Abstract
The recently recognised protein-coding genes MCM7 and TSR1 have shown significant promise for phylogenetic resolution within the Ascomycota and Basidiomycota, but have remained unexamined within other fungal groups (except for Mucorales). We designed and tested primers to amplify these genes across early-diverging fungal clades, with emphasis on the Kickxellomycotina, zygomycetous fungi with characteristic flared septal walls forming pores with lenticular plugs. Phylogenetic tree resolution and congruence with MCM7 and TSR1 were compared against those inferred with nuclear small (SSU) and large subunit (LSU) rRNA genes. We also combined MCM7 and TSR1 data with the rDNA data to create 3- and 4-gene trees of the Kickxellomycotina that help to resolve evolutionary relationships among and within the core clades of this subphylum. Phylogenetic inference suggests that Barbatospora, Orphella, Ramicandelaber and Spiromyces may represent unique lineages. It is suggested that these markers may be more broadly useful for phylogenetic studies among other groups of early-diverging fungi.
Keywords: DNA replication licensing factor, Harpellales, Kickxellomycotina, MCM7, MS277, MS456, ribosomal biogenesis protein, Trichomycetes, TSR1, Zygomycota
INTRODUCTION
The molecular revolution has transformed our understanding of the evolutionary relationships between groups of fungi – with examples of both artificial and natural clades being refuted or recognised, respectively. However, in the early-diverging fungi, the process has been only partially successful. Some monophyletic groups have been broken up. For example, the Chytridiomycota had two other phyla, the Blastocladiomycota and the Neocallimastigomycota, created for certain taxa previously belonging to it (James et al. 2006b, Hibbett et al. 2007). The Zygomycota has been split into numerous subphyla (Hibbett et al. 2007). However, the relationships between and sometimes within these groups have resisted efforts with existing phylogenetic techniques for genes in broad usage. James et al. (2006a) were unable to define well-supported relationships between most of the basal groups, leading them to be regarded as incertae sedis within the most recent reclassification of Fungi (Hibbett et al. 2007). Additional genes might provide better support for phylogenetic analyses and understanding of these evolutionary relationships, especially when combined with increased taxon sampling. Ultimately, well-supported phylogenies (depicted as trees) allow one to (re-)evaluate and hopefully improve classification systems, as well as understand the ancient environmental pressures that have guided and shaped fungal diversity.
While improving molecular techniques and phylogenomics undoubtedly will provide better evidence to address these questions, the results may not be evident for some time. Firstly, only a limited number of early-diverging taxa have been genome sequenced. Efforts with additional taxa are in progress, but currently only three species of Chytridiomycota, one each of Blastocladiomycota and Kickxellomycotina, as well as four species of Mucoromycotina have their genomes available (based on available online searches and the list at http://www.fungalgenomes.org). Furthermore, many early-diverging fungi will prove difficult to genome-sequence as they have not yet been cultured axenically and offer genomic DNA samples that are low in concentration and potentially contaminated with host DNA. This is particularly true of the symbiotic members of the Entomophthoromycotina, Kickxellomycotina and Zoopagomycotina, each of which has at least one major clade with no member species yet successfully cultured. For this reason, finding powerful single-copy nuclear genes that can be amplified and sequenced using current techniques (for available samples) remains a reasonable phylogenetic option in pursuit of answers to critical evolutionary questions while also considering project timeframes and budgets.
Fortunately, the wealth of information emerging from genomic sequencing projects can be utilized concurrently to discover candidate single-copy genes for such purposes. Using a bioinformatics approach, Aguileta et al. (2008) mined genomic sequences among Fungi to identify clusters of orthologous single-copy genes. Individual phylogenetic trees, inferred from the predicted protein sequences, were compared to a phylogenetic tree based on a concatenated alignment of protein sequences. Two genes, MS456 and MS277, demonstrated high topological congruence with the overall consensus tree using all of the genes in the study (Aguileta et al. 2008). MS456 corresponds to the MCM7 gene, a DNA replication licensing factor that forms part of a hexameric protein complex required for DNA replication (Moir et al. 1982, Kearsey & Labib 1998). MS277 corresponds to the TSR1 gene, a ribosome biogenesis protein (Gelperin et al. 2001).
Although Aguileta et al. (2008) demonstrated the utility and power of these two genes for phylogenetic analysis, neither primer sequences nor PCR protocols were provided. Schmitt et al. (2009) aligned amino acid sequences (from GenBank) to design new degenerate primers to amplify regions of both MCM7 and TSR1. With these primers, they were able to sequence MCM7 and TSR1 for 42 species of lichenised ascomycetes. The resulting phylogeny was well-resolved and demonstrated the potential use of these genes for other taxa. Raja et al. (2011) performed additional testing of MCM7 among the Ascomycota and found that it resolved relationships more strongly than the ribosomal large subunit (LSU), one of the most commonly used genes within the ascomycetes. Morgenstern et al. (2012) generated a phylogeny using MCM7 sequences from genome-sequenced fungi, which included some early-diverging taxa. Hermet et al. (2012) utilized both MCM7 and TSR1 in a study of Mucor, demonstrating the potential utility of the MCM7 and TSR1 genes outside of the Dikarya. Despite the apparent phylogenetic potential, beyond the Mucorales (Hermet et al. 2012) these genes have not yet been investigated for their power to resolve relationships among the early-diverging fungi.
To address this and potentially improve our understanding of evolution within this section of the fungal tree of life, we attempted to amplify and sequence the MCM7 and TSR1 genes for putative species within the Kickxellomycotina. This subphylum is a diverse group, among which members may be saprotrophic, mycoparasitic, or obligate symbionts of arthropods. Natural affinities among its members have long been suspected on morphological grounds (Moss & Young 1978). Some molecular-based studies (James et al. 2006a, Sekimoto et al. 2011) have suggested it is monophyletic, whereas others (White et al. 2006a) have suggested the relationship between the Kickxellomycotina and closely related taxa may be more complex. Some studies, such as Tanabe et al. (2004), have been inconclusive, with different genes disagreeing on the monophyly of the clade. Furthermore, the relationships between the four orders (Asellariales, Dimargaritales, Harpellales and Kickxellales) that comprise the subphylum are not fully substantiated.
Our primary goal was to assess the phylogenetic utility of MCM7 and TSR1 for these early-diverging fungal taxa. In so doing, we compared these genes against a combined nuclear 18S and 28S rDNA phylogeny, with attention to tree congruence and resolution. Additionally, 3-gene (18S+28S+MCM7) and 4-gene (18S+28S+MCM7+TSR1) phylogenies were examined to assess their use in combination. These data provide an opportunity to assess the inferred evolutionary relationships and history among members of the Kickxellomycotina, one of the first multi-gene phylogenies with such a focus (but also see Wang 2012).
MATERIALS AND METHODS
DNA samples used for this study were extracted according to White (2006). Some samples were prepared from axenic cultures, whereas others were prepared from the dissection of host arthropods (Table 2).
Table 2.
Fungal species, isolate number, and source, amplified with specific primer combinations.
