Abstract
Huntington's disease (HD) is an autosomal dominant neurodegenerative disease caused by a triplet (CAG) expansion mutation. The length of the triplet repeat is the most important factor in determining age of onset of HD, although substantial variability remains after controlling for repeat length. The Venezuelan HD kindreds encompass 18,149 individuals spanning 10 generations, 15,409 of whom are living. Of the 4,384 immortalized lymphocyte lines collected, 3,989 DNAs were genotyped for their HD alleles, representing a subset of the population at greatest genetic risk. There are 938 heterozygotes, 80 people with variably penetrant alleles, and 18 homozygotes. Analysis of the 83 kindreds that comprise the Venezuelan HD kindreds demonstrates that residual variability in age of onset has both genetic and environmental components. We created a residual age of onset phenotype from a regression analysis of the log of age of onset on repeat length. Familial correlations (correlation ± SE) were estimated for sibling (0.40 ± 0.09), parent-offspring (0.10 ± 0.11), avuncular (0.07 ± 0.11), and cousin (0.15 ± 0.10) pairs, suggesting a familial origin for the residual variance in onset. By using a variance-components approach with all available familial relationships, the additive genetic heritability of this residual age of onset trait is 38%. A model, including shared sibling environmental effects, estimated the components of additive genetic (0.37), shared environment (0.22), and nonshared environment (0.41) variances, confirming that ≈40% of the variance remaining in onset age is attributable to genes other than the HD gene and 60% is environmental.
Huntington's disease (HD) is a fatal, autosomal dominant neurological illness that causes involuntary movements, severe emotional disturbance, and cognitive decline. There is currently no treatment to slow the progression of the disease, which leads without remission to death, ≈20 years after onset. The prevalence of HD is ≈1 in 10,000 people in the Americas, Europe, and Australasia (1).
Genetic and clinical data from the Venezuelan kindreds described in this article were responsible for localizing the HD gene on chromosome 4p16.3 (2) and for subsequently discovering the defective gene and identifying the nature of its mutation (3). The huntingtin mutation is an unstable trinucleotide (CAG) repeat expansion in the ORF of exon 1 of the gene. The mutation leads to the expression of an expanded polyglutamine repeat in the huntingtin protein (3). Although ubiquitously expressed, this protein causes neuronal dysfunction and death, particularly in the striatum and cortex. The pathophysiological processes involved, from expanded repeat to cell death, are currently under intense investigation (4–6).
The expanded HD allele is autosomal dominant, and symptoms inevitably will appear in a gene carrier who lives a normal life span. Phenotypic expression, such as age of onset, disease duration, and the concatenation of physical and mental symptoms, however, are variable (1). The most critical determinant of age of onset is the number of CAG repeats in the HD allele. Despite this fact, family members and even individuals with identical repeat lengths have significant variability in their ages of onset, suggesting that modifying factors exist (7–9). Previous studies have indicated that some of these modifying factors may be familial, but they involved only small sample sizes (10) or sibling pairs (11). Such methodologies cannot reliably separate the influences of genes and shared (family) environment, and it is unclear whether the age of onset of HD is influenced primarily by additional genetic or environmental factors or by a combination of both. Our study, using data collected from the Venezuelan HD kindreds, was undertaken to identify genetic and environmental factors modifying the age of onset of the disease.
The Venezuelan HD kindreds first were described by Negrette, Avila-Giron, and Bonilla (12). These kindreds now represent the largest and best characterized HD population in the world. Twenty-three years of prospective genetic, neurological, and cognitive studies have been carried out with kindred members. The families primarily live in the region of Lake Maracaibo in Zulia state. Some dwell in the urban city of Maracaibo; others live in tiny, rural fishing villages around the lake. The Venezuelan kindreds are highly heterogeneous. The majority are Hispanic, but their genetic and phenotypic heterogeneity results from matings with European sailors and traders and some Native American admixture. Most members of the Venezuelan kindreds are descendents of a woman living in the early nineteenth century in a stilt village on Lake Maracaibo. She died of HD and passed her abnormal allele through 10 generations.
Methods
Sample. Since 1979, an international, interdisciplinary team has traveled annually to Maracaibo, Venezuela, to identify HD families and document the extent and progression of their disease. Genetic and clinical data from these kindreds have been used in this investigation.
Phenotypic and Genetic Measures. A critical component of the study has been to develop and validate assessment tools that are specific and sensitive to HD so as to characterize its natural history. A protocol of neurological, cognitive, and psychiatric assessments was designed to specifically meet the cultural and educational requirements of this mainly uneducated community (13, 14). We have created a large prospective database containing clinical and genetic information, spanning 23 years on more than 2,547 people who have been examined almost annually.
Age of onset of HD was determined either prospectively or retrospectively. Both involved the examination of individuals by experienced neurologists using the Venezuelan HD Neurological Examination (13, 14). This scale assesses oculomotor function, dysarthria, dysdiakokinesis, chorea, dystonia, parkinsonism, gait and postural stability, and functional capacity.
