Abstract
RNA interference technology, silencing targeted genes in mammalian cells, has become a powerful tool for studying gene function. For the first time in cancer research, we show that direct injection of a pool of naked small interfering RNA (siRNA) duplexes can prevent tumorigenesis in an animal model, suggesting a novel preventive and therapeutic strategy for cancer management. As a model system, we used siRNA duplexes of CXCR4 to block breast cancer metastasis. Here, we show that blocking CXCR4 expression at the mRNA level by a combination of two siRNAs impairs invasion of breast cancer cells in Matrigel invasion assay and inhibits breast cancer metastasis in an animal model. Targeting more than one site of the target gene may be important to overcome the functional redundancy of other variants of a single gene, especially in in vivo experiments. Moreover, our studies confirm the necessity of CXCR4 in breast cancer metastasis.
Introduction
RNA interference is a cellular mechanism by which double-stranded RNA triggers the silencing of the corresponding cellular gene (1–3). The double-stranded RNA in the cell is processed into short, 21- to 22-nucleotide double-stranded RNAs termed small interfering RNAs (siRNA; refs. 4, 5). A major breakthrough in the application of RNA interference technology in mammalian cells came from the development of a 21- to 22-nucleotide synthetic siRNA to silence targeted genes in mammalian cells (6, 7). Sorensen et al. showed that cationic liposome-based i.v. injection of mice with a plasmid encoding the green fluorescent protein with its cognate siRNA had the effect of inhibiting green fluorescent protein expression in various organs (8). Recently, it has also been shown that duplex siRNA can be effectively delivered and subsequently detected 24 hours post injection in the liver, lung, kidney, spleen, and pancreas of postnatal mice by the tail vein injection without a carrier (9). Song et al. also reported that silencing Fas expression using a single-dose i.v. injection of siRNA duplexes protected mice from autoimmune hepatitis for up to 10 days of observation (10).
Breast cancer cells usually metastasize to the regional lymph nodes, bone marrow, lungs, and the liver in an organ-selective process. The chemokine receptor, CXCR4, has shown to be one of the critical factors for breast cancer metastasis through interaction with its ligand, stromal cell–derived factor-1 (SDF-1; ref. 11). Here, we successfully blocked the in vitro invasion and in vivo metastasis of breast cancer cells in our animal model by silencing of CXCR4 gene expression with siRNAs. The siRNAs directed to CXCR4 will be useful for both the study of CXCR4 gene function and therapeutic applications for breast cancer metastasis.
Materials and Methods
Construction of siRNAs and Transfection
We designed and purchased two different siRNA duplexes of CXCR4 (Genbank accession no. NM_003467), siRNA1(sense, 5′-UAAAAUCUUCCUGCCCACCdTdT-3′), and siRNA2 (sense, 5′-GGAAGCUGUUGGCUGAAAAdTdT-3′) from Dharmacon (Lafayette, CO). The nonspecific control siRNA duplexes were purchased from Dhamacon with the same GC content as CXCR4 siRNAs (42%, D001206-10). Human breast carcinoma cell line MDA-MB-231 was cultured in 5% CO2 at 37°C in RPMI 1640 (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Sigma). The siRNAs were transfected into MDA-MB-231 cells at a final concentration of 120 nmol/L using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) in vitro.
Detection of siRNA Efficiency
To determine the efficiency of siRNA, at 48 hours post transfection, the transfected cells were collected to measure the mRNA levels and protein levels of CXCR4. At the same time point, cells were also immunostained with the biotinylated CXCR4 antagonist to measure CXCR4 protein levels as previously described (12). For Western blot analysis, a polyclonal rabbit anti-CXCR4 antibody (Ab-2) and a monoclonal mouse anti-β-actin were obtained from EMD Biosciences (San Diego, CA) and Sigma, respectively.
Tumor Cell Invasion Assay
The invasion assay was done by using a Matrigel invasion chamber from BD Biocoat Cellware (San Jose, CA) as previously described (12).
Cytotoxicity
The cells at 48 hours post transfection with siRNAs of CXCR4 were seeded in quadruplicate in 96-well plates (3,000 cells per well in 100 µL of medium). Twenty-four hours later, the cell proliferation was measured by the Cell Titer 96 AQ (Promega, Madison, WI) according to the manufacturer’s instruction. These experiments were repeated twice.
