurofibromatosis 1
Neurofibromatosis type 1 (NF1) or von Recklin-ghausen neurofibromatosis is one the most common autosomal dominant disorders primarily affecting the nervous system [Friedman, 1999]. While NF1 is inherited in 50–70% of cases, the remainder of affected patients represent de novo mutations. Common manifestations of the syndrome include, but are not limited to, café-au-lait spots, skinfold freckling, Lisch nodules, optic gliomas, and neurofibromas [Williams et al. 2009]. Neurofibromas are benign tumors of the peripheral nervous system that can eventually transform into malignant peripheral nerve sheath tumors (MPNSTs) [Rasmussen et al. 2001].
The NF1 gene was discovered by linkage analysis after it was localized to 17q11.2 [Cawthon et al. 1990]. It encodes for the protein neurofibromin which functions as a GTPase-activating protein and is mutated in NF1 [Xu et al. 1990; Wallace et al. 1990]. The Ras family of proteins normally function to control cell differentiation and growth. Neurofibromin functions by hydrolyzing the active Ras-guanidine triphosphate to the inactive Ras-guanidine diphosphate as seen in Figure 2. The most frequent malignancies seen in patients with NF1 are optic gliomas, MPNSTs, and astrocytomas [Korf, 2000]. Leukemia is another cancer that is seen at a higher frequency among patients with NF1 [Stiller et al. 1994]. In contrast to the general population in which 80% of all childhood leukemias are of the lymphoid variety, almost two out of three leukemias diagnosed in patients with NF1 are myeloid in nature. In particular, juvenile myelomonocytic leukemia (JMML) is a rare RAS-driven malignancy which is seen at a substantially higher rate (200–500 fold higher) in patients with NF1 compared with the general population [Side et al. 1998].
Figure 2.
Germline syndromes associated with the Ras signaling pathway.
Schematic diagram showing the GM-CSF receptor spanning the phospholipid bilayer. After stimulation with a cytokine, the receptor propagates a signaling cascade through the Ras-MAPK, CBL, and JAK-STAT pathways. Neurofibromin, encoded by the gene NF1 is a negative regulator of the Ras pathway by binding to activated Ras and catalyzing GTP hydrolysis, thereby returning Ras to the inactive GDP-bound state. Overall, 1 in 10,000 individuals are affected with an inherited syndrome that is caused by a mutation in the Ras pathway [Tidyman and Rauen, 2012]. Color coding indicates a specific association between a mutated gene and a syndrome that predisposes children to leukemia. Syndromes include: Noonan syndrome (OMIM 163950); NF1 (OMIM 162200), and CBL syndrome (OMIM 613563). Illustration courtesy of Alessandro Baliani. Copyright © 2013. GDP, guanidine diphosphate; GM-CSF, granulocyte macrophage colony-stimulating factor; GTP, guanidine triphosphate; JAK-STAT, Janus kinase signal transducers and activators of transcription; MAPK, mitogen-activated protein kinase; NF1, neurofibromatosis type 1.
Noonan syndrome
In 1968 Noonan reported on 19 patients who shared similar features including dysmorphic facies, short stature, webbed neck, cryptorchidism and congenital heart abnormalities among other characteristic findings [Noonan, 1968]. Prior to this report the majority of these patients who were men were diagnosed with ‘the male Turner syndrome‘ due to several overlapping findings between the two syndromes [Avin, 1956]. In the 1990s the locus for the genetic lesion underlying Noonan syndrome (NS) was mapped to chromosome 12q24.1 and in 2001 it was shown that missense mutations in the gene PTPN11 are responsible for nearly 50% of cases [Tartaglia et al. 2001]. Since then, several other genes, including SOS1, RAF1, KRAS, HRAS, BRAF, MEK1/MAP2K1, MEK2/MAP2K2, NRAS, SHOC2, CBL, and SPRED1 have been reported in the group of germline disorders now known as ‘Rasopathies’ [Aoki et al. 2005; Schubbert et al. 2006; Rodriguez-Viciana et al. 2006; Roberts et al. 2006; Razzaque et al. 2007; Brems et al. 2007; Cirstea et al. 2009; Sarkozy et al. 2009; Niemeyer et al. 2010; Komatsuzaki et al. 2010]. All of the genes signal in the Ras/Raf/MEK/ERK pathway [Roberts et al. 2013].
One particularly fascinating hematologic consequence of NS is the occurrence in some patients of a transient myeloproliferative neoplasm (MPN) in the first year of life that has many characteristics of JMML. This finding is of particular interest because the association of germline PTPN11 mutations in the context of NS and MPN led to the discovery that somatic mutations in PTPN11 are found in 35% of cases of nonsyndromic JMML [Tartaglia et al. 2003; Loh et al. 2004]. However, the spectrum of germline mutations in PTPN11 is different from the somatic lesions seen in JMML, leading investigators to study and conclude that the biochemical sequelae of the somatic alterations are more severe than the germline alterations. Similar to the transient myeloproliferative disorder (TMD) seen in Down syndrome, most NS/MPN cases do not require any treatment and resolve without intervention [Kratz, 2005; Tartaglia et al. 2006], though some patients do become symptomatic and require low-dose chemotherapy. In contradistinction to TMD of trisomy 21, there is not an increased risk of developing myeloid neoplasms at a later age as a result of the prior TMD [Jongmans et al. 2011].
CBL syndrome or Noonan syndrome-like disorder
The CBL gene located at 11q23.3 encodes for an E3 ubiquitin ligase and multifunction adapter protein of the same name. There are three members of the Cbl family of proteins, CBL (c-CBL), CBL-b, and CBL-c (Cbl-3). Cbl is involved in trafficking and degradation of tyrosine kinases via a ubiquitin ligase function and is involved in many cellular pathways [Keane et al. 1999; Schmidt and Dikic, 2005]. Missense mutations in the linker region or zinc finger of the CBL gene lead to decreased ubiquitination of receptor and nonreceptor tyrosine kinases [Caligiuri et al. 2007; Sanada et al. 2009; Niemeyer et al. 2010].
A germline syndrome was identified in 2010 characterized by impaired growth, developmental delay, cryptorchidism, deafness and a predisposition to JMML in patients with germline mutations in CBL [Martinelli et al. 2010; Niemeyer et al. 2010]. As is seen in NF1, 50% of cases are autosomally inherited and 50% occur as de novo germline events. Like patients with NS who have germline PTPN11 mutations predisposing them to the development of transient JMML, patients with germline CBL mutations have an increased risk of developing JMML that is frequently, but not uniformly, self-resolving. Unfortunately, even in spontaneously resolving cases, it appears that many of these patients will go on to develop life-threatening vasculitides in the second generation of life [Niemeyer et al. 2010]. Due to the rarity of the reports thus far in the literature, the frequency of malignancies in patients with CBL is not yet established.