A diagnosis of HD among the Venezuelan family members was based on the appearance of an appropriate motor abnormality characteristic of adult or juvenile onset HD. Although cognitive abnormalities frequently precede the onset of motor symptoms by years, cognitive impairment was found to be too variable to be used in assessing age of onset. Three independent diagnoses, given by three separate examiners simultaneously or by one or more examiners in subsequent years, were required; age of onset was designated by the first of three consecutive diagnoses.
In some instances family members were symptomatic at our first encounter. Their ages of onset were established retrospectively by assessing the clinical severity of their symptoms and from family history.
Transformed lymphocyte lines were created as described (15) from the 4,384 blood samples collected. A subset of 3,989 individuals at highest genetic risk was genotyped for the size of its HD allele. An additional 2,743 people at high genetic risk in these kindreds have not yet had their DNA collected or genotyped.
Statistical Analyses. We performed both genetic and phenotypic statistical analyses of these Venezuelan kindreds. By using the statistical software package sas 8 (16), we evaluated the characteristics of repeat length and age of onset and estimated the relationship between these two variables. Pearson product-moment correlations were estimated in the full sample as well as in subsamples conditioned on the number of repeats in the longer allele. Natural logarithmic transformations of the raw data were performed in the presence of nonnormal distributions. Simple and multiple regression analyses were performed to assess the overall and independent predictive power of the longer allele, as well as of other covariates such as the individual's shorter allele, gender, and paternal and maternal longer and shorter alleles.
We also used linear regression to compute a residual age of onset for subsequent genetic analyses. Because the relationship between repeat length and age of onset is curvilinear, we analyzed the log-transform of age of onset (LAO). We fitted a simple linear regression predicting a subject's LAO from that subject's repeat length of the longer allele [E (LAO) = α + β* (repeat length)]. Subtracting the predicted LAO from the actual LAO produces a residual age of onset score that is independent of the effect of the repeat length of the HD gene.
By using statistical and genetic software packages, we estimated the familial relationships and the genetic component of this residual age of onset phenotype. We used the general linear model and the variance-components procedure of sas to estimate the between and within sibship variances. Sibships were defined with a family code formed from the parental codes. A model that predicts the residual age of onset from this family code then was fitted to the data, allowing for partitioning of the variance into between and within sibships. The sibling intraclass correlation then was calculated from these variance components.
We used the family correlations (fcor) program of the sage genetic software package (17) to form familial relationships (sibling, parent-offspring, avuncular, and cousin) from which familial correlations were estimated in our sample. This program produces pedigree statistics, including asymptotic standard errors.
These familial correlations then were used as input to the mx structural equation modeling software (18), which was used to perform variance-components analysis of the residual age of onset trait. Variance-components analysis in a genetically informative data set can be used to partition the phenotypic variance observed in a trait into its genetic and environmental components. More sophisticated analysis can further decompose the environmental variance into shared (family) and nonshared components, and the genetic variance into additive and nonadditive variances attributable to polygenic factors or even due to a single major gene. Depending on the data available, all or some of these variance components can be estimated simultaneously.
Finally, we used the qtdt genetic software package (19, 20) to confirm the heritability estimate for the residual age of onset trait. qtdt uses all available familial relationships in a sample, taking full advantage of the depth of the Venezuelan kindreds, and is more informative than mx.
Results
Analyses of the Venezuelan HD Kindreds. The Venezuelan HD kindreds comprise 18,149 individuals. There are 9,162 males, 8,256 females, and 731 individuals of unknown gender. Of these, 15,409 are living, and 78% are younger than age 40 years. There are 83 independent kindreds with HD. The majority, 14,761 individuals, belong to the main kindred, tracing their origin to a single founder living in the early 1800s. The remaining 3,883 individuals form 82 kindreds of varying size. Most are quite large: three comprise between 200 and 600 people, and nine other kindreds each include between 100 and 199 individuals. Descriptive statistics of the kindreds are shown in Table 1.
Table 1. Venezuelan HD kindred statistics.
| Numbers of kindreds by size
|
|||
|---|---|---|---|
| Number of individuals | Number of kindreds | Number of individuals, sibships, and relative pairs | |
| Total number of individuals in Venezuelan HD kindreds | 18,149 | ||
| 14,761 | 1 | Male | 9,162 |
| 200–600 | 3 | Female | 8,256 |
| 100–200 | 9 | Unknown gender | 731 |
| 51–99 | 17 | Living | 15,409 |
| 10–50 | 20 | Immortalized lymphocyte lines | 4,384 |
| 4–9 | 7 | HD genotype completed | 3,989 |
| 3 | 26 | Heterozygotes (≥40 CAGs) | 938 |
| Variably penetrant alleles (35–39 CAGs) | 80 | ||
| Homozygotes | 18 | ||
| Completely penetrant alleles (≥40 CAGs on both chromosomes) | 15 | ||
| Compound alleles (≥40 CAGs on one chromosome; 35–39 CAGs on other chromosome) | 3 | ||
| Founders | 3,783 | ||
| Nonfounders | 12,626 | ||
| Unconnected singletons | 134 | ||
| Sibships | 4,502 | ||
| Pairs | |||
| Sibling | 26,330 | ||
| Parent-offspring | 25,252 | ||
| Grandparental | 33,534 | ||
| Avuncular | 97,870 | ||
| Half-sibling | 8,181 | ||
| Cousin | 180,494 | ||
The full Venezuelan HD kindreds encompass 10 generations and 4,502 sibships, with an average size of 4.998 siblings and a range of 1 to 21 siblings. The kindreds contain the following pairs: 26,330 sibling, 25,252 parent-offspring, 33,534 grandparental, 97,870 avuncular, 8181 half-sibling, and 180,494 cousin. Additionally, 3,783 individuals are founders, 12,626 people are considered nonfounders, and 134 persons are unconnected to any kindred.