Animal Experiments
Animal experiments were done on 6- to 8-week-old CB-17 severe combined immunodeficient (SCID) female mice (Taconic Farms, Germantown, NY) divided into six groups with six mice per group. Group 1 mice were given injections of 2 × 106 MDA-MB-231 tumor cells transfected with nonspecific control siRNA duplexes and later were given injections of the control siRNA twice weekly. Groups 2 and 3 animals were given i.v. injections through the tail vein of 2 × 106 MDA-MB-231 tumor cells transfected with CXCR4 siRNA1+2 at 48 hours before the animal injection. The mice in group 2 were treated with CXCR4 siRNA1+2 twice weekly (0.5 µg/g body weight) whereas the mice in group 3 were not treated after administration of tumor cells. Mice in group 4 were given injections of 2 × 106 MDA-MB-231 cells without prior siRNA1+2 transfection and were treated only with CXCR4 siRNA1+2 twice weekly (0.5 µg/g body weight) post injection. Groups 5 and 6 animals received i.v. injections through the tail vein of 2 × 106 MDA-MB-231 tumor cells transfected with CXCR4 siRNA1 or 2, respectively, at 48 hours before the animal injection and were treated with siRNAs twice weekly. (For clarification, refer to Table 1.) The animals were sacrificed at 45 days after the tumor cell injection. Mice in groups 1 and 2 were imaged on the positron emission tomography (microPET) scanner with [18F]fluorodeoxyglucose (FDG), prior to sacrifice. Whole lung tissues were harvested in optimum cutting temperature (OCT, Fisher Scientific, Suwanee, GA) compound and snap-frozen in liquid nitrogen. The frozen lung tissues were sectioned, fixed in ice-cold acetone, and subjected to H&E histostaining to evaluate the presence or absence of tumors. These experiments were repeated once more to confirm the results. All protocols for animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at Emory University.
Table 1.
| Group | siRNAs | Pretransfection of siRNAs |
Posttreatment of naked siRNAs |
|---|---|---|---|
| 1 | Control siRNA |
Yes | Yes |
| 2 | siRNA1+ siRNA2 |
Yes | Yes |
| 3 | siRNA1+ siRNA2 |
Yes | No |
| 4 | siRNA1+ siRNA2 |
No | Yes |
| 5 | siRNA1 | Yes | Yes |
| 6 | siRNA2 | Yes | Yes |
Real-time Reverse Transcription–PCR Analyses
For reverse transcription–PCR (RT-PCR), total RNA was isolated from five 20-µm frozen sections of animal lungs by using Trizol (Invitrogen). The PCR primers of a human housekeeping gene, hypoxanthine-guanine-phosphoribosyltransferase (hHPRT), were designed according to a previous report (11). The human CXCR4-specific primers and the primers for β-actin are from our previous report (12). First-strand cDNA synthesis and amplification were done using a GeneAmp Gold RNA PCR Reagent Kit (Applied Biosystems, Foster City, CA). RT-PCR analysis and SYBR Green quantitative PCR amplifications (Applied Biosystems) were done in an iCycler with a multicolor real-time PCR detection system (Bio-Rad, Hercules, CA).
MicroPET Imaging of Lung Metastasis
Mouse microPET images were acquired on the Concord P4 microPET scanner that has 26-cm transaxial and 8-cm axial fields of view. Quality-control scans were done before scanning any animals and the scanner was calibrated by scanning a uniform phantom with the similar activity concentration as in the animals. Images were reconstructed with measured attenuation correction (20-minute scan with a Ge-68 point source that spirals through the field of view). The attenuation data were reconstructed into an image that was further segmented into tissue, air, and bone regions to which known attenuation coefficients were assigned. The resulting images were quantitatively calibrated and had 2-mm isotropic resolution. [18F]FDG was synthesized by the method described by Hamacher et al. (13) at specific activities of ~5,000 Ci/mmol. MicroPET studies were done using six mice from two groups, control siRNA (group 1) and CXCR4 siRNA1+2 treated (group 2). One hour after tail vein injections of 200 µCi [18F]FDG in a volume of 0.1 mL, the animals were anesthetized and placed on a platform inside of microPET scanner. Fifteen-minute images were acquired using microPET with the long axis of the mouse parallel to the long axis of the scanner. Data acquisition and processing, including image reconstruction, image display, and analyses were done by using the ASIPro program provided by Concorde Microsystems (Knoxville, TN). A pixel region of interest was outlined in the regions of increased FDG uptake, and after correction for radioactive decay, the maximal standard uptake value (SUVmax) was semiquantitatively calculated according to Truong et al. (14).
