Abstract
Several subsets of dendritic cells have been shown to produce type I IFN in response to viral infections, thereby assisting the natural killer cell-dependent response that eliminates the pathogen. Type I IFN production can be induced both by unmethylated CpG-oligodeoxynucleotide and by double-stranded RNA. Here, we describe a codominant CpG-ODN unresponsive phenotype that results from an N-ethyl-N-nitrosourea-induced missense mutation in the Tlr9 gene (Tlr9CpG1). Mice homozygous for the Tlr9CpG1 allele are highly susceptible to mouse cytomegalovirus infection and show impaired infection-induced secretion of IFN-α/β and natural killer cell activation. We also demonstrate that both the Toll-like receptor (TLR) 9 → MyD88 and TLR3 → Trif signaling pathways are activated in vivo on viral inoculation, and that each pathway contributes to innate defense against systemic viral infection. Whereas both pathways lead to type I IFN production, neither pathway offers full protection against mouse cytomegalovirus infection in the absence of the other. The Tlr9CpG1 mutation alters a leucine-rich repeat motif and lies within a receptor domain that is conserved within the evolutionary cluster encompassing TLRs 7, 8, and 9. In other TLRs, including three mouse-specific TLRs described in this paper, the affected region is not represented. The phenotypic effect of the Tlr9CpG1 allele thus points to a critical role for TLR9 in viral sensing and identifies a vulnerable amino acid within the ectodomain of three TLR proteins, essential for a ligand response.
Natural killer cells (NK) make an essential contribution to mammalian innate immune defense against viral infections (1). NK can directly sense viral pathogens through a specific and complex set of inhibitory and activating receptors (2–4). The molecular interactions that are required for viral sensing and response have been best documented in the case of mouse cytomegalovirus (MCMV) infection. The MCMV-encoded protein m157 interacts directly with the NK membrane protein LY49H, which contains an immunoreceptor tyrosine-based activation motif (ITAM) (5, 6). On ligand binding, LY49H causes NK activation, manifested in part by IFN-γ and perforin production.
However, an effective NK response also depends on extrinsic signals. Once MCMV infection is established, NK are activated by cytokines and chemokines, including type I IFN and IL-12, which are secreted by dendritic cells (DC). These molecules are known to influence the outcome of MCMV infection (7–9). Indeed, during MCMV infection, bidirectional communication between DC and NK is established. Both DC-derived cytokines and direct pathogen recognition are essential for NK activation. In turn, NK participate in the maintenance of the DC population (10). However, little is known about the earliest events that lead to type I IFN synthesis, which is known to occur during viral infection.
The host sensors that initially detect viral pathogens and cause cytokine production by myeloid cells have been investigated by several groups, some of which have pointed to possible involvement of Toll-like receptors (TLRs) (11, 12). The TLRs constitute a family of ligand-binding molecules that engage a variety of microbial products (13). On binding, they activate signaling cascades leading to the synthesis of proinflammatory molecules. TLR3, which recognizes double-stranded RNA (dsRNA) (14), and TLR9, which recognizes double-stranded DNA unmethylated at CpG motifs (15, 16), are likely to play a key role in viral infections (17). Indeed, two groups have reported that herpes simplex virus (HSV)-1- and HSV-2-induced IFN-β production is abrogated in cells harvested from TLR9 knockout mice (11, 12). A role for TLR3 in viral detection has been suggested by in vivo and ex vivo analyses that showed that this receptor is involved in the recognition of poly I:C and dsRNA from Lang reovirus (14). However, none of these studies has demonstrated that these TLRs are crucial for an efficient antiviral response in vivo.
We previously described the phenotype of Lps2 mice, which carry a nonsense mutation in the Trif/Ticam-1 gene (18, 19). The Trif gene encodes an essential adaptor molecule for MyD88-independent signaling downstream from TLRs 3 and 4 (18, 20). In macrophages harvested from homozygous Lps2 mutant mice, neither LPS nor dsRNA is capable of triggering phosphorylation or dimerization of IFN response factor (IRF)-3. Moreover, the animals display impaired responses to MCMV, fail to produce type I IFN when infected, and exhibit an abnormally elevated splenic viral titer during infection.