Repeat Length Distribution in the Venezuelan HD Kindreds. The HD mutation is an unstable triplet repeat (CAG) expansion, acting with a dominant gene action. Alleles with 34 CAG repeats or fewer do not produce symptoms. Alleles containing 35–39 repeats produce incomplete penetrance, while a repeat of ≥40 CAGs is considered fully penetrant. In our analyses, both in the normal and pathological ranges, the allele containing the most repeats is designated the “longer” allele and the other, the “shorter” allele. Unless otherwise noted, repeat length refers to the longer allele.
Those at highest genetic risk, 3,989 individuals, were genotyped for the length of their CAG repeats; these sizes range from 14 to 86 repeats. Kindred members can be separated into three groups according to the size of their repeat length: 2,953 people have normal sized alleles (9–34 CAGs) on both chromosomes; another 80 individuals possess alleles that fall into the variably penetrant range (35–39 CAGs); and 956 people have fully penetrant alleles (≥40 CAGs). These 956 individuals include 938 heterozygotes. They also include 18 homozygotes. Of these, 15 homozygotes are “true” homozygotes and have alleles containing ≥40 CAGs on both copies of their chromosome 4s. Three individuals are “compound homozygotes,” people with a fully penetrant allele on one chromosome 4 and an allele in the variably penetrant range on the other.
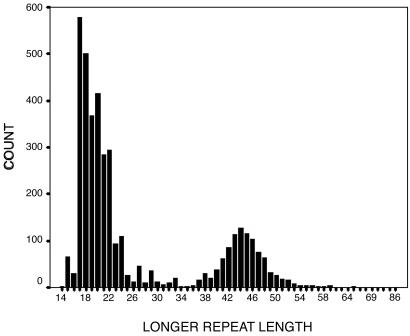
The distribution of CAG repeat length of the longer allele in the full sample is bimodal (Fig. 1), representing the combination of separate curves, one for those with normal alleles (mode = 17) and one for those with expanded alleles (mode = 44). These two distributions overlap slightly for repeat lengths in the low 30s.
Fig. 1.
Histogram of the longer allele repeat length in the Venezuelan HD kindreds. Repeat length ranges are defined as normal (14–34 CAGs), incompletely penetrant (35–39 CAGs), and fully penetrant (≥40 CAGs).
The range of repeat lengths for those with normal alleles is distributed continuously but with kurtosis and positive skew. The peak at ≈17 reveals that this is the most common length for the normal allele, a finding confirmed by examination of the shorter allele of heterozygotes, which also has a mode at 17. The short negative tail is probably attributable to a floor effect, because very few alleles are smaller than 17 repeats. The smallest repeat lengths observed in this entire sample were 14 in the longer allele and 9 in the shorter allele. The long positive tail within the normal range is probably a result of expanded alleles that have not reached the penetrance threshold.
The second smaller peak represents the fully penetrant range of repeats. This second population, involving 956 individuals, has a peak at 44 and is positively skewed. The mode repeat length is 44 (median 45) with 90% of alleles falling between 40 and 50 CAG repeats. The low tail of this distribution includes the variably penetrant range. The long positive tail of the distribution extends as far as 86 repeats. Alleles in this extreme of the distribution probably have mutated several times because, for all individuals with >65 repeats, at least one parent also had an allele in the fully penetrant range, although never with precisely the same repeat length as their offspring. As long as alleles continue mutating, there is no a priori limit to the maximum number of repeats an allele can have, although the longest repeats may not be viable.
Determination of HD Age of Onset. During our 23 years of investigation, the age of onset of HD has been determined for 458 people (Table 2). More than half, 285 individuals, were diagnosed prospectively; 173 people were diagnosed retrospectively. Of those 458 people, 214 were male and 244 were female. Ages of onset range from 2 to 69 years with an approximately normal distribution and a mean age of onset of 34.35 (± 10.07) years. There were no significant sex differences (t = 1.13, P = 0.26). Of the 18 homozygotes, 12 of the 15 true homozygotes have age of onset data. All three compound homozygotes have been diagnosed with HD. Analyses in this article only pertain to heterozygotes.