| Species | Isolate # | Source / Host | Culture | Country | Coll. | Primer combination |
GenBank accession or genome identifier |
||||||
| 18S | 28S | MCM7 | TSR1 | 18S | 28S | MCM7 | TSR1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batrachochytrium dendrobatidis8 | JEL423 | Broad Institute sequencing project3 | – | – | – | – | – | – | Supercontig 14 188674-190464 | NG_027619 | BDEG_04439.1 | BDEG_02071.1 | |
| Spizellomyces punctatus8 | DAOM BR117 | Broad Institute sequencing project3 | – | – | – | – | – | – | Supercontig 32 115-2731 | NG_027618.1 | SPPG_04788.3 | SPPG_02880.2 | |
| Rhizophlyctis rosea | JEL318 | AFTOL DNA sample | – | – | – | – | 7f-16r | – | AY635829.1 | DQ273787.1 | KC297581 | – | |
| Allomyces macrogynus8 | ATCC 38327 | Broad Institute sequencing project3 | – | – | – | – | – | – | Supercontig 63 186963-188875 | Supercontig 63 164434-171606 | AMAG_00422.2 | AMAG_17353.1 | |
| Allomyces arbuscula | Brazil 2 | AFTOL DNA sample | – | – | – | – | 7f-16r | 1018f-2356r | NG_017166.1 | DQ273806.1 | KC297582 | KC297629 | |
| Coelomomyces stegomyiae | DUH0008925 | AFTOL DNA sample | – | – | – | – | 7f-16r | 1018f-2356r | NG_017164 | NG_027644.1 | KC297583 | KC297630 | |
| Mucor circinelloides8 | CBS277.49 | JGI sequencing project5 | – | – | – | – | – | – | Scaffold 11 800000-801950 | Scaffold 11 804000-809666 | Scaffold 14 106477-109288 | Scaffold 1 4335584-4338189 | |
| Phycomyces blakesleeanus8 | NRRL1555 | JGI sequencing project5 | – | – | – | – | – | – | NG_017190.1 | NG_027559.1 | Scaffold 5 753037-756512 | Scaffold 24 361829-364983 | |
| Rhizopus oryzae8 | 99-880 | Broad Institute sequencing project4 | – | – | – | – | – | – | Supercontig 6 2079534-2081357 | Supercontig 6 2074749-2079337 | RO3G_11608 | RO3G_12091.3 | |
| Coprinopsis cinerea8 | okayama7#1306 | GenBank | – | – | – | – | – | – | M92991 | AF041494 | AACS02000002.1 | AACS02000003.1 | |
| Cryptococcus neoformans8 | B-3501A | GenBank | – | – | – | – | – | – | BR000310.1 | BR000310.1 | AAEY01000032.1 | AAEY01000024.1 | |
| Ustilago maydis8 | 5217 | GenBank | – | – | – | – | – | – | X62396 | AF453938 | AACP01000247.1 | AACP01000184.1 | |
| Aspergillus nidulans8 | FGSC A4 | GenBank | – | – | – | – | – | – | U77377.1 | EU840227.1 | AACD01000102.1 | AACD01000107.1 | |
| Saccharomyces cerevisiae8 | S288c | GenBank | – | – | – | – | – | – | NC_001144.5 | NC_001144.5 | BK006936.2 | BK006938.2 | |
| Schizosaccharomyces pombe8 | 972h- | GenBank | – | – | – | – | – | – | CU329672.1 | NC_003421.2 | CU329671.1 | CU329670.1 | |
| Rhopalomyces elegans | NRRL A-10835 | AFTOL DNA sample | – | – | – | 7f-16r | 1018f-2356r | NG_017191.1 | NG_027654.1 | KC297584 | KC297631 | ||
| Conidiobolus coronatus | NRRL28638 | AFTOL DNA sample | – | – | – | – | 7f-16r | – | NG_017182.1 | NG_027617.1 | KC297585 | – | |
| Entomophaga conglomerata | ARS-2273 | Adult Chironomidae | yes | – | – | – | 7f-16r | – | AF368509.1 | – | KC297586 | – | |
| Entomophthora muscae | ARSEF3074 | AFTOL DNA sample | – | – | – | – | 7f-16r | 1018f-2356r | NG_017183.1 | NG_027647.1 | KC297587 | KC297632 | |
| Barbatospora ambicaudata | TN-49-W4a | Simulium vandalicum | yes | USA | MMW | SR1R-NS8 | ITS1F-NL4 | 8bf-16r | 1492f-2356r | KC297614 | KC297566 | KC297588 | KC297633 |
| Coemansia reversa | NRRL1564 | JGI sequencing project5 | yes | – | – | – | – | – | Scaffold 121 3595-5518 | Scaffold 121 7595-11423 | Scaffold 81 3398-5725 | Scaffold 24 40229-42782 | |
| Coemansia braziliensis | NRRL-1566 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007532.1 | AF031069.1 | KC297589 | KC297634 | |
| Kickxella alabastrina | NRRL-2693 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007537.1 | AF031064.1 | JX155485 | KC297635 | |
| Dipsacomyces acuminosporus | NRRL-2925 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007534.1 | AF031065.1 | KC297590 | KC297636 | |
| Martensiomyces pterosporus | NRRL-2642 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007539.1 | AF031066.1 | KC297591 | KC297637 | |
| Linderina pennispora | NRRL-3781 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007538.1 | FJ517544.1 | JX155486 | KC297638 | |
| Spirodactylon aureum | NRRL-2810 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007541.1 | AF031068.1 | KC297592 | KC297639 | |
| Spiromyces minutus | NRRL-3067 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007542.1 | DQ273810.1 | KC297593 | KC297640 | |
| Spiromyces aspiralis | NRRL-22631 | ARS Culture Collection | yes | – | – | – | 8bf-16r | 1492f-2356r | AF007543.1 | NG_027560.1 | KC297594 | KC297641 | |
| Orphella catalaunica | NOR-40-W10 + W12 | Leuctrid | no | Norway | MMW | NS1AA-NS8AA | ITS1F-LR5 | 8bf-16r | – | KC297617 | KC297569 | KC297598 | – |
| Orphella dalhousiensis | NS-34-W16 | Paracapnia sp. | no | Canada | GenBank/MMW | – | – | 8bf-16r | – | DQ322626.1 | DQ273830.1 | KC297599 | – |
| Ramicandelaber longisporus | ARSEF 6175 | Eggs/cysts, Heterodera glycines | yes | China | – | SR1R-NS8 | NL1-LR11 | 7f-16r | 1492f-2356r | KC297615 | KC297567 | KC297595 | KC297642 |
| Ramicandelaber longisporus | ARSEF 6176 | Eggs/cysts, Heterodera glycines | yes | China | – | SR1R-NS8 | – | 7f-16r | – | KC297616 | – | KC297596 | – |
| Dimargaris bacillispora | NRRL2808 | AFTOL DNA sample | yes | – | – | – | 7f-16r | 1018f-2356r | NG_017180.1 | NG_027650.1 | KC297597 | KC4564209 | |
| Harpellomyces montanus | TN-22-W5B | Thaumaleidae | no | USA | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297618 | KC297570 | KC297600 | – |
| Harpellomyces eccentricus | NOR-56-W1 | Thaumaleidae | no | Norway | MMW | – | – | 8bf-16r | – | – | – | KC297601 | – |
| Bojamyces sp. | CA-18-W17 | Ephemeroptera | no | USA | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | JX155619 | JX155645 | JX155471 | – |
| Capniomyces sasquatchoides | ID-130-N5 | Plecoptera | no | USA | NKR | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297619 | KC297571 | KC297602 | – |
| Capniomyces stellatus | MIS-21-127 | Allocapnia sp. | yes | USA | GenBank/RWL | – | – | 8bf-16r | 1492f-2356r | DQ367451.1 | EF396194.1 | JX155472 | KC297643 |
| Caudomyces sp. | OR-8-W10 | Tipulidae | no | USA | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | 1492f-2356r | KC297620 | KC297572 | KC297603 | KC297649 |
| Caudomyces sp. | UT-1-W16a | Antocha sp. | no | USA | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | 1492f-2356r | JX155620 | JX155646 | JX155473 | KC297650 |
| Furculomyces boomerangus | AUS-77-4 | Tanytarsus nr. inextentus | no | Australia | RWL | – | – | 8bf-16r | – | AF277013.1 | AF031074.1 | JX155466 | – |
| Genistelloides hibernus | KS-19-M23 | Capniidae | no | USA | GenBank/JKM | – | – | 8bf-16r | 1492f-2356r | DQ367456.1 | JQ302921 | JX155474 | KC297651 |
| Graminella microspora | NOR-35-1 | Baetis rhodani | no | Norway | RWL | – | – | 8af-16r | – | JQ302867 | JQ302945 | KC297604 | – |
