Abstract
Siah proteins are E3 ubiquitin ligases. They are homologues of the Drosophila seven in absentia (Sina), a protein required for the R7 photoreceptor development. We have previously found that the expression of human siah-1 and its mouse homologue siah-1b are induced by p53 during apoptosis and tumor reversion. So far, no evidence that the siah-1b gene is a direct transcriptional target of p53 has been provided. In the present study we investigate this issue. Northern blot analysis with a specific probe demonstrates an increase in siah-1b transcription on activation of endogenous and inducible exogenous p53. To explore whether this effect is directly mediated by p53 we analyzed 20 kb of chromosome X DNA, containing the siah-1b locus. A p53-binding site was identified in the siah-1b promoter, located at nucleotides -2155/-2103 relative to the translational start site. This site is composed of two half-sites, conforming to the p53-binding consensus sequence but separated by a nonclassical 33-bp spacer. In luciferase assays, p53 induces a substantial increase in siah-1b promoter activity. Gel shift and DNase-I-footprinting studies, combined with mutational analysis and chromatin immunoprecipitation, indicate that p53 effectively binds the siah-1b promoter in vitro and in vivo. Thus, the siah-1b gene is a direct transcriptional target of p53.
The Drosophila seven in absentia (Sina) is a protein required for neuronal differentiation of the R7 photoreceptor cells in the eye of Drosophila melanogaster (1). It acts downstream of the Sevenless tyrosine kinase receptor to degrade the transcriptional repressor Tramtrak (2, 3). The Drosophila Sina protein and its mammalian Siah homologues are phylogenetically conserved E3 ligases, enzymes involved in ubiquitination and proteasome-mediated degradation of protein substrates. This function is due to their N-terminal RING-finger domain, which recruits ubiquitin-conjugating E2 enzymes and promotes the ligation of ubiquitin to the substrate (4). The two functional human genes, siah-1 and siah-2, are responsible for degradation of Kid, BOB/OBF1, synaptophysin, synphilin, transforming growth factor-β-induced early gene, and TRAF2 (5–10). Siah-1, through the binding and down-modulation of Numb, is a positive regulator of Notch activity (11).
We previously cloned the siah-1 gene, one of the human homologues of Sina, and showed its overexpression in the epithelium of the small intestine, a well established example of physiological programmed cell death (12). Moreover, in our studies of tumor reversion, we found siah-1 overexpressed at the mRNA level (12) and at the protein level (13, 14) in cells with a suppressed malignant phenotype. In a 3D basement membrane reconstituted in matrigel, Siah-1 is able to reorganize MCF7 cells in structures similar to those observed with normal breast cells (14, 15). Cells overexpressing Siah-1 showed an increase in apoptosis and gave rise to significantly fewer tumors than the parental cells when injected into scid/scid mice (13, 14). We previously demonstrated that cells transfected with the cyclin-dependent kinase inhibitor p21 exhibit high levels of Siah-1 (16) and that Siah-1 has common downstream effectors with p21 and the tumor suppressor p53 (13). Moreover, Siah-1 acts in a complex with Skp1, Eb1, SIP (Siah interacting protein), and adenomatous polyposis coli protein (pAPC) to facilitate, in a p53-dependent manner, the degradation of β-catenin, thus inducing apoptosis and inhibiting cell proliferation and transformation (17, 18). Pw1/Peg3 is another p53-inducible gene product that cooperates with Siah-1 in promoting cell death (19), whereas BAG-1, an antiapoptotic protein, antagonizes the effect of Siah-1 on apoptosis (20).
The human siah-1 gene has two murine homologues, siah-1a and siah-1b, which are widely expressed in various tissues of the embryo and adult (21). The RNA sequences of siah-1a and siah-1b are 95% homologues with each other, 90% with their human counterpart, and 72% with Drosophila Sina. Siah-1a drives the degradation of c-myb, a protooncogene involved in cellular proliferation and apoptosis (22), and is necessary for progression past metaphase during meiosis I of spermatogenesis (23). We previously identified siah-1b by cDNA differential display as a gene induced by p53 in murine M1 myeloid leukemia cells (24). This finding suggests a function for Siah-1b in apoptosis, as observed for its human homologue. Gene expression analysis revealed that human siah-1 transcription was significantly correlated with the dosage of p53 (25).
p53 is a homotetrameric transcription factor that can activate or repress the transcription of a series of genes controlling cell cycle progression, apoptosis, DNA repair, and other types of stress response. These genes include p21, MDM2, cyclin G, BAX, noxa, puma, TSAP6, and many others (26–32). The p53 protein is normally short-lived and present at low levels, but in response to stress it accumulates in the nucleus, where it binds to specific DNA sequences within chromatin (33). The consensus p53-binding site is composed of two 10-base half-sites, each conforming to the sequence 5′-PuPuPuC(A/T)(T/A)GPy-PyPy-3′ (where Pu and Py represent purines and pyrimidines, respectively) and separated by a spacer of 0–13 bp (34–37). Based on this consensus sequence, a computer algorithm developed to identify p53-binding sites in the human genome disclosed siah-1 as a possible downstream target of p53 (38).
In the present study we show that the siah-1b promoter contains a functional p53 responsive element (p53RE), which is able to bind p53 in vitro and in vivo. Activation of p53 in different cell systems leads to a significant increase in siah-1b transcription. Thus, the siah-1b gene is a direct transcriptional target of p53.