Table 2. Age of onset by gender and HD genotype.
| Age of onset, years
|
|||||
|---|---|---|---|---|---|
| n | Mean | SD | Range | Median | |
| Total | 458 | 34.35 | 10.07 | 2–69 | 34 |
| Sex* | |||||
| Male | 214 | 33.77 | 9.74 | 2–67 | 34 |
| Female | 244 | 34.86 | 10.34 | 8–69 | 34 |
| Genotype | |||||
| Homozygote | 15 | 30.07 | 6.69 | 19–40 | 30 |
| Completely penetrant alleles | 12 | 28.75 | 6.17 | 19–37 | 30 |
| Compound alleles | 3 | 35.33 | 7.23 | 27–40 | 39 |
| Heterozygote | 443 | 34.49 | 10.14 | 2–69 | 34 |
t test for gender differences = 1.13; P = 0.26
We have divided kindred members into three groups differentiated by age of onset: juvenile, typical, and late (Table 3). The juvenile form of HD (age of onset ≤20 years) is characterized by parkinsonism, tremor, seizures, and a more fulminant course (21). The juvenile repeat length varies from 43 to 86 CAGs (mean = 60.15). Anyone with ≥60 CAGs in their longer HD allele invariably will show symptoms by 20 years of age.
Table 3. Repeat length of longer allele by age of onset class.
| Age of onset, years
|
Repeat length
|
||||
|---|---|---|---|---|---|
| n | Mean | SD | Range | Median | |
| Juvenile, 2–20 | 40 | 60.15 | 9.32 | 43–86 | 60 |
| Typical, 21–50 | 377 | 45.72 | 2.97 | 40–58 | 45 |
| Late, >50 | 26 | 41.85 | 1.56 | 40–45 | 42 |
The typical ages of onset fall between 21 and 50 years. Allele lengths among this group range from 40 to 58 CAG repeats (mean = 45.72). Those with late onset become symptomatic at ≥51 years. Their repeat lengths extend from 40 to 45 CAGs (mean = 41.85). Individuals with allele lengths of between 40 and 50 CAG repeats (the most commonly found repeat sizes) can manifest symptoms as children, adults, or elderly. Factors in addition to the size of the CAG repeat clearly play a role in determining the age of HD onset.
In the 443 heterozygous individuals for whom we have age of onset and repeat length data, the negative correlation between the log of the CAG repeat and log age of onset of symptomatic HD was highly significant (r = -0.85, P < 0.0001). The size of the repeat in the longer allele accounts for 72% of the variance in age of onset. Most of this relationship can be attributed to 75 individuals with ≥50 CAG repeats whose early age of onset correlates strongly and inversely with the size of their repeat length (r = -0.81), and accounts for 66% of the variance.
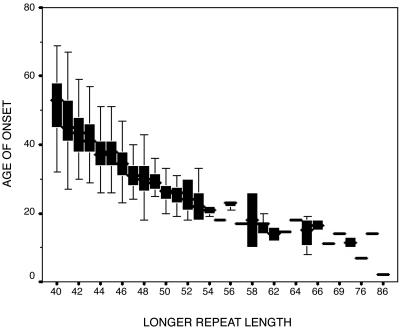
Of the members of the Venezuelan kindreds for whom we have confirmed ages of onset, 87% have repeat lengths between 40 and 50 CAGs in their HD allele. In this group the size of the HD allele is less strongly correlated with age of onset (r =-0.66, P < 0.0001), accounting for only 44% of the variance. This extensive variability exists within each repeat length (see Fig. 2).
Fig. 2.
Box plot of age of onset and repeat length of the longer allele. The curvilinear relationship between the two variables can be observed. It also is important to note the large variability of age of onset values, even within each repeat length.
After controlling for the effect of the longer allele, not even the size of the shorter allele, any of the parental alleles, or gender accounted significantly for any remaining variance in age of onset.
Familial Correlations for Residual Age of Onset. A linear regression model predicting LAO from the longer HD allele produced a residual age of onset score. By using sas, a variance-components analysis was performed to estimate the between and within sibling-pair variances of this phenotype, allowing for the computation of the sibling intraclass correlation.
The 443 heterozygous members of the Venezuelan kindreds, with CAG repeat lengths ranging from 40 to 86 and for whom we have age of onset data, form 223 sibships. The estimated sibling correlation for the residual age of onset was 0.42 (P < 0.0001). Twice the value of this sibling correlation represents familiality (22). Our analysis suggests that as much as 84% of the variance in residual age of onset is familial; that is, it is not attributable to the HD gene itself but rather to other genetic and/or family environmental factors.
Familial relationships among these 443 heterozygotes, determined by using the fcor program from the sage package, formed the following familial relationships: 441 sibling pairs, 202 parent-offspring pairs, 729 avuncular pairs, and 1,331 cousin pairs (Table 4). The sibling correlation for residual age of onset was estimated at 0.40 ± 0.09, which is remarkably consistent with the variance-component estimate. The lesser correlations also computed by sage were: parent-offspring (0.10 ± 0.11), avuncular (0.07 ± 0.11), and cousin (0.15 ± 0.10) correlations. These results are evidence of heritable influences, but they also highlight the possibility of nonadditive genetic or environmental modifiers of the age of onset of HD.