Statistical Analysis
Statistical significance was determined by Student’s t test. The correlation coefficient between CXCR4 and hHPRT was calculated by using Microsoft Excel.
Results
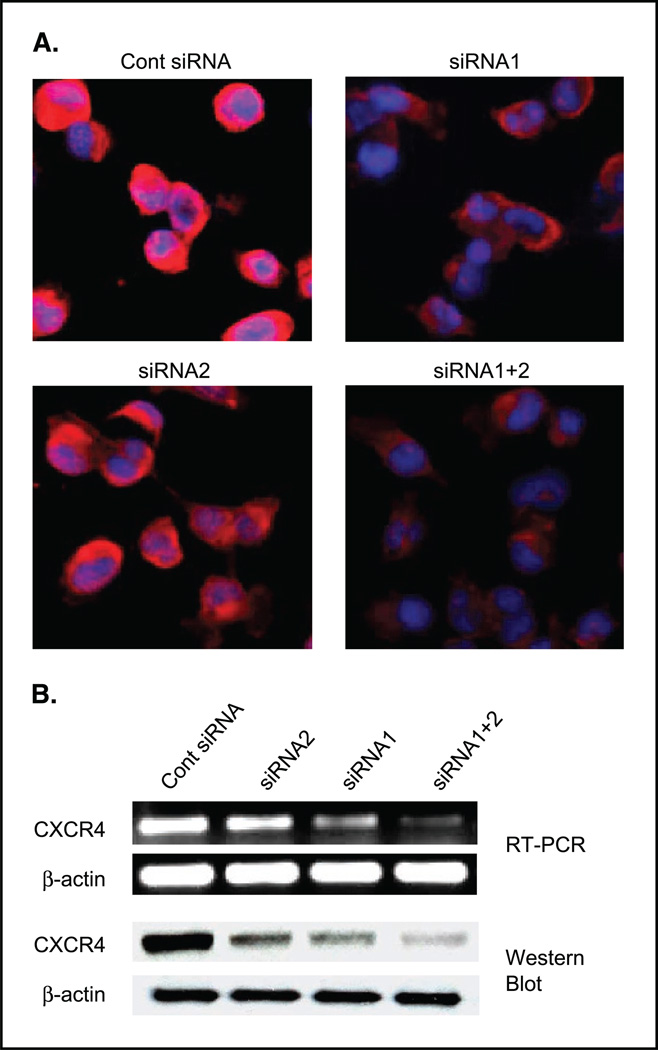
Inhibition of CXCR4/SDF-1 interaction by selective antagonists or anti-CXCR4 antibody blocks breast cancer metastasis in vivo (11, 12, 15, 16). Thus, we sought to determine whether lowering CXCR4 mRNA levels by using siRNA might inhibit breast cancer metastasis. Preventing expression of CXCR4 by RNA interference would likely be more efficient in preventing cancer metastasis mediated by the CXCR4/SDF-1 interaction. Two siRNAs were developed specific for the CXCR4 mRNA. The siRNA1 was more efficient in reduction of CXCR4 expression than siRNA2 (Fig. 1). The combination of siRNA1 and siRNA2 (siRNA1+2) achieved even better suppression of CXCR4 expression at the mRNA and protein level (Fig. 1).
Figure 1.
CXCR4 expression levels of MDA-MB-231 cells at 48 hours posttransfection of CXCR4 siRNA-transfected MDA-MB-231 cells were stained with immunofluorescence by using biotinylated CXCR4 antagonistic peptide and streptavidin-phycoerythrin. Red, CXCR4 phycoerythrin staining; blue, counterstaining of nuclei. B, RT-PCR analysis of CXCR4 of the siRNA-transfected MDA-MB-231 cells showed that siRNA1+2 effectively blocked the expression of CXCR4 mRNA. Western blot results of the siRNA-transfected MDA-MB-231 cells by using anti-CXCR4 antibody Ab-2 (1:1,000) showed that siRNA1+2 blocked the expression of CXCR4 protein almost completely. β-Actin (Sigma, 1:2,500) was used as a loading control. Cont, control.