DNA repair defects
Mismatch repair deficiency syndrome
Lynch syndrome or Hereditary nonpolyposis colon cancer is characterized by early onset colonic and extracolonic malignancies which result from inherited or sporadic mutations in genes responsible for DNA repair [Lynch et al. 2006]. Heterozygous mutations in the mismatch repair genes MSH2 and MLH1 account for nearly 90% of cases in Lynch syndrome [Peltomaki and Vasen, 2004]. While leukemia is not a typical malignancy seen in Lynch syndrome [Cohen, 1992], there is a variant of this disorder that presents with similar features to NF1 called mismatch repair deficiency syndrome, which is caused by homozygous mutations in one of four mismatch repair genes: MLH1, MSH2, MSH6, or PMS2 [Ricciardone et al. 1999; Wang et al. 1999; Wimmer and Etzler, 2008]. In 1999, three families were reported in which children displayed homozygous mutations in the MLH1 gene causing a phenotype similar to NF1 as well as a predisposition to develop hematologic malignancies. In another case report, a child born to consanguineous parents was found to have multiple café au lait spots at the time he was diagnosed with ALL at age 24 months. Although he did not fulfill all the criteria for diagnosis of NF1, his peculiar phenotype prompted a genetic workup which revealed homozygosity for a MSH2 mutation which he inherited from each of his parents who were heterozygous and asymptomatic [Whiteside et al. 2002]. Since then, more than 70 patients from over 40 families have been characterized with homozygous or compound heterozygous mutations in one of the four MMR genes. Biallelic mutations in PMS2 account for more than half of all documented cases [Wimmer and Etzler, 2008]. This syndrome has also been referred to by the acronym CoLoN, alluding to the Colonic tumors, Leukemia/lymphomas, and features of Neurofibromatosis that these patients frequently exhibit [Bandipalliam, 2005].
Fanconi anemia
Fanconi anemia (FA) is an autosomal recessive disorder characterized by clinical manifestations, including congenital abnormalities, progressive bone marrow failure and propensity to both hematologic and solid cancers [Fanconi, 1967]. Initial testing for diagnosis involves analysis of chromosomal breakage induced by diepoxybutane (DEB) or other crosslinking agents such as mitomycin C [Auerbach, 2009]. FA is also categorized by FA complementation status, with eight groups, A–G, including D1 and D2, accounting for more the 90% of all cases (Figure 3) [Kee and D’Andrea, 2012].
Figure 3.
Germline syndromes associated with the DNA repair pathway.
Schematic diagram depicting aspects of the DNA repair pathway. Depending on the type of DNA damage that occurs, various pathways including mismatch repair, nucleotide excision repair, base excision repair, homologous recombination repair, and nonhomologous end joining can be employed by cells to repair damaged DNA [Branzei and Foiani, 2008]. Color coding indicates a specific association between a mutated gene and a syndrome that predisposes children to leukemia. Syndromes include Fanconi anemia (OMIM 227650 and 227645 are the two most common types); ataxia telangiectasia (OMIM 208900); Bloom syndrome (OMIM 210900); Nijmegen breakage syndrome (OMIM 251260); and Werner syndrome (OMIM 277700). Illustration courtesy of Alessandro Baliani. Copyright © 2013. Adapted with permissions from Ahmed and Dokal [2009].
Information from the largest FA registry in the United States suggests that roughly 33% of patients with FA will develop a hematologic malignancy by the age of 40, with AML being the most common disease followed by MDS and ALL [Kutler, 2003]. Patients with FA are also at risk of developing solid cancers such as squamous cell carcinoma (SCC) of the head and neck, vulvar SCC and various types of liver tumors among others. The risk of developing bone marrow failure by the age of 40 approaches 90% without bone marrow transplantation [Kutler, 2003].
Of particular interest is the occurrence of germline BRCA2 mutations (Fanconi complementation type D1) in FA. A report detailing six children from five families with FA who developed leukemia at a particularly young age revealed biallelic BRCA2 mutations [Wagner, 2004]. These patients had a 40% chance of developing leukemia by the age of 5 compared with patients with non-BRCA2 FA who carry a 1% risk at the same age. Thus, the occurrence of leukemia in patients with FA at less than 5 years of age should prompt an investigation into the patient’s BRCA2 locus. Mutations in BRIP1 and PALB2 which encode for nuclear binding proteins of BRCA1/2 have also been shown to cause FA-J and FA-N, respectively [Levran et al. 2005; Reid et al. 2006]. Most recently, a whole exome analysis of patients with FA without known mutations revealed that several patients harbored biallelic germline mutations in ERCC4, previously associated with xeroderma pigmentosum [Bogliolo et al. 2013].
Ataxia telangiectasia
Ataxia telangiectasia (AT) is an autosomal recessive neurodegenerative disorder characterized by progressive ataxia, ocular telangiectasias, immune dysregulation, and a predisposition to lymphoreticular malignancies, in particular after exposure to ionizing radiation [Gatti et al. 2001]. Patients with AT are either homozygous or compound heterozygotes for mutations in the gene ataxia telangiectasia mutated (ATM) located on 11q22.3 that results in truncated proteins in the majority of families with AT [Gumy-Pause et al. 2003]. Wild-type ATM protein kinases are responsible for repairing double-stranded DNA breaks and coordinating the ensuing cellular response by interacting with p53, BRCA1 and the checkpoint kinase protein (CHK2). Patients with heterozygous germline mutations appear to have a slightly increased risk of developing malignancies [Thompson et al. 2005], although there is considerable debate regarding the exact incidence rates [Ahmed and Rahman, 2006]. Fifteen percent of 57 sporadic childhood leukemias harbored heterozygous ATM mutations in one study [Pause et al. 2003], indicating ATM also plays a role as a somatic event. The greatest risk, however, is in patients with biallelic germline mutations who are at increased risk of developing lymphoma and leukemia with observed/expected ratios of between 50 and 750 [Morrell et al. 1986].
Of note, α fetoprotein (AFP) levels are elevated in patients with AT and rise with increasing age [Stray-Pedersen et al. 2007]. AFP can therefore be helpful as an adjunctive diagnostic test of AT in the correct clinical context. When treating leukemia in patients with AT, it is important to remember that ionizing radiation can carry exquisite toxicity in these patients owing to their impaired DNA repair pathway [Pollard and Gatti, 2009].
Nijmegen breakage syndrome
Nijmegen breakage syndrome (NBS) is another autosomal recessive neurodegenerative disorder with striking similarities to AT, including a strong predisposition to lymphoma and leukemia [Weemaes et al. 1981]. The defining clinical characteristics of NBS include microcephaly, ‘birdlike’ facies, intellectual impairment and growth retardation [Chrzanowska et al. 2012]. In one study of 26 patients with NBS, nearly 50% had developed a malignancy, with lymphoma being the most highly represented [Krüger et al. 2007]. The underlying mutation in NBS1 is distinct from AT [Varon et al. 1998], although the mutated proteins in both disorders interact with each other as part of the double-stranded breakage repair complex called MRN (Mre11, Rad50, and Nbs1 proteins) [Carney et al. 1998; Lee and Paull, 2005]. The degree of truncation in the Nbs1 protein inversely correlates with the risk of developing cancer [Krüger et al. 2007]. NBS1 functions as a hypomorphic protein and even in its truncated form has been shown to bind to MRE11 and RAD50 to stimulate ATM and its downstream targets. Therefore, patients with the lowest expression of NBS1 will be at increased risk of potential progression to malignancy via downregulation of double-strand DNA breaks and decreased interaction with p53 and CHK2 [Krüger et al. 2007].