Table 4. Familial correlations for residual age of onset.
| Familial pairs | Gender | Number of pairs | Correlation (SE) |
|---|---|---|---|
| Sibling | 441 | 0.40 (0.09) | |
| Same sex | 220 | 0.34 (0.16) | |
| Brother-brother | 108 | 0.18 (0.20) | |
| Sister-sister | 113 | 0.49 (0.13) | |
| Different sex | 220 | 0.40 (0.11) | |
| Parent-offspring | 202 | 0.10 (0.11) | |
| Same sex | 93 | 0.15 (0.15) | |
| Different sex | 109 | 0.06 (0.19) | |
| Avuncular | 729 | 0.07 (0.11) | |
| Cousin | 1,319 | 0.15 (0.10) | |
| Great-avuncular | 213 | 0.14 (0.15) | |
| First cousin once-removed | 2,615 | 0.06 (0.08) | |
| First half-cousin once-removed | 305 | 0.04 (0.09) | |
| Second cousin | 3,443 | 0.17 (0.06) | |
| Second half-cousin | 1,282 | -0.08 (0.13) | |
| First cousin twice-removed | 749 | 0.06 (0.08) | |
| Second cousin once-removed | 5,970 | 0.09 (0.04) | |
| Second half-cousin once-removed | 2,340 | 0.03 (0.11) | |
| Third cousin | 6,160 | 0.08 (0.04) | |
| Third half-cousin | 654 | -0.08 (0.12) |
The parent-offspring correlation differs between same-sex (0.15) and different-sex (0.06) pairings. These differences are in the opposite direction and not as large as those between same-sex (0.34) and sister-brother (0.40) sibling pairs. Moreover, the brother-brother correlation is much lower (0.18) than the sister-sister (0.49) or the sister-brother (0.40) correlations. This possible sex effect on parent-offspring and sibling-pair correlations is intriguing, although it is only suggestive, given the large standard errors associated with these estimates.
These familial correlations suggest that genes influence this residual age of onset phenotype, at least partially, although the balance between genetic and environmental influences remains unclear.
Heritability Estimate for Residual Age of Onset. It is possible to separate genetic and environmental influences on the residual age of onset phenotype by applying variance-components methodology to a genetically informative sample such as our Venezuelan kindreds. We used mx (18) to fit a genetic and environmental model to the familial correlation data obtained from sage. For this analysis, we used the familial correlations for the closest relationships in Table 4, yielding sufficient degrees of freedom to estimate simultaneously several variance components. The additive genetic (0.37), shared sibling environmental (0.22), and nonshared environmental (0.41) variances were estimated. The total environmental effect (shared and nonshared) accounts for 63% of the variance.
With qtdt, formal estimates of heritability, obtained by fitting a model with additive polygenic genetic and nonshared environmental effects, indicate that residual age of onset has an additive genetic heritability of 38% (P = 0.0001), very similar to the result obtained with mx. This degree of genetic influence is remarkably similar to that encountered for most other common multifactorial traits of biomedical interest and is encouraging for the linkage mapping that is in progress (23). Our results represent direct estimates of genetic and environmental effects for residual age of onset of HD and provide the most compelling evidence so far for genetic modifiers of HD in humans.
Discussion
HD is a devastating, fatal disorder without remission. Notwithstanding its relentless progression, significant variability exists in symptoms and age of onset, even within families and, most surprisingly, among people sharing the identical CAG repeat length in their HD gene. Careful study of this variability should illuminate factors that modify age of onset, symptoms, and even new biological disease mechanisms and therapeutic targets. The Venezuelan kindreds are unique in that they comprise the world's largest genetically related HD community and have already provided a wealth of genetically and phenotypically informative data. The sheer magnitude of the overall kindreds, the size of individual nuclear families (from 1 to 21 children), the interrelationships among the branches, the high level of genetic heterogeneity, and the relative immobility of family members, coupled with their extraordinary cooperation, has taught us immensely and promises to reveal even more in the future.
The growth of communities at risk, compressed logistically by poverty and stigma, has increased the number of families both with the disease and in which both parents are affected or at risk. All children of homozygotes are destined to inherit the disease, which raises the genetic hazard in these communities to the extreme.
Although huntingtin is expressed ubiquitously, the gene is subject to some somatic mosaicism (24–27). In seminal fluid, as well as in testicular and brain tissue from some kindred members, the repeat size can expand dramatically. We do not know, however, whether these expansions in gametic and somatic cells have functional consequences. Do the sequelae of instability produce an appreciable, deleterious change in the protein itself? Does increased allelic instability translate to greater impaired neural activity?