Materials and Methods
Cell Culture and p53 Activation. The M1 murine myeloid leukemia cell line and its derivative LTR6 clone, expressing a temperature-sensitive p53, have been described (24, 39). In mouse NIH 3T3 cells p53 was activated by incubation with 15 nM Actinomycin D (ActD) (Sigma) for 16–24 h. Human p53-null H1299 cells were grown according to ATCC recommendations.
Cloning and Mutagenesis. The siah-1b promoter region (nucleotides -2613/-1694) was cloned into the pGL3-enhancer vector (Promega). The siah-1b p53-binding element was mutated (CATG to TATA) either within the first half-site (siah-1b mut1) or within the second half-site (siah-1b mut2) by using the QuikChange Multi SiteDirected Mutagenesis kit (Stratagene). The p53RE (nucleotides -2160/-2098) was deleted in siah-1b Δ with the following primers: forward, 5′-CTAAAATGGGTCTCAAGACCTCCCCTGAGA-3′, and reverse, 5′-TCTCAGGGGAGGTCTTGAGACCCATTTTAG-3′. The distal region of the p21 promoter (nucleotides -7615/-7354 from ATG) containing the p53RE was cloned into the pGL3 enhancer vector by using the following primers: forward, 5′-CTAGGTACCCCAGAGGATACCTTGCAAGGCTGCA-3′, and reverse, 5′-TATAGATCTTCTCTGTCTCCATTCATGCTCCTCC-3′. The mouse p53 coding sequence (CDS) was cloned in a pCMV vector.
Proteins and Antibodies. The human p53 protein was purified from insect cells infected with recombinant baculovirus. The following antibodies were used: anti-p53 FL-393 (Santa Cruz Biotechnology) and PAb421 (Oncogene Research Products); anti-clathrin heavy chain H-300 (Santa Cruz Biotechnology); anti-α-tubulin B-5-1-2 (Sigma).
Northern Blot Analysis. Northern blot analyses were performed as described (24). The following probes were used: mouse p21 nucleotides 111/340 (accession no. NM_007669), cyclin G1 nucleotides 202/443 (accession no. BC005534), and human GAPDH (Clontech). The probe CDS corresponds to the region nucleotides 251/896 of the human siah-1 cDNA (accession no. U63295). A specific probe for siah-1a (probe a) was designed from the 3′ UTR (nucleotides 1676/1934) of siah-1a mRNA (accession no. Z19579), whereas the specific probe for siah-1b (probe b) corresponds to nucleotides 1/230 in the 5′ UTR of the mRNA (accession no. Z19580).
Rapid Amplification of cDNA Ends (RACE)-PCR. The RACE-PCR was performed by using the Marathon cDNA Amplification kit (Clontech) following the manufacturer's instructions. In brief, mRNA derived from mouse cells was subjected to reverse transcription by using a modified oligo(dT) primer. The resulting cDNAs were ligated to an adaptor and used as templates in 5′ or 3′ RACE-PCR by using the adaptor-specific primer with siah-1 primers designed to amplify both siah-1a and siah-1b cDNA: siah-1 forward, 5′-CCCCTTGTGAGTCAACACATAGTGCTGC-3′, and siah-1 reverse, 5′-TGGGGCGACAGTTGCTACAAACAAG-3′. For the 5′ amplification we performed a nested PCR by using an internal adaptor-specific primer and the reverse siah-1b primer 5′-AGACTCGCCAAGTCATTGTTGGATGC-3′.
Luciferase Assays. Cells were transfected with 300 ng of the different siah-1b promoter constructs (see Cloning and Mutagenesis) and 300 ng of pCMV vector, either empty or expressing mouse p53. Transfection was performed by using LipofectAMINE PLUS reagent (Life Technologies, Grand Island, NY) according to the manufacturer's recommendations. Twenty-four hours after transfection the cells were washed and cell extracts were prepared by using a reporter lysis reagent (25 mM Tris/8 mM MgCl2/1mMDTT/1% Triton X-100/15% glycerol). After normalization of each extract for protein content, luciferase activity was measured by using the Victor Luminometer (Perkin–Elmer) after addition of 20 nmol of Luciferin (Roche Diagnostics) and 100 nmol of ATP (Sigma).
Electrophoretic Mobility-Shift Assay. Synthetic oligonucleotides containing the p53RE of siah-1b, either wild type (5′-TCTCAAGACATGTCCAGACCTCCCCTGATCACATTCAAAAGGGTCTCAAGACATGTCCAGACC-3′) or double mutant (5′-TCTCAAGATATATCCAGACCTCCCCTGATCACATTCA AAAGGGTCTCAAGATATATCCAGACC-3′) were radiolabeled at their 5′ end by using T4 Polynucleotide Kinase (Biolabs) and [γ-32P]ATP (Amersham Pharmacia Biosciences). The complementary oligonucleotides were then annealed and purified on a polyacrylamide gel. The probes were incubated for 20 min at room temperature with 18 ng of recombinant p53 in a buffer containing 20 mM Tris·HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, and 0.5 mg/ml BSA; then 20 ng of DNA competitor poly(dI-dC) (Amersham Pharmacia) was added to a final volume of 30 μl. To induce a supershift, 300 ng of p53-specific PAb421 antibody was included in the mixture, and the incubation was continued for 20 min at room temperature. The reaction products were analyzed by electrophoresis on a 4% nondenaturing polyacrylamide gel.
