Abstract
Sample preparation remains a challenge in untargeted metabolomics studies and no method currently results in complete extraction of all metabolite classes in human plasma. Because a large variety of molecules, with vast differences in dynamic range, could be involved in human disease, there is an urgent need to develop analytical techniques that result in comprehensive coverage of metabolites. Furthermore, analysis of more focused molecular classes could be necessary to more fully interrogate markers of human disease. However, such techniques, which generally involve multiple steps, often result in high variability. We have optimized a combined liquid-liquid and solid phase extraction method for plasma and have compared that to traditional methanol precipitation using spiked internal standards as controls. This method, based largely on previously published methods, results in 5 separate fractions enriched for aqueous species, phospholipids, fatty acids, neutral lipids, and hydrophobic lipids. Using liquid chromatography mass spectrometry as the analytical method, we detect over 3,806 metabolites using the new method versus 1,851 metabolites using methanol alone. Qualitative analysis and quantitative analysis of both internal standards (ISTDs) and endogenous metabolites demonstrate excellent reproducibility with CV’s below 15% for the combined method compared to 30% using the methanol method. While both methods have applications for clinical metabolomics, fractionation resulted in greater overall coverage and can be used for initial classification of molecular species.
Keywords: Metabolomics, profiling, liquid chromatography, mass spectrometry, plasma
1. Introduction
Advances in liquid chromatography mass spectrometry techniques have enabled metabolomics, or metabolite profiling, to become an effective means of measuring a large number of small molecule metabolites (molecular weight less than 1,500 Da) from biological samples without bias [1–5]. In spite of advances in column chromatography, instrumentation, and data analysis software, sample preparation is still an immense challenge in the rapidly growing field of clinical metabolomics. The choice of sample preparation method affects the observed metabolite profile and data quality, and can ultimately affect reported results. Without a deep understanding of the capabilities and limitations of the sample preparation method used in a given study, the accuracy of biological interpretation of collected data may be compromised [6].
Recently, several sample preparation methods have been applied to clinical samples with the goal of improving overall metabolite coverage, including organic solvent precipitation, liquid-liquid extraction (LLE), and solid-phase extraction (SPE)[7]. Polson et al. compared the effectiveness of 4 categories of protein removal techniques (organic solvent, acid, salt, and metal) in plasma for LC-MS studies [8]. Acetonitrile was found to be the most efficient organic solvent with 93.2% protein removal versus 88.6% by ethanol and 88.7% by methanol. Although it is presumed that improved protein removal results in improved extraction, no metabolite coverage was reported in this study. Want and colleagues also compared protein precipitation methods (organic precipitation, acid precipitation, and heat denaturation) in human serum for LC-MS [9]. A comparison of 14 protein precipitation methods (2:1 solvent to serum ratio), demonstrated that methanol and ethanol removed 98% and 96% of protein, respectively. Overall, 2,136 metabolite features were found when acetone-methanol (30:70) was used, 2,056 features when methanol alone was used, and 1,919 or 1,606 features when ethanol or acetonitrile were used, respectively. The average percent relative standard deviation (RSD) for the intensity of all detected metabolite features from each method was also calculated. It was determined that samples extracted with acetonitrile had the highest variation at 36% RSD, compared to 24% for acetone, 25% for methanol, and 27% for ethanol. Overall, methanol precipitation was recommended as the first choice for large scale metabolite profiling studies due to its straightforward preparation, low protein interference, comprehensive metabolite profile, and reproducible results. Bruce et al. did a comprehensive study on the performance of four different solvents (methanol, ethanol, acetonitrile, and acetone) and combinations of these solvents for sample preparation in human plasma [10]. Ethanol, methanol, and acetone all gave very good coefficient of variation (%CV) values of under 20% for nearly all labeled internal standards and endogenous compounds. Methanol-ethanol (1:1) and methanol-acetonitrile-acetone (1:1:1) at a 4:1 ratio to plasma were found to be the optimal methods as measured by the number of extracted molecular features, data quality, and column lifetime.
Solid phase extraction (SPE) has also been used for metabolomic profiling and enables fractionation of compounds in addition to protein removal. Michopoulos and colleagues fractionated human plasma using SPE composed of a C18 bonded phase with methanol as the eluting solvent[7]. Over 1,500 molecular features were detected and good repeatability was achieved when the SPE method was used compared to 1,300 features when the methanol precipitation was used. Excellent retention time stability over the entire run, with CV% values of less than 1%, was obtained. The reproducibility of the abundance of selected ions was found to be between 5 to 20 %CV.
Liquid-liquid extraction (LLE) is a very common sample preparation method used primarily in targeted metabolomics analysis. According to Folch et al [11] and Bligh-Dyer [12], chloroform-methanol extraction can be used to recover all major classes of lipids, including phosphatidylcholine (PC), sphingomyelin (SM), phosphatidylethanolamine (PE), cholesteryl ester (CE), ceramide (Cer), and lysophosphatidylcholine (LPC). However, this method can be inconvenient due to the high density of chloroform and its toxicity. Matyash reported that substituting chloroform with methyl tert-butyl ether (MTBE) also recovers most major classes of lipids with the same or better coverage than the Folch method and with reduced toxic effects [13].
Recently, Ferreiro-Vera et al. compared deproteinization and metabolome coverage when LLE and SPE were used to extract molecules from human serum samples [14]. In all, 1,685 molecular features were found when the tetrahydrofuran (THF) precipitation method was used, 1,911 features when the ethanol-chloroform (2:1) mixture extraction method was used, and 2,667 features by SPE. Fractionation may increase metabolite coverage in part due to a reduction or elimination of ion suppression for many metabolite species. SPE also provided better recovery of phospholipids than LLE and can be considered an alternate approach for metabolite profiling.
Each of the previously mentioned techniques, including organic precipitation, LLE, and SPE, has advantages and disadvantages for metabolite profiling of plasma. While straightforward, organic precipitation can be variable and results in a relatively low number of metabolites. LLE and SPE can improve sample quality, but additional fractions must be subsequently analyzed which is a disadvantage for global metabolite profiling. Any single method will necessarily bias the results towards specific classes of molecules. We sought to determine if combining methods could result in dramatically improved coverage and decreased variability when applied to human plasma. Differences in molecular features, reproducibility, and data quality were compared following extraction using various precipitation, LLE, and SPE strategies. Qualitative and quantitative measurements of internal standards were employed to evaluate the performance of each method.
2. Experimental
2.1 Reagents
All chemicals and solvents were of analytical or HPLC grade. Acetonitrile, methanol, ethanol, chloroform, acetic acid, and formic acid were purchased from Fisher (Fair Lawn, New Jersey). Methyl tert-butyl ether was purchased from J.T. Baker (Center Valley, Pennsylvania); isopropanol and water were from Burdick and Jackson (Morristown, New Jersey); and ammonium acetate was from Sigma-Aldrich (St. Louis, Missouri).
Internal standard (ISTD) compounds: creatinine-d3 (hydrophilic) was purchased from CDN Isotopes (Pointe-Claire, Canada); lysine-d4 (hydrophilic) and testosterone-d2 (hydrophobic) were purchased from Cambridge Isotope Laboratories (Andover, Massachusetts); heptadecanoic acid (17:0 FA) and cis-10-nonadecenoic acid (19:1 FA) were purchased from Sigma-Aldrich (St. Louis, Missouri). Lipids, 1,2-diheptadecanoyl-sn-glycero-3-phosphoethanolamine (17:0 PE), 1,2-dipentadecanoyl-sn-glycero-3-phosphocholine (15:0 PC), and N-heptadecanoyl-D-erythro-sphingosine (C17 Cer(d18:1/17:0)), were purchased from Avanti Polar Lipid Inc. (Alabaster, Alabama).
2.2 Sample Preparation
2.2.1 Instrumental
Centrifugation was carried out with a thermostatted centrifuge Microfuge 22R from Beckman Coulter (Palo Alto, California). A 12-port Phenomenex vacuum manifold was used for SPE fractionation.