Bloom syndrome
Bloom syndrome (BS) is an autosomal recessive disorder most common among Ashkenazi Jews [Roa et al. 1999] which is characterized by growth retardation, photosensitivity rashes and predisposition to malignancy at an early age [German, 1995]. The genetic defect is in the BLM gene which resides at 15q26.1 [Straughen et al. 1996] and encodes a protein in the RecQ helicase family which is postulated to support genomic stability in the face of DNA damage [Cheok et al. 2005; Chu and Hickson, 2009]. Lymphoma and leukemia are the two most common types of cancers seen in patients with BS, although in contrast to other DNA repair defect syndromes, patients with BS develop a wide spectrum of malignancies similar to the general population but at an earlier age [German, 1997]. In a report summarizing malignancies detected in a BS registry, there were 21 instances of leukemia documented in 168 patients [German, 1997].
Werner syndrome and Rothmund–Thomson syndrome (RTS) are similar disorders caused by mutations in WRN [Gray et al. 1997] and RECQL4 [Furuichi et al. 1999], respectively, which are also members of the RecQ helicase family. Werner syndrome is remarkable for premature aging in addition to overlapping features with BS. Of note, soft tissue sarcomas are the most common malignancy in Werner syndrome, while osteosarcoma is the most common in RTS [Goto et al. 1996; Wang et al. 2001]. RAPADILINO syndrome is another disorder related to the RecQ family, characterized by mutations in RECQL4 and is inherited in an autosomal dominant fashion. Typical features include short stature, infantile diarrhea, patellar hypoplasia, and radial ray anomalies, but not poikiloderma as is common in RTS. In one study of 15 patients with RAPADILINO syndrome, four had developed lymphoma and two had developed osteosarcoma [Siitonen et al. 2008].
Immunodeficiency syndromes
Patients with primary inherited immunodeficiencies are at risk of developing malignancies at significantly higher rates than the general population [Shapiro, 2010]. Risks vary depending on the type of immunodeficiency and malignancy but the lifetime incidence has been reported in the 4–25% range [Mueller and Pizzo, 1995]. In addition to the increased risk of infection, Epstein Barr virus in particular predisposes patients who are immunocompromised to develop lymphoma, which is the most common type of malignancy seen in this population. Other viral infections could activate proto-oncogenes in the host via transduction or stimulate transcription factors that induce oncogenesis.
Wiskott–Aldrich syndrome
Wiskott–Aldrich syndrome (WAS) is an X-linked disorder characterized by the triad of thrombocytopenia, primary immunodeficiency, and eczema along with a predisposition to autoimmunity and malignancy [Albert et al. 2011]. There is tremendous clinical variability in this disorder that is closely related to the type of underlying mutation in the WAS protein which functions in regulating actin polymerization in hematopoietic cells [Ochs and Thrasher, 2006]. Classic WAS that presents with the clinical triad is most often seen when mutations in the WASP gene lead to an absent or truncated form of the protein. If the WAS protein is normal in size despite the underlying mutation, it more frequently results in other phenotypes, including X-linked thrombocytopenia without immunodeficiency or X-linked neutropenia [Villa et al. 1995; Jin et al. 2004].
Approximately 70% of patients with WAS will develop at least one autoimmune disorder, most commonly, hemolytic anemia [Dupuis-Girod et al. 2003]. Roughly 13% of patients with WAS will develop at least one malignancy, with lymphoma being the most prevalent subtype. Autoimmunity appears to be the single most important risk factor for developing a malignancy in this patient population [Dupuis-Girod et al. 2003]. Based on a seminal survey, 25% of patients with WAS who had an autoimmune component to their disorder developed a malignancy, while only 5% of patients who did not display autoimmune phenomenon developed a malignancy [Sullivan et al. 1994]. The mechanism for this association has yet to be elucidated.
Bruton agammaglobulinemia
Bruton agammaglobulinemia, also referred to as X-linked agammaglobulinemia (XLA), is a primary immunodeficiency caused by mutations in the Bruton tyrosine kinase (BTK) gene at Xq21.3-22 [Bruton, 1952; Väliaho et al. 2006]. The mutated gene, normally responsible for B-cell maturation, causes arrest of B cells before maturing into plasma cells capable of producing immunoglobulins [Rawlings and Witte, 1994]. There are hundreds of reported mutations in the BTK gene leading to XLA. Most are missense, resulting in varying degrees of enzymatic function [Conley et al. 2005]. As early as 1963, it was recognized that leukemia and lymphoma occurred more frequently in patients with XLA than the population at large [Page et al. 1963]. However, surveying a more recent registry of 201 patients with XLA in the United States, only four patients were noted to have a history of malignancy, with one case each of osteosarcoma, lymphoma, adenocarcinoma of the lung, and reticulum cell sarcoma [Winkelstein et al. 2006]. Thus, the exact incidence of leukemia and lymphoma in this condition is not clear at this time.
Down syndrome and hematologic malignancies
Constitutional trisomy of chromosome 21 carries the risk of several unique hematologic sequelae [Ravindranath, 2004]. First, approximately 10% of patients with Down syndrome will develop a TMD that frequently self resolves [Massey, 2004]. However, TMD in patients with risk factors for adverse outcomes such as premature infants, those with high white blood cell counts and those with visceromegaly often require AML like chemotherapy [Gamis, 2004]. Approximately 30% of babies with Down syndrome and a history of TMD will develop MDS or AML. De novo AML is treated less aggressively in patients with Down syndrome due to their exquisite sensitivity to chemotherapy, both in terms of response and potential for toxicity. Bone marrow transplant is rarely employed in patients with trisomy 21, except in the most aggressive of all cases. The overall incidence of leukemia in Down syndrome is roughly 2%, which is 10–100 times higher than the general population [Hasle et al. 2000]. Interestingly the incidence of almost all other malignancies with the exception of retinoblastoma and germ cell tumors is markedly lower in the Down syndrome population. In general, the distribution of leukemia between ALL and AML in people with Down syndrome is similar to that of the general population (85% ALL), except in the first 3 years of life when AML predominates, with amegakaryoblastic leukemia (AMKL) presenting as the most common form. AMKL is characterized by GATA1 mutations, which normally encodes for a transcription factor important in erythroid and megakaryocyte development [Wechsler et al. 2002].
Patients with trisomy 21 are also at increased risk of developing ALL. Recently, JAK2 mutations as well high expression levels of the cytokine receptor CRLF2 have been found in 19% and 66% of patients with Down syndrome and ALL, respectively [Mullighan et al. 2009; Hertzberg et al. 2010]. Overexpression of CRLF2 is associated with genomic lesions of CRLF2, most notably in people with Down’s syndrome, of a PAR1 deletion that fuses the P2RY8 gene located on the pseudo autosomal regions of the sex chromosomes with the CRFL2 gene.
Proposed mechanisms for the increased incidence of leukemia in patients with trisomy 21 involve genes on chromosome 21 including cystathionine β synthase (CBS), which is thought to act on the folate pathway and superoxide dismutase (SOD1), which is suspected to increase DNA damage and lead to the aforementioned GATA1 mutations [Cabelof et al. 2009]. While a putative tumor suppressor gene on chromosome 21 has been postulated to increase the risk of leukemia, a specific disomic homozygous mutation has yet to be elucidated [Taub, 2004]. The genetic basis for the increased predisposition to leukemia but protection from solid malignancies is not well understood at this time [Malinge et al. 2009].