In Vitro DNase I Footprinting. The radiolabeled forward primer, 5′-CCATGGAGCCACCTCAGCTC-3′, and the reverse primer, 5′-ACTAGTAATGAGTTTCCTCTCCTACATGAA-3′, were used in PCR reactions with 25 ng of DNA of the following constructs: siah-1b, siah-1b mut1, or siah-1b mut2. The 410-bp labeled PCR product containing the p53RE in its center was used in DNase-I-footprinting experiments according to the Sure Core Footprinting protocol (Promega) with minor modifications. For each reaction, 500 ng or 1 μg of p53 or 1 μg of irrelevant protein were mixed with 500 ng of PAb421 antibody. After 30 min of preincubation on ice, 10 ng (30,000 cpm) of 5′ end-labeled DNA was added, and incubation was carried out for another 30 min on ice. After adjusting the concentration to 5 mM CaCl2 and 10 mM MgCl2, samples were digested with DNase I (0.15 unit) for 1 min at room temperature and resolved on a sequencing gel. DNA-sequencing reactions were done by using the sequenase 2.0 DNA Sequencing Kit (USB).
Chromatin Immunoprecipitation and Western Blot Analysis. Experiments were performed with the chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's instructions. Samples were immunoprecipitated with antibodies against clathrin heavy chain or against p53 (FL-393). PCR analysis of the isolated DNA fragments used the following primer pairs: siah-1b forward, 5′-CCATTGTGTGCATCTTCCTGAGCCC-3′, and reverse, 5′-GAACTAACCTAGCACTAGTAATGAG-3′; β2 microglobulin forward, 5′-GCTCTGAAGATTCATTTGAACCTGC-3′, and reverse, 5′-ATCCAAGTAATGAGAGTACAGAGG-3′; p21 forward, 5′-CCAGAGGATACCTTGCAAGGC-3′, and reverse, 5′-TCTCTGTCTCCATTCATGCTCCTCC-3′. Specificity of the primers was checked by blast analysis by using the Ensembl mouse genomic database. PCR reactions with the different primers gave rise to a single specific product of the expected size. The linear range for each primer pair was determined empirically by using increasing amounts of LTR6 genomic DNA. PCR products were resolved on 2% agarose gels.
Whole-cell extracts were generated by using standard conditions. Extracts containing 20 μg of total protein were subjected to Western blot analysis by using the FL-393 and B-5-1-2 antibodies.
Results
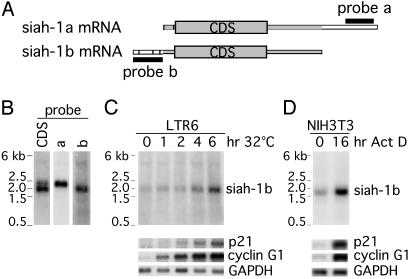
p53-Dependent Induction of siah-1b mRNA. To confirm and extend previous studies showing that siah-1b is activated during p53-induced apoptosis, we used the information available in the National Center for Biotechnology Information mRNA database to analyze the sequences of siah-1a and siah-1b, whose alignment is schematically shown in Fig. 1A. This analysis confirmed that the siah-1 CDS probe used previously (23) recognizes both siah-1a and siah-1b. In Northern blot analysis this probe hybridizes to two transcripts of 2.3 and 1.9 kb, respectively (Fig. 1B). To study more specifically siah-1b transcription, we designed a probe in the 5′ UTR (accession no. Z19580) of its mRNA (nucleotides 1/230) (Fig. 1 A). In the same manner we designed a probe specific for siah-1a (accession no. Z19579) in the 3′ UTR of its mRNA (nucleotides 1676/1934) (Fig. 1 A). On mRNA from mouse cells the siah-1a probe (probe a) recognizes the 2.3-kb band, whereas the siah-1b probe (probe b) recognizes only the 1.9-kb band (Fig. 1B). These results disagree with the suggestion (40) that the band at 2.3 kb includes both siah-1a and siah-1b transcripts.
Fig. 1.
Activation of p53 induces increased siah-1b mRNA levels. (A) Schematic alignment of siah-1a and siah-1b mRNAs. Exons are represented by bars and the CDSs are represented by large rectangles. Homologous sequences are represented by the gray segments. White segments identify sequences specific to each mRNA. The relative positions of the siah-1a and siah-1b probes (black bars) used for Northern blot analysis are also indicated. (B) Northern blots of mouse mRNA hybridized with the siah-1 CDS (probe CDS), the siah-1a probe (probe a), or the siah-1b probe (probe b). (C) Northern blot analysis of mRNA isolated from LTR6 cells after shifting to 32°C for 1, 2, 4, and 6 h. LTR6 cells at 38°C are used as negative controls. (D) Northern blot analysis of mRNA extracted from NIH 3T3 cells after 16 h of incubation with 15 nM ActD; untreated cells (0 h) were used as control. (C and D) Blots were hybridized with a siah-1b-specific probe. p21 and cyclin G1 were used as positive controls for p53 activation. GAPDH was used as control for equal loading.
According to the published sequences of siah-1b mRNA (accession nos. Z19580 and BC052887) the expected size of the siah-1b transcript is 1.7 kb. Because our siah-1b-specific probe recognizes a band of bigger size, we performed RACE-PCR experiments to determine more precisely the 5′ and 3′ ends of siah-1b mRNA. The 3′ RACE-PCR confirmed the published sequences, but the preliminary 5′ RACE analysis revealed a possible alternative splicing of the siah-1b first exon. The two different first exons (NM009173 and BC052887) described by Della et al. (21) or sequenced by the I.M.A.G.E. Consortium (http://image.llnl.gov/), respectively. Each represent 1/7 of the clones analyzed by us. The remaining 5/7 of the clones were found to contain a first exon of 231 bp, which so far has not been described (accession no. AY495086). Transcripts containing this longer exon are expected to have a size of 1.9 kb, confirming the results of our Northern blot analysis.