2.2.2. Human plasma
Human plasma powder, purchased from Sigma-Aldrich, was reconstituted in 5mL water. This plasma sample was aliquoted to 100 µL aliquots and stored at −80°C.
2.2.3. Internal standards
Hydrophilic ISTDs were dissolved in 50% methanol-water to form a solution at 40 µg/mL; hydrophobic and lipid ISTDs were dissolved in chloroform-methanol (1:1) at 40 µg/mL.
2.2.4. Sample preparation protocols
Frozen plasma aliquots (100 µL) were thawed on ice, and 10 µL of each ISTD solution was added.
2.2.4.1. Precipitation
A volume of 400 µL ice-cold methanol or methanol-ethanol (1:1) was added to plasma sample aliquots containing internal standards, and vigorously vortexed for 15 min at room temperature. The samples were then centrifuged at 18000 × g0°C for 15 min. The supernatant was transferred to a new tube and dried by SpeedVac at 45°C. The dried supernatant residue was reconstituted in 80 µL of methanol followed by 20 µL of water, then centrifuged again at 18000 × g0°C for 15 min. The supernatant was collected and stored prior to analysis at −80°C after dilution with 100 µL 80% methanol in water.
2.2.4.2.MTBE extraction
A volume of 400 µL ice-cold methanol was added to plasma sample aliquots containing internal standards, then the mixture was vortexed vigorously for 30 sec and centrifuged at 18000 × g0°C for 15 min.
The supernatant was transferred to a glass culture tube followed by drying under N2 at 35°C. 2.5 mL of MTBE was added to the dried residue followed by vortexing vigorously for 10 sec, then 625 µL of water was added to the mixture and vortexed vigorously for 30 sec. The upper layer was removed to a new glass culture tube, and the lower layer was re-extracted with 2.5 mL of MTBE. Combined MTBE solution was dried under N2 at 35°C, and then dissolved in 200 µL methanol. This solution represents the MTBE lipid fraction used for analysis.
The lower layer was dried by N2, and then reconstituted in 100 µL water followed by 400 µL methanol. This mixture was frozen at −80°C for 30 min and then centrifuged at 18000 × g0°C for 15 min. The supernatant was removed and transferred into new vials, dried by SpeedVac at 40°C, and re-suspended into 5% ACN in water (aqueous fraction) for analysis.
2.2.4.3.MTBE extraction and SPE fractionation
The same procedure as outlined above for MTBE extraction was first used to obtain an aqueous fraction and a lipid fraction, followed by fractionation of the lipids using SPE. SPE fractionation of lipids was performed on 12-port vacuum manifold with Phenomenex Strata NH2 (55 µm, 70 Å) cartridges (100 mg/1mL). The MTBE-extracted lipid fraction was dried by N2, and dried lipids were dissolved into 500 µL chloroform. Cartridges were primed with two 500 µL volumes of hexane, then the lipid-containing chloroform solution was loaded and the flow through collected. The column was eluted with 800 µL chloroform-isopropanol (2:1) to the same flow through tube. The resulting sample was dried and dissolved in 200 µL methanol, resulting in a neutral lipid fraction. Next, the column was eluted with 800 µL 2% acetic acid in diethyl ether to a new tube, dried by N2, and dissolved in 200 µL methanol, resulting in a fatty acids fraction. The column was eluted with 500 µL methanol to new tube, dried by SpeedVac at 35°C, and dissolved in 200 µL methanol, resulting in a phospholipids fraction. All fractions were stored at −80°C prior to analysis.
Following precipitation with methanol, a white pellet remains. Metabolites in this pellet were extracted with 1.0 mL MTBE, vortexed vigorously for 30 sec, and centrifuged at 18000 × g0°C for 15 min. The supernatant was transferred to a new tube and the pellet re-extracted with 1.0 mL MTBE. The combined MTBE solution was dried by N2 at 35°C, reconstituted in 200 µL methanol, and stored at −80°C prior to analysis. This solution represents the pellet sample used for analysis.
2.3. Liquid Chromatography Time-of-Flight (LC-TOF) analysis
All samples were analyzed using an Agilent 1200 series Rapid Resolution system (Agilent Technologies, Santa Clara, CA, USA) containing a binary pump, degasser, well-plate autosampler with thermostat, and thermostatted column compartment. Because lipids and hydrophilic compounds have varying chemical properties, both reverse phase chromatography and hydrophilic interaction chromatography (HILIC) were applied to achieve better resolution for all metabolites. All lipid fractions were analyzed using a C18 column, and the aqueous fractions were analyzed using a HILIC column. Samples from organic precipitation were analyzed using both C18 and HILIC columns. All samples were analyzed in triplicate.
2.3.1. Reverse phase chromatography
An Agilent ZORBAX SB-C18 column 2.1 mm × 50 mm, 1.8 µm was used for samples from organic precipitation and for all lipid fractions, and an Agilent ZORBAX SB-C8, 2.1 mm × 12.5 mm, 5 µm guard column was placed in series in front of the analytical column. LC parameters: autosampler temperature, 4°C; injection volume, 2 µL; column temperature, 60°C; and flow rate, 0.25 mL/min. Mobile phases were 0.1% formic acid in water (phase A) and 0.1% formic acid in isopropanol:acetonitrile:water (60:36:4) (phase B). The gradient started at 30%B, and increased to 70%B from 0 to 1 min, then increased to 100%B from 1 to 15 min and held for 5 min, followed by 5 min 10%B washing and 5 min post run.
2.3.2. Hydrophilic interaction chromatography
A Phenomenex Kinetex HILIC column 2.1 mm × 50 mm, 2.6 µm was used for samples from organic precipitation and for all aqueous fractions, again with a guard column. LC parameters: autosampler temperature, 4°C; injection volume, 2 µL; column temperature, 20°C; and flow rate, 0.5 mL/min. Mobile phases were 10 mM ammonium acetate in 50% acetonitrile, pH 5.8 (phase A) and 10 mM ammonium acetate buffer in 90% acetonitrile, pH 5.8 (phase B). The gradient started at 100%B from 0 to 2 min, then decreased to 50%B from 2 to 15 min, followed by 5 min 0%B washing and 10 min post run.
2.3.3. TOF parameters
An Agilent 6510 TOF mass spectrometer equipped with an electrospray ionization (ESI) source was used. Mass spectra were acquired in extended dynamic range mode (2 GHz) using the following settings: ESI capillary voltage, 4000V; fragmentor, 170V; nebulizer gas, 50 psig; drying gas, 7 L/min; drying temperature, 300°C. Data were acquired at a rate of 2.0 spectra/sec in the mass range of m/z 50–1700, and stored in centroid mode. Mass accuracy was maintained by continually sprayed internal reference ions, m/z 121.0509 and 922.0098 in positive mode, m/z 112.9856 and 1033.9881 in negative mode.
2.4. Data processing
MassHunter workstation software (Agilent Technologies) was used for qualitative and quantitative analysis of raw data, including molecular feature extraction, formula generation, and concentration determination. Mass Profiler Professional (Agilent Technologies, MPP version 12.1) was used for molecular feature normalization and alignment, statistical analysis, and compound identification.
The molecular feature extraction algorithm in Mass Hunter extracted ions above 600 counts with a single charge state. Other extraction parameters were as follows: m/z range, 100 to 1100; peak spacing tolerance, 0.0025 m/z, plus 7.0 ppm; adducts species in positive mode, +H, +Na, and adducts in negative mode, −H, +HCOO; compound ion count threshold, two or more ions. All the related ions (including isotopes and dimers) were treated as a single feature. The term ‘features’, refers to compounds which have been extracted from the chromatogram using the ‘Molecular Feature Extraction’ algorithm. These features are then aligned based on retention time, mass-to-charge, abundance and molecular weight using Mass Profiler Professional. The data then undergoes filtering and re-analysis using the ‘Find by Formula’ algorithm in Qualitative Analysis. This allows isotope patterns and retention times to be distinguished to prevent duplicate features due to adducts, thus resulting in improved reliability of compounds identified.