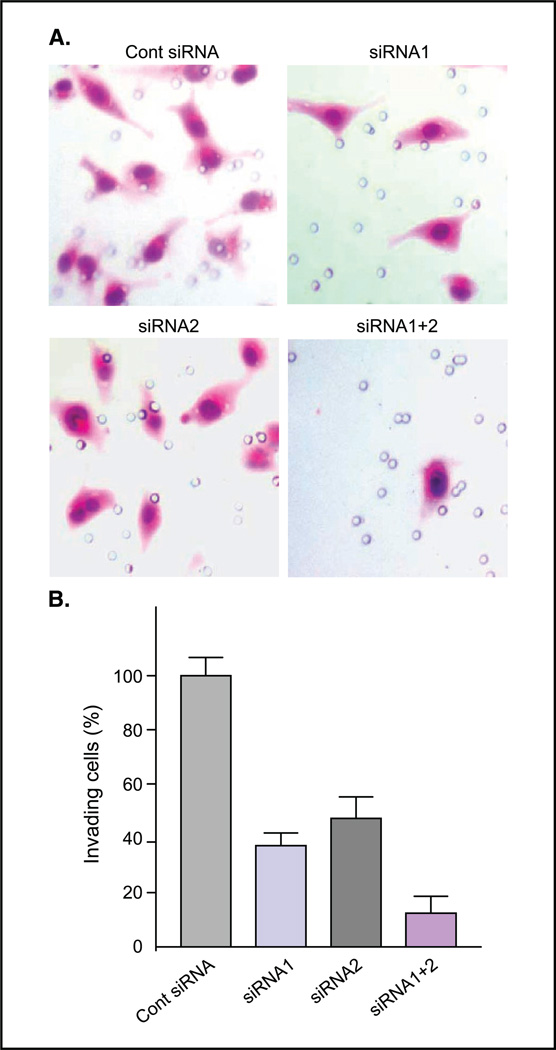
Lowering CXCR4 mRNA levels by siRNAs inhibited CXCR4/SDF-1-mediated invasion as measured by the Matrigel invasion assay. The invasion of MDA-MB-231 cells transfected with siRNA1 decreased to 39% of the control cells, to 51% with siRNA2, and to only 16% with both siRNA1+2 (Fig. 2).
Figure 2.
Invasion of MDA-MB-231 cells transfected with CXCR4 siRNAs. A, H&E staining showing the invasion of MDA-MB-231 cells transfected with control siRNA (Cont siRNA), siRNA1, siRNA2, or siRNA1+2 in a Matrigel invasion assay. B, invasiveness of MDA-MB-231 cells transfected with siRNA1+2, siRNA1, and siRNA2 relative to the control are 16% (P < 0.0003), 39% (P < 0.0014), and 51% (P < 0.0026), respectively. Average of three experiments.
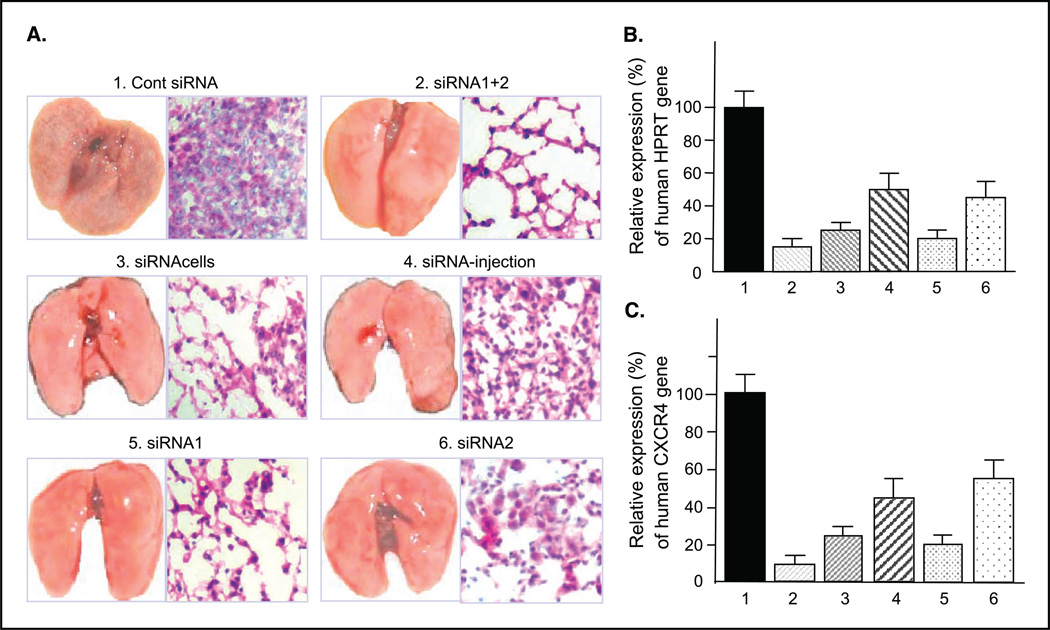
MDA-MB-231 cells transfected with CXCR4 siRNA1+2 were i.v injected through the tail vein of female SCID mice. Synthetic siRNA-mediated RNA interference in human cells is transitory with cells recovering from a single treatment in 4 to 6 days (7). Because of this, the CXCR4-silencing siRNAs were i.v. injected twice weekly by themselves (without liposome) after the injection of tumor cells. Six groups of animals used are described in Table 1. Forty-five days after the tumor cell injection, all animals in the control group (group 1) developed lung metastases. In contrast, only one animal in group 2 developed metastases and these were barely visible. The crude estimation of the average area of micrometastasis on the lung surface from six animals in group 1 was 54.2 ± 6.9%, whereas that from six animals in group 2 was 4.2 ± 1.7%. A representative picture of lungs in Fig. 3A shows grossly cystic lung micrometastasis in the control group. On the other hand, three representative pictures of lungs from five treated groups show significantly fewer visible lung metastases, most notably decreased in groups 2 and 5. The H&E staining of the lung tissues from group 2 shows the morphology of normal lung, whereas that from the control group shows invading tumor cells (Fig. 3A). These results were further confirmed by semiquantitative real-time RT-PCR using primers for the human housekeeping gene hHPRT that do not cross-react with its mouse counterpart (Fig. 3B). MDA-MB-231 cells in the lung were detected by RT-PCR using hHPRT rather than CXCR4 because