| Harpella or Pennella sp. | NF-MC-15 | Adult Prosimulium mixtum ovarian cysts | no | Canada | MCB | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297621 | KC297573 | KC297605 | – |
| Harpella melusinae(cysts) | NF-15-5A | Adult Prosimulium mixtum ovarian cysts | no | Canada | RWL | SR1R-NS8 | NL1AA-LR7AA | 8bf-16r | 1492f-2356r | JX155621 | JX155647 | JX155475 | KC297652 |
| Harpella sp. | NF-MC-18 | Adult Prosimulium mixtum ovarian cysts | no | Canada | MCB | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | KC297622 | KC297574 | KC297606 | – | |
| Lancisporomyces falcatus | NS-X-2 | Paracapnia angulata | no | Canada | DBS | – | – | 8bf-16r | – | JQ302865 | JQ302943 | JX155477 | – |
| Lancisporomyces vernalis | SPA-X-40 | Nemoura | no | Spain | LGV | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297623 | KC297575 | KC297607 | – |
| Legerioides tumidus | NH-1-M869a | Isopoda | no | USA | JKM | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297576 | KC297608 | – | |
| Legeriomyces minae | PEI-X-6 | Ephemeroptera | no | Canada | DBS | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | JX155622 | JX155648 | JX155478 | – |
| Legeriosimilis sp. | CA-10-W15 | Ephemeroptera | no | USA | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297625 | KC297577 | KC297609 | – |
| Legeriomyces sp. nov. | PEI-X-4 | Ephemerellidae | no | Canada | DBS | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297626 | KC297578 | KC297610 | – |
| Legeriosimilis sp. | NOR-31-2 | Siphloneuridae | no | Norway | RWL | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297627 | KC297579 | KC297611 | – |
| Pennella sp. | NOR-7-W12 | Simuliidae | no | Norway | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | KC297628 | KC297580 | KC297612 | – |
| Pteromaktron sp. | OR-11-W8 | Ephemeroptera | no | USA | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | JX155623 | JX155649 | JX155479 | – |
| Smittium culisetae | COL-18-3 | Culiseta impatiens | yes | USA | GenBank/RWL | – | – | 8bf-16r | 1492f-2356r | NG_017185.1 | NG_027648.1 | AEW26363.1 | KC297644 |
| Smittium culisetae | MAL-X-1 | Aedes crinifer | yes | Malaysia | CLL | – | – | 8bf-16r | 1492f-2356r | JQ302897 | JQ302835 | AEW26370.1 | KC297645 |
| Unnamed Trichopteran tricho | ALG-13-W1 | Trichoptera | no | Canada | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | 1492f-2356r | JX155625 | JX155651 | JX155481 | KC297653 |
| Unnamed Trichopteran tricho | ALG-10-W3 | Trichoptera | no | Canada | MMW | NS1AA-NS8AA | NL1AA-LR7AA | 8bf-16r | – | JX155624 | JX155650 | JX155480 | – |
| Smittium culicis | 43-1-2 | Chironomus sp. | yes | Australia | LCF/BH | – | – | 8bf-16r | 1492f-2356r | JQ302893 | DQ367512.1 | AEW26366.1 | KC297646 |
| Smittium mucronatum | FRA-12-3 | Psectrocladius sordidellus | yes | France | KUMYCOL/RWL | – | – | 8bf-16r | 1492f-2356r | AF277030.1 | JQ302833 | AEW26371.1 | KC297647 |
| Smittium simulii | 41-1-6 | Orthocladius sp. | yes | Australia | LCF/BH | – | – | 8bf-16r | 1492f-2356r | JQ302861 | JQ302939 | AEW26374.1 | KC297648 |
| Austrosmittium sp. | 32-1-8 | Orthocladiinae | no | Australia | KUMYCOL | – | – | 8bf-16r | – | – | DQ367494.1 | KC297613 | – |
| Coleopteromyces amnicus | ARG-15-6F | Scirtidae | no | Argentina | LCF | – | – | 8bf-16r | – | JQ302853 | JQ302931 | JX155465 | – |
| Pseudoharpella arcomylica | LCF#3 | Dixidae | no | USA | LCF | – | – | 8bf-16r | – | JQ302882 | JQ302956 | JX155467 | – |
| Smittium angustum | AUS-126-30 | Tanytarsus sp. | yes | Australia | RWL | – | – | 8bf-16r | – | AF277005.1 | JQ302822 | JX155420 | – |
| Smittium annulatum | CR-143-8 | Simuliidae | yes | Costa Rica | RWL | – | – | 8bf-16r | – | AF277024.1 | JQ302832 | JX155421 | – |
| Smittium caudatum | KS-1-2 | Chironomidae | yes | USA | KUMYCOL/RWL | – | – | 8bf-16r | – | AF277031.1 | JQ302948 | JX155422 | – |
| Smittium coloradense | RMBL-13-41 | Cricotopus sp. | yes | USA | RWL | – | – | 8bf-16r | – | AF277041.1 | JQ302912 | JX155423 | – |
| Smittium commune | KS-6-6 | Chironomidae | yes | USA | RWL | – | – | 8bf-16r | – | AF277034.1 | JQ302901 | JX155426 | – |
| Smittium cylindrosporum | CHI-27-1 | Cricotopus sp. | yes | Chile | RWL | – | – | 8bf-16r | – | AF277018.1 | JQ302828 | JX155433 | – |
| Smittium gravimetallum | KS-F1-3 | Dicrotendipes fumidus | yes | USA | LCF | – | NL1AA-LR7AA | 8bf-16r | – | AF277037.1 | JX155634 | JX155437 | – |
| Smittium megazygosporum | SC-DP-2 | Simulium vittatum | yes | USA | KUMYCOL/CEB | – | – | 8bf-16r | – | AF277045.1 | JQ302823 | JX155442 | – |
| Smittium morbosum | AUS-X-1 | Anopheles hilli | yes | Australia | KUMYCOL/RWL | – | – | 8bf-16r | – | AF277014.1 | JQ302913 | JX155443 | – |
| Smittium orthocladii | LCF-BT-1 | Corynoneura sp. | yes | USA | LCF/MMW | – | – | 8bf-16r | – | DQ367446.1 | JQ302900 | AEW26378.1 | – |
| Smittium tipulidarum | RMBL-31-1 | Elliptera astigmatica | yes | USA | KUMYCOL/RWL | – | – | 8bf-16r | – | AF277043.1 | JQ302836 | JX155452 | – |
| Smittium tronadorium | ARG-24-2F | Paraheptagyia sp. | yes | Argentina | LCF | – | – | 8bf-16r | – | AF277004.1 | JQ302904 | JX155454 | – |
| Stachylina lentica | NOR-58-10 | Chironomus sp. | no | Norway | RWL | – | NL1AA-LR7AA | 8bf-16r | – | JQ302874 | JX155628 | JX155468 | – |
| Stachylina sp. | NS-X-10 | Chironomidae | no | Canada | DBS | – | – | 8bf-16r | – | – | – | JX155469 | – |
| Trichozygospora chironomidarum | TN-3-16 | Chironomidae | yes | USA | RWL | – | NL1AA-LR7AA | 8bf-16r | – | JQ302841 | KC297568 | JX155470 | – |
1AS, Amy Slaymaker; AR, Alen Rizzo; BH, Barb Hayford; CEB, Charles ‘Eddie’ Beard; CLL, Claudia Lopez Lastra; DBS, Douglas B. Strongman; GM, Maria Gabriela Mazzucchelli; JKM, JK Misra; JL, Joyce Longcore; LCF, Leonard C. Ferrington, Jr.; LGV, Laia Guàrdia Valle; MCB, Murray Colbo; MJC, Matías J. Cafaro; MMW, Merlin White; NKR, Nicole Reynolds; PVC, Paula Clarke; RWL, Robert W. Lichtwardt; SM, Steve Moss; WKR, Will K. Reeves. Some of the sequences were generated from culturable isolates from the University of Kansas Mycological Culture Collection, represented as KUMYCOL.
2Accession numbers in bold were generated for this study (or as joint effort with Wang 2012 study).
3Data derived from Origins of Multicellularity Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/).
4Data derived from Rhizopus oryzae Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/).
5These sequence data were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community.
6rDNA was not available from the genome sequencing project, so data from other isolates was used. The isolate used for the SSU rDNA was not specified in GenBank. The LSU rDNA was taken from isolate C13.
7rDNA was not available from the genome sequencing project, so data from other isolates was used. The SSU rDNA was taken from isolate “MUCL 30488, CBS 445.63”. The LSU rDNA was taken from isolate MS 115.
8Species used for initial primer design and in silico testing.