The calculated neutral mass, retention time (RT), and summed abundances of each feature were exported to a compound exchange format (.cef file) for MPP analysis. MPP normalization and alignment parameters were as follows: abundance filter, >5000 counts; number of ions, 2; alignment RT window, 0.15 min intercept and 0.2% slope; alignment mass window, 2 mDa intercept and 5 ppm slope. The normalized features were filtered again, and only masses appearing in two of three samples were accepted. Background noise was removed by subtracting masses found in blank runs from filtered masses. Venn diagrams were used to compare masses from each method.
The extracted ion chromatogram (EIC) of each ISTD or endogenous metabolite was extracted with ±20 ppm single ion expansion using MassHunter software. Statistical comparisons were conducted using 2-tailed T-test with unequal variance.
2.5. Compound identification
An in-house database comprising data from METLIN and Lipid Maps was employed to tentatively identify compounds. All features were searched using the Agilent Personalized METLIN software and empirical formulas were generated for unknown compounds with the following parameters: ppm limit = 5, isotope model = common organic molecules, limit charge state to +1, and use +H or −H. Additional searches were conducted using sodium and potassium adducts but that data is not included in the current study. The hits with top scores were selected. Because the project goal was to compare the overall performance of sample preparation methods, no further confirmation of molecular species was performed.
3. Results and discussion
3.1. Protein precipitation with organic solvents
Protein precipitation is straightforward, inexpensive, and can result in good removal of proteins. Bruce et al. reported methanol-ethanol (1:1) was found to be an optimal solvent in precipitation method for global metabolic profiling analysis. However, the number of metabolites and the recovery of individual classes of metabolites were not reported in this study. Therefore we compared methanol and methanol-ethanol (1:1) as initial protein precipitation methods. Metabolites resulting from these two methods were analyzed in both positive and negative ionization mode using either SB-C18 or HILIC columns.
3.1.1. Reverse phase chromatography (RPC)-Positive ionization mode
As shown in Table 1, methanol-ethanol (1:1) precipitation results in a higher number of molecular features (1,620) compared to methanol alone (1,410), with over 70% features (1,138) in common. This result is consistent with Ferreiro-Vera’s results, where 1,325 features were detected using ethanol precipitation versus 851 features using methanol alone[14]. This could be due to the fact that the relatively hydrophobic solvent ethanol provides better solubilization of hydrophobic lipids compared to methanol. To better understand the differences between these two methods, tentative identification of common and unique features in each method was performed (Supplementary S1). Biologically relevant metabolites including free fatty acid, monoacylglycerol (MG), diacylglycerol (DG), short chain phosphatidylcholines (PC), glycerophosphatidylcholines (GPC), and glycerophosphatidylethanolamine (GPE) were found in both methods. Vitamin-D3 derivatives, sphingolipids, PC, and phosphatidylethanolamines (PE) were only found in methanol-ethanol extracted samples, while hydroxyl- or bromo-modified fatty acid and carnitine analogs were found only when the methanol-only method was used.
Table 1.
Comparison of molecular features detected by LC-TOF on RPC when methanol (MeOH) or methanol-ethanol (MeOH-EtOH) precipitation methods are used. RPC = reverse phase chromatography
| MeOH | MeOH-EtOH | Common features (%)a |
Unique in MeOH |
Unique in MeOH-EtOH |
|
|---|---|---|---|---|---|
| RPC-Positive Mode | 1410 | 1620 | 1138 (70%) | 272 | 482 |
| RPC-Negative Mode | 512 | 526 | 448 (85%) | 64 | 78 |
| Common in RPC | 71 | 73 | |||
| Total | 1851 | 2073 |
The percentage of common is calculated by the number in common vs numbers in MeOH-EtOH precipitation.
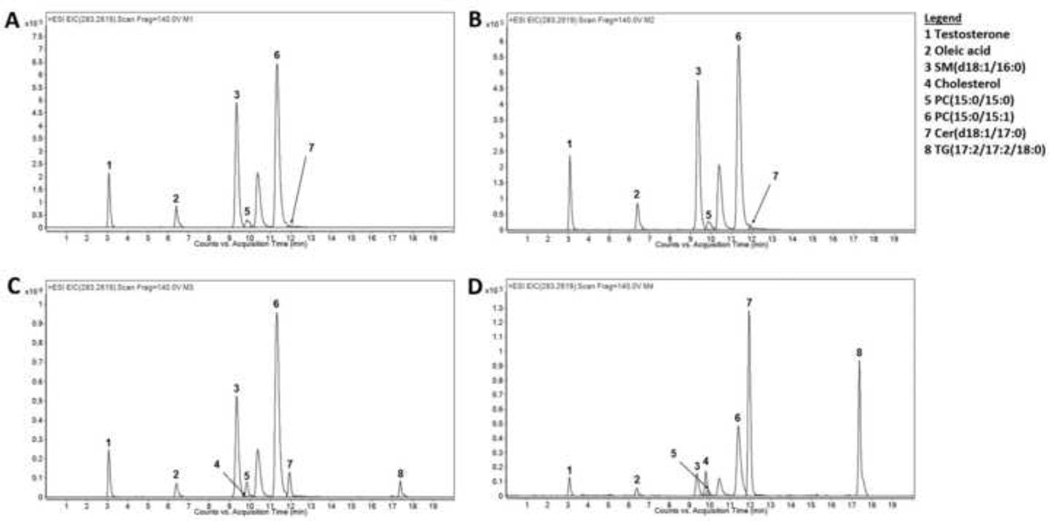
The recovery of 2 ISTDs and 6 endogenous metabolites was compared when various methods were used. As shown in Fig. 1A (Methanol precipitation) and 1B (Methanol-ethanol precipitation), while most compounds were recovered and resolved using RPC, cholesterol and TG(17:2/17:2/18:0) were not seen when either precipitation method was used. Figure 1 also includes data resulting from MTBE extraction (Fig. 1C) and MTBE:SPE extraction (Fig. 1D), as discussed in detail later.
Fig. 1.
The extracted ion chromatograms of representative ISTDs and endogenous metabolites on RPC with Methanol precipitation (A), Methanol-Ethanol (1:1) precipitation (B), MTBE extraction (C), and MTBE-SPE fractionation (D) methods are used. Extraction was performed and resulting samples were separated using RPC and analyzed using LCMS in positive mode as described in the text.
The chromatographic reproducibility of the organic precipitation methods was also evaluated by ISTDs. The retention time (RT) CV% values of all lipid ISTDs on reverse phase chromatography in both methanol and methanol-ethanol precipitation methods are below 1% (Shown in Supplementary S4).