An essential component of our Venezuelan HD research is to investigate genotype/phenotype relationships: How does the study of the natural history of HD in this genetically related community give us evidence of genotypic or environmental modifiers? Age of onset is one component, among many, to parse the disease and search for modifiers.
In the Venezuelan kindreds, the repeat size of the longer allele varies from 14 to 86 repeats, showing peaks at 17 (normal alleles) and 44 (expanded alleles) repeats (Fig. 1). How the two distributions of normal and expanded alleles evolved is not evident from this analysis.
Once the longer allele has been controlled for, the shorter allele does not account for much variance in age of onset in people heterozygous for the HD gene, suggesting that variability within the normal allele range has no observable effects. Whatever the pathophysiological mechanism by which the CAG expansion in the huntingtin allele is translated into neurodegeneration leading to death, we know from studying the Venezuelan kindreds that the larger the size of the repeat length, the earlier the age of onset.
Members of the Venezuelan kindreds also manifest a statistically significantly earlier age of onset in comparison to American and Canadian populations. The Venezuelans' average age of onset is 34.35 (± 10.07) years in contrast to the mean age of onset for Americans (37.47 ± 13.28) and Canadians (40.36 ± 12.97) (28). Variability in age of onset is quite appreciable among the Venezuelan kindreds: from 2 to 69 years in individuals for whom we have DNA samples and from 2 to 84 years from family history information.
What accounts for this earlier age of onset is a subject of exploration. It could be that other deleterious modifying genes not associated with the HD gene also are segregating in these kindreds. Because kindred members are interrelated, many of them may inherit both the HD gene and these pernicious modifiers. There also could be shared environmental modifiers that are influencing age of onset. Although the families are scattered widely in both urban and rural environments and across all socioeconomic strata, the majority live in extreme poverty and their diet is marginal. Because most live along the edges of Lake Maracaibo and fish for a living, they are exposed to potential pollutants from the oil industry that also occupies much of the lake. Effluvia from the lake and poor sanitation create a toxic brew in which people eke out a meager survival. All of these environmental factors and more could contribute to an earlier age of onset. Life expectancy, in general, in Venezuela is not decreased. Although census statistics do not exist for the communities in which families with HD live, the average Venezuelan life expectancy at birth for males is 70.7 years and females is 76.5 years (www.who.int/country/ven/en/).
Members of the Venezuelan kindreds with variably penetrant alleles, between 35 and 39 CAGs, represent the largest such family group in the world. Of the 80 persons with variably penetrant alleles, 78 are members of the main kindred. One individual has 21 descendents and another has 24. Although these descendents exhibited minor expansions and contractions of their variably penetrant alleles, in no instance did an allele contract to <35 or expand to >40 CAG repeats. None of those with variably penetrant alleles have yet been diagnosed with HD (Table 1).
Our analysis of the Venezuelan kindred confirms the strong, inverse correlation between repeat length and age of onset (Fig. 2). As this relationship is exponential, the LAO was analyzed. This significant, negative correlation weakens, however, for individuals whose repeat lengths fall between 40 and 50 CAGs, the majority of individuals in the Venezuelan kindreds and elsewhere. Within this group there is great variability in age of onset, even among those with identical length repeats. We have demonstrated in our analysis of the Venezuelan kindred that this variability is attributable to genetic and environmental modifiers acting in concert with the HD gene.
Analysis of the Venezuelan kindreds present familial correlations that are estimated from a larger sample and for more familial relationships but are in general accord with data reported for epidemiological samples from Italy (10) and small nuclear families in North America (11). Other studies also have found variation in several genes associated with age of onset but were able to explain only a small proportion of the total variance (29–32). A genome scan using sibling pairs in North America recently has been completed and is suggestive of linkage in three genomic loci (11).
By using variance-components analysis, we have been able to obtain a more precise estimate of the familiality than previous studies have reported. Surprisingly, even controlling for the effect of allele length, our results indicate that fully 59% of the variability in residual age of onset is familial and attributable to either genetic or shared environmental factors.
There is an intriguing sex effect in the pattern of familial correlations, although not statistically significant. For sibling pairs, the sister correlation (0.49) is larger than the sister-brother correlation (0.40) and even the brother correlation (0.18). For parent-offspring pairs, the same-sex correlation (0.15) is larger than the different-sex correlation (0.06). These gender differences imply a sex-limitation effect or favored same-sex transmission, but the large standard errors associated with these correlations make these suggestions tentative. Finally, the heritability estimate of 38% demonstrates that additive genetic effects importantly influence age of onset, in addition to the HD gene. Our analysis of the Venezuelan kindreds provides the strongest evidence to date for genetic factors influencing age of onset of HD that are statistically independent of the mutation causing the disease. We also confirm the role of environmental modifiers in modulating age of onset.
With virtually complete penetrance after 40 or more CAG repeats and with its invariably fatal course, the HD gene would seem to be the epitome of genetic determinism. And yet our results demonstrate that genetics as well as environmental factors modulate its impact. Identifying these modifiers, particularly those that lead to postponing age of onset until much later in life and prolonging productive years, would be most advantageous.