In the present study, we have identified a codominant N-ethyl-N-nitrosourea (ENU)-induced phenotype in which homozygous animals fail to respond to CpG-oligodeoxynucleotide (ODN) and also show severely impaired responses to MCMV infection as evidenced by viral titers measurement and survival analysis. This immunodeficiency phenotype is caused by a hypomorphic Tlr9 allele (Tlr9CpG1), encoding a structurally aberrant receptor. Tlr9CpG1 homozygotes display a low level of cytokine induction and NK activation on viral infection. Mice homozygous for a null allele of MyD88 show a similar phenotype, indicating that the TLR9 → MyD88 axis is essential for effective innate antiviral defense. We also tested the response to MCMV infection in mice homozygous for a null allele of Tlr3. Our results indicate that TLR3 plays a significant role, although a less crucial role than TLR9, in defense against systemic MCMV infection.
Materials and Methods
Mice. C57BL/6, BALB/c, Tlr3-/- mice (14), MyD88-/- mice (provided by S. Akira, Osaka University, Osaka), and TLR9CpG1/CpG1 mice were maintained under pathogen-free conditions in The Scripps Research Institute animal care facility. All mice used in the experiments were 6–9 weeks in age. All experimental procedures were conducted in accordance with institutional guidelines for animal care and use.
ENU Mutagenesis and in Vitro Stimulation of Thioglycolate-Elicited Macrophages. Germ-line mutagenesis was performed in mice by using ENU (19). Ex vivo stimulation of peritoneal macrophages with multiple microbial inducers and measurement of secreted tumor necrosis factor (TNF)-α activity were conducted as described in refs. 18 and 19. For analysis of the Tlr9CpG1/CpG1 phenotype, CpG-ODN (5′-tccatgacgttcctgatgct-3′) was used at the stated concentrations. Secreted TNF-α activity was measured 4 hours after induction by using the L-929 cell bioassay.
Virus Culture and Assay. MCMV (Smith strain) was prepared by in vivo propagation in 3-week-old BALB/c mice infected with 1 × 104 plaque-forming units (pfu) by an i.p. route. Salivary gland homogenates were prepared at day 14 postinfection, and viral titer was determined by plaque assay (21). Splenic viral titers were determined by plaque assay 4 days after MCMV infection.
Cytokine Measurements. IL-12 and IFN-γ were measured in serum harvested 36 h after MCMV infection by using an ELISA (R & D Systems). Type I IFN titers were determined with a bioassay, measuring the protection conferred by serially diluted serum against the cytopathic effect of the vesicular stomatitis virus in L929 cells (22). Purified IFN-α (R & D Systems) was used as a standard.
Fluorescence-Activated Cell Sorting Assay. Splenocytes were harvested and passed through a cell strainer 36 h postinfection, into Dulbecco's modified Eagle's medium containing 10% FBS and 2% penicillin/streptomycin solution (GIBCO). After fixation and permeabilization, cells were stained for the surface markers NK1.1 and T cell antigen receptor (TCR) β as well as intracellular IFN-γ by using antibodies from BD Bioscience. Flow cytometry was performed by using a FACSCalibur cell sorter and analyzed with cellquest software.
Transfection of HEK 293 Cells and Assay of TLR9 Signaling Activity. Full-length muTLR9 cDNAs (amplified from C57BL6/J and Tlr9CpG1/CpG1 mutant peritoneal macrophage mRNA) were cloned into pFlagCMV3 mammalian expression vector (Sigma). Plasmids were isolated by using the Endo-free Maxi prep kit (Qiagen, Valencia, CA). HEK293 cells (2 × 105) were transfected by using Lipofectamine (Invitrogen) with 0.4 μg of each expression vector and endothelial leukocyte adhesion molecule 1 (ELAM-1) luciferase reporter vector (23). After 24 h, cells were stimulated with 0.1 and 1.0 μM CpG-ODN for 5 h. Luciferase assays were performed by using the Steady-Glo luciferase assay system (Promega).
Identification and cDNA Cloning of TLRs 11, 12, and 13. All murine and human ESTs were downloaded from the National Center for Biotechnology Information database, translated in all six reading frames by using the program pepdata, and searched for Toll/IL-1R homology (TIR) domain homology (hmmersearch, E < 10) by using a hidden Markov model based on representatives of all known animal TIR domains. Three hits were acquired among the mouse ESTs. The corresponding cDNAs were amplified from macrophage mRNA by using primers based on genomic sequence to capture complete coding regions. Alignment of all human and mouse TLR protein sequences was performed by using the clustalw program (gap opening penalty = 10; gap extension penalty = 0.20; PAM series as the protein weight matrix). The TLR coding sequences were submitted to GenBank (accession nos. AY510704–AY510706).