Germline polymorphisms
There has been an explosion of genome-wide association studies that identify inherited single nucleotide polymorphisms (SNPs) that are associated with the development of human diseases, including childhood leukemia, as well as predicting response to therapy and/or relapse [Yang, 2009; Orsi et al. 2012; Yang et al. 2012]. Polymorphisms in IKZF1, CDKN2A, ARID5B and CEBPE have all been shown to influence the risk of developing ALL in children [Treviño et al. 2009; Sherborne et al. 2010; Orsi et al. 2012]. While these associations are statistically significant, they are still modest in terms of relative risk, confirming that risk factors for developing ALL are polygenic in nature. SNPs in the genes PYGL and PDE4B have also been shown to influence risk of response to therapy [Yang et al. 2012]. These genes are responsible for metabolism of several chemotherapeutics used in treatment of childhood ALL, and are therefore logical determinants of outcome [Zaza et al. 2005; Kim et al. 2011].
Future directions
Cancer predisposition syndromes have become increasingly recognized through thoughtful collaborations between clinicians and scientists. Through advances in next-generation sequencing technologies, new germline mutations that alter proteins responsible for critical cellular functions have been identified. These discoveries are critical to facilitate early diagnoses for families and clinicians, which have obvious implications for cancer surveillance and genetic counseling. In addition, the discovery of these alterations has revealed commonly altered pathways in a number of genes, yielding new insights into the biological mechanisms underpinning these disorders. Ultimately, we hope and anticipate that novel therapies will arise for these patients as continued progress is made.
Footnotes
Funding: This work was supported by the Leukemia and Lymphoma Society (grant number 6059-09 to MLL), NIH (grant number T32 CA128583-6 to ES), the Frank A. Campini Foundation (to MLL), and the Team Conner Foundation.
Conflict of interest statement: The authors declare no conflicts of interest in preparing this article.
Contributor Information
Elliot Stieglitz, Department of Pediatrics, Benioff Children’s Hospital, University of California San Francisco, USA.
Mignon L. Loh, Department of Pediatrics, Benioff Children’s Hospital, 513 Parnassus Ave, HSE-302, Box 0519, University of California San Francisco/ Helen Diller Family Comprehensive Cancer Center, San Francisco, CA 94143, USA
References
- Ahmed M., Dokal I. (2009) Understanding aplastic anaemia/bone-marrow failure syndromes. Paediatr Child Health 19: 351–357 [Google Scholar]
- Ahmed M., Rahman N. (2006) ATM and breast cancer susceptibility. Oncogene 25: 5906–5911 [DOI] [PubMed] [Google Scholar]
- Albers C., Paul D., Schulze H., Freson K., Stephens J., Smethurst P., et al. (2012) Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet 44: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M., Notarangelo L., Ochs H. (2011) Clinical spectrum, pathophysiology and treatment of the Wiskott–Aldrich syndrome. Curr Opin Hematol 18: 42–48 [DOI] [PubMed] [Google Scholar]
- Alter B. (2007) Diagnosis, genetics, and management of inherited bone marrow failure syndromes. Hematology Am Soc Hematol Educ Program 2007: 29–39 [DOI] [PubMed] [Google Scholar]
- Alter B., Giri N., Savage S., Peters J., Loud J., Leathwood L., et al. (2010) Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol 150: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., et al. (2005) Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet 37: 1038–1040 [DOI] [PubMed] [Google Scholar]
- Aubert G., Hills M., Lansdorp P. (2012) Telomere length measurement—caveats and a critical assessment of the available technologies and tools. Mutat Res 730: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A. (2009) Fanconi anemia and its diagnosis. Mutat Res 668: 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin K., Gupta M., Jr, Coats S., Tulpule A., Mostoslavsky G., Balazs A., et al. (2008) Mitotic spindle destabilization and genomic instability in Shwachman–Diamond syndrome. J Clin Invest 118: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin J. (1956) The male Turner syndrome. Arch Pediatr Adolesc Med 91: 630. [DOI] [PubMed] [Google Scholar]
- Ball S. (2011) Diamond Blackfan anemia. Hematology Am Soc Hematol Educ Program 2011: 487–491 [DOI] [PubMed] [Google Scholar]
- Ballmaier M., Germeshausen M. (2011) Congenital amegakaryocytic thrombocytopenia: clinical presentation, diagnosis, and treatment. Semin Thromb Hemost 37: 673–681 [DOI] [PubMed] [Google Scholar]
- Bandipalliam P. (2005) Syndrome of early onset colon cancers, hematologic malignancies & features of neurofibromatosis in HNPCC families with homozygous mismatch repair gene mutations. Fam Cancer 4: 323–333 [DOI] [PubMed] [Google Scholar]
- Baranger L., Baruchel A., Leverger G., Schaison G., Berger C. (1990) Monosomy-7 in childhood hemopoietic disorders. Leukemia 4: 345–349 [PubMed] [Google Scholar]
- Bodian M., Sheldon W., Lightwood R. (1964) Congenital hypoplasia of the exocrine pancreas. Acta Paediatr 53: 282–293 [DOI] [PubMed] [Google Scholar]
- Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., et al. (2013) Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Med Genet 92: 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock G., Morrison J., Popovic M., Richards N., Ellis L., Durie P., et al. (2003) Mutations in SBDS are associated with Shwachman–Diamond syndrome. Nat Genet 33: 97–101 [DOI] [PubMed] [Google Scholar]
- Boria I., Garelli E., Gazda H., Aspesi A., Quarello P., Pavesi E., et al. (2010) The ribosomal basis of Diamond–Blackfan anemia: mutation and database update. Hum Mutat 31: 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. (2008) Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9: 297–308 [DOI] [PubMed] [Google Scholar]
- Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., et al. (2007) Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1–like phenotype. Nat Genet 39: 1120–1126 [DOI] [PubMed] [Google Scholar]
- Bruton O. (1952) Agammaglobulinemia. Pediatrics 9: 722–728 [PubMed] [Google Scholar]
- Burwick N., Coats S., Nakamura T., Shimamura A. (2012) Impaired ribosomal subunit association in Shwachman–Diamond syndrome. Blood 120: 5143–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof D., Patel H., Chen Q., van Remmen H., Matherly L., Ge Y., et al. (2009) Mutational spectrum at GATA1 provides insights into mutagenesis and leukemogenesis in Down syndrome. Blood 114: 2753–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri M., Briesewitz R., Yu J., Wang L., Wei M., Arnoczky K., et al. (2007) Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood 110: 1022–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo K., Vinh D., Maric I., Wang W., Noel P., Stetler-Stevenson M., et al. (2011) Myelodysplasia in autosomal dominant and sporadic monocytopenia immunodeficiency syndrome: diagnostic features and clinical implications. Haematologica 96: 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney J., Maser R., Olivares H., Davis E., Le Beau M., Yates J., 3rd, et al. (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486 [DOI] [PubMed] [Google Scholar]