To confirm that siah-1b is activated in the process of p53-induced apoptosis, we used the LTR6 clone derived from M1 mouse myeloid leukemia cells. LTR6 cells are stably transfected with a temperature-sensitive p53 mutant (val135) that gains wild-type function on shifting the temperature from 38 to 32°C, leading cells into programmed cell death (39). As expected, Northern blot analysis confirmed the transcriptional activation of mouse p21 and mouse cyclin G1, both of which are well established p53 target genes, on shift of LTR6 cells to 32°C (Fig. 1C). A parallel, albeit more modest, increase in siah-1b mRNA was also observed under the same conditions (Fig. 1C), consistent with the conclusion that siah-1b is a p53-inducible gene.
Activated p53 can trigger different effector pathways, depending on cell type and mode of activation. To assess the generality of p53-mediated siah-1b induction, we therefore subjected another cell line, NIH 3T3, to treatment with the transcription inhibitor ActD, a well documented activator of the p53 pathway (41, 42). As seen in Fig. 1D, this also resulted in a marked increase in siah-1b mRNA.
Identification of a Putative p53 Consensus Site in the siah-1b Gene. To determine whether the siah-1b gene is a direct transcriptional target of p53, 20 kb of the region of murine chromosome X containing the siah-1b locus (nucleotides -18621/+1379 relative to the initiator ATG) were analyzed by using the patch software (www.gene-regulation.com/cgibin/pub/programs/patch/bin/patch.cgi). A putative p53 consensus binding site was identified in the intronic region comprising nucleotides -2155/-2103, between exons 2 and 3. This DNA element consists of two typical half-sites, 5′-AGACATGTCC-3′. The distance between the two half-sites is unusual: whereas the consensus specifies a spacer of 0–13 bp (33), the two half sites found in siah-1b are 33 bp apart. Moreover, from the analysis of many confirmed p53 target sites, it seems that the majority of the physiologically relevant ones have no spacer at all (34, 38). It was therefore of particular interest to test the interaction of p53 with this putative binding site.
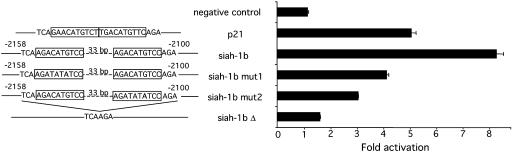
Functionality of the p53 Responsive Element in the siah-1b Gene. The region nucleotides -2613/-1694 of the siah-1b gene, containing the putative p53RE located at nucleotides -2155/-2103, was subcloned into the pGL3-enhancer vector and tested for p53-dependent transcriptional activity in a luciferase assay. The different constructs represented on Fig. 2 Left were transiently transfected into p53-null H1299 lung adenocarcinoma cells with or without a plasmid driving the expression of the mouse p53 protein. Luciferase activity in the presence of p53 was normalized for the activity in the absence of p53 to obtain a fold of activation. The empty luciferase vector (negative control) was not affected by the presence of p53, whereas inclusion of siah-1b sequences led to an 8-fold increase in luciferase activity when p53 was present (Fig. 2). To verify that the positive effect of p53 on the transcriptional activity of the reporter plasmid is mediated by the consensus site identified within the siah-1b gene, we mutated nucleotides within this site that are known to be necessary for direct binding of p53. Mutation of CATG to TATA in the first half-site (siah-1b mut1; Fig. 2) led to a 2-fold reduction in the activating effect of p53, and a similar mutation in the second half-site (siah-1b mut2) resulted in a 3-fold reduction. Deletion of the entire putative p53RE (64 bp; siah-1b Δ) abrogated almost completely the effect of p53. Similar results were obtained in mouse NIH 3T3 cells transfected with the same set of reporter plasmids (Fig. 5A, which is published as supporting information on the PNAS web site). Moreover, the extent of activation was p53-dose-dependent (Fig. 5B). These observations imply that p53 responsiveness of the reporter plasmid is indeed due to the presence of the putative intronic p53-binding site identified by us in the siah-1b gene.
Fig. 2.
DNA derived from the siah-1b gene confers transcriptional activation by p53. Promoter activity was measured in luciferase assays with reporter plasmids comprising the region nucleotides -2613/-1694 of siah-1b or its three mutants (siah-1b mut1, siah-1b mut2, and siah-1b Δ) after transient transfection into H1299 cells. (Left) Schematic representation of different constructs. (Right) Corresponding promoter activity of the constructs. A p21 promoter fragment containing the distal p53RE was used as positive control and the empty pGL3 vector was used as negative control. Luciferase activity is reported as fold activation, representing the ratio of the values standardized by protein concentration measured in the presence or absence of p53. Values shown are mean ± SD (n = 3).