CXCR4 levels in these cells would be expected low by the siRNA and might not indicate the presence of metastasis in the lungs. Real-time RT-PCR analyses showed high expression of hHPRT mRNA in metastasis-infiltrated lungs of the SCID mice in the control group. In contrast, there were significantly fewer metastases in the lungs of groups 2, 3, and 5 based on the expression levels of hHPRT in the lungs (Fig. 3B). As anticipated, real-time RT-PCR for human-specific CXCR4 gene of these lung tissues showed the high CXCR4 expression in the control group mouse lungs and much lower CXCR4 expression in the lungs of the treated group (Fig. 3C).
Figure 3.
Effect of CXCR4 siRNAs on inhibition of breast cancer metastasis in vivo. A, representative photographs of lungs and their H&E stainings (original magnification ×100) of each group from two independent experiments. B, average of real-time RT-PCR of hHPRT from siRNA-treated groups relative to that of control group. 1, group 1; 2, group 2; 3, group 3; 4, group 4; 5, group 5; 6, group 6 in Table 1. The average expression levels of hHPRT in groups 2, 3, 4, 5, and 6 were 12.5% (P < 0.0040), 21.5% (P < 0.0064), 47% (P < 0.0447), 25.3% (P < 0.0063), and 42.1% (P < 0.031), respectively, compared with that of group 1. C, percentage of human CXCR4 average expression level of each treated group is relative to that of the control group. The average expression levels of human CXCR4 in groups 2, 3, 4, 5, and 6 were 8.1% (P < 0.0006), 22.9% (P < 0.0022), 40.1% (P < 0.0157), 23.8% (P < 0.0074), and 59.1% (P < 0.152), respectively, compared with that of control group.
The potential cytotoxicity/clonogenicity of CXCR4 siRNA was determined by using the Cell Titer AQ96 Assay kit (Promega) on the reseeded cells transfected with either nonspecific control siRNA or the CXCR4 siRNA1+2, 48 hours post transfection. The growth of siRNA1+2 transfected cells was 85.9 ± 15.7% (P = 0.139) of control cell growth over 24 hours, indicating a relatively minor effect of siRNA treatment on adherent cell growth/clonogenicity. It is therefore unlikely that the absence of the lung metastasis in the siRNA-treated animals was due to the cytotoxic effect of CXCR4 siRNA on MDA-MB-231 cells, but rather due to inhibition of the metastatic process. This lack of cytotoxicity also implies a limited systemic effect on normal cells.