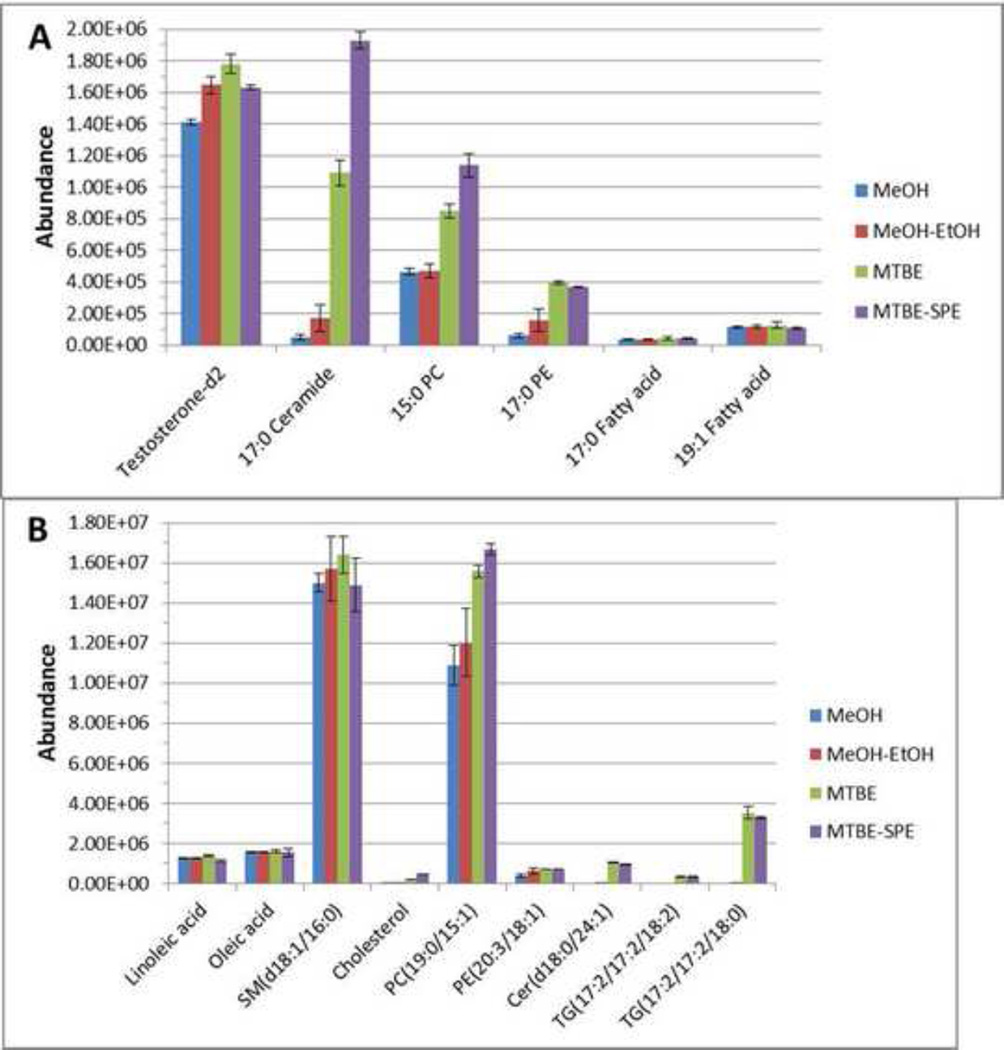
Internal standards were also used to compare the extraction efficiency of the organic precipitation methods (Fig. 2A). Similar amounts of 15:0 PC (p=0.89), 17:0 fatty acid (p=0.74), and 19:1 fatty acid (p=0.69) were extracted in both precipitation methods, with CV% values less than 15%. There is a significant increase of testosterone-d2 (p=0.011) when the methanol-ethanol method is used. Over 3 times more 17:0 Cer and 17:0 PE were extracted using the methanol-ethanol method compared to the methanol method. However, the CV% values of 17:0 Cer and 17:0 PE are significantly lower when the methanol method is used (25% and 30% versus 47% and 49%). For the endogenous metabolites (Fig. 2B), similar amounts of oleic acid (p=0.33), linoleic acid (p=0.49) SM(d18:0/16:0) (p=0.53) and PC(19:0/15:1) (p=0.38) were extracted using either method., A higher amount of PE(20:3/18:1) (p=0.094) was extracted with the methanol-ethanol precipitation compared to methanol only precipitation. Only trace amounts of Cer(18:0/24:1), cholesterol, and TG(17:2/17:2/18:0) were found in methanol-ethanol samples and these compounds were not detected when only methanol was used. This further confirms an increase in the extraction of hydrophobic lipids when ethanol-methanol is used.
Fig. 2.
The abundance of lipid ISTDs (A) and endogenous metabolites (B) following extraction and reverse phase chromatography and analyzed in positive ionization mode. Extraction was performed and resulting samples were separated using RPC and analyzed using LCMS in positive mode as described in the text.
3.1.2 Reverse phase chromatography (RPC)-Negative ionization mode
Overall, 512 features were detected in negative mode when the methanol supernatant was analyzed versus 526 features detected in the methanol-ethanol (1:1) supernatant (Table 1). Over 85% (448) features were found in common between the methods and there were 64 features unique to the methanol method and 78 features unique to the methanol-ethanol method.
A comparison of metabolites found only in negative ionization mode was performed based on a compound’s neutral mass and retention time. When methanol was used for extraction, an additional 441 (512–71) features were found compared to positive mode (Table 1). These included steroid lipids, diacylglycerol, free fatty acids, glucose, glycine derivative, and some glycophospholipids. When methanol-ethanol was used, an additional 453 (526–73) features were found compared to positive mode (Table 1). These included the same types of molecules listed above for the methanol method.
In total, 1,851 molecular features were extracted and detected in negative and positive mode when the methanol precipitation method was used versus 2,073 features when methanol-ethanol (1:1) precipitation was used.
3.1.3. Hydrophilic Interaction Chromatography (HILIC) - Positive ionization mode
Compared to reverse phase chromatography, HILIC offers improved retention of the polar/ionic analytes that elute very early or are not retained on reverse phase columns. Several groups have already reported that metabolite profiling studies utilizing HILIC offer a complementary approach to reverse phase LC-MS analysis for polar metabolites [15–17]. Here, the supernatant from methanol versus methanol:ethanol precipitation methods was analyzed using HILIC in positive and negative ionization modes.
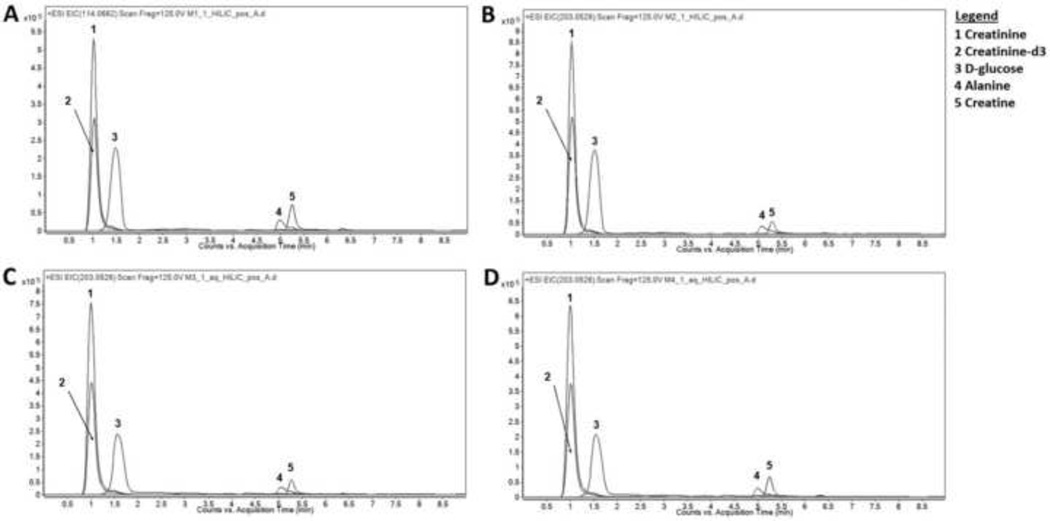
As shown in Table 2, 1,669 features were found when methanol alone was used to precipitate proteins compared to 1,831 features when methanol-ethanol was used. There were 1,278 (over 70%) common features, with 391 features unique to the methanol method and 553 features unique to methanol-ethanol method. Tentative identification of features was performed and results are listed in Supplementary Table S2. Dimethylglycine, ubiquinone, methionine, histidine, PEs, PCs, CERs, and others acids were found only in the methanol-ethanol method. The chromatographic behaviors of selected ISTDs and endogenous metabolites are shown in Fig. 3A and 3B. Regardless of the method, all hydrophilic compounds were detected and had excellent resolution on the HILIC column. Figure 3 also includes data resulting from MTBE extraction (Fig. 3C) and MTBE:SPE extraction (Fig. 3D), as discussed in detail later.
Table 2.