- Cawthon R., Weiss R., Xu G., Viskochil D., Culver M., Stevens J., et al. (1990) A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 62: 193–201 [DOI] [PubMed] [Google Scholar]
- Cheok C., Bachrati C., Chan K., Ralf C., Wu L., Hickson I. (2005) Roles of the Bloom’s syndrome helicase in the maintenance of genome stability. Biochem Soc Trans 33: 1456–1459 [DOI] [PubMed] [Google Scholar]
- Chitambar C., Robinson W., Glode L. (1983) Familial leukemia and aplastic anemia associated with monosomy 7. Am J Med 75: 756–762 [DOI] [PubMed] [Google Scholar]
- Chrzanowska K., Gregorek H., Dembowska-Bagińska B., Kalina M., Digweed M. (2012) Nijmegen breakage syndrome (NBS). Orphanet J Rare Dis 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Hickson I. (2009) RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Cirstea I., Kutsche K., Dvorsky R., Gremer L., Carta C., Horn D., et al. (2009) A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet 42: 27–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. (1992) Muir–Torre syndrome in patients with hematologic malignancies. Am J Hematol 40: 64–65 [DOI] [PubMed] [Google Scholar]
- Conley M., Broides A., Hernandez-Trujillo V., Howard V., Kanegane H., Miyawaki T., et al. (2005) Genetic analysis of patients with defects in early B-cell development. Immunol Rev 203: 216–234 [DOI] [PubMed] [Google Scholar]
- Cuellar-Rodriguez J., Gea-Banacloche J., Freeman A., Hsu A., Zerbe C., Calvo K., et al. (2011) Successful allogeneic hematopoietic stem cell transplantation for GATA2 deficiency. Blood 118: 3715–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale D., Person R., Bolyard A., Aprikyan A., Bos C., Bonilla M., et al. (2000) Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood 96: 2317–2322 [PubMed] [Google Scholar]
- Diamond L., Blackfan K. (1938) Hypoplastic Anemia. Am J Dis Child 464–467 [Google Scholar]
- Dokal I. (2011) Dyskeratosis congenita. Hematology Am Soc Hematol Educ Program 2011: 480–486 [DOI] [PubMed] [Google Scholar]
- Donadieu J., Leblanc T., Meunier B., Barkaoui M., Fenneteau O., Bertrand Y., et al. (2005) Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica 90: 45–53 [PubMed] [Google Scholar]
- Draptchinskaia N., Gustavsson P., Andersson B., Pettersson M. (1999) The gene encoding ribosomal protein S19 is mutated in Diamond–Blackfan anaemia. Nat Genet 21: 169–175 [DOI] [PubMed] [Google Scholar]
- Dupuis-Girod S., Medioni J., Haddad E., Quartier P., Cavazzana-Calvo M., Le Deist F., et al. (2003) Autoimmunity in Wiskott–Aldrich syndrome: risk factors, clinical features, and outcome in a single-center cohort of 55 patients. Pediatrics 111: e622–e627 [DOI] [PubMed] [Google Scholar]
- Dutt S., Narla A., Lin K., Mullally A., Abayasekara N., Megerdichian C., et al. (2011) Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117: 2567–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S., Gleizes P. (2011) Diamond Blackfan anemia: ribosomal proteins going rogue. Semin Hematol 48: 89–96 [DOI] [PubMed] [Google Scholar]
- Fanconi G. (1967) Familial constitutional panmyelocytopathy, Fanconi’s anemia (F.A.). I. Clinical aspects. Semin Hematol 4: 233–240 [PubMed] [Google Scholar]
- Friedman J. (1999) Epidemiology of neurofibromatosis type 1. Am J Med Genet 89: 1–6 [PubMed] [Google Scholar]
- Furuichi Y., Kitao S., Shimamoto A., Goto M., Miller R., Smithson W., et al. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nat Genet 22: 82–84 [DOI] [PubMed] [Google Scholar]
- Gaidzik V., Bullinger L., Schlenk R., Zimmermann A., Rock J., Paschka P., et al. (2011) RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML Study Group. J Clin Oncol 29: 1364–1372 [DOI] [PubMed] [Google Scholar]
- Gaitonde S., Boumendjel R., Angeles R., Rondelli D. (2010) Familial childhood monosomy 7 and associated myelodysplasia. J Pediat Hematol Oncol 32: e236–e237 [DOI] [PubMed] [Google Scholar]
- Gamis A. (2004) Acute myeloid leukemia and Down Syndrome evolution of modern therapy? State of the art review. Pediatr Blood Cancer 44: 13–20 [DOI] [PubMed] [Google Scholar]
- Gatti R., Becker-Catania S., Chun H., Sun X., Mitui M., Lai C., et al. (2001) The pathogenesis of ataxia-telangiectasia. Clinic Rev Allerg Immunol 20: 87–108 [DOI] [PubMed] [Google Scholar]
- Geddis A. (2011) Congenital amegakaryocytic thrombocytopenia. Pediatr Blood Cancer 57: 199–203 [DOI] [PubMed] [Google Scholar]
- German J. (1995) Bloom’s syndrome. Dermatol Clin 13: 7–18 [PubMed] [Google Scholar]
- German J. (1997) Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet 93: 100–106 [DOI] [PubMed] [Google Scholar]
- Germeshausen M., Ballmaier M., Welte K. (2006) MPL mutations in 23 patients suffering from congenital amegakaryocytic thrombocytopenia: the type of mutation predicts the course of the disease. Hum Mutat 27: 296–296 [DOI] [PubMed] [Google Scholar]
- Goldgar D., Easton D., Cannon-Albright L., Skolnick M. (1994) Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer I 86: 1600–1608 [DOI] [PubMed] [Google Scholar]
- Goto M., Miller R., Ishikawa Y., Sugano H. (1996) Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol Biomarkers Prev 5: 239–246 [PubMed] [Google Scholar]
- Gray M., Shen J., Kamath-Loeb A., Blank A., Sopher B., Martin G., et al. (1997) The Werner syndrome protein is a DNA helicase. Nat Genet 17: 100–103 [DOI] [PubMed] [Google Scholar]
- Gumy-Pause F., Wacker P., Sappino A. (2003) ATM gene and lymphoid malignancies. Leukemia 18: 238–242 [DOI] [PubMed] [Google Scholar]
- Gunz F., Gunz J., Chapman C., Houston I. (2009) Familial leukaemia: a study of 909 families. Scand J Haematol 15: 117–131 [DOI] [PubMed] [Google Scholar]
- Gustavsson P., Willig T., Haeringen A., Tchernia G., Dianzani I., Donnér M., et al. (1997) Diamond–Blackfan anaemia: genetic homogeneity for a gene on chromosome 19q13 restricted to 1.8 Mb. Nat Genet 16: 368–371 [DOI] [PubMed] [Google Scholar]
- Hahn C., Chong C., Carmichael C., Wilkins E., Brautigan P., Li X., et al. (2011) Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 43: 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle H., Clemmensen I., Mikkelsen M. (2000) Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet 355: 165–169 [DOI] [PubMed] [Google Scholar]
- Hertzberg L., Vendramini E., Ganmore I., Cazzaniga G., Schmitz M., Chalker J., et al. (2010) Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood 115: 1006–1017 [DOI] [PubMed] [Google Scholar]
- Holmfeldt L., Wei L., Diaz-Flores E., Walsh M., Zhang J., Ding L., et al. (2013) The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 45: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. (1997) The genetics of familial leukemia. Leukemia 11: 1347–1359 [DOI] [PubMed] [Google Scholar]
- Hsu A., Sampaio E., Khan J., Calvo K., Lemieux J., Patel S., et al. (2011) Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118: 2653–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K., Ishii E., Eguchi M., Takada H., Suminoe A., Good R., et al. (1999) Identification of mutations in the c-mpl gene in congenital amegakaryocytic thrombocytopenia. Proc Natl Acad Sci U S A 96: 3132–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Mazza C., Christie J., Giliani S., Fiorini M., Mella P., et al. (2004) Mutations of the Wiskott–Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood 104: 4010–4019 [DOI] [PubMed] [Google Scholar]