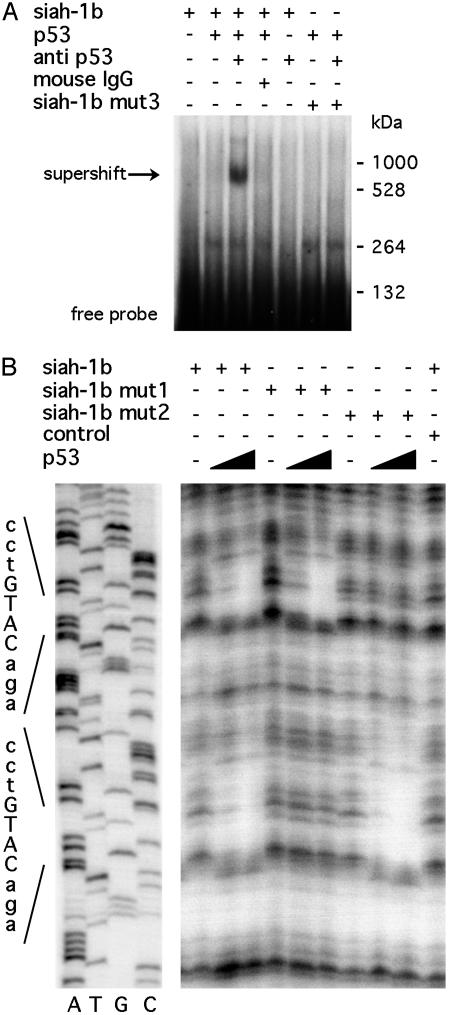
p53 Binds to the siah-1b Promoter in Vitro. Electrophoretic mobility-shift assays were used to analyze the binding and affinity of p53 to the p53RE identified in the siah-1b gene. A 63-bp radiolabeled probe including this element was incubated with purified baculovirus-derived p53 protein, in the presence of the p53-specific monoclonal antibody PAb421, which stimulates the formation of p53-DNA complexes by imparting a conformational change in p53 (43, 44). As seen in Fig. 3A, such incubation resulted in specific gel retardation (lane 3). A similar effect was seen when in vitro translated p53 was used in the reaction (Fig. 6, lane 3, which is published as supporting information on the PNAS web site). The specificity of the binding was verified by competition with an excess of unlabeled DNA containing a p53 consensus site derived from the p21 gene (data not shown). In contrast, no shift was observed when the p53 consensus binding site was mutated (siah-1b mut3, Fig. 3A, lanes 6 and 7). Comparison with the positions of molecular weight markers run on the same gel suggests that the supershifting complex has an approximate size of 850 kDa, corresponding to a p53 tetramer plus four antibody molecules and the probe (45). Thus, despite the nonconventional nature of the consensus sequence, the mode of binding of p53 appears to be similar to that occurring with more typical p53-binding sites.
Fig. 3.
p53 binds to the siah-1b promoter in vitro.(A) Electrophoretic mobility-shift assay was performed with a probe corresponding to the p53RE of the siah-1b promoter (nucleotides -2160/-2098). This probe was incubated with recombinant p53 protein (lanes 2, 3, 4, 6, and 7) in the absence (lanes 1, 2, 4, and 6) or in the presence (lanes 3, 5, and 7) of the PAb421 antibody. In lanes 6 and 7, the probe contained a double mutation in the p53 consensus site (siah-1b mut3). A pool of mouse IgGs was used as control for the PAb421 antibody specificity (lane 4). (B) In vitro DNase-I-footprinting experiment with labeled probes designed to have the p53RE placed in their center. Probes containing the p53RE of siah-1b (lanes 1–3) or its mutants (lanes 4–6 for siah-1b mut1 and lanes 7–9 for siah-1b mut2) were incubated with either 0.5 (lanes 2, 5, and 8) or 1 μg of purified p53 (lanes 3, 6, and 9) or with an irrelevant protein (lane 10) before digestion with DNase I. The positions of the p53-binding sites were identified by running DNA-sequencing reactions next to the footprinting experiment (Left).
The use of short double-stranded oligonucleotide probes for p53 bandshift assays has been questioned (46). Therefore, we also performed DNase-I-footprinting studies by using a 410-bp labeled probe corresponding to nucleotides -2290/-1880 of the siah-1b gene, centered around the p53RE. As shown in Fig. 3B, p53 protected the two half-sites from DNase I cleavage but did not protect the spacer DNA between the two half-sites. The latter may be because of the relatively long distance between the two half-sites. A similar footprint was obtained in the absence of antibody (data not shown). Remarkably, mutation of either of the two half-sites from CATG to TATA abrogated p53-mediated protection of that particular half-site, but not affecting the protection of the other one, whose sequence remained wild type (Fig. 3B, lanes 4, 5, and 6 marked for siah-1b mut1 and lanes 7, 8, and 9 marked for siah-1b mut2). These results demonstrate that p53 can bind to either one or both of the consensus half-sites found in the siah-1b gene. In conclusion, electrophoretic mobility-shift assay and DNase I footprinting demonstrate that p53 is able to associate in vitro with the p53 consensus located at nucleotides -2155/-2112 of the siah-1b gene.