Comparison of molecular features detected by LC-TOF on HILIC when methanol (MeOH) or methanol-ethanol (MeOH-EtOH) precipitation methods were used. HILIC = Hydrophilic interaction chromatography.
| MeOH | MeOH-EtOH | Common features (%)a |
unique in MeOH |
unique in MeOH-EtOH |
|
|---|---|---|---|---|---|
| HILIC-Positive Mode | 1669 | 1831 | 1278 (70%) | 391 | 553 |
| HILIC-Negative Mode | 467 | 461 | 386 (84%) | 81 | 75 |
| Common in HILIC | 47 | 59 | |||
| Total | 2089 | 2233 |
The percentage of common is calculated by the number in common vs numbers in MeOH-EtOH precipitation.
Fig. 3.
The extracted ion chromatograms of representative ISTDs and some endogenous metabolites when HILIC is used following Methanol precipitation (A), Methanol-Ethanol (1:1) precipitation (B), MTBE extraction (C), and MTBE-SPE fractionation (D) methods. Extraction was performed and resulting samples were separated using HILIC and analyzed using LCMS in positive mode as described in the text.
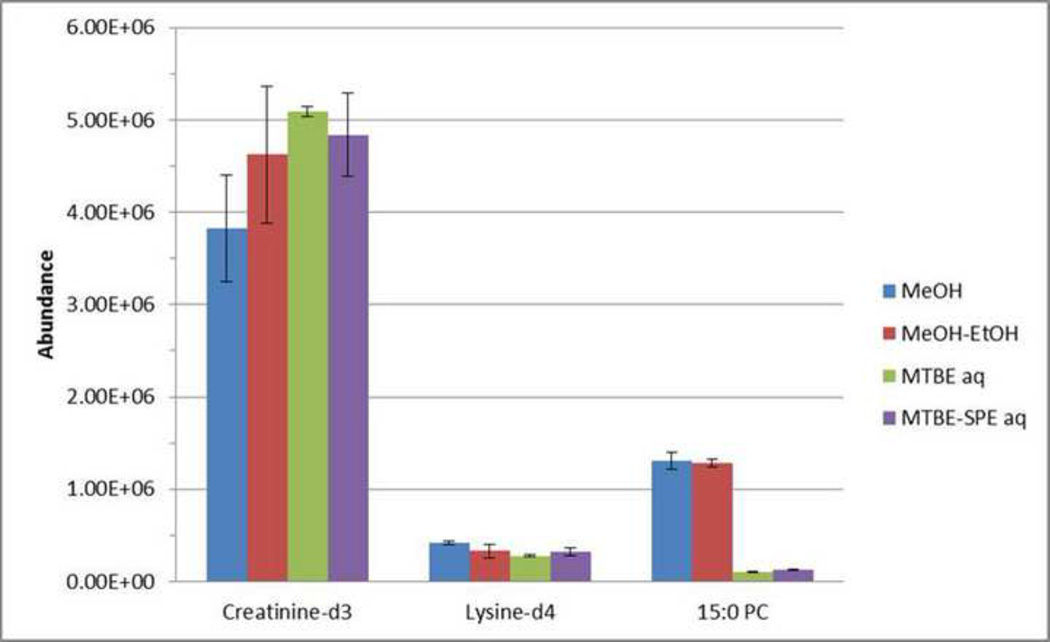
The retention time reproducibility of spiked internal standards is very good with CV% values less than 1% (Shown in Supplementary S6). As shown in Fig. 4, the recovery of creatinine-d3 is about 10% higher when methanol-ethanol is used compared to methanol alone, with %CV around 20% for both methods. Similar amounts of lysine-d4 (p=0.15) and 15:0 PC (p=0.62) are extracted using either method, with lower variation. These results are similar to what was obtained using reverse phase chromatography.
Fig. 4.
The abundance of hydrophilic spikes following extraction and separation by HILIC. Extraction was performed and resulting samples were separated using HILIC and analyzed using LCMS in positive mode as described in the text.
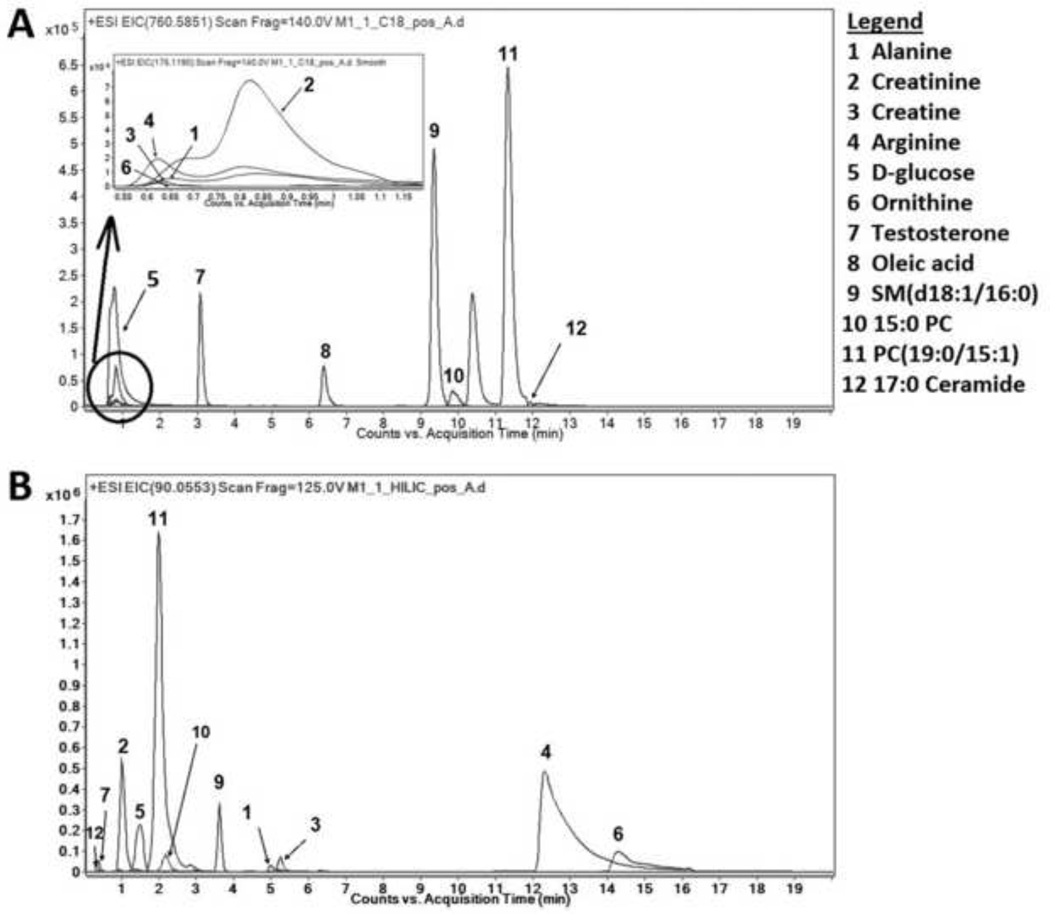
The elution profile of selected ISTDs and endogenous metabolites when RPC or HILIC are used are compared in Fig. 5A and 5B, respectively. Although lipids are well-resolved when RPC is used (compounds 9–12), co-elution of hydrophilic compounds results in ion-suppression (compounds 1–6). On the other hand, hydrophilic compounds have improved resolution when HILIC is used (Fig 5B). However, there is also co-elution of neutral and lipid compounds; testosterone-d2 co-eluted with 17:0 Cer, and PC(19:0/15:1) co-eluted with an unknown compound with a similar m/z. These could be separated using RPC. Oleic acid was not detected when HILIC was used but was detected using RPC. This comparison clearly shows advantages and disadvantages of these widely-used chromatographic methods. Based on our results, RPC was more appropriate for resolving lipids while HILIC resulted in better separation and detection of hydrophilic compounds.
Fig. 5.
The comparison of selected ion chromatograms of representative ISTDs and endogenous metabolites in RPC (A) and HILIC (B). Extraction was performed and resulting samples were separated using either RPC or HILIC and analyzed using LCMS in positive mode as described in the text.