- Jongmans M., Kuiper R., Carmichael C., Wilkins E., Dors N., Carmagnac A., et al. (2009) Novel RUNX1 mutations in familial platelet disorder with enhanced risk for acute myeloid leukemia: clues for improved identification of the FPD/AML syndrome. Leukemia 24: 242–246 [DOI] [PubMed] [Google Scholar]
- Jongmans M., van der Burgt I., Hoogerbrugge P., Noordam K., Yntema H., Nillesen W., et al. (2011) Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur J Hum Genet 19: 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos G., Baumann I., Passmore S., Locatelli F., Hasle H., Schultz K., et al. (2003) Refractory anemia in childhood: a retrospective analysis of 67 patients with particular reference to monosomy 7. Blood 102: 1997–2003 [DOI] [PubMed] [Google Scholar]
- Kazenwadel J., Secker G., Liu Y., Rosenfeld J., Wildin R., Cuellar-Rodriguez J., et al. (2012) Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 119: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M., Ettenberg S., Nau M., Banerjee P., Cuello M., Penninger J., et al. (1999) cbl-3: a new mammalian cbl family protein. Oncogene 18: 3365. [DOI] [PubMed] [Google Scholar]
- Kee Y., D’Andrea A.D. (2012) Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest 122: 3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.W., Rai D., Aguiar R.C.T. (2011) Gene set enrichment analysis unveils the mechanism for the phosphodiesterase 4B control of glucocorticoid response in B-cell lymphoma. Clin Cancer Res 17: 6723–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleihues P., Schäuble B., Hausen Zur A., Estève J., Ohgaki H. (1997) Tumors associated with p53 germline mutations: a synopsis of 91 families. Am J Pathol 150: 1–13 [PMC free article] [PubMed] [Google Scholar]
- Klein C. (2011) Genetic defects in severe congenital neutropenia: emerging insights into life and death of human neutrophil granulocytes. Annu Rev Immunol 29: 399–413 [DOI] [PubMed] [Google Scholar]
- Klopocki E., Schulze H., Strauß G., Ott C., Hall J., Trotier F., et al. (2007) Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia–absent radius syndrome. Am J Med Genet 80: 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuzaki S., Aoki Y., Niihori T., Okamoto N., Hennekam R., Hopman S., et al. (2010) Mutation analysis of the SHOC2 gene in Noonan-like syndrome and in hematologic malignancies. J Hum Genet 55: 801–809 [DOI] [PubMed] [Google Scholar]
- Korf B. (2000) Malignancy in neurofibromatosis type 1. Oncologist 5: 477–485 [DOI] [PubMed] [Google Scholar]
- Kostmann R. (1956) Infantile genetic agranulocytosis; agranulocytosis infantilis hereditaria. Acta Paediatr Suppl 45: 1–78 [PubMed] [Google Scholar]
- Kratz C. (2005) The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood 106: 2183–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz C., Emerling B., Donovan S., Laig-Webster M., Taylor B., Thompson P., et al. (2001) Candidate gene isolation and comparative analysis of a commonly deleted segment of 7q22 implicated in myeloid malignancies. Genomics 77: 171–180 [DOI] [PubMed] [Google Scholar]
- Krüger L., Demuth I., Neitzel H., Varon R., Sperling K., Chrzanowska K., et al. (2007) Cancer incidence in Nijmegen breakage syndrome is modulated by the amount of a variant NBS protein. Carcinogenesis 28: 107–111 [DOI] [PubMed] [Google Scholar]
- Kutler D. (2003) A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101: 1249–1256 [DOI] [PubMed] [Google Scholar]
- Lee J., Paull T. (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Levran O., Attwooll C., Henry R., Milton K., Neveling K., Rio P., et al. (2005) The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet 37: 931–933 [DOI] [PubMed] [Google Scholar]
- Li F. (1969) Soft-tissue sarcomas, breast cancer, and other neoplasms : a familial syndrome? Ann Intern Med 71: 747–752 [DOI] [PubMed] [Google Scholar]
- Lipton J., Atsidaftos E., Zyskind I., Vlachos A. (2006) Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr Blood Cancer 46: 558–564 [DOI] [PubMed] [Google Scholar]
- Lipton J., Federman N., Khabbaze Y., Schwartz C., Hilliard L., Clark J., et al. (2001) Osteogenic sarcoma associated with Diamond–Blackfan anemia: a report from the Diamond–Blackfan Anemia Registry. J Pediat Hematol Oncol 23: 39–44 [DOI] [PubMed] [Google Scholar]
- Loh M., Vattikuti S., Schubbert S., Reynolds M., Carlson E., Lieuw K., et al. (2004) Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood 103: 2325–2331 [DOI] [PubMed] [Google Scholar]
- Lynch H., Boland C., Gong G., Shaw T., Lynch P., Fodde R., et al. (2006) Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet 14: 390–402 [DOI] [PubMed] [Google Scholar]
- Malinge S., Izraeli S., Crispino J. (2009) Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood 113: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D., Li F., Strong L., Fraumeni J., Nelson C., Kim D., et al. (1990) Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250: 1233–1238 [DOI] [PubMed] [Google Scholar]
- Martinelli S., De Luca A., Stellacci E., Rossi C., Checquolo S., Lepri F., et al. (2010) Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Med Genet 87: 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey G. (2004) Transient leukemia in newborns with Down syndrome. Pediatr Blood Cancer 44: 29–32 [DOI] [PubMed] [Google Scholar]
- McLean T., Ringold S., Neuberg D., Stegmaier K., Tantravahi R., Ritz J., et al. (1996) TEL/AML-1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood 88: 4252–4258 [PubMed] [Google Scholar]
- Minelli A., Maserati E., Giudici G., Tosi S., Olivieri C., Bonvini L., et al. (2001) Familial partial monosomy 7 and myelodysplasia: different parental origin of the monosomy 7 suggests action of a mutator gene. Cancer Genet Cytogenet 124: 147–151 [DOI] [PubMed] [Google Scholar]
- Morrell D., Cromartie E., Swift M. (1986) Mortality and cancer incidence in 263 patients with ataxia-telangiectasia. J Natl Cancer I 77: 89–92 [PubMed] [Google Scholar]
- Mueller B., Pizzo P. (1995) Cancer in children with primary or secondary immunodeficiencies. J Pediatr 126: 1–10 [PubMed] [Google Scholar]
- Mullighan C., Collins-Underwood J., Phillips L., Loudin M., Liu W., Zhang J., et al. (2009) Rearrangement of CRLF2 in B-progenitor–and Down syndrome–associated acute lymphoblastic leukemia. Nat Genet 41: 1243–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K., Davies S., Shimamura A. (2013) Clinical and molecular pathophysiology of Shwachman–Diamond syndrome: an update. Hematol Oncol Clin North Am 27: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A., Ebert B. (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer C., Kang M., Shin D., Furlan I., Erlacher M., Bunin N., et al. (2010) Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet 42: 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro J., Baker S., Preisinger A., Jessup J., Hosteller R., Cleary K., et al. (1989) Mutations in the p53 gene occur in diverse human tumour types. Nature 342: 705–708 [DOI] [PubMed] [Google Scholar]