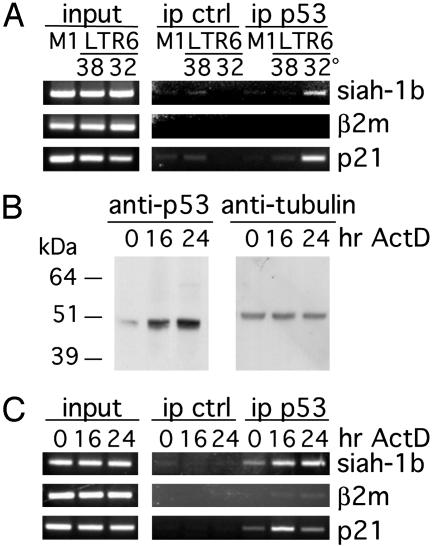
p53 Associates with siah-1b Chromatin in Vivo. To determine whether p53 could bind the intronic siah-1b p53RE in vivo, we performed ChIP experiments. The LTR6 cell system was used for this purpose, under the same conditions where endogenous siah-1b transcription is efficiently activated by p53 (see Fig. 1C). M1 and LTR6 cells were maintained at 38°C or 32°C for 6 h and treated with formaldehyde to generate covalently cross-linked DNA–protein complexes within the cells. Cross-linked chromatin was then immunoprecipitated with antibodies against clathrin heavy chain for negative control or against p53. PCR amplification was then performed on the immunoprecipitated DNA and on total input DNA after fragmentation (input). Primers specific for siah-1b, corresponding to positions -2208/-2184 (forward) and -1891/-1967 (reverse) were designed in the genomic sequence containing the p53RE, thus avoiding cross-reactivity with the other siah homologues. As seen in Fig. 4A, siah-1b and p21 chromatin were specifically immunoprecipitated with anti-p53 antibodies only from LTR6 cells maintained at 32°C; hence, like the corresponding elements in the p21 gene promoter, the p53RE within the siah-1b intron is indeed occupied by wild-type p53 within living cells.
Fig. 4.
p53 binds to siah-1b chromatin in vivo. ChIP was performed after p53 activation in vivo with antibodies against clathrin heavy chain (ip ctrl) or p53 (ip p53). PCR was performed with gene-specific primers. Total lysate was used as a control for PCR amplification (input). p53 binding was tested by using siah-1b-specific primers; p21 primers and β2 microglobulin primers were used as positive and negative controls, respectively. (A) Results of ChIP analysis done on LTR6 cells, activated by temperature shift to 32°C for 6 h. M1 and LTR6 cells maintained at 38°C served as negative controls. (B) Western blot analysis of p53 in NIH 3T3 cells activated with 15 nM ActD for 16 or 24 h. Untreated cells were used as negative control (0 h). α-Tubulin served as an equal loading control. (C) Results of ChIP analysis done on NIH 3T3 cells treated as in B.
To rule out the possibility that the in vivo binding of p53 to the siah-1b genomic DNA was due to a p53 overexpression artefact, we induced activation of endogenous p53 in NIH 3T3 cells by treatment with ActD, which led to a substantial increase in the steady-state levels of p53 (Fig. 4B). When such ActD-treated NIH 3T3 cells were used as the starting material for ChIP analysis, a clear association was observed between p53 and siah-1b chromatin and with p21 chromatin (Fig. 4C). p53 binding to the siah-1b chromatin remained maximal after 24 h of ActD treatment, whereas binding to the p21 promoter region decreased between 16 and 24 h of treatment. Thus, both transfected and endogenous p53 are found specifically associated with the siah-1b gene in vivo, further confirming that this is a bona fide direct p53 target gene.
Discussion
In the present study we show that the mouse siah-1b gene, shown previously to be up-regulated during p53-mediated apoptosis (24), contains a functional p53RE within its second intron. This p53RE enables the transactivation of the siah-1b gene by p53 in response to anticancer agents such as ActD. Through the use of suitable molecular tools, which discern between the different highly homologous mouse siah-1 transcripts, we show that siah-1b mRNA is increased as a result of p53 activation, thus resolving previous ambiguities stemming from the use of nonspecific probes (12, 16, 21, 24, 40).
Basal levels of siah-1b mRNA are also present before p53 activation, suggesting that the protein has a role in nonstressed cells. Moreover, siah-1b expression can be induced by p53-independent mechanisms (13), although expression is maximal when p53 is activated.
A recent report, with siah-1b-null mouse embryo fibroblasts, has concluded that Siah-1b is not involved in the p53 pathway (40). However, it is possible that in such cells p53 activates multiple downstream effectors. The role of Siah-1b could thus be substituted by proteins encoded by one or more of the other p53-responsive genes that are transactivated simultaneously with siah-1b. Moreover, the apparent discrepancy between our data and those of Bowtell and coworkers (40) may also be due to different experimental conditions; it is conceivable that the contribution of Siah-1b to the p53 response may vary as a function of the cellular context. In this regard, it is noteworthy that the effect of p53 itself is also greatly cell-context-dependent; thus, activation of the same temperature-sensitive p53 mutant can induce growth arrest, apoptosis, or differentiation in a context-dependent manner (47).
The pattern and extent of induction of different p53 target genes greatly depends on the concentration of active p53 protein within a given cell (48). In this regard, it is of note that the siah-1b gene seems to require higher levels of p53 activity for maximum activation than the p21 gene (Fig. 1C), and this requirement is also seen when the corresponding p53REs are compared in luciferase assays (see Fig. 5B). This finding is most likely because of differences in the affinity of p53 for the corresponding p53REs, as seen also with other p53 target genes (49). This difference of affinity is also suggested by the fact that although for p21 10 min of crosslinking were sufficient to obtain an optimal PCR signal in the ChIP analysis, 15 min were required for siah-1b (data not shown).