3.1.4 Hydrophilic Interaction Chromatography (HILIC)-Negative ionization mode
Overall, similar numbers of metabolites were found when negative mode was used in conjunction with HILIC to compare methanol (467 features) and methanol-ethanol (461 features) extraction methods (Table 2). These methods shared 386, or 84%, features in common. In total, 2,089 molecular features were detected when methanol supernatants were analyzed using HILIC, and 2,233 features in methanol-ethanol supernatants.
In summary, both methanol-only and methanol-ethanol precipitation methods were very reproducible and resulted in fairly similar number of features when either RPC or HILIC was used. The addition of ethanol into methanol results in the extraction of additional hydrophobic compounds such as creatinine, 17:0 Cer, and 17:0 PE.
3.2 Comparison of MTBE extraction and methanol precipitation
Liquid-liquid extraction is generally used in targeted analyses, especially when lipids are the focus of the study. Here we combined methanol precipitation with MTBE extraction in order to achieve a higher degree of extraction for lipids. We hypothesized that a less complex sample would also result in few overlapping compounds, and reduced ion suppression of metabolites when RPC was used for lipids and HILIC was used for hydrophilic compounds. Therefore we performed multiple comparisons following methanol-MTBE extraction or methanol-only extraction. Because they are comparable methods, the methanol-only method was chosen over methanol-ethanol (1:1) for this comparison. Supernatants from methanol and the MTBE aqueous fraction were compared using HILIC chromatography in both positive and negative ionization mode. Methanol supernatants and the MTBE lipid fraction were resolved using RPC and analyzed in both ionization modes.
3.2.1. Reverse phase chromatography (RPC) – Positive ionization mode
In all, 1,828 molecular features were found in the MTBE extracted lipid fraction, and 1,410 features were found when methanol alone was used in conjunction with RPC (Table 3). Surprisingly, there were only 560 shared features between these two methods, and 1,268 unique features were detected in the lipid MTBE fraction. This difference could be due to compound overlap and/or high ion suppression when thousands of metabolites are being separated in a single LC-MS run. These results were obtained using HPLC with capillary flow, and it is acknowledged that UPLC would likely result in improved resolution. Tentative identification of the metabolites found to be unique to the MTBE lipid fraction was performed (Supplementary S3). Not surprisingly, several lipid classes including free fatty acids, sphingosines, diacylglycerols, ceramides, glycerophosphoethanolamine, glycerophosphocholine, sphingomyelin, and triacylglycerols were detected only in MTBE lipid fraction. Therefore separating lipids and hydrophilic metabolites results in improved detection of lipid species.
Table 3.
Comparison of metabolites from methanol precipitation (MeOH) and MTBE extraction (MTBE) on RPC and HILIC column in both positive and negative ionization mode.
| MeOH | MTBE | Common features (%)a |
Unique in MeOH |
Unique in MTBE |
|
|---|---|---|---|---|---|
| RP-Positive Mode | 1410 | 1828 | 560 (31%) | 850 | 1268 |
| RP-Negative Mode | 512 | 619 | 325 (53%) | 187 | 294 |
| HILIC-Positive Mode | 1669 | 1297 | 685 (53%) | 984 | 612 |
| HILIC-Negative Mode | 467 | 271 | 203 (75%) | 264 | 68 |
The percentage of common is calculated by the number in common vs numbers in MTBE extraction.
The recovery of 2 ISTDs and 6 endogenous metabolites was compared when various methods were used. All of these compounds were recovered and had good peak shape when MTBE extraction was performed. Significant amounts of cholesterol and TG(17:2/17:2/18:0) were detected following MTBE extraction, and these could not be detected when methanol alone was used (Fig. 1C). MTBE extraction also resulted in improved recovery of 15:0 PC (p=0.00136), PC(19:0/15:1) (p<0.001) and 17:0 Cer (p=0.00173) compared to methanol. The retention time (RT) CV% values of all lipid ISTDs on reverse phase chromatography are below 1% (Shown in Supplementary S4).
Internal standards were used to compare the extraction efficiency of the methods. Overall the MTBE method resulted in dramatic increases in lipid recoveries compared to methanol precipitation (Fig. 2A). For example, recovery of Testosterone-d2 (p=0.00611) was increased by 26%, 17:0 Cer (p=0.00173) was increased almost 200%, 15:0 PC (p=0.00136) was increased by 100%, and 17:0 PE (p<0.001) was increased by approximately 400%. Similar amounts of fatty acids were extracted using either method. The CV% values of ISTD abundances are generally less than 8%, except for fatty acids, which were approximately 20%.
MTBE also resulted in improved extraction for several endogenous metabolites (Fig. 2B). For example, recovery of PC(19:0/15:1) (p<0.001) increased by 40%, and PE(20:3/18:1) (p=0.012) increased by 58%. Similarly, cholesterol, Cer(d18:0/24:1) (p<0.001), TG(17:2/17:2/18:2) (p=0.00261), and TG(17:2/17:2/18:0) (p=0.00291) were detected when MTBE was used but were not observed when methanol alone was used. Recovery of linoleic acid and oleic acid was similar for either of the four methods.
3.2.2. Reverse phase chromatography (RPC) - Negative ionization mode
Metabolites extracted using MTBE or methanol, were also analyzed in negative ionization mode. In all, 619 features were found in the MTBE lipid fraction and 512 were found in the methanol supernatant, with 325 features in common (Table 3). Similar to the results from positive ionization, more lipids were extracted when MTBE was used compared to methanol precipitation alone.
3.2.3. HILIC
In positive ionization mode (Table 3), there were 1,297 features detected in the aqueous fraction following MTBE extraction, compared to 1,669 features detected in the methanol supernatant. Only 685 features were in common between these two methods; 612 features were only found in MTBE extracted samples and 984 features were only found in methanol supernatants. This could be predicted since the methanol method results in both aqueous and lipid species. Tentative identification of compounds showed that species such as homoserine, histidine, uric acid, and glycophosphocholine could be recovered in MTBE aqueous fraction but were not detected in methanol supernatants (Shown in Supplementary S5).
Internal standards and endogenous metabolites (Fig. 3A and 3C) had similar chromatographic behavior regardless of the method used with ISTD retention time CV values less than 2% (Data are shown in Supplementary S6). The separation of hydrophilic metabolites from lipids by MTBE also resulted in a slight increase of creatinine-d3 (p=0.063) with 1.04% CV versus 15.2% when no separation occurs (Fig. 4). Slightly less lysine-d4 was recovered from the MTBE aqueous fraction, and trace amounts of 15:0 PC was detected in the MTBE aqueous fraction, while most of it was extracted into the lipid fraction.
An additional 68 unique features were found when negative mode was used to analyze the MTBE aqueous fraction (Table 3).
Overall, the separation of metabolites into lipid and hydrophilic fractions using MTBE prior to analysis significantly increases coverage by 1,268 lipids and 612 hydrophilic compounds compared to the methanol precipitation method in positive mode. Reproducibility is improved when MTBE is used with CV’s less than 8% for lipids and 5% for hydrophilic compounds versus 29% and 15% for the methanol method. Since it entails a second extraction step, liquid-liquid extraction also resulted in cleaner samples, thereby presumably prolonging column lifetime.
3.3. MTBE extraction with SPE fractionation
Methanol precipitation combined with MTBE extraction significantly increases the metabolite coverage by separating lipids from hydrophilic compounds prior to analysis by RPC. However, our results showed that co-eluting lipids could still result in ion suppression. To address this issue, SPE fractionation was employed following MTBE extraction in order to further separate lipid classes. For these experiments, the supernatant resulting from methanol precipitation was separated by MTBE into aqueous and lipid fractions as described above. The lipid fraction was then separated using SPE. Finally, the pellet resulting from the initial methanol precipitation step was also extracted with MTBE. Although time consuming, this is a very comprehensive method that is expected to extract a very high number of metabolites. In addition, some information regarding the chemistry of the molecules can be elucidated based on their elution profile.