The ultimate aim of these studies is to develop new therapeutic agents that are available and accessible to everyone, including the Venezuelans, at an affordable price. In our Venezuelan investigations, we have attempted to conduct our research in a manner that is culturally sensitive as well as ethically, sociologically, and psychologically protective of the specific needs, constraints, and priorities of these individuals, families, and communities. It would be fitting if they, who have given so much, could be the first to reap the benefits.
Acknowledgments
We are indebted to the Venezuelan families who have participated in this project throughout all these years. Their generous collaboration has been critical to the success of this project and many others. We also acknowledge grants from the National Institute of Neurological Disorders and Stroke, the National Institutes of Health, the W. M. Keck Foundation, and the Hereditary Disease Foundation.
Abbreviations: HD, Huntington's disease; LAO, log-transform of age of onset.
Columbia University, 1051 Riverside Drive, New York, NY 10032
Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, United Kingdom
Indiana University, 975 West Walnut Street, Indianapolis, IN 46202
Deceased September 29, 2001.
Massachusetts General Hospital, 114 16th Street, Charlestown, MA 02129
Deceased January 31, 1999.
Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139
Universidad del Zulia, AP 1151, Maracaibo, Venezuela
Deceased September 13, 2003.
Mattel Children's Hospital at University of California, Los Angeles, CA 90095
Hospital Virgen del Camino, Irunlarrea, 4, 31008 Pamplona, Spain
University of Texas, 1515 Holcombe Boulevard, Houston, TX 77030
University of South Florida, 12901 Bruce B. Downs Boulevard, Tampa, FL 33612
University of Rochester, 1351 Mt. Hope Avenue, Rochester, NY 14620
Miami Children's Hospital, 3100 SW 62nd Avenue, Miami, FL 33155
North Shore University Hospital, 444 Community Drive, Manhasset, NY 11030
University of Southern California, 835 West 37th Street, Los Angeles, CA 90089
Mt. Sinai Medical Center, 4300 Alton Road, Miami Beach, FL 33140
Health and Human Services Agency, 4588 Market Street, San Diego, CA 92102
New York University, 550 1st Avenue, New York, NY 10016
National Institute of Neurological Disorders and Stroke, National Institutes of Health, 10 Center Drive, Bethesda, MD 20892
University of California, 2121 Gillespie, Irvine, CA 92697
University of Alabama, Birmingham, AL 35294
Thomas Jefferson University, 233 South 10th Street, Philadelphia, PA 19107
University of Iowa, 200 Hawkins Drive, Iowa City, IA 52242
City University of New York, 365 5th Avenue, New York, NY 10016
Oregon Health Sciences University, 3181 SW Sam Jackson Park Road, Portland, OR 97201
Albany Medical College, 47 New Scotland Avenue, Albany, NY 12208
University of Ulm, 89069 Ulm, Germany.
References
- 1.Bates, G., Harper, P. & Jones, L. (2002) Huntington's Disease (Oxford Univ. Press, Oxford), 3rd Ed.
- 2.Gusella, J. F., Wexler, N. S., Conneally, P. M., Naylor, S. L., Anderson, M. A., Tanzi, R. E., Watkins, P. C., Ottina, K., Wallace, M. R., Sakaguchi, A. Y., et al. (1983) Nature 306, 234-238. [DOI] [PubMed] [Google Scholar]
- 3.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 4.Feigin, A. & Zgaljardic, D. (2002) Curr. Opin. Neurol. 15, 483-489. [DOI] [PubMed] [Google Scholar]
- 5.Ross, C. A. (2002) Neuron 35, 819-822. [DOI] [PubMed] [Google Scholar]
- 6.Tobin, A. J. & Signer, E. R. (2000) Trends Cell. Biol. 10, 531-536. [DOI] [PubMed] [Google Scholar]
- 7.Snell, R. G., MacMillan, J. C., Cheadle, J. P., Fenton, I., Lazarou, L. P., Davies, P., MacDonald, M. E., Gusella, J. F., Harper, P. S. & Shaw, D. J. (1993) Nat. Genet. 4, 393-397. [DOI] [PubMed] [Google Scholar]
- 8.Andrew, S. E., Goldberg, Y. P., Kremer, B., Telenius, H., Theilmann, J., Adam, S., Starr, E., Squitieri, F., Lin, B., Kalchman, M. A., et al. (1993) Nat. Genet. 4, 398-403. [DOI] [PubMed] [Google Scholar]
- 9.Duyao, M., Ambrose, C., Myers, R., Novelletto, A., Persichetti, F., Frontali, M., Folstein, S., Ross, C., Franz, M., Abbott, M., et al. (1993) Nat. Genet. 4, 387-392. [DOI] [PubMed] [Google Scholar]
- 10.Squitieri, F., Sabbadini, G., Mandich, P., Gellera, C., Di Maria, E., Bellone, E., Castellotti, B., Nargi, E., de Grazia, U., Frontali, M., et al. (2000) Am. J. Med. Genet. 95, 366-373. [DOI] [PubMed] [Google Scholar]
- 11.Li, J. L., Hayden, M. R., Almqvist, E. W., Brinkman, R. R., Durr, A., Dode, C., Morrison, P. J., Suchowersky, O., Ross, C. A., Margolis, et al. (2003) Am. J. Hum. Genet. 73, 682-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negrette, A. (1955) Corea de Huntington: Estudio de Una Sola Familia a Traves de Varias Genereaciones (Universidad de Zulia, Maracaibo, Venezuela).