The identified p53RE is functional, despite its unusual structure. So far, no functional p53RE with such a long spacer between the two half-sites has been described. It is possible that the sequence of the spacer also has a role in enabling such an unusual p53RE to function properly. The footprinting analysis suggests that the spacer is not directly involved in interactions with p53; nevertheless, this region may be recognized in vivo by other proteins required for optimal p53-induced transcription of siah-1b. Also, each of the two half-sites seems to function even on its own, when the other half-site is mutant and incapable of binding p53; this independent functioning is seen both in DNA-binding analysis (Fig. 3B) and in functional assays (Fig. 2). Several studies have proposed a role for the Siah proteins in p53-mediated responses (11, 13, 19, 50–52). It is also noteworthy that siah-1b knockout mice are not viable, emphasizing the importance of this gene in embryonic development. The exact way in which Siah contributes to p53-mediated apoptosis or cell cycle arrest is still not clearly defined; one possibility is that Siah's contribution may have to do with the proposed role of Siah-1 in mediating p53-dependent degradation of β-catenin in response to DNA damage (51). By establishing a direct link between p53 and transcriptional regulation of the siah-1b gene, our study further supports the importance of the latter in the p53 response.
Supplementary Material
Acknowledgments
We thank Pierre Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for his advice and support. This work was supported in part by European Commission Grant QLK6-CT-2000-00159.
Abbreviations: Sina, seven in absentia; p53RE, p53 responsive element; CDS, coding sequence; ChIP, chromatin immunoprecipitation; ActD, Actinomycin D.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY495086).
References
- 1.Carthew, R. W. & Rubin, G. M. (1990) Cell 63, 561-577. [DOI] [PubMed] [Google Scholar]
- 2.Li, S. C., Songyang, Z., Vincent, S. J., Zwahlen, C., Wiley, S., Cantley, L., Kay, L. E., Forman-Kay, J. & Pawson, T. (1997) Proc. Natl. Acad. Sci. USA 94, 7204-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang, A. H., Neufeld, T. P., Kwan, E. & Rubin, G. M. (1997) Cell 90, 459-467. [DOI] [PubMed] [Google Scholar]
- 4.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germani, A., Bruzzoni-Giovanelli, H., Fellous, A., Gisselbrecht, S., Varin-Blank, N. & Calvo, F. (2000) Oncogene 19, 5997-6006. [DOI] [PubMed] [Google Scholar]
- 6.Tiedt, R., Bartholdy, B. A., Matthias, G., Newell, J. W. & Matthias, P. (2001) EMBO J. 20, 4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler, T. C., Chin, L. S., Li, Y., Roudabush, F. L. & Li, L. (2002) J. Biol. Chem. 277, 10273-10282. [DOI] [PubMed] [Google Scholar]
- 8.Nagano, Y., Yamashita, H., Takahashi, T., Kishida, S., Nakamura, T., Iseki, E., Hattori, N., Mizuno, Y., Kikuchi, A. & Matsumoto, M. (2003) J. Biol. Chem. 278, 51504-51514. [DOI] [PubMed] [Google Scholar]
- 9.Johnsen, S. A., Subramaniam, M., Monroe, D. G., Janknecht, R. & Spelsberg, T. C. (2002) J. Biol. Chem. 277, 30754-30759. [DOI] [PubMed] [Google Scholar]
- 10.Habelhah, H., Frew, I. J., Laine, A., Janes, P. W., Relaix, F., Sassoon, D., Bowtell, D. D. & Ronai, Z. (2002) EMBO J. 21, 5756-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Susini, L., Passer, B. J., Amzallag-Elbaz, N., Juven-Gershon, T., Prieur, S., Privat, N., Tuynder, M., Gendron, M. C., Israel, A., Amson, R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 15067-15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemani, M., Linares-Cruz, G., Bruzzoni-Giovanelli, H., Roperch, J. P., Tuynder, M., Bougueleret, L., Cherif, D., Medhioub, M., Pasturaud, P., Alvaro, V., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 9039-9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roperch, J. P., Lethrone, F., Prieur, S., Piouffre, L., Israeli, D., Tuynder, M., Nemani, M., Pasturaud, P., Gendron, M. C., Dausset, J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 8070-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuynder, M., Susini, L., Prieur, S., Besse, S., Fiucci, G., Amson, R. & Telerman, A. (2002) Proc. Natl. Acad. Sci. USA 99, 14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruzzoni-Giovanelli, H., Faille, A., Linares-Cruz, G., Nemani, M., Le Deist, F., Germani, A., Chassoux, D., Millot, G., Roperch, J. P., Amson, R., et al. (1999) Oncogene 18, 7101-7109. [DOI] [PubMed] [Google Scholar]
- 16.Linares-Cruz, G., Bruzzoni-Giovanelli, H., Alvaro, V., Roperch, J. P., Tuynder, M., Schoevaert, D., Nemani, M., Prieur, S., Lethrosne, F., Piouffre, L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuzawa, S. I. & Reed, J. C. (2001) Mol. Cell 7, 915-926. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzawa, S., Li, C., Ni, C. Z., Takayama, S., Reed, J. C. & Ely, K. R. (2003) J. Biol. Chem. 278, 1837-1840. [DOI] [PubMed] [Google Scholar]