Results
Identification of an ENU-Induced CpG-ODN Non-Responder Phenotype and Determination of the Causative Mutation Within the Tlr9 Locus. Among >11,000 mice examined to date, we identified an F3 mutant male in which thioglycolate-elicited peritoneal macrophages were markedly hyporesponsive to stimulation with CpG-ODN. Normal TNF-α production was observed when macrophages were stimulated with all other TLR agonists (data not shown). This finding suggested that the mutation, termed CpG1, specifically affected TLR9 signaling.
On sequencing the Tlr9 coding region, a T → C base transition was observed at position 1496 (gi:13626029), predicting the amino acid substitution L499P (Fig. 1A). To examine the affected residue with reference to all other mammalian TLRs, a hmmersearch of all human and mouse ESTs was performed to generated an updated set of TIR domain proteins for alignment. Three mouse TLRs were identified (TLRs 11, 12, and 13). They have not previously been reported and lack human orthologs. The total complement of mouse TLR proteins now numbers 12 because the mouse ortholog of human TLR10 is a degenerate pseudogene.
Fig. 1.
Isolation and identification of CpG1, an ENU-induced point mutation in TLR9. (A) Trace file of amplified genomic DNA from homozygous mutant mice (upper chromatogram) and normal animals (lower chromatogram). (Left) Location of the mutation within the 11th LRR in the TLR9 ectodomain. (Lower) Multiple alignment of all human and mouse TLR protein sequences reveals that the mutation resides in a region shared only by TLRs 7, 8, and 9. At left, the identity of each sequence is indicated. H, human paralog; M, mouse paralog. Note that TLRs 11 and 12 are nearest homologs, and TLR13 is most closely related to TLR3. (B) Transfection-based assay of TLR9 signaling activity. Error bars indicate the SD of duplicate transfections.
All 10 human TLRs, and all 12 mouse TLRs, were optimally aligned. Residue 499 of the mouse TLR9 protein lies within a part of the receptor ectodomain that, on comparison with all other TLRs in the multiple alignment plot, is represented only in TLRs 7, 8, and 9 [known to represent an evolutionary cluster (24)]. The leucine in question is located near the beginning of the eleventh leucine-rich repeat (LRR) motif predicted by simple modular research tool (smart) analysis. In TLR8, an isoleucine is represented at this position.
When reconstructed and overexpressed in transfected HEK 293 cells, the mutant TLR9 protein was insensitive to stimulation with CpG-ODN whereas the wild-type protein, expressed in the same system, was strongly CpG-ODN responsive (Fig. 1B). In addition, homozygous mutant mice were bred to normal C57BL/6 mice and then backcrossed to the mutant stock so that Tlr9CpG1 homozygotes and heterozygotes were obtained. Cells from Tlr9CpG1/CpG1 mice were uniformly unresponsive to CpG-ODN (0.1 μM concentration) stimulation whereas cells from Tlr9+/+ mice were uniformly responsive. Heterozygotes Tlr9CpG1/+ exhibited an intermediate phenotype. These data establish that the CpG1 phenotype is linked to the observed mutation in Tlr9 (P < 0.0001) and also reveal that the mutant allele is codominant, or alternatively, that the wild-type allele is haploinsufficient (Fig. 2A).
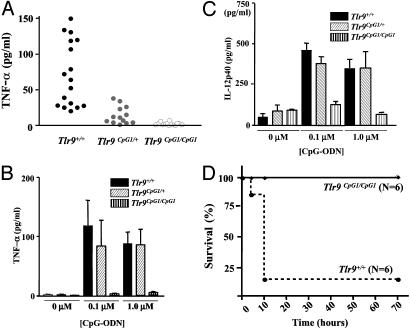
Fig. 2.