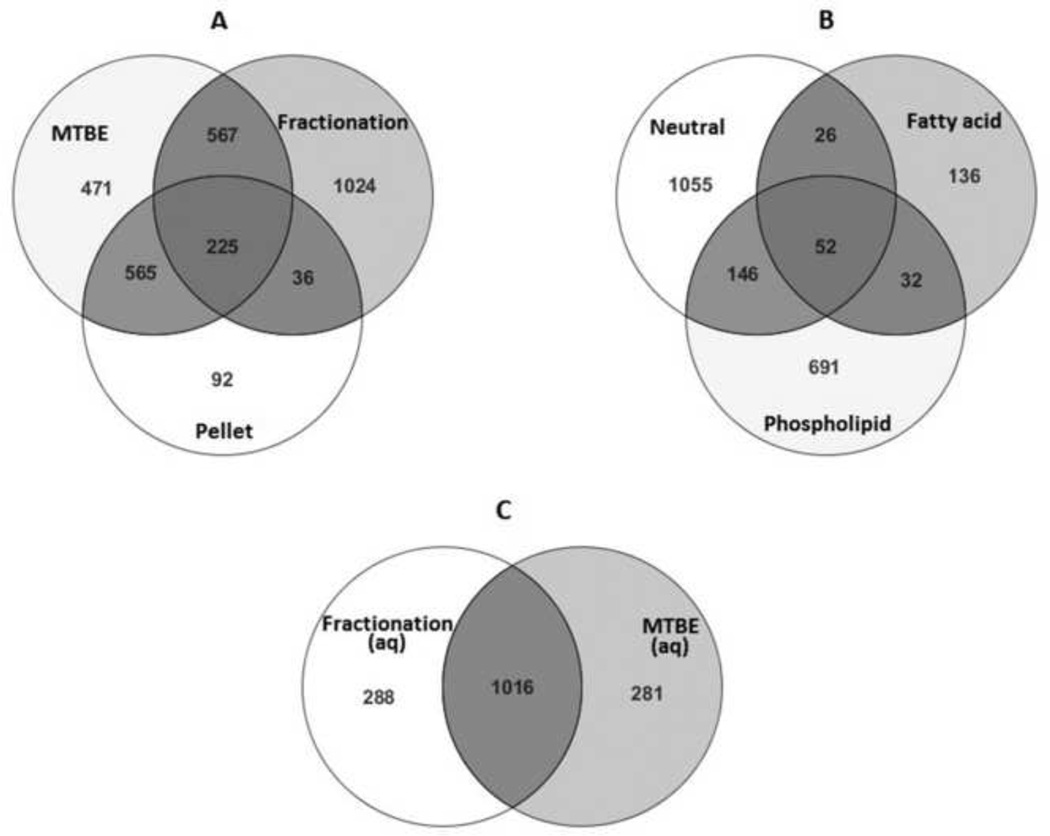
This new, combined method fractionates the MTBE lipid fraction into three fractions which are enriched for neutral, fatty acid, and phospholipid compounds respectively. A separate step can be utilized to extract hydrophobic lipids that are found in the pellet following methanol precipitation and would otherwise be lost. As shown in Fig. 6A, a total of 1852 (1024+36+225+567) features were found in all three SPE lipid fractions, compared to 1828 (471+567+225+565) features found in the MTBE-only lipid fraction, and 918 (565+225+36+92) features in the methanol pellet. Comparing the effect of SPE fractionation, 1060 (1024+36) unique features were recovered when SPE fractionation was employed; however, there were 1036 (471+565) features unique to the MTBE-only lipid fraction which were therefore lost when the SPE column was used. Although SPE fractionation may result in decreased ion suppression and increased resolution, many features are also lost due to inefficient transfer or poor NH2 column absorption. When features found in the methanol pellet fraction were compared to MTBE only lipids without SPE, 790 (565 + 225) out of 918 (86%) features were common. Therefore, 565 out of 1036 lost features would be recovered with the extraction of the pellet. Metabolites found to be unique in the MTBE only lipids without SPE (Supplementary S7), included highly hydrophobic compounds such as ceramides, sphingomyelins, and triglycoceramides.
Fig. 6.
A comparison of MTBE extraction vs MTBE-SPE fractionation methods using Venn Diagrams. Extraction was performed and resulting samples were separated using RPC (lipids) or HILIC (aqueous) and analyzed using LCMS in positive mode as described in the text. Mass Profiler Professional was used to analyze resulting data. A, Comparison of lipid fraction, fractionated lipid fractions, and pellet fraction; B, Comparison of fractionated fractions; C, Comparison of the aqueous fractions.
The chromatography of several representative ISTDs and endogenous metabolites were compared between the precipitation and extraction methods (Fig. 1D). Similar to MTBE extraction, all lipid ISTDs were recovered, including cholesterol and triacylglycerol. The reproducibility of the method was excellent with CV% values less than 0.5% (Shown in Supplementary S4). Comparing the recovery of each ISTDs and endogenous metabolites (Fig. 2A and 2B), the MTBE-SPE extraction method resulted in improved extraction for metabolites such as 17:0 Cer (p<0.001) and 15:0 PC (p=0.00384), which were not extracted using methanol precipitation alone.
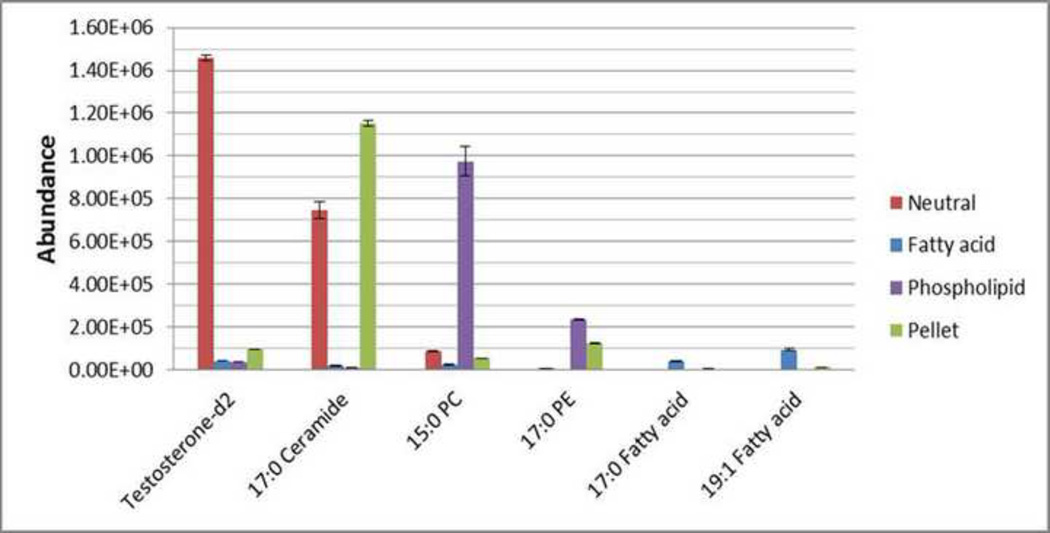
All three SPE lipid fractions, neutral, fatty acid, and phospholipid were analyzed using the same LC-MS parameters and the resulting chromatograms overlapped very well (Data not shown). Overall, there were 1279 features found in the neutral fraction, 246 features found in the fatty acid fraction, and 921 features found in the phospholipid fraction (Fig. 6B). There were 52 features common to all fractions, 146 features shared between neutral and phospholipid fractions, 26 features shared between neutral and fatty acid fractions, and 32 features shared between phospholipid and fatty acid fractions. However, most features were unique to a single fraction; for example, 1,055 out of 1,279 (82%) were unique to the neutral fraction and 691 out of 921 (75%) were unique to the phospholipid fraction. This confirms a relatively high specificity for fractionation based on chemical properties. Tentative identification of unique features in each fraction was performed (Supplementary S8). Compounds including sphingosine, cholesterol, glycoceramides, triglycoceramides were found in the neutral fraction, while most phosphocholines, phosphoethanolamines were found in the phospholipid fraction, and fatty acids such as palmitoleic acid, heptadecylenic acid, hexadecanoic acid, 3-hydroxy-eicosanoic acid were found only in the fatty acid fraction. This is further confirmed by comparing ISTDs (Fig. 7) where the majority of testosterone-d2 and ceramide were found in the neutral fraction, phospholipids were eluted to the phospholipid fraction, while fatty acids were fractionated to the fatty acids fraction. Therefore, the SPE fractionation did efficiently separate the lipids into different classes. The extraction of the pellet resulted in additional recovery of ceramide, testosterone-d2, 15:0 PC, and 17:0 PE.