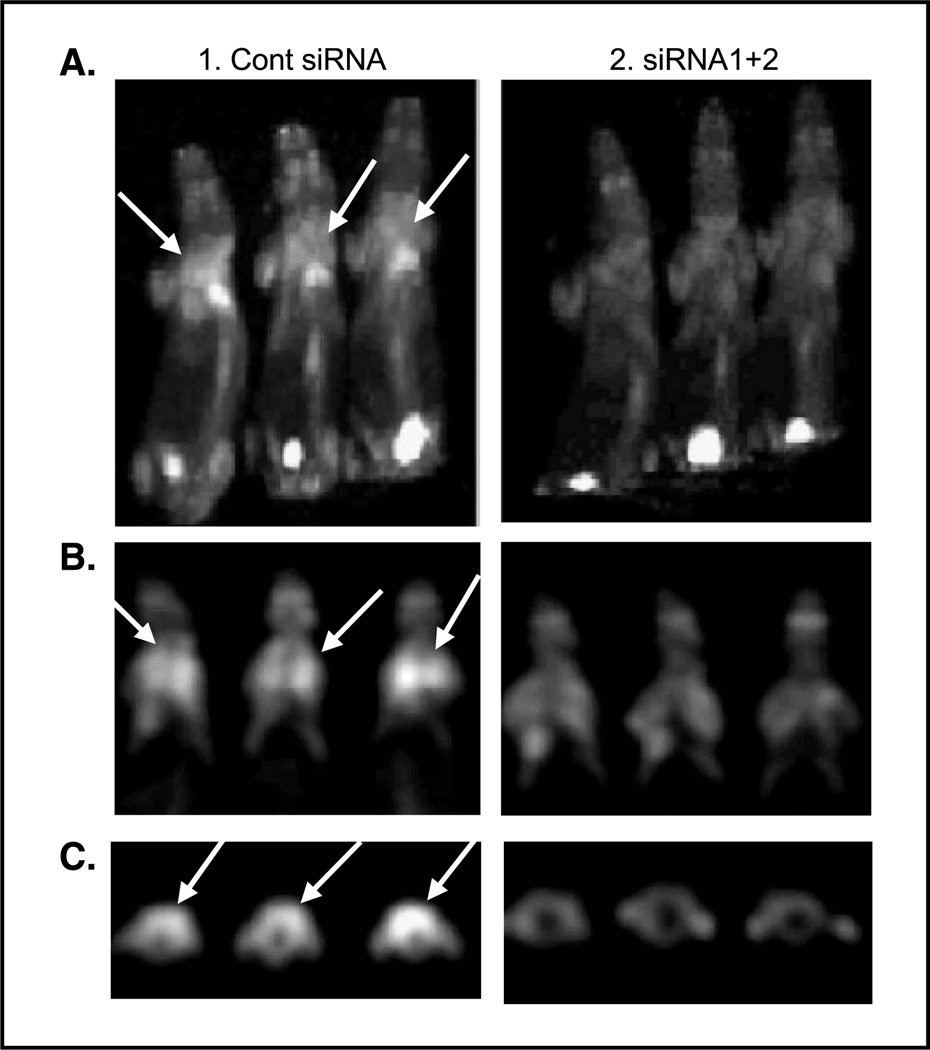
We utilized microPET imaging with FDG to detect lung metastases in mice in groups 1 and 2. Figure 4 shows representative FDG-PET images confirming lung metastasis in the control group and significantly fewer lung metastases in group 2. Figure 4A is a maximum-intensity projection generated from three representative mice in group 1 (control). The chest area is significantly brighter in each mouse of the control group (left) than any of the mice in the siRNA1+2-treated group (right). The high FDG uptake can also be seen in the bladder due to the secretion of FDG. Figure 4B and C are selected coronal and transaxial section images, respectively. The SUVmax of the lung area in Fig. 4C was 8.6, 7.1, 9.3, 2.2, 2.5, and 2.1. The crude estimation of the average area of micrometastasis on the lung surface from those six animals were 54%, 48%, 57%, 0%, 6%, and 1%. The relative expression levels of hHPRT in mouse 2, 3, 4, 5, and 6 were 0.82, 1.07, 0.02, 0.07, and 0.02, respectively, compared with that of mouse 1. Collectively, these images clearly show that FDG uptake is much higher in lungs from the control group (left) than those from the siRNA1+2-treated group (right ), which indicates significantly more lung metastases in the control group compared with those in the siRNA1+2-treated group.