PCR amplification
MCM7
Initial attempts to amplify MCM7 were conducted using the primers MCM7-709for and MCM7-1348rev of Schmitt et al. (2009). PCR products from three taxa (Coemansia braziliensis, Dipsacomyces acuminosporus and Smittium culisetae) were amplified successfully and sequenced using the same primers. However, further attempts using these primers with other taxa were unsuccessful. We subsequently designed new primers, assuming that the taxa of interest had primer sites that were not well-matched to the originals of Schmitt et al. (2009).
Specifically, using those three sequences (above) with others from GenBank and several from genome-sequencing projects published online (see Table 2), a reference alignment of MCM7 protein sequences was compiled, spanning the Dikarya and several groups of early-diverging fungi, that was used to design six new degenerate primers (Table 1). Two sets of primers were used for the majority of our data collection. One set uses the Schmitt et al. (2009) primer MCM7-709for but with our reverse primer (MCM7-16r). The latter appeared to be more conserved amongst a greater diversity of taxa and worked well on the majority of early-diverging fungi tested, except for members of the Harpellales where a second set, MCM7-8bf and MCM7-16r, was compatible with the majority of the taxa tested from that order. Both primer combinations amplified a region of approximately 850 base pairs.
Table 1.
Primers used to amplify nuclear (SSU and LSU) rDNA or protein-coding genes (MCM7; TSR1), among the Kickxellomycotina and some other early-diverging fungi.
| Primer name | Gene | Source | Direction | Sequence (5’–3’) | Translated amino acid acid sequence (5’–3’) | Length | Degeneracy |
|---|---|---|---|---|---|---|---|
| MCM7-709for | MCM7 | Schmitt et al. 2009 | For | ACIMGIGTITCVGAYGTHAARCC | TRVSDVKP | 23 bp | 481 |
| MCM7-8bf | MCM7 | New for this study | For | GTIGCIGCITAYYTITGYGAY | VAAYLCD | 21 bp | 16 |
| MCM7-8af | MCM7 | New for this study | For | TGYGGIWSIGARGTITTYCARGA | CGSEVFQ | 23 bp | 64 |
| MCM7-1348rev | MCM7 | Schmitt et al. 2009 | Rev | GATTTDGCIACICCIGGRTCWCCCAT | MGDPGVAKS | 26 bp | 242 |
| MCM7-16r | MCM7 | New for this study | Rev | GTYTGYTGYTCCATIACYTCRTG | HEVMEQQT | 23 bp | 32 |
| TSR1-1018f | TSR1 | New for this study | For | AAYGARCARACITGGCCIACIGA | NEQTWPT(D/E) | 23 bp | 8 |
| TSR1-1492f | TSR1 | New for this study | For | TGGGAYCCITWYGARAAYYTICC | WDP(Y/F)ENLP | 23 bp | 64 |
| TSR1-2356r | TSR1 | New for this study | Rev | CAYTTCATRTAICCRTGIGTICC | GTHGYMKC | 23 bp | 8 |
| NS1AA | SSU rDNA | Wang 2012 | For | AAGCCATGCATGTCTAAGTATAA | – | 23 bp | – |
| SR1R | SSU rDNA | Vilgalys & Hester 1990 | For | TACCTGGTTGATYCTGCCAGT | – | 21 bp | 2 |
| NS8AA | SSU rDNA | Wang 2012 | Rev | TACTTCCTCTAAATGACCAAGTTTG | – | 25 bp | – |
| NS8 | SSU rDNA | White et al. 1990 | Rev | TCCGCAGGTTCACCTACGGA | – | 20 bp | – |
| ITS1F | LSU rDNA | Gardes & Bruns 1993 | For | CTTGGTCATTTAGAGGAAGTAA | – | 22 bp | – |
| ITS3 | LSU rDNA | White et al. 1990 | For | GCATCGATGAAGAACGCAGC | – | 20 bp | – |
| NL1 | LSU rDNA | O’Donnell 1993 | For | GCATATCAATAAGCGGAGGAAAAG | – | 24 bp | – |
| NL1AA | LSU rDNA | Wang 2012 | For | GAGTGAAGCGGGAAIAGCTCAAG | – | 23 bp | – |
| NL4 | LSU rDNA | O’Donnell 1993 | Rev | GGTCCGTGTTTCAAGACGG | – | 19 bp | – |
| LR5 | LSU rDNA | Vilgalys & Hester 1990 | Rev | TCCTGAGGGAAACTTCG | – | 17 bp | – |
| LR7AA | LSU rDNA | Wang 2012 | Rev | CCACCAAGATCTGCACTAGA | – | 20 bp | – |
| LR11 | LSU rDNA | Vilgalys lab page3 | Rev | GCCAGTTATCCCTGTGGTAA | – | 20 bp | – |
1Degeneracy given by Schmitt et al. (2009) as 32 (three-fold degeneracies calculated as two-fold).
2Degeneracy given by Schmitt et al. (2009) as 16 (three-fold degeneracies calculated as two-fold).
3Available at http://www.biology.duke.edu/fungi/mycolab/primers.htm.
The PCR reagents used for the MCM7-709for and MCM7-1348rev primer combination included 11 μL of Promega Go-Taq Green Hot Master Mix, 2.20 μL of each primer at 10 μM concentration, 0.44 μL of 25 mM MgCl2 (to a total concentration of 2.5 mM), 4.16 μL dH2O, and 2 μL of genomic DNA. Cycling conditions used an initial denaturation step of 95 °C for 2 min, 45 cycles of 95 °C for 30 s, annealing at 56 °C for 45 s, and extension at 72 °C for 1 min 15 s, a final extension at 72 °C for 10 min, followed with a final hold step at 4 °C. Reagents for the MCM7-8bf and MCM7-16r primer were identical except that 0.35 μL of 50 μg/μL BSA was added (while reducing the water by an equal amount). Cycling conditions included an initial denaturation step of 95 °C for 2 min, with 45 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 45 s, and extension at 72 °C for 1.5 min, followed by a final extension step at 72 °C for 10 min, before a min, before a final hold at 4 °C.
TSR1
As with the MCM7, except with greater sequence variation of the TSR1 gene, a reference alignment was prepared but used first to conduct in silico testing before attempting amplifications of it. Specifically, the translated protein sequences of both primers were compared visually to the translated protein sequences in the alignment to assess their conservation and putative compatibility. Again, sequences from GenBank and various genome sequencing projects (see Table 2) were used to make this initial assessment. When published primers (Schmitt et al. 2009) did not appear compatible with the early-diverging fungi, based on estimated compatibility with the Blastocladiomycota, Chytridiomycota and Mucoromycotina, the closest relatives to the Kickxellomycotina for which we had data, we considered the development of new primers. Ultimately, three new primers were developed and tested (Table 1). One set (TSR1-1018f with TSR1-2356r) successfully amplified products 1250–1300 bp for most non-harpellid Kickxellomycotina. The other set (TSR1-1492f to TSR1-2356r) generated fragments from 700–800 bp but was more broadly compatible within the Kickxellomycotina. Since the latter products were generated entirely from within the range of the gene region amplified by the other set, only this shorter region was used within the analysis (longer sequences were truncated accordingly).
rRNA genes
Ribosomal RNA gene sequences were amplified and sequenced as well as obtained from GenBank (Table 2). Wang et al. (2013) developed primers for both the small rDNA subunit (18S), specifically primers NS1AA and NS8AA, and the large subunit (28S), with primers NL1AA and LR7AA. Those primers were specifically designed to avoid amplification of host DNA from mixed genomic DNA samples, a situation that is not uncommon when fungi are prepared as micro-dissections from arthropod digestive tracts. PCR reagents used for the NS1AA and NS8AA primer combination included 11 μL of Promega Go-Taq Green Master Mix, 0.66 μL of each primer at 10 μM concentration, 0.88 μL of 25 mM MgCl2 (to a final concentration of 2.5 mM), 0.35 μL of 50 μg/μL BSA, 6.45 μL dH2O, and 2 μL of genomic DNA. Cycling conditions included an initial denaturation step of 95 °C for 2 min, 45 cycles of denaturation at 95 °C for 30 s, annealing at 62 °C for 45 s, and extension at 72 °C for 3 min, with a final extension step at 72 °C for 10 min and a final hold at 4 °C. The PCR cocktail used for the NL1AA and LR7AA primer combination included 11 μL of Promega Go-Taq Green Hot Master Mix, 0.66 μL of each primer at 10 μM concentration, 0.44 μL of 25 mM MgCl2 (to a total concentration of 2.5 mM), 2.20 μL of 5M Betaine, 0.35 μL of 50 μg/μL BSA, 4.69 μL dH2O, and 2 μL of genomic DNA. Cycling conditions included an initial denaturation step of 95 °C for 2 min, 45 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 45 s, and extension at 72 °C for 3 min, with a min, with a final extension at 72 °C for 10 min followed by a final hold at 4 °C.