In vivo effects of the Tlr9CpG1 mutation. (A) TNF-α secretion by macrophages from controls C57BL/6 (Tlr9+/+, filled dots), heterozygotes (Tlr9CpG1/+, gray dots) and homozygotes (Tlr9CpG1/CpG1, open dots) animals after CpG-ODN induction (0.1 μM). Each dot represents the result of a duplicate induction assay performed on cells from a single animal. (B) TNF-α production or (C) IL-12p40 production by peritoneal macrophages at low (0.1 μM) CpG ODN concentration, as influenced by Tlr9 genotype. Error bars indicate SD; n = 5 mice. (D) Kaplan–Meier survival curves for Tlr9CpG1/CpG1 mice and Tlr9+/+ mice, after sensitization with d-galactosamine and challenge with CpG-ODN. Mice were monitored for a 3-day period, at which time all survivors seemed healthy.
The effect of heterozygosity for the Tlr9CpG1 allele on TNF-α and IL-12 secretion was further examined by varying the concentration of CpG-ODN used as a stimulus. A high CpG-ODN concentration (1 μM) allows a clear-cut distinction between Tlr9CpG1/+ and Tlr9CpG1/CpG1 genotypes but fails to resolve Tlr9CpG1/+ and Tlr9+/+ genotypes (Fig. 2 B and C). Hence, the codominant character of the Tlr9CpG1 allele is observed only at low CpG-ODN concentrations (<0.1 μM).
The Tlr9CpG1/CpG1 genotype was found to protect d-galactosamine-sensitized animals against the lethal effect of CpG-ODN-induced TNF-α in vivo. Homozygous mutants and wild-type controls were sensitized with d-galactosamine (20 mg per mouse) and then injected with CpG-ODN (20 nmol per mouse). One hundred percent of the homozygous mutants survived the injection whereas 83% of wild-type C57BL/6 mice succumbed within 10 h (Fig. 2D; P = 0.0025).
The TLR9 → MyD88 Signaling Axis Is Critical to Control MCMV Infection. Tlr9CpG1/CpG1 mice were examined in a second screen, designed to identify mutations that impair MCMV resistance. In this screen, a systemic infection is induced by i.p. injection of MCMV, by using an inoculum (5 × 105 pfu) that is well tolerated by normal C57BL/6 mice. This protocol has been established to distinguish between sensitive strains (such as BALB/c), which succumb 4–5 days after the inoculation, and resistant strains (such as C57BL/6), which survive even 8 days after the infection.
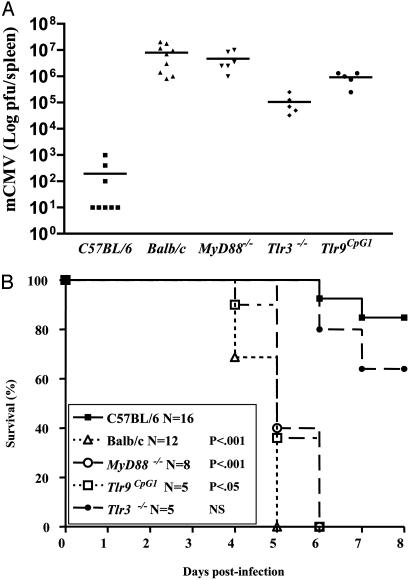
As shown in Fig. 3A, Tlr9CpG1/CpG1 mice accumulate exaggerated viral loads 4 days after infection, with titers approaching those observed in the BALB/c strain. Because TLR9 signals through MyD88 (25), we determined the viral load in MyD88-/- mice and observed that these animals also show high viral titers.
Fig. 3.
Tlr9CpG1/CpG1, MyD88-/-, and Tlr3-/- mice are hypersusceptible to viral infections. (A) Viral titers, expressed as log pfu per spleen, were determined in mice 4 days after i.p. inoculation with 5 × 105 pfu of MCMV. (B) Mice of the indicated genotype were infected i.p. with 5 × 105 pfu of MCMV, and survival was monitored for a period of 7 days. P values indicate comparisons with the survival curve of C57BL/6 control animals.
We have previously reported that the Lps2 allele of the Trif/Ticam-1 gene impairs macrophage IFN-β production in response to dsRNA. As a result, these mice are also permissive for MCMV infection (18). Because Trif is necessary for TLR3 signaling (18, 20), which is MyD88-independent (18), we reasoned that either component of the Trif → TLR3 axis might impair the immune response to MCMV. Accordingly, Tlr3-/- mice were examined for MCMV susceptibility. The average viral titer in the spleen of infected Tlr3-/- mice never reached that observed in Tlr9CpG1/CpG1 or MyD88-/- mice but showed a highly significant increase compared with wild-type controls (Fig. 3A), comparable to that previously reported in TrifLps2/Lps2 mutant mice (19). In the course of our infection studies, we repeatedly observed that, as in BALB/c mice, MyD88-/- and Tlr9CpG1/CpG1 mice showed obvious signs of sickness 4 days after MCMV inoculation. As shown in Fig. 3B, death follows viral infection of Tlr9CpG1/CpG1 and MyD88-/- mice almost as rapidly as it does in BALB/c animals. Tlr3-/- mutants showed no significant survival difference compared with C57BL/6 controls although, at a subjective level, the Tlr3-/- animals seemed sicker than controls, and a trend toward higher mortality was observed.