Figure 4.
Effect of CXCR4 siRNAs on inhibition of breast cancer metastasis in vivo confirmed by FDG-PET. Representative images of FDG-PET of group 1 (control siRNA) and group 2 (siRNA1+2). A, maximum-intensity projection of six representative mice from group 1 (left, three mice) and group 2 (right, three mice). B, coronal sectional images from the lung area from the same animals in A. C, transaxial sectional images from the lung area from the same animals in A.
Discussion
Two siRNAs were developed specifically for the CXCR4 mRNA. The combination of siRNA1 and siRNA2 (siRNA1+2) achieved better suppression of CXCR4 expression in MDA-MB-231 cells than either one alone. Lowering CXCR4 mRNA levels by siRNAs inhibits CXCR4/SDF-1-mediated invasion as measured in the Matrigel invasion assay. The enhanced gene silencing with a combination of two different siRNAs not only resulted in a decrease of the protein and mRNA expression levels but also in the functional level.
siRNA directed against CXCR4 was shown to inhibit in vitro T-cell tropic HIV-1 infection of cells in vitro (17–19). An inducible RNAi system that targets a different sequence of CXCR4 also inhibited breast cancer cell migration in vitro (20). Previous work including our own had shown that blocking CXCR4/SDF-1 interaction by using anti-CXCR4 antibody or antagonist reduced metastatic burden in the lungs (11, 12). Our current work is different from previous work because we reduced CXCR4 levels by using antisense technology, rather than blocking receptor/ligand interaction. Prevention of metastasis by using CXCR4 antagonist (inhibition of protein interaction) does not guarantee the prevention of metastasis by using CXCR4 siRNA (inhibition of mRNA expression). Both are needed to confirm that CXCR4 is indeed a target for cancer prevention.
Various treatment combinations of siRNAs of CXCR4 were compared in experimental metastatic animal models of MDA-MB-231 cells. Groups 1 to 4 were compared to determine (a) whether lowering CXCR4 levels can block breast cancer metastasis to the lung and (b) what is the efficiency of the pretreatment or posttreatment of siRNAs. Our results showed that the most effective treatment was the injection of the CXCR4 siRNA1+2 transfected cells plus twice weekly i.v. injection of siRNA1+2. Direct injection of naked siRNAs resulted in 53% reduction in metastasis derived from untransfected cells and 78% reduction in metastasis in cells pretreated with siRNA determined by HPRT levels. The delivery of siRNA may be improved by delivering it in liposome or by conjugating to the ligands of surface receptors to be endocytosed. Nevertheless, as long as effective siRNA concentration was maintained, we found that cancer metastasis could be suppressed. Although the effect of siRNA in human cells is only transitory, mice injected with the siRNA-tranfected tumor cells without the maintaining siRNA injection formed 21.5% of lung metastases compared with the control group. One probable explanation is that tumor cells transfected with CXCR4 siRNA1+2 could not attach to lung tissues because the CXCR4 levels of these cells were substantially lowered. As a result, SDF-1 in lungs could not attract these tumor cells expressing inadequate CXCR4 protein on their cell surface. Groups 1, 2, 5, and 6 were compared to determine whether different combinations of siRNAs would result in differing degrees of efficiency in preventing metastasis. siRNA1 was more efficient in lowering CXCR4 expression than siRNA2 under both in vitro and in vivo conditions (group 5 versus group 6). The combination of siRNA1 and siRNA2 (siRNA1+2) achieved even more suppression of CXCR4 expression in vitro than either one alone. Similarly, the combination of siRNA1+2 in vivo prevented lung metastasis slightly more than siRNA1 alone (group 2 versus group 5). CXCR4 siRNA treatment only suppressed CXCR4 levels in MDA-MB-231 cells without affecting hHPRT levels in vitro. However, CXCR4 siRNA treatment significantly lowered lung metastasis reflected as hHPRT levels in the animal lungs. The reduced metastasis to the lungs in CXCR4 siRNA-treated animals was due to inhibited metastatic capability rather than cytotoxicity due to the siRNA. The overall correlation coefficient between hHPRT and CXCR4 (Fig. 3B and C) was 0.9674, demonstrating that lowered CXCR4 expression levels blocked breast cancer cells from forming lung metastasis. These findings were further supported by detection of lung metastasis using noninvasive FDG-PET, which allows sensitive detection of tumor cells regardless of depth, whereas most optical imaging methods are limited to the detection of s.c. tumors. Thus, CXCR4 is required and critical for lung metastasis of breast cancer cells.