Electrophoresis and sequencing
For all amplified sequences, the PCR product was electrophoresed in 1 % Lonza Seaplaque GTG agarose (low EDTA 1X TAE buffer), stained with Gelstar nucleic acid stain (Cambrex), and visualized on a Clare Chemical DR46B transilluminator. Bands of the appropriate size were excised with medium sized pipet tips (pre-cut by a few mm to increase the bore necessary for bands being cut) and DNA was extracted using a ‘freeze and squeeze’ method. Briefly, pipet tips with excised gel cores were placed in a 1.5 mL microcentrifuge tube, frozen at -20 °C, spun at 14500′G for 10 min, frozen again at -20 °C again, and similarly centrifuged once more. Cycle sequencing reactions were set up using the Applied Biosystems BigDye v. 3.1 kit for bidirectional sequencing. The resulting products were sent to the University of Wisconsin Madison Biotechnology Centre for capillary electrophoresis.
Phylogenetic analyses
DNA sequences were first aligned using the MUSCLE algorithm (Edgar 2004) and then imported into Mesquite (Maddison & Maddison 2011) for final manual adjustment. Introns were removed from the MCM7 sequences via visual inspection for translation into hypothetical proteins. For the MCM7 protein alignment, the reading frame was designated and set, and the nucleotide sequences translated into proteins. This protein alignment was then re-aligned with MUSCLE (Edgar 2004). Regions of poor or ambiguous alignment were manually removed.
Each of the alignments was tested using an appropriate model selection program. The 18S and 28S nucleotide sequences, as well as each of the three individual codon positions of the MCM7 nucleotide alignment, were tested with jModelTest (Guindon & Gascuel 2003, Posada 2008). Model selection was based on the corrected AIC (AICc) score. For all sequences tested, except for the 2nd codon position of the MCM7 nucleotide alignment, the GTR+ Γ+I method had the highest AICc score. For the 2nd codon position, the GTR+ Γ model was slightly higher; however, for simplicity of analysis, the GTR+ Γ+I model was used in all cases. The ProtTest programme (Drummond & Strimmer 2001, Guindon & Gascuel 2003, Abascal et al. 2005) was used on preliminary MCM7 and TSR1 datasets to determine the best model of amino acid evolution for these genes. The LG+ Γ+I model, described by Le & Gascuel (2008), consistently received the highest score and was used.
Phylogenetic inference was conducted through both the Maximum-Likelihood (ML) method and Bayesian inference (BI). The Bayesian tree was used as the primary tree for all analyses, with the Maximum-Likelihood results offered as well, considering possible Bayesian overestimation of branch supports (Suzuki et al. 2002). MrBayes v. 3.1.2 was used for Bayesian inference (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003, Altekar et al. 2004). The LG+ Γ+I model of protein evolution, mentioned above, is not implemented natively in MrBayes v.3.1.2 and was done by setting a fixed GTR model and using the LG exchange matrix and equilibrium frequencies as a dirichlet prior. The online version of AWTY was used to assess tree convergence (Wilgenbusch et al. 2004). GARLI v. 2.0 was used for maximum-likelihood calculations (Zwickl 2006).
Nine analyses were performed: MCM7 nucleotide, MCM7 protein, TSR1 protein, 18S, 28S, nuclear 18S + 28S, 3-gene (18S + 28S + MCM7 protein), 4 gene (18S + 28S + MCM7 protein + TSR1 protein) and 18S + 28S for the taxa in the TSR1 protein alignment only. For the MCM7 nucleotide tree, each codon position was treated as an independent partition. For all trees, different genes were always treated as unlinked partitions. For all trees, 10 mil generations (BI) and 100 bootstrap replicates (ML) were performed, with half of the BI generations treated as burn-in.
A total of nine topologies were produced by our analyses (Fig. 1–5 and S1–S4). Six of the analyses used a large number of taxa (76–81) and were primarily intended to investigate the use of MCM7, whereas three of the analyses used a smaller number (38–39) and were intended to evaluate the use of TSR1 and the combined four-gene analysis (see Table 2). For all of the analyses, branches were considered well-supported (and shown in figures with heavy bold lines) if they had a Bayesian posterior probability (BPP) of > 0.95 and a maximum-likelihood bootstrap proportion (MLBP) of > 0.70.
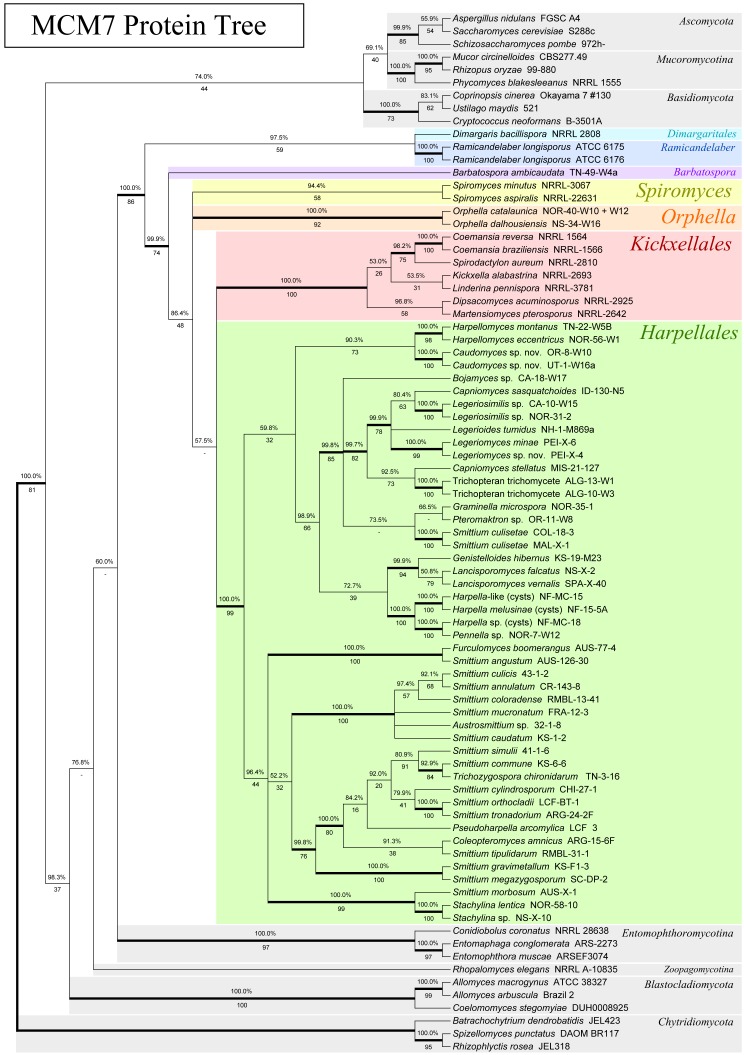
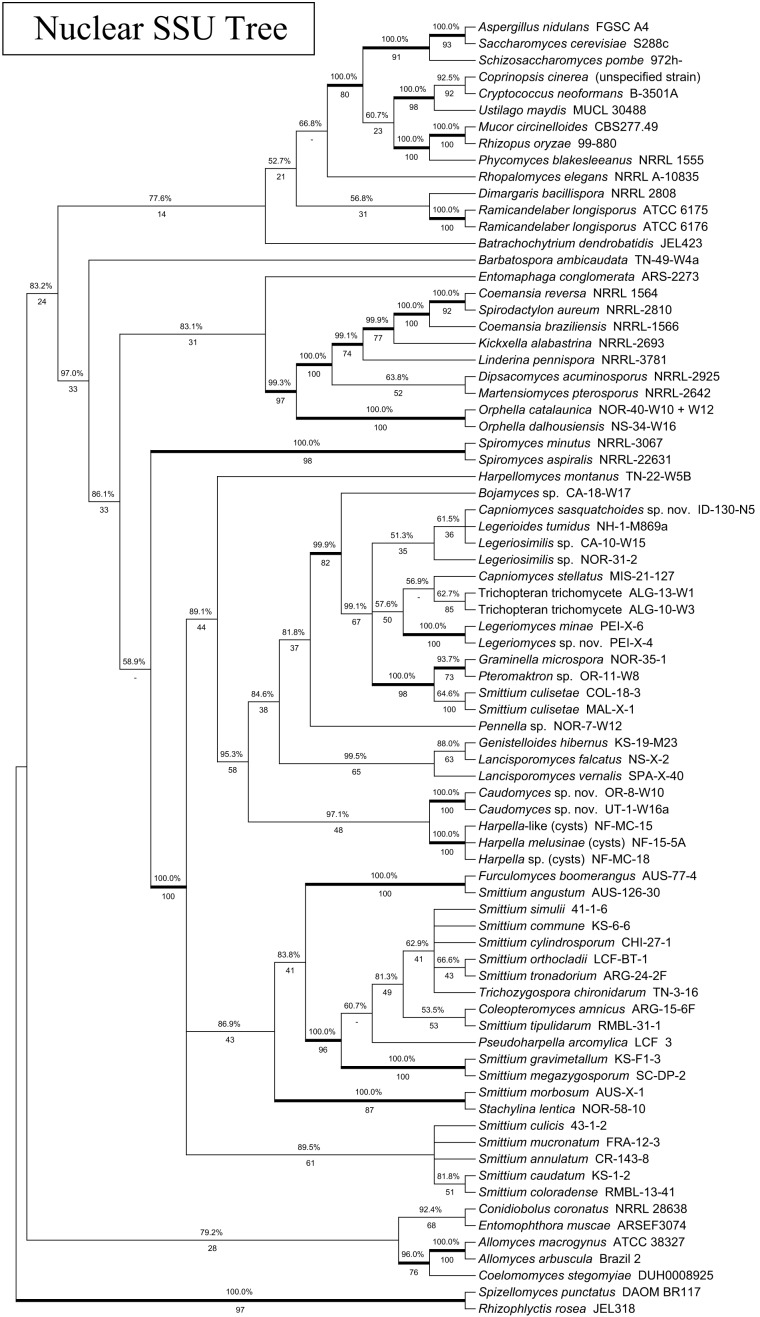
Fig. 1.