- 19.Relaix, F., Wei, X., Li, W., Pan, J., Lin, Y., Bowtell, D. D., Sassoon, D. A. & Wu, X. (2000) Proc. Natl. Acad. Sci. USA 97, 2105-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuzawa, S., Takayama, S., Froesch, B. A., Zapata, J. M. & Reed, J. C. (1998) EMBO J. 17, 2736-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Della, N. G., Senior, P. V. & Bowtell, D. D. (1993) Development (Cambridge, U.K.) 117, 1333-1343. [DOI] [PubMed] [Google Scholar]
- 22.Weston, K. (1998) Curr. Opin. Genet. Dev. 8, 76-81. [DOI] [PubMed] [Google Scholar]
- 23.Dickins, R. A., Frew, I. J., House, C. M., O'Bryan, M. K., Holloway, A. J., Haviv, I., Traficante, N., de Kretser, D. M. & Bowtell, D. D. (2002) Mol. Cell. Biol. 22, 2294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amson, R. B., Nemani, M., Roperch, J. P., Israeli, D., Bougueleret, L., Le Gall, I., Medhioub, M., Linares-Cruz, G., Lethrosne, F., Pasturaud, P., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon, H., Liyanarachchi, S., Wright, F. A., Davuluri, R., Lockman, J. C., de la Chapelle, A. & Pellegata, N. S. (2002) Proc. Natl. Acad. Sci. USA 99, 15632-15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.el-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W. & Vogelstein, B. (1993) Cell 75, 817-825. [DOI] [PubMed] [Google Scholar]
- 27.Wu, X., Bayle, J. H., Olson, D. & Levine, A. J. (1993) Genes Dev. 7, 1126-1132. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto, K. & Beach, D. (1994) EMBO J. 13, 4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita, T., Krajewski, S., Krajewska, M., Wang, H. G., Lin, H. K., Liebermann, D. A., Hoffman, B. & Reed, J. C. (1994) Oncogene 9, 1799-1805. [PubMed] [Google Scholar]
- 30.Oda, E., Ohki, R., Murasawa, H., Nemoto, J., Shibue, T., Yamashita, T., Tokino, T., Taniguchi, T. & Tanaka, N. (2000) Science 288, 1053-1058. [DOI] [PubMed] [Google Scholar]
- 31.Yu, J., Zhang, L., Hwang, P. M., Kinzler, K. W. & Vogelstein, B. (2001) Mol. Cell 7, 673-682. [DOI] [PubMed] [Google Scholar]
- 32.Passer, B. J., Nancy-Portebois, V., Amzallag, N., Prieur, S., Cans, C., Roborel De Climens, A., Fiucci, G., Bouvard, V., Tuynder, M., Susini, L., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 2284-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W. & Vogelstein, B. (1992) Nat. Genet. 1, 45-49. [DOI] [PubMed] [Google Scholar]
- 34.Tokino, T., Thiagalingam, S., el-Deiry, W. S., Waldman, T., Kinzler, K. W. & Vogelstein, B. (1994) Hum. Mol. Genet. 3, 1537-1542. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Y., Farmer, G., Soussi, T. & Prives, C. (1995) Oncogene 10, 779-784. [PubMed] [Google Scholar]
- 36.Waterman, J. L., Shenk, J. L. & Halazonetis, T. D. (1995) EMBO J. 14, 512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funk, W. D., Pak, D. T., Karas, R. H., Wright, W. E. & Shay, J. W. (1992) Mol. Cell. Biol. 12, 2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoh, J., Jin, S., Parrado, T., Edington, J., Levine, A. J. & Ott, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8467-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonish-Rouach, E., Resnitzky, D., Lotem, J., Sachs, L., Kimchi, A. & Oren, M. (1991) Nature 352, 345-347. [DOI] [PubMed] [Google Scholar]
- 40.Frew, I. J., Dickins, R. A., Cuddihy, A. R., Del Rosario, M., Reinhard, C., O'Connell, M. J. & Bowtell, D. D. (2002) Mol. Cell. Biol. 22, 8155-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leveillard, T., Andera, L., Bissonnette, N., Schaeffer, L., Bracco, L., Egly, J. M. & Wasylyk, B. (1996) EMBO J. 15, 1615-1624. [PMC free article] [PubMed] [Google Scholar]
- 42.Andera, L. & Wasylyk, B. (1997) Mol. Med. 3, 852-863. [PMC free article] [PubMed] [Google Scholar]
- 43.Hupp, T. R., Meek, D. W., Midgley, C. A. & Lane, D. P. (1992) Cell 71, 875-886. [DOI] [PubMed] [Google Scholar]
- 44.Halazonetis, T. D., Davis, L. J. & Kandil, A. N. (1993) EMBO J. 12, 1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagaich, A. K., Zhurkin, V. B., Sakamoto, H., Gorin, A. A., Clore, G. M., Gronenborn, A. M., Appella, E. & Harrington, R. E. (1997) J. Biol. Chem. 272, 14830-14841. [DOI] [PubMed] [Google Scholar]
- 46.McKinney, K. & Prives, C. (2002) Mol. Cell. Biol. 22, 6797-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo, P. K., Chen, J. Y., Tang, P. P., Lin, J., Lin, C. H., Su, L. T., Wu, C. H., Chen, T. L., Yang, Y. & Wang, F. F. (2001) J. Biol. Chem. 276, 37186-37193. [DOI] [PubMed] [Google Scholar]
- 48.Inga, A., Storici, F., Darden, T. A. & Resnick, M. A. (2002) Mol. Cell. Biol. 22, 8612-8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szak, S. T., Mays, D. & Pietenpol, J. A. (2001) Mol. Cell. Biol. 21, 3375-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu, G., Chung, Y. L., Glover, T., Valentine, V., Look, A. T. & Fearon, E. R. (1997) Genomics 46, 103-111. [DOI] [PubMed] [Google Scholar]
- 51.Liu, J., Stevens, J., Rote, C. A., Yost, H. J., Hu, Y., Neufeld, K. L., White, R. L. & Matsunami, N. (2001) Mol. Cell 7, 927-936. [DOI] [PubMed] [Google Scholar]
- 52.Maeda, A., Yoshida, T., Kusuzaki, K. & Sakai, T. (2002) FEBS Lett. 512, 223-226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.