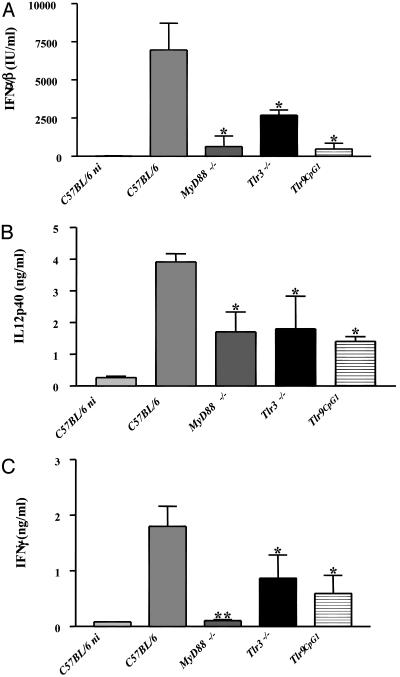
Cytokine Induction and NK Activation Are Impaired in Tlr9CpG1/CpG1 and MyD88 -/- Mice. Because the secretion of certain cytokines is a well known and essential response for the clearance of viral infections, we infected C57BL/6 control mice and mutant mice with an inoculum of virus that was sublethal during the term of the experiment (5 × 104 pfu per mouse) and monitored their cytokine levels in the serum at a time (36 h postinfection) when this response is at its maximum (26). As shown in Fig. 4A, type I IFN secretion is dramatically reduced in MyD88-/- mice as well as in Tlr9CpG1/CpG1 mice. IL-12 p40 levels (Fig. 4B) and IFN-γ levels (Fig. 4C) are also diminished in the homozygous mutants. We also note that IFN-γ production is strongly inhibited in MyD88-/- animals, which might be attributable to additional signaling defects in these mice (see Discussion). The decrease in serum cytokine levels is less pronounced in Tlr3-/- mice but is statistically significant.
Fig. 4.
In vivo impairment of cytokine production after MCMV infection in mice lacking the TLR9 → MyD88 signaling pathway. (A) IFN-α/β activity was measured in the serum of noninfected (ni) controls and MCMV-infected animals 36 h postinoculation. Values are expressed as international units (IU)/ml of serum. IL-12p40 (B) and IFN-γ (C) concentrations were measured in the serum of the same animals as those shown in A by ELISA and are expressed in ng/ml. Data represent mean values with SD (n = 4 mice). Statistical analysis was performed by using the ANOVA test with prism software (GraphPad, San Diego) (*, P < 0.05, **, P < 0.001, with respect to the C57BL/6 control values).
A clear link has been established between cytokine induction and NK activation during MCMV infection (8–10). We analyzed splenic NK and NKT cell populations 36 h postinfection by gating on NK1.1+/TCRβ- (for NK) and NK1.1+/TCRβ+ (for NKT cells) and then measuring intracellular IFN-γ. Fig. 5 shows the results of this analysis. The values (mean ± SD, n = 4 mice) indicate the proportion of IFN-γ+ NK or NKT cells among the total population. These data illustrate the pronounced effect (a 4-fold reduction) of both the Tlr9CpG1/CpG1 and the MyD88-/- genotypes on NK and NKT cell activation that occurs in the course of MCMV infection. The Tlr3-/- genotype was associated with a smaller, but still significant, decrease in the proportion of activated NK and NKT cells.
Fig. 5.
Impairment of NK and NKT cell activation after MCMV inoculation in TLR9 and MyD88 mutant mice. Purified splenic cells from uninfected or MCMV-infected animals were recovered 36 h postinoculation and gated against NK1.1 and TCRβ surface markers. (Left) IFN-γ intracellular staining (and isotype antibody as control) for NK (NK1.1+TCRβ-). (Right) Results for NKT cells (NK1.1+TCRβ+). Mean values ± SD are indicated (n = 4 mice).