- 13.Young, A. B., Shoulson, I., Penney, J. B., Starosta-Rubinstein, S., Gomez, F., Travers, H., Ramos-Arroyo, M. A., Snodgrass, S. R., Bonilla, E., Moreno, H., et al. (1986) Neurology 36, 244-249. [DOI] [PubMed] [Google Scholar]
- 14.Penney, J. B., Jr., Young, A. B., Shoulson, I., Starosta-Rubenstein, S., Snodgrass, S. R., Sanchez-Ramos, J., Ramos-Arroyo, M., Gomez, F., Penchaszadeh, G., Alvir, J., et al. (1990) Movement Disorders 5, 93-99. [DOI] [PubMed] [Google Scholar]
- 15.Bond, C. E. & Hodes, M. E. (1996) Clin. Chem. 42, 773-774. [PubMed] [Google Scholar]
- 16.SAS Institute (2000) sas (SAS Institute, Cary, NC), Version 8.
- 17.Statistical Solutions (2002) sage, Statistical Analysis for Genetic Epidemiology (Statistical Solutions, Cork, Ireland).
- 18.Neale, M. C., Boker, S. M., Xie, G., Maes, H. H. (2002) mx: Statistical Modeling (Virginia Commonwealth Univ., Richmond, VA).
- 19.Abecasis, G. R., Cookson, W. O. & Cardon, L. R. (2000) Eur. J. Hum. Genet. 8, 545-551. [DOI] [PubMed] [Google Scholar]
- 20.Abecasis, G. R., Cardon, L. R. & Cookson, W. O. (2000) Am. J. Hum. Genet. 66, 279-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper, P. (1991) Huntington's Disease (Saunders, London).
- 22.Falconer, D. (1989) Introduction to Quantitative Genetics (Wiley, New York).
- 23.Neale, M. & Cardon, L. (1992) Methodology for Genetic Studies of Twins and Families (Kluwer Academic, Boston).
- 24.Yoon, S. R., Dubeau, L., de Young, M., Wexler, N. S. & Arnheim, N. (2003) Proc. Natl. Acad. Sci. USA 100, 8834-8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nenguke, T., Aladjem, M. I., Gusella, J. F., Wexler, N. S. & Arnheim, N. (2003) Hum. Mol. Genet. 12, 1021-1028. [DOI] [PubMed] [Google Scholar]
- 26.Leeflang, E. P., Tavare, S., Marjoram, P., Neal, C. O., Srinidhi, J., MacFarlane, H., MacDonald, M. E., Gusella, J. F., de Young, M., Wexler, N. S., et al. (1999) Hum. Mol. Genet. 8, 173-183. [DOI] [PubMed] [Google Scholar]
- 27.Leeflang, E. P., Zhang, L., Tavare, S., Hubert, R., Srinidhi, J., MacDonald, M. E., Myers, R. H., de Young, M., Wexler, N. S., Gusella, J. F., et al. (1995) Hum. Mol. Genet. 4, 1519-1526. [DOI] [PubMed] [Google Scholar]
- 28.Rosenblatt, A., Brinkman, R. R., Liang, K. Y., Almqvist, E. W., Margolis, R. L., Huang, C. Y., Sherr, M., Franz, M. L., Abbott, M. H., Hayden, M. R., et al. (2001) Am. J. Med. Genet. 105, 399-403. [PubMed] [Google Scholar]
- 29.Chattopadhyay, B., Ghosh, S., Gangopadhyay, P. K., Das, S. K., Roy, T., Sinha, K. K., Jha, D. K., Mukherjee, S. C., Chakraborty, A., Singhal, B. S., et al. (2003) Neurosci. Lett. 345, 93-96. [DOI] [PubMed] [Google Scholar]
- 30.Squitieri, F., Cannella, M., Giallonardo, P., Maglione, V., Mariotti, C. & Hayden, M. R. (2001) Brain Res. Bull. 56, 233-238. [DOI] [PubMed] [Google Scholar]
- 31.Naze, P., Vuillaume, I., Destee, A., Pasquier, F. & Sablonniere, B. (2002) Neurosci. Lett. 328, 1-4. [DOI] [PubMed] [Google Scholar]
- 32.Djousse, L., Knowlton, B., Hayden, M., Almqvist, E. W., Brinkman, R., Ross, C., Margolis, R., Rosenblatt, A., Durr, A., Dode, C., et al. (2003) Am. J. Med. Genet. 119A, 279-282. [DOI] [PubMed] [Google Scholar]