Fig. 7.
The recovery of ISTDs in fractions using the MTBE-SPE method. Extraction was performed and resulting samples were separated using RPC and analyzed using LCMS in positive mode as described in the text.
Following MTBE-SPE fractionation and analysis, the resulting aqueous fraction was analyzed on a HILIC column using both positive and negative ionization modes. The comparison of aqueous fractions when MTBE-only and MTBE-SPE extraction methods are used is shown in Fig. 6C. As expected, there was almost identical numbers of features found in both methods (1,304 and 1,297, for MTBE-only and MTBE-SPE, respectively) and 1,016 features (78%) were in common.
As the extracted ion chromatograms of ISTDs and endogenous metabolites show (Fig. 3D), the hydrophilic compounds had similar recoveries and all were reproducible. As depicted in Fig. 4, creatinine-d3 (p=0.079) had 26% increase in abundance using the MTBE-SPE method compared to methanol precipitation, however lysine-d4 (p=0.037) was 20% lower. All internal standards were extracted with CV% values less than 2% in both MTBE-only and MTBE-SPE extraction methods (Shown in Supplementary S6).
In total, 3806 molecular features (1,852 lipids + 918 pellet lipids − 261 common compounds + 1,297 hydrophilic compounds) were found when the MTBE-SPE method was used and samples analyzed in positive ionization mode. This includes compounds found in aqueous, neutral, fatty acid, phospholipid, and pellet fractions.
4. Conclusions
Sample preparation is crucial in order to obtain good metabolite coverage and reproducibility when performing metabolomic profiling studies. Recently, Vuckovic reviewed commonly used sample preparation methods for untargeted metabolomics using LC-MS [6]. He pointed out the ideal method should be non-selective to ensure adequate depth of metabolite coverage, simple and fast to avoid metabolite loss or degradation, reproducible, and should incorporate a metabolism-quenching step to represent true metabolome composition at the time of sampling. To our knowledge, no reported method can obtain full coverage of all 4,651 metabolites that have so far been found in human serum (www.serummetabolome.ca). In order to improve metabolite coverage and enable more emphasis on lipid classes, methanol precipitation was followed by LLE or by LLE and SPE. These methods were compared with organic precipitation using methanol or methanol-ethanol (1:1). Overall, 1,851 metabolites could be detected when methanol precipitation was used with CV% values less than 30% (Table 1). In comparison, the methanol-ethanol (1:1) method increased the metabolite coverage to 2,073 compounds (Table 1). The presence of ethanol improved the extraction of sphingolipids and phospholipids, and increased the recovery of testosterone-d2, 17:0 Cer, 17:0 PE, and creatinine-d3. Separation of lipids from hydrophilic metabolites by MTBE increased metabolite coverage to 3,125 metabolites (1,828 lipids plus 1,297 hydrophilic compounds, Table 3). This was likely due to a decrease in ion suppression and improved peak separation that results from a less complex sample. Important metabolites such as cholesterol and triacylglycerol were lost when precipitation was used but these were extracted by MTBE. Dramatic increases in recovery of compounds such as 17:0 Cer, Cer(d18:0/24:1), 15:0 PC, and 17:0 PE were also found when the MTBE method was used. Further separation of the MTBE lipid fraction into three fractions by SPE was very efficient, and the extraction of lipids from the methanol pellet recovered more than 50% of the metabolites lost on SPE. Overall, the combined MTBE-SPE method increased metabolite coverage to 3,806, and it has advantages including low protein interference, low ion-suppression, better resolution and peak separation, and high reproducible data. However, the method is time consuming and there is loss when combined with further fractionation using an SPE column. While our MTBE-SPE method results in a higher number and more classes of metabolites compared to MeOH precipitation, we are unable to perform a direct comparison with previously published methods due to differences in data processing. During data processing using our workflow, isotope clusters, adducts, and multimers are merged into single compounds and our methods therefore result in a substantially smaller number of compounds compared to if merging were not performed. Therefore we are only able to compare extraction methods performed using the same data processing methods. Overall, we found the MTBE and MTBE-SPE methods to be appropriate when a high level of metabolome coverage, increased representation of lipid species, and a priori knowledge of molecular class and species are desired.
Supplementary Material
Highlights.
Efficiency of sample preparation methods were compared for metabolite profiling
Compared number of molecules, reproducibility, recovery of spiked standards
Methanol-ethanol extraction increased recovery of hydrophobic molecules vs methanol
MTBE increased extraction of hydrophobic lipids such as ceramides and triglycerols
MTBE+SPE recovered more molecules than MTBE alone
Acknowledgements
The NJH MS facility is supported in part by CCSTI UL1 TR000154. Funding from NIH grants P20 HL-113445 and R01 HL-095432 also supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data. All the tentative identification results are listed in supplementary tables.
References
- 1.Becker S, Kortz L, Helmschrodt C, Thiery J, Ceglarek U. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2012;883–884:68. doi: 10.1016/j.jchromb.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Metz TO, Zhang Q, Page JS, Shen Y, Callister SJ, Jacobs JM, Smith RD. Biomark. Med. 2007;1:159. doi: 10.2217/17520363.1.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roux A, Lison D, Junot C, Heilier JF. Clin. Biochem. 2011;44:119. doi: 10.1016/j.clinbiochem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Zhang A, Sun H, Wang P, Han Y, Wang X. Analyst. 2012;137:293. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- 5.Zhou B, Xiao JF, Tuli L, Ressom HW. Mol. Biosyst. 2012;8:470. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuckovic D. Anal. Bioanal. Chem. 2012;403:1523. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- 7.Michopoulos F, Lai L, Gika H, Theodoridis G, Wilson I. J. Proteome Res. 2009;8:2114. doi: 10.1021/pr801045q. [DOI] [PubMed] [Google Scholar]
- 8.Polson C, Sarkar P, Incledon B, Raguvaran V, Grant R. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2003;785:263. doi: 10.1016/s1570-0232(02)00914-5. [DOI] [PubMed] [Google Scholar]
- 9.Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, Trauger SA, Siuzdak G. Anal. Chem. 2006;78:743. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 10.Bruce SJ, Tavazzi I, Parisod V, Rezzi S, Kochhar S, Guy PA. Anal. Chem. 2009;81:3285. doi: 10.1021/ac8024569. [DOI] [PubMed] [Google Scholar]
- 11.Folch J, Lees M, Sloane Stanley GH. J. Biol. Chem. 1957;226:497. [PubMed] [Google Scholar]
- 12.Bligh EG, Dyer WJ. Can J. Biochem. Physiol. 1959;37:911. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 13.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. J. Lipid Res. 2008;49:1137. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreiro-Vera C, Priego-Capote F, Luque de Castro MD. J. Chromatogr. A. 2012;1240:21. doi: 10.1016/j.chroma.2012.03.074. [DOI] [PubMed] [Google Scholar]
- 15.Gika HG, Theodoridis GA, Wilson ID. J. Sep. Sci. 2008;31:1598. doi: 10.1002/jssc.200700644. [DOI] [PubMed] [Google Scholar]
- 16.Spagou K, Tsoukali H, Raikos N, Gika H, Wilson ID, Theodoridis G. J. Sep. Sci. 2010;33:716. doi: 10.1002/jssc.200900803. [DOI] [PubMed] [Google Scholar]
- 17.Spagou K, Wilson ID, Masson P, Theodoridis G, Raikos N, Coen M, Holmes E, Lindon JC, Plumb RS, Nicholson JK, Want EJ. Anal. Chem. 2011;83:382. doi: 10.1021/ac102523q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.