Phylogeny of the Kickxellomycotina and other fungal taxa based on an alignment of MCM7 translated protein sequences. Tree is based on a 50 % majority-rules consensus of 10k trees produced with Bayesian inference (5k used as burn-in). Numbers above branches are Bayesian posterior probabilities. Numbers below branches are maximum-likelihood bootstrap supports produced from 100 bootstrap replicates. Bold branches are highly supported (> 95 % BPP and > .70 MLBP).
Fig. 5.
Phylogeny of the Kickxellomycotina based on a concatenated alignment of SSU and LSU rDNA as well as MCM7 and TSR1 translated protein sequences. The method used for tree inference and the format of the tree are the same as for Fig. 3.
Fig. S1.
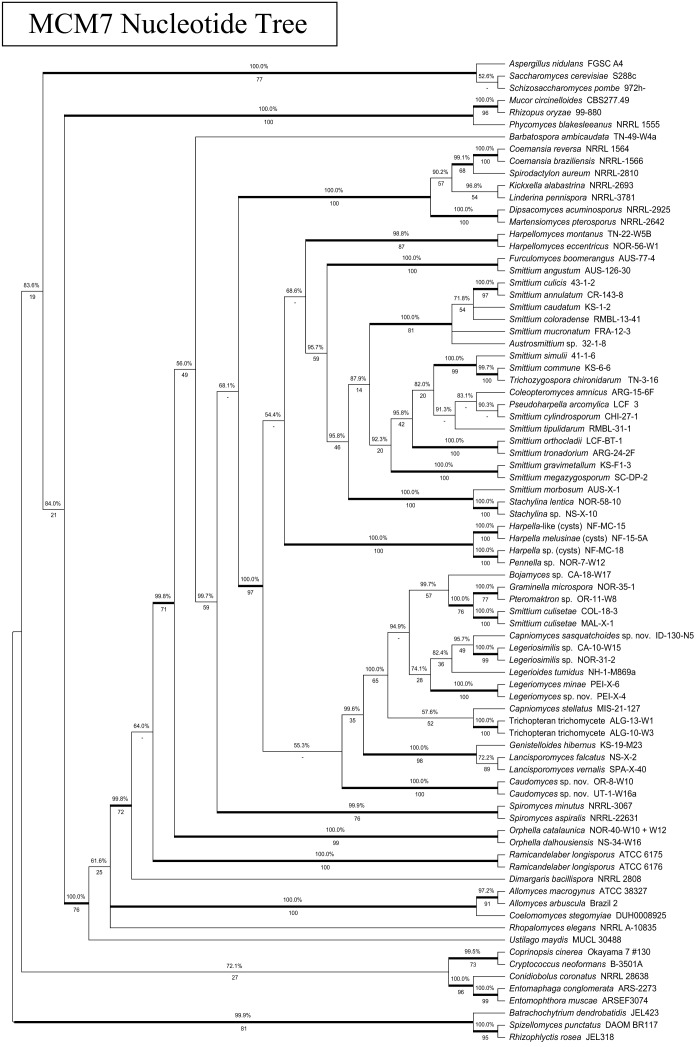
Phylogeny of the Kickxellomycotina based on an alignment of MCM7 nucleotide sequences. Tree is based on a 50 % majority-rules consensus of 10k trees produced with Bayesian inference (5k used as burn-in). The three codon positions were all considered to be on different, unlinked partitions during tree calculation. Numbers above branches are Bayesian posterior probabilities. Numbers below branches are maximum-likelihood bootstrap supports produced from 100 bootstrap replicates. Bold branches are highly supported (> 95 % BPP and > .70 MLBP).
Fig. S4.
Phylogeny of the Kickxellomycotina based on a concatenated alignment of nuclear small subunit (SSU) and nuclear large subunit (LSU) rDNA. For this tree, only taxa for which we had TSR1 were included in the alignment, to provide a basis for comparison to the TSR1 protein tree. Tree is based on a 50 % majority-rules consensus of 10k trees produced with Bayesian inference (5k used as burn-in). Numbers above branches are Bayesian posterior probabilities. Numbers below branches are maximum-likelihood bootstrap supports produced from 100 bootstrap replicates. Bold branches are highly supported (> 95 % BPP and > .70 MLBP).
RESULTS
We report 68 new MCM7 sequences, 26 new TSR1 sequences, and 46 new rDNA sequences (Table 2) for a variety of taxa within the early-diverging fungal lineages. We amplified most of the lineages tested with at least one primer combination for each gene. Recommended primer combinations for MCM7 (Table 3) and TSR1 (Table 4) are provided for each primer and clade tested.
Table 3.
MCM7 protein-coding gene testing status among early-diverging fungal groups with notes on earlier and newly established primer combinations.
| Clade tested | Recommended primers | Notes |
|---|---|---|
| Chytridiomycota | MCM7-709f, MCM7-16r | |
| Blastocladiomycota | MCM7-709f, MCM7-16r | |
| Zoopagales | MCM7-709f, MCM7-16r | |
| Entomophthorales | MCM7-709f, MCM7-8af, MCM7-16r | MCM7-709f preferred over MCM7-8af |
| Kickxellomycotina | ||
| Harpellales | MCM7-8bf, MCM7-16r | MCM7-709f works for a couple of species |
| Kickxellales | MCM7-8bf, MCM7-16r | MCM7-709f works for some but not all species |
| Asellariales | – | Attempted unsuccessfully |
| Dimargaritales | MCM7-709f, MCM7-16r | MCM7-8bf not tested |
| Orphella clade | MCM7-8bf, MCM7-16r | MCM7-709f may work, but not as well as 8bf |
| Barbatospora clade | MCM7-8bf, MCM7-16r | MCM7-709f not tested |
| Spiromyces clade | MCM7-8bf, MCM7-16r | MCM7-709f amplified an incorrect gene when attempted |
| Ramicandelaber clade | MCM7-709f, MCM7-8bf, MCM7-16r | MCM7-709f seemed to sequence better |
Table 4.