We used siRNAs to inhibit CXCR4 gene expression and determined the effects of these siRNAs on breast cancer metastasis. Inhibition of CXCR4 expression at the mRNA level by a combination of two siRNAs significantly impaired the invasion of the breast cancer cell line MDA-MB-231 in a Matrigel invasion assay and blocked the lung metastasis of MDA-MB-231 cells in an animal model. These data are consistent with the requirement of CXCR4 in metastasis shown by using anti-CXCR4 antibody (11, 12) and suggest CXCR4 as a novel target for prevention of metastasis. In contrast to cytotoxic therapy, the metastatic program is specifically targeted by targeting CXCR4, one of the critical factors of metastasis. Takei et al. (21) reported that intratumoral injection of vascular endothelial growth factor siRNA suppressed tumor in an animal model. However, intratumoral injection lacks the merit of clinical implication. In our study, we used i.v. injection of a pool of siRNAs to prevent lung metastasis of breast cancer. This is the first demonstration that a pool of naked siRNA duplexes given through i.v. injection of animals inhibits breast cancer metastasis, opening a new avenue of cancer metastasis prevention.
Acknowledgments
Grant support: Georgia Cancer Coalition Distinguished Cancer Scientist Development Fund (H. Shim) and the AACR-Cancer Research and Prevention Foundation Fellowship in Cancer Prevention Research (Z. Liang).
We thank Drs. Jay Umbreit, Georgia Chen, and Daniel Brat at Emory University for critical reading of the manuscript.
References
- 1.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. RNA interference—2001. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 3.Plasterk RH. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM, Martinez J, Patkaniowska A, et al. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 8.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 9.Lewis DL, Hagstrom JE, Loomis AG, et al. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 10.Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 11.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 12.Liang Z, Wu T, Lou H, et al. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004;64:4302–4308. doi: 10.1158/0008-5472.CAN-03-3958. [DOI] [PubMed] [Google Scholar]
- 13.Hamacher K, Coenen HH, Stocklin G. Efficient stereospecific synthesis of no-carrier-added 2-[18F]-fluoro-2-deoxy-D-glucose using aminopolyether supported nucleophilic substitution. J Nucl Med. 1986;27:235–238. [PubMed] [Google Scholar]
- 14.Truong MT, Erasmus JJ, Munden RF, et al. Focal FDG uptake in mediastinal brown fat mimicking malignancy: a potential pitfall resolved on PET/CT. AJR Am J Roentgenol. 2004;183:1127–1132. doi: 10.2214/ajr.183.4.1831127. [DOI] [PubMed] [Google Scholar]
- 15.Tamamura H, Hori A, Kanzaki N, et al. T140 analogs as CXCR4 antagonists identified as anti-metastatic agents in the treatment of breast cancer. FEBS Lett. 2003;550:79–83. doi: 10.1016/s0014-5793(03)00824-x. [DOI] [PubMed] [Google Scholar]
- 16.Hatse S, Princen K, Bridger G, De Clercq E, Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002;527:255–262. doi: 10.1016/s0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- 17.Anderson J, Banerjea A, Planelles V, Akkina R. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res Hum Retroviruses. 2003;19:699–706. doi: 10.1089/088922203322280928. [DOI] [PubMed] [Google Scholar]
- 18.Martinez MA, Gutierrez A, Armand-Ugon M, et al. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16:2385–2390. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- 19.Ji J, Wernli M, Klimkait T, Erb P. Enhanced gene silencing by the application of multiple specific small interfering RNAs. FEBS Lett. 2003;552:247–252. doi: 10.1016/s0014-5793(03)00893-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4804. [PubMed] [Google Scholar]
- 21.Takei Y, Kadomatsu K, Yuzawa Y, et al. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]