- Noonan J. (1968) Hypertelorism with Turner phenotype: a new syndrome with associated congenital heart disease. Arch Pediatr Adolesc Med 116: 373–380 [DOI] [PubMed] [Google Scholar]
- Ochs H., Thrasher A. (2006) The Wiskott–Aldrich syndrome. J Allergy Clin Immunol 117: 725–738 [DOI] [PubMed] [Google Scholar]
- Orsi L., Rudant J., Bonaventure A., Goujon-Bellec S., Corda E., Evans T., et al. (2012) Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE). Leukemia 26: 2561–2564 [DOI] [PubMed] [Google Scholar]
- Osato M. (2004) Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 23: 4284–4296 [DOI] [PubMed] [Google Scholar]
- Ostergaard P., Simpson M., Connell F., Steward C., Brice G., Woollard W., et al. (2011) Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 43: 929–931 [DOI] [PubMed] [Google Scholar]
- Owen C., Barnett M., Fitzgibbon J. (2007) Familial myelodysplasia and acute myeloid leukaemia - a review. Br J Haematol 140: 123–132 [DOI] [PubMed] [Google Scholar]
- Owen C., Toze C., Koochin A., Forrest D., Smith C., Stevens J., et al. (2008) Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 112: 4639–4645 [DOI] [PubMed] [Google Scholar]
- Pabst T., Eyholzer M., Haefliger S., Schardt J., Mueller B. (2008) Somatic CEBPA mutations are a frequent second event in families with germline CEBPA mutations and familial acute myeloid leukemia. J Clin Oncol 26: 5088–5093 [DOI] [PubMed] [Google Scholar]
- Pabst T., Mueller B., Zhang P., Radomska H., Narravula S., Schnittger S., et al. (2001) Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat Genet 27: 263–270 [DOI] [PubMed] [Google Scholar]
- Page A., Hansen A., Good R. (1963) Occurrence of leukemia and lymphoma in patients with agammaglobulinemia. Blood 21: 197–206 [PubMed] [Google Scholar]
- Parikh S., Bessler M. (2012) Recent insights into inherited bone marrow failure syndromes. Curr Opin Pediatr 24: 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquet M., Bellanné-Chantelot C., Tavitian S., Prade N., Beaupain B., LaRochelle O., et al. (2013) High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood 121: 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause F., Wacker P., Maillet P., Betts D., Sappino A. (2003) ATM gene alterations in childhood acute lymphoblastic leukemias. Hum Mutat 21: 554. [DOI] [PubMed] [Google Scholar]
- Peltomaki P., Vasen H. (2004) Mutations associated with HNPCC predisposition – update of ICG-HNPCC/INSiGHT mutation database. Dis Markers 20: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard J., Gatti R. (2009) Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol 74: 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell B., Jiang L., Muzny D., Treviño L., Dreyer Z., Strong L., et al. (2012) Identification of TP53 as an acute lymphocytic leukemia susceptibility gene through exome sequencing. Pediatr Blood Cancer 60: E1–E3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preudhomme C. (2002) Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 100: 2717–2723 [DOI] [PubMed] [Google Scholar]
- Rasmussen S., Yang W., Friedman J. (2001) Mortality in neurofibromatosis 1: an analysis using U.S. death certificates. Am J Hum Genet 68: 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath Y. (2004) Down syndrome and leukemia: new insights into the epidemiology, pathogenesis, and treatment. Pediatr Blood Cancer 44: 1–7 [DOI] [PubMed] [Google Scholar]
- Rawlings D., Witte O. (1994) Bruton’s tyrosine kinase is a key regulator in B-cell development. Immunol Rev 138: 105–119 [DOI] [PubMed] [Google Scholar]
- Razzaque M., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., et al. (2007) Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet 39: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Reid S., Schindler D., Hanenberg H., Barker K., Hanks S., Kalb R., et al. (2006) Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39: 162–164 [DOI] [PubMed] [Google Scholar]
- Renneville A., Mialou V., Philippe N., Kagialis-Girard S., Biggio V., Zabot M., et al. (2008) Another pedigree with familial acute myeloid leukemia and germline CEBPA mutation. Leukemia 23: 804–806 [DOI] [PubMed] [Google Scholar]
- Rey M., Duffy S., Brown J., Kennedy J., Dick J., Dror Y., et al. (2008) Enhanced alternative splicing of the FLVCR1 gene in Diamond Blackfan anemia disrupts FLVCR1 expression and function that are critical for erythropoiesis. Haematologica 93: 1617–1626 [DOI] [PubMed] [Google Scholar]
- Ricciardone M., Özçelik T., Cevher B., Özdağ H., Tuncer M., Gürgey A., et al. (1999) Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res 59: 290–293 [PubMed] [Google Scholar]
- Roa B., Savino C., Richards C. (1999) Ashkenazi Jewish population frequency of the Bloom syndrome gene 2281Δ6ins7 mutation. Genet Test 3: 219–221 [DOI] [PubMed] [Google Scholar]
- Roberts A., Allanson J., Tartaglia M., Gelb B. (2013) Noonan syndrome. Lancet 381: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Araki T., Swanson K., Montgomery K., Schiripo T., Joshi V., et al. (2006) Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet 39: 70–74 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Tetsu O., Tidyman W., Estep A., Conger B., Cruz M., et al. (2006) Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311: 1287–1290 [DOI] [PubMed] [Google Scholar]
- Rosenberg P., Zeidler C., Bolyard A., Alter B., Bonilla M., Boxer L., et al. (2010) Stable long-term risk of leukaemia in patients with severe congenital neutropenia maintained on G-CSF therapy. Br J Haematol 150: 196–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada M., Suzuki T., Shih L., Otsu M., Kato M., Yamazaki S., et al. (2009) Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460: 904–908 [DOI] [PubMed] [Google Scholar]
- Sankaran V., Ghazvinian R., Do R., Thiru P., Vergilio J., Beggs A., et al. (2012) Exome sequencing identifies GATA1 mutations resulting in Diamond–Blackfan anemia. J Clin Invest 122: 2439–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozy A., Carta C., Moretti S., Zampino G., Digilio M., Pantaleoni F., et al. (2009) Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat 30: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S., Bertuch A. (2010) The genetics and clinical manifestations of telomere biology disorders. Genet Med 12: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Dikic I. (2005) The Cbl interactome and its functions. Nat Rev Mol Cell Biol 6: 907–918 [DOI] [PubMed] [Google Scholar]
- Schubbert S., Zenker M., Rowe S., Böll S., Klein C., Bollag G., et al. (2006) Germline KRAS mutations cause Noonan syndrome. Nat Genet 38: 331–336 [DOI] [PubMed] [Google Scholar]
- Sellick G., Spendlove H., Catovsky D., Pritchard-Jones K., Houlston R. (2005) Further evidence that germline CEBPA mutations cause dominant inheritance of acute myeloid leukaemia. Leukemia 19: 1276–1278 [DOI] [PubMed] [Google Scholar]
- Shannon K., Turhan A., Rogers P., Kan Y. (1992) Evidence implicating heterozygous deletion of chromosome 7 in the pathogenesis of familial leukemia associated with monosomy 7. Genomics 14: 121–125 [DOI] [PubMed] [Google Scholar]
- Shapiro R. (2010) Malignancies in the setting of primary immunodeficiency: implications for hematologists/oncologists. Am J Hematol 86: 48–55 [DOI] [PubMed] [Google Scholar]