TSR1 protein-coding gene testing status among early-diverging fungal groups with notes on earlier and newly established primer combinations.
| Clade tested | Recommended primers | Notes |
|---|---|---|
| Chytridiomycota | TSR1-1492f, TSR1-2356r | Not sequenced, but amplification product noted. |
| Blastocladiomycota | TSR1-1018f, TSR1-2356r | TSR1-1492f not tested. |
| Zoopagomycotina | TSR1-1018f, TSR1-2356r | TSR1-1492f not tested. |
| Entomophthoromycotina | TSR1-1018f, TSR1-2356r | TSR1-1492f not tested. |
| Kickxellomycotina | ||
| Harpellales | TSR1-1492f, TSR1-2356r | TSR1-1018f does not appear to work. |
| Kickxellales | TSR1-1018f, TSR1-1492f, TSR1-2356r | TSR1-1018f and TSR-1492f both work well. |
| Asellariales | - | Attempted unsuccessfully. |
| Dimargaritales | TSR1-1018f, TSR1-2356r | TSR1-1492f not tested. |
| Orphella clade | TSR1-1492f, TSR1-2356r | PCR product did not sequence cleanly but was identifiable as fungal TSR1. |
| Barbatospora clade | TSR1-1492f, TSR1-2356r | TSR1-1018f amplified but would not sequence. |
| Spiromyces clade | TSR1-1018f, TSR1-1492f, TSR1-2356r | TSR1-1018f and TSR1-1492f both work well. |
| Ramicandelaber clade | TSR1-1018f, TSR1-1492f, TSR1-2356r | TSR1-1018f and TSR1-1492f both work well. |
For MCM7, primer combination MCM7-709f and MCM7-16r was effective for most taxa, with the exception of the Harpellales. MCM7-16r appeared to be more conserved than MCM7-1348rev, amplifies a larger region, and is highly conserved and therefore useful for placing distantly related taxa. Fewer spurious bands were noted in PCR attempts with MCM7-16r. We were unable to develop a primer closer to the beginning (on the 5’ end) of the MCM7 gene. Several clades within the Kickxellomycotina appear to be variable at the MCM7-709f priming site. For these orders, the MCM7-8bf forward primer, which is further downstream, appears to have a higher success rate but is also specific to the Kickxellomycotina and is not recommended for use with other clades. Both primer combinations appear to work well for genomic samples derived from axenic cultures, but sometimes amplify bacterial genes in vouchers prepared from dissected insect guts. Additionally they occasionally amplify host MCM genes, although not MCM7. For example, the MCM2 gene for the host arthropod was amplified in a few of our attempts from mixed genomic samples. These issues are similar to those we observe for primers used to amplify other genes used for phylogenetic studies, such as RPB1 and RPB2, when used under similar conditions. Schmitt et al. (2009) did not report any size variation or introns within their MCM7 dataset. However, we observed spliceosomal introns for some species within the Blastocladiomycota, Chytridiomycota, Entomophthoromycotina, Kickxellomycotina and Zoopagomycotina. No pattern was observed regarding intron position or presence. The largest fragment sequenced in this study was 1139 bp, about 300 bp larger than the observed average size, within our study, of 850 bp (for primers MCM7-709 and MCM7-16r). We also experienced minor size variation (~10 aa) within the translated amino acid sequences. Our alignment also had a small ambiguously aligned region (approx. 15 aa) which was excluded from analysis.
For TSR1, primer combination TSR1-1018f–TSR1-2356r worked best for members of the Blastocladiomycota, Entomophthoromycotina and Zoopagomycotina. In our view, it is preferable over TSR1-1492f–TSR1-2356r because it amplifies a larger region. This region appears to be more conserved and is recommended for the Chytridiomycota, which we found did not have the correct primer site for TSR1-1018f. Within the Kickxellomycotina, TSR1-1492f and TSR1-2356r appears to work for most clades. TSR1-1018f and TSR1-2356r works for many groups, except for the Harpellales.
Schmitt et al. (2009) did report the presence of both introns and hypervariable regions within TSR1, and both of these phenomena were observed within our sample set as well. Introns often occurred within highly variable sections of the gene that were not well aligned, making them difficult to precisely locate. At this scale of phylogenetic comparison, the introns could not be reliably aligned between various taxa and were thus excluded from further consideration. Introns are listed and positions given within Fig. 6. Several hypervariable regions could not be aligned and also needed to be excluded within the dataset. Of the 9 253 characters in the complete alignment, 1 226 of them were excluded in the final analysis. The overall rate of success for amplifications seemed to be lower with TSR1 than with MCM7, but no host insect sequences were observed for TSR1 during the course of this study.
Fig. 6.
Map of the genes MS456 (MCM7) and MS277 (TSR1). 5’ end is at left. Forward primers are marked with blue arrows, reverse primers with red arrows. Introns are labelled in green. Red numbers designate the position of the feature on a reference sequence from C. reversa. Blue numbers designate the position of features on a reference sequence from A. nidulans. Intron locations are given by the position in the alignment in which those introns would be present, if they existed in the reference species,
To assess the congruence of the MCM7 gene to the accepted phylogeny of the trichomycete fungi, the topology of the MCM7 nucleotide and protein trees was compared to a tree based on 18S and 28S rDNA as well as to existing analyses. The MCM7 nucleotide tree had one significant and well-supported incongruity with the rDNA tree as well as the accepted phylogeny: the basidiomycete Ustilago maydis was placed in a well-supported group including the Kickxellomycotina, the Zoopagomycotina and the Blastocladiomycota, instead of with the other basidiomycetes. In general, the MCM7 nucleotide tree failed to recover a higher-level classification of the fungi that was congruent with the accepted phylogeny (James et al. 2006a, White et al. 2006a, Hibbett et al. 2007). We suspect that the third codon base is saturated at this level of taxon selection and is introducing noise into the analysis. It is recommended that future studies utilizing MCM7 to study the entire tree of Fungi or large clades either use the amino acid translation, or at least consider excluding the third codon base from analysis, if it is not otherwise down-weighted.
The MCM7 protein analysis was, in general, more congruent with both the rDNA and the accepted phylogeny. No well-supported incongruities between the MCM7 protein analysis and the accepted phylogeny were apparent. The MCM7 protein analysis did have one incongruity involving Coemansia braziliensis, Coemansia reversa and Spirodactylon aureum that was also shown by the MCM7 nucleotide analysis. This is discussed more in-depth in the section on Kickxellales below.
To assess the congruence of TSR1, we compared its topology to a smaller rDNA analysis containing only the taxa for which we had data on TSR1. A few well-supported incongruities were noted. Dimargaris bacillospora placed within the Mucoromycotina; this species is an obligate mycoparasite of Mucorales that is often cultured with Cokeromyces recurvatus (Benny 2005), and it is likely that our DNA isolate was derived from such a mixed culture. As this may indicate that our sequence is derived from the host, not from Dimargaris, we removed this taxon from the four-gene analysis. This analysis also placed Coprinopsis cinerea with Ustilago maydis, instead of with Cryptococcus neoformans, placed C. reversa with C. braziliensis similar to the MCM7 analysis, and was incongruent in several places within the order Harpellales. This may be due to significant sequence length variation and the difficulty in accurately identifying and removing introns within this group.
DISCUSSION
Overall assessment of MCM7 and TSR1
We developed and tested, along with those from Schmitt et al. (2009), primers for MCM7 and TSR1 that amplify regions of these genes suitable for phylogenetic reconstruction among the early-diverging fungi. Within the Kickxellomycotina we were able to sequence three of the four orders for MCM7 as well as four other genera that may represent new orders; for TSR1, we were able to sequence two of these orders and three of these other genera. Finally, we also amplified and sequenced other groups of early-diverging fungi, including members of the Blastocladiomycota, Chytridiomycota, Entomophthoromycotina and Zoopagomycotina for comparative purposes. The Glomeromycota, Mucoromycotina or the Neocallimastigomycota were not tested. Our assessment of each primer (and combinations of them) for both MCM7 and TSR1 among the clades tested, are detailed in Table 3 and 4.