Discussion
The L499P substitution specified by Tlr9CpG1 falls within a centrally placed LRR motif: 1 of 19 such motifs found in the TLR9 ectodomain. Within this LRR, the mutation would be expected to disrupt an alpha helix that normally contributes to the single loop formed by all LRRs (27). The region of the receptor within which the mutation occurs is represented only in a subset of TLR proteins. Neither the previously published TLRs (TLRs 1–6 and TLR10) nor the three mouse TLRs reported herein (TLRs 11, 12, and 13) exhibit a homologous region. It might therefore be imagined that the residue in question has a specialized function, related to the types of ligands that are engaged by TLRs 7, 8, and 9 (nucleotide-based molecules).
The Tlr9CpG1 allele was identified in two independent screening procedures: an ex vivo assay designed to identify new components of the TLR signaling pathways, and an in vivo protocol used to recover mutations that affect the innate antiviral response. The mutation markedly impairs the antiviral response, and the magnitude of the impairment is very similar to that associated with the MyD88-/- genotype, and indeed, to that associated with the BALB/c genotype. Hence, absent input from the TLR9 → MyD88 signaling axis, the normal m157 → LY49H sensing apparatus within the NK of the C57BL/6 mouse is inadequate to contain an MCMV infection.
Although MyD88 serves most of the TIR domain receptors (excluding TLR3), it is logical to suppose that most of the protection that MyD88 affords during MCMV infection results from its interaction with TLR9. Consistent with this conclusion, TLR2 and TLR4 deficiencies have no influence on the course of MCMV infection (P.G., unpublished results). On the other hand, the viral resistance phenocopy observed in MyD88-/- and Tlr9CpG1/CpG1 is imperfect. The exceptionally low level of serum IFN-γ observed in infected MyD88-/- animals (Fig. 4C) might reflect a requirement for MyD88 in certain TLR-independent signaling pathways for NK activation. IL-18, which signals through MyD88 (28) but not Trif (P.G., E.J., and K.H., unpublished results) and is essential for NK expansion after mCMV infection (10), may account for the discrepancy.
Because the DNA of herpesviruses such as MCMV is G/C-rich and has immunostimulatory activities (29), the MCMV-sensitivity phenotype that we have observed is likely due to a lack of TLR9-mediated viral DNA recognition and resulting impairment of IFN-α/β secretion. Two independent studies have recently identified plasmacytoid dendritic cells (pDC) and/or IFN producing cells (IPC) as the source of type I IFN production during HSV-1 and HSV-2 infections (11, 12) and have shown that IFN production is TLR9-dependent. Our data do not permit discrimination between these two categories of DCs as principal sources of type I IFNs in MCMV infection. We have examined the splenic pDC (CD8+/CD11c+/CD11b-) population in MCMV-infected MyD88-/- mice and observed that they undergo a reduction in numbers at day 1.5 after infection, and an expansion at day 4 after infection: a pattern also observed in wild-type control animals (data not shown). This finding, however, does not rule out their involvement in viral DNA recognition because this pattern of response is itself dependent on NK activation (10). It has also been suggested that IFN-α/β produced by IPC on viral challenge can activate immature DC (30), which in turn secrete type I IFN. Alternatively, both types of DC, as well as macrophages [which also express TLR9 (31)], may be activated by MCMV in vivo and synergize in resisting the viral pathogen.
Our data also implicate TLR3 as a key participant in the antiviral response. Approximately 1,000-fold augmentation of viral load in the spleen was observed in Tlr3-/- mice inoculated with MCMV, a finding consistent with earlier studies that showed a comparable increase in TrifLps2/Lps2 mice (18). TLR3 is known to serve as a ligand for dsRNA, and we presume that dsRNA may be produced as a consequence of bidirectional transcription from the MCMV genome in the course of infection (32).
The fact that impairment of either TLR3 or TLR9 signaling pathways has a dramatic effect on the course of disease is somewhat surprising. The TLR3 → Trif signaling axis is believed to be independent of the TLR9 → MyD88 signaling axis. Yet both signals lead to the induction of type I IFN (12, 18, 20). Abrogation of TLR3 signaling causes a >60% decrement, and abrogation of TLR9 signaling causes a >90% decrement in the amount of type I IFN that is measured in serum after infection. Hence, the two pathways seem to elicit the production of type I IFN in a superadditive or codependent manner.