- Sherborne A., Hosking F., Prasad R., Kumar R., Koehler R., Vijayakrishnan J., et al. (2010) Variation in CDKN2A at 9p21. 3 influences childhood acute lymphoblastic leukemia risk. Nat Genet 42: 492–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Side L., Emanuel P., Taylor B., Franklin J., Thompson P., Castleberry R., et al. (1998) Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood 92: 267–272 [PubMed] [Google Scholar]
- Siitonen H., Sotkasiira J., Biervliet M., Benmansour A., Capri Y., Cormier-Daire V., et al. (2008) The mutation spectrum in RECQL4 diseases. Eur J Hum Genet 17: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Cavenagh J., Lister T., Fitzgibbon J. (2004) Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med 351: 2403–2407 [DOI] [PubMed] [Google Scholar]
- Song W., Sullivan M., Legare R., Hutchings S., Tan X., Kufrin D., et al. (1999) Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 23: 166–175 [DOI] [PubMed] [Google Scholar]
- Stiller C., Chessells J., Fitchett M. (1994) Neurofibromatosis and childhood leukaemia/lymphoma: a population-based UKCCSG study. Br J Cancer 70: 969–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straughen J., Ciocci S., Ye T., Lennon D., Proytcheva M., Alhadeff B., et al. (1996) Physical mapping of the Bloom syndrome region by the identification of YAC and P1 clones from human chromosome 15 band q26. 1. Genomics 35: 118–128 [DOI] [PubMed] [Google Scholar]
- Stray-Pedersen A., Borresen-Dale A., Paus E., Lindman C., Burgers T., Abrahamsen T. (2007) Alpha fetoprotein is increasing with age in ataxia–telangiectasia. Eur J Paediatr Neurol 11: 375–380 [DOI] [PubMed] [Google Scholar]
- Sullivan K., Mullen C., Blaese R., Winkelstein J. (1994) A multiinstitutional survey of the Wiskott–Aldrich syndrome. J Pediatr 125: 876–885 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., et al. (2006) Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Med Genet 78: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M., Mehler E., Goldberg R., Zampino G., Brunner H., Kremer H., et al. (2001) Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 29: 465–468 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Niemeyer C., Fragale A., Song X., Buechner J., Jung A., et al. (2003) Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet 34: 148–150 [DOI] [PubMed] [Google Scholar]
- Taub J. (2004) Prenatal origin of GATA1 mutations may be an initiating step in the development of megakaryocytic leukemia in Down syndrome. Blood 104: 1588–1589 [DOI] [PubMed] [Google Scholar]
- Thompson D., Duedal S., Kirner J., McGuffog L., Last J., Reiman A., et al. (2005) Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer I 97: 813–822 [DOI] [PubMed] [Google Scholar]
- Tidyman W., Rauen K. (2012) The RASopathies: Syndromes of Ras/MAPK Pathway Dysregulation. Berlin: Springer [Google Scholar]
- Treviño L., Yang W., French D., Hunger S., Carroll W., Devidas M., et al. (2009) Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 41: 1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S., Martin D., Zon L., D’Andrea A., Wong G., Orkin S. (1989) Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339: 446–451 [DOI] [PubMed] [Google Scholar]
- Tsai F., Orkin S. (1997) Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89: 3636–3643 [PubMed] [Google Scholar]
- Väliaho J., Smith C., Vihinen M. (2006) BTKbase: the mutation database for X-linked agammaglobulinemia. Hum Mutat 27: 1209–1217 [DOI] [PubMed] [Google Scholar]
- Varon R., Vissinga C., Platzer M., Cerosaletti K., Chrzanowska K., Saar K., et al. (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93: 467–476 [DOI] [PubMed] [Google Scholar]
- Villa A., Notarangelo L., Macchi P., Mantuano E., Cavagni G., Brugnoni D., et al. (1995) X–linked thrombocytopenia and Wiskott–Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet 9: 414–417 [DOI] [PubMed] [Google Scholar]
- Vinh D., Patel S., Uzel G., Anderson V., Freeman A., Olivier K., et al. (2010) Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood 115: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A., Ball S., Dahl N., Alter B., Sheth S., Ramenghi U., et al. (2008) Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol 142: 859–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A., Rosenberg P., Atsidaftos E., Alter B., Lipton J. (2012) Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood 119: 3815–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. (2004) Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood 103: 3226–3229 [DOI] [PubMed] [Google Scholar]
- Wallace M., Marchuk D., Andersen L., Letcher R., Odeh H., Saulino A., et al. (1990) Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science 249: 181–186 [DOI] [PubMed] [Google Scholar]
- Walne A., Dokal I. (2009) Advances in the understanding of dyskeratosis congenita. Br J Haematol 145: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Lasset C., Desseigne F., Frappaz D., Bergeron C., Navarro C., et al. (1999) Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res 59: 294–297 [PubMed] [Google Scholar]
- Wang L., Levy M., Lewis R., Chintagumpala M., Lev D., Rogers M., Plon S. (2001) Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Med Genet 102: 11–17 [DOI] [PubMed] [Google Scholar]
- Wechsler J., Greene M., McDevitt M., Anastasi J., Karp J., Le Beau M., et al. (2002) Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet 32: 148–152 [DOI] [PubMed] [Google Scholar]
- Weemaes C., Hustinx T., Scheres J., van Munster P., Bakkeren J., Taalman R. (1981) A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr Scand 70: 557–564 [DOI] [PubMed] [Google Scholar]
- Whiteside D., McLeod R., Graham G., Steckley J., Booth K., Somerville M., et al. (2002) A homozygous germ-line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple café-au-lait spots. Cancer Res 62: 359–362 [PubMed] [Google Scholar]
- Williams V., Lucas J., Babcock M., Gutmann D., Korf B., Maria B. (2009) Neurofibromatosis type 1 revisited. Pediatrics 123: 124–133 [DOI] [PubMed] [Google Scholar]
- Willig T., Gazda H., Sieff C. (2000) Diamond–Blackfan anemia. Curr Opin Hematol 7: 85–94 [DOI] [PubMed] [Google Scholar]
- Wimmer K., Etzler J. (2008) Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum Genet 124: 105–122 [DOI] [PubMed] [Google Scholar]
- Winkelstein J., Marino M., Lederman H., Jones S., Sullivan K., Burks A., et al. (2006) X-linked agammaglobulinemia. Medicine 85: 193–202 [DOI] [PubMed] [Google Scholar]
- Xu G., O’Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., et al. (1990) The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell 62: 599–608 [DOI] [PubMed] [Google Scholar]
- Yang J. (2009) Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA 301: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Cheng C., Devidas M., Cao X., Campana D., Yang W., et al. (2012) Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood 120: 4197–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaza G., Cheok M., Yang W., Panetta J., Pui C., Relling M., et al. (2005) Gene expression and thioguanine nucleotide disposition in acute lymphoblastic leukemia after in vivo mercaptopurine treatment. Blood 106: 1778–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]