It is possible that this relatively modest superadditivity results from signal transducer and activator of transcription (STAT)-1-mediated induction of additional type I IFN in response to IFN that is produced (33). However, when an infectious endpoint is examined, the effect of mutational inactivation of either the TLR3 or TLR9 pathway is even more pronounced, suggesting that numerous functional defects (rather than the relatively modest observed defect of type I IFN production) contribute to immunocompromise. MCMV titer is elevated by three orders of magnitude as the result of Tlr3 mutation, and by four orders of magnitude as the result of Tlr9 mutation. Whereas it might have been supposed that either pathway would complement the loss of the other, it seems, to the contrary, that both pathways are essential for containment of infection. Are these the only two pathways that allow DC to sense MCMV infection? It is possible that dsRNA sensing may also occur by means of protein kinase R (PKR), which may prompt the production of type I IFN (34). The relative importance of this pathway in DC remains to be established. Compound homozygous mutants (Tlr3-/-;Tlr9CpG1/CpG1) are currently being established and should clarify whether residual awareness of infection exists, absent both receptors for viral sensing.
In the past few years, a large volume of data has demonstrated the involvement of TLRs in the defense against bacterial infections, but their role in viral infection has remained unclear. In contrast to previous reports (11, 12), our in vivo infectious model establishes a prominent and essential function for TLR9 in MCMV resistance because mice devoid of CpG-mediated signaling quickly die after viral inoculation. Furthermore, our data suggest that both MyD88-dependent and -independent signals are essential for cytokine responses. TLR3- and TLR9-dependent type I IFN production activates NK, which in turn confine the viral pathogen and prevent its rapid spread during the critical interval before activation of an adaptive immune response.
Acknowledgments
We thank Marc Dalod (Centre d'Immunologie de Marseille-Luminy, Marseille, France) for helpful advice. This work was supported by National Institutes of Health Grant U54A154523 and by the Fondation Philippe (to P.G.).
Abbreviations: DC, dendritic cells; dsRNA, double-stranded RNA; ENU, N-ethyl-N-nitrosourea; LRR, leucine-rich repeat; MCMV, mouse cytomegalovirus; NK, natural killer cells; ODN, oligodeoxynucleotide; pfu, plaque-forming units; TLR, Toll-like receptor; HSV, herpes simplex virus; TNF, tumor necrosis factor; TCR, T cell antigen receptor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY510704–AY510706).
References
- 1.French, A. R. & Yokoyama, W. M. (2003) Curr. Opin. Immunol. 15, 45-51. [DOI] [PubMed] [Google Scholar]
- 2.Brown, M. G., Dokun, A. O., Heusel, J. W., Smith, H. R., Beckman, D. L., Blattenberger, E. A., Dubbelde, C. E., Stone, L. R., Scalzo, A. A. & Yokoyama, W. M. (2001) Science 292, 934-937. [DOI] [PubMed] [Google Scholar]
- 3.Daniels, K. A., Devora, G., Lai, W. C., O'Donnell, C. L., Bennett, M. & Welsh, R. M. (2001) J. Exp. Med. 194, 29-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, S. H., Girard, S., Macina, D., Busa, M., Zafer, A., Belouchi, A., Gros, P. & Vidal, S. M. (2001) Nat. Genet. 28, 42-45. [DOI] [PubMed] [Google Scholar]
- 5.Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B. & Lanier, L. L. (2002) Science 296, 1323-1326. [DOI] [PubMed] [Google Scholar]
- 6.Smith, H. R., Heusel, J. W., Mehta, I. K., Kim, S., Dorner, B. G., Naidenko, O. V., Iizuka, K., Furukawa, H., Beckman, D. L., Pingel, J. T., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 8826-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asselin-Paturel, C., Boonstra, A., Dalod, M., Durand, I., Yessaad, N., Dezutter-Dambuyant, C., Vicari, A., O'Garra, A., Biron, C., Briere, F. & Trinchieri, G. (2001) Nat. Immunol. 2, 1144-1150. [DOI] [PubMed] [Google Scholar]
- 8.Dalod, M., Salazar-Mather, T. P., Malmgaard, L., Lewis, C., Asselin-Paturel, C., Briere, F., Trinchieri, G. & Biron, C. A. (2002) J. Exp. Med. 195, 517-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalod, M., Hamilton, T., Salomon, R., Salazar-Mather, T. P., Henry, S. C., Hamilton, J. D. & Biron, C. A. (2003) J. Exp. Med. 197, 885-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews, D. M., Scalzo, A. A., Yokoyama, W. M., Smyth, M. J. & Degli-Esposti, M. A. (2003) Nat. Immunol. 4, 175-181. [DOI] [PubMed] [Google Scholar]
- 11.Krug, A., Luker, G. D., Barchet, W., Leib, D. A., Akira, S. & Colonna, M. (2003) Blood 103, 1433-1437. [DOI] [PubMed] [Google Scholar]
- 12.Lund, J., Sato, A., Akira, S., Medzhitov, R. & Iwasaki, A. (2003) J. Exp. Med. 198, 513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda, K., Kaisho, T. & Akira, S. (2003) Annu. Rev. Immunol. 21, 335-376. [DOI] [PubMed] [Google Scholar]
- 14.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732-738. [DOI] [PubMed] [Google Scholar]
- 15.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K. & Akira, S. (2000) Nature 408, 740-745. [DOI] [PubMed] [Google Scholar]
- 16.Ahmad-Nejad, P., Hacker, H., Rutz, M., Bauer, S., Vabulas, R. M. & Wagner, H. (2002) Eur. J. Immunol. 32, 1958-1968. [DOI] [PubMed] [Google Scholar]
- 17.Vaidya, S. A. & Cheng, G. (2003) Curr. Opin. Immunol. 15, 402-407. [DOI] [PubMed] [Google Scholar]
- 18.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., et al. (2003) Nature 424, 743-748. [DOI] [PubMed] [Google Scholar]
- 19.Hoebe, K., Du, X., Goode, J., Mann, N. & Beutler, B. (2003) J. Endotoxin Res. 9, 250-255. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K. & Akira, S. (2003) Science 301, 640-643. [DOI] [PubMed] [Google Scholar]
- 21.Orange, J. S., Wang, B., Terhorst, C. & Biron, C. A. (1995) J. Exp. Med. 182, 1045-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orange, J. S. & Biron, C. A. (1996) J. Immunol. 156, 4746-4756. [PubMed] [Google Scholar]
- 23.Schindler, U. & Baichwal, V. R. (1994) Mol. Cell. Biol. 14, 5820-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du, X., Poltorak, A., Wei, Y. & Beutler, B. (2000) Eur. Cytokine Netw. 11, 362-371. [PubMed] [Google Scholar]
- 25.Schnare, M., Holt, A. C., Takeda, K., Akira, S. & Medzhitov, R. (2000) Curr. Biol. 10, 1139-1142. [DOI] [PubMed] [Google Scholar]
- 26.Ruzek, M. C., Miller, A. H., Opal, S. M., Pearce, B. D. & Biron, C. A. (1997) J. Exp. Med. 185, 1185-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobe, B. & Deisenhofer, J. (1995) Curr. Opin. Struct. Biol. 5, 409-416. [DOI] [PubMed] [Google Scholar]
- 28.Adachi, O., Kawai, T., Takeda, K., Matsumoto, M., Tsutsui, H., Sakagami, M., Nakanishi, K. & Akira, S. (1998) Immunity 9, 143-150. [DOI] [PubMed] [Google Scholar]
- 29.Lundberg, P., Welander, P., Han, X. & Cantin, E. (2003) J. Virol. 77, 11158-11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Bon, A. & Tough, D. F. (2002) Curr. Opin. Immunol. 14, 432-436. [DOI] [PubMed] [Google Scholar]
- 31.Gao, J. J., Diesl, V., Wittmann, T., Morrison, D. C., Ryan, J. L., Vogel, S. N. & Follettie, M. T. (2002) J. Leukocyte Biol. 72, 1234-1245. [PubMed] [Google Scholar]
- 32.Jacobs, B. L. & Langland, J. O. (1996) Virology 219, 339-349. [DOI] [PubMed] [Google Scholar]
- 33.Marie, I., Durbin, J. E. & Levy, D. E. (1998) EMBO J. 17, 6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P. & Reis e Sousa, C. (2003) Nature 424, 324-328. [DOI] [PubMed] [Google Scholar]