Phylogenetic utility for the genes varied. The translated MCM7 protein sequences appear to be similar in resolving power to the SSU rDNA (see Table 5), potentially making it a valuable single copy protein-coding gene contribution to multi-gene studies. Congruity with earlier multi-gene trees (Aguileta et al. 2008) suggests that it is resistant to environmental selective pressures and long-branch attraction. Whereas MCM7 analyses were generally congruent to those from rDNA, without having phylogenies based on whole-genomes for comparison, it is difficult to estimate whether the MCM7 protein or the rDNA tree better reflects the evolutionary history in the few cases where they disagree.
Table 5.
Comparative analysis of phylogenetic trees.
| Alignment | Figure | Treebase # | ML score (RAxML) | # Taxa in Alignment | # Char in Alignment | # Interior Branches Total | # Interior Branches Supported | % Interior Branches Supported |
|---|---|---|---|---|---|---|---|---|
| MCM7 protein | Fig. 1 | 13444 | -12111.24453 | 81 | 266 | 72 | 39 | 54.17 % |
| TSR1 protein | Fig. 2 | 13444 | -8224.179649 | 39 | 207 | 33 | 21 | 63.64 % |
| Nuclear SSU + LSU | Fig. 3 | 13444 | -25369.28207 | 76 | 2492 | 67 | 40 | 59.70 % |
| Nuclear SSU+LSU+MCM7 protein | Fig. 4 | 13444 | -38458.25623 | 76 | 2758 | 73 | 51 | 69.86 % |
| Nuclear SSU+LSU+MCM7 protein+TSR1 protein | Fig. 5 | 13444 | -35502.47807 | 38 | 2965 | 35 | 29 | 82.86 % |
| MCM7 nucleotide1 | Fig. S1 | 13444 | -34531.25508 | 81 | 780 | 75 | 41 | 54.67 % |
| SSU rDNA1 | Fig. S2 | 13444 | -14149.10549 | 78 | 1414 | 66 | 28 | 42.42 % |
| LSU rDNA1 | Fig. S3 | 13444 | -11824.38916 | 77 | 1078 | 65 | 26 | 40.00 % |
| SSU+LSU (TSR1 taxa)1 | Fig. S4 | 13444 | -20628.4317 | 39 | 2492 | 35 | 25 | 71.43 % |
1Not presented in main body of document – see supplementary materials.
In our view, TSR1 was more challenging (and perhaps less useful) when compared to MCM7, at least at the taxonomic scale of this study. While it was possible to reconstruct a phylogeny of fungi with TSR1 that was congruent on the large scale with previous analyses (James et al. 2006a, White et al. 2006a, Liu et al. 2009) and with the combined rDNA analysis here, it did present more hindrances in this regard than MCM7. Furthermore, introns needed to be removed from further consideration in preparing the alignment file. Nonetheless, at this time, these issues do not seem severe enough to suggest eliminating TSR1 from future consideration. They should, however, be taken into account by those considering its potential utility. TSR1 is likely to be more useful in studies on clades of more closely related species, where its greater variability may be considered an asset.
Combined analyses using both the 3- and 4-gene datasets had greater resolving power than any single-gene analysis. The 3-gene analysis utilizing the rDNA (SSU and LSU) along with MCM7 protein sequences yielded high resolving power across the greatest number of taxa, whereas the four-gene analysis utilizing these genes along with the TSR1 protein sequences had the highest proportion of fully-supported branches of any analysis (noting also the differences in taxon number between them). Since this, along with Wang (2012), represent the first multi-gene studies primarily concentrating on the Kickxellomycotina that includes sequences from both rDNA and protein-coding genes, we also present a clade-based phylogenetic perspective on the various clades presented (Fig. 1–5).
Phylogenetic analyses
Kingdom Fungi
Except for the Kickxellomycotina, taxon sampling limits our commentary about the other fungal groups. However, by comparing our trees to evolutionary hypotheses presented by others, we offer our assessment of the power of these genes for large-scale phylogenetic reconstruction. The relationships within the early-diverging fungal lineages are still in need of refinement. Hibbett et al. (2007) could not distinguish between early-diverging fungal clades at higher taxonomic levels due to limited molecular phylogenetic support (and to some extent taxon sampling). However, existing analyses do offer hints. James et al. (2006a) used a combination of three rDNA genes and three protein-coding genes to place the Entomophthoromycotina, Kickxellomycotina and Zoopagomycotina on an unsupported branch along with the Dikarya, the Glomeromycota, and the Mucoromycotina. However, our MCM7 protein tree (Fig. 1) placed the Entomophthoromycotina, the Kickxellomycotina and the Zoopagomycotina in a group together with Blastocladiomycota, and separate from the Dikarya and the Mucoromycotina. That branch was supported by the Bayesian but not by the maximum-likelihood analysis (BPP: 98.3 %, MLBP: 37/100). The four-gene tree (Fig. 5) placed representatives of the Entomophthoromycotina together with Blastocladiomycota in a well-supported group (BPP: 100.0 %, MLBP: 80/100); the Dikarya, Kickxellomycotina, Mucoromycotina and Zoopagomycotina were placed in another well-supported group (BPP: 99.9 %, MLBP: 78/100).
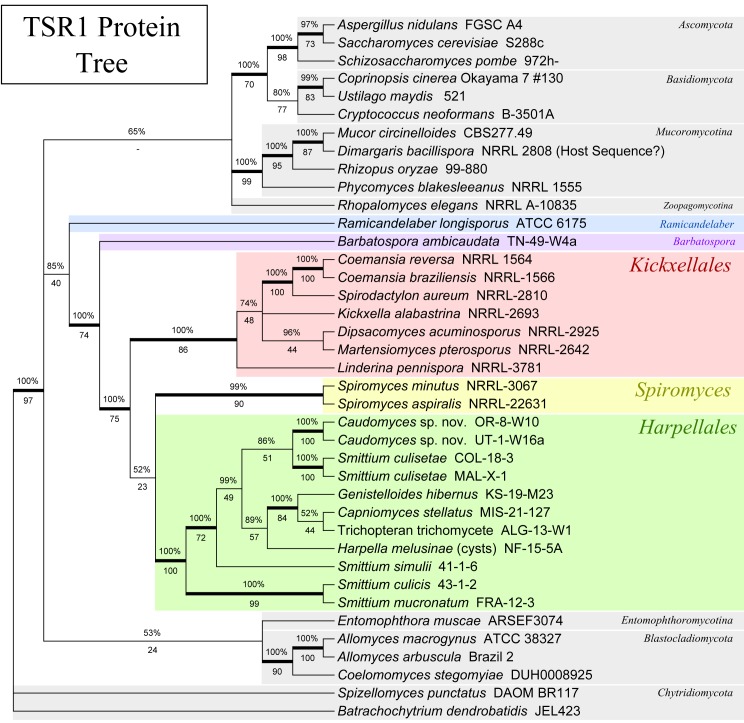
With regard to the later-diverging fungi, the TSR1 protein tree (Fig. 2) placed the Dikarya on a well-supported branch (BPP: 100.0 %, MLBP: 70/100). The MCM7 protein tree (Fig. 1) placed the Ascomycota together with the Mucoromycotina, but was not well supported (BPP: 69.1 %, MLBP: 40/100). Multi-gene analyses (Fig. 4, 5) recovered a well-supported Dikarya (3-gene: BPP: 98.7 %, MLBP: 72/100; 4-gene: BPP: 99.9 %, MLBP: 94/100) as well as a well-supported Dikarya+Mucoromycotina clade (3-gene: BPP: 100.0 %, MLBP: 90/100; 4-gene: BPP: 100.0 %, MLBP: 98/100).
Fig. 2.
Phylogeny of the Kickxellomycotina based on an alignment of TSR1 translated protein sequences. The method of tree calculation and the tree format are the same as Fig. 1.
Fig. 4.
Phylogeny of the Kickxellomycotina based on a concatenated alignment of SSU and LSU rDNA as well as MCM7 translated protein sequences. The method used for tree inference and the format of the tree are the same as for Fig. 3.
Kickxellomycotina
The Kickxellomycotina, the primary focus of this study, are a subphylum of fungi previously placed within the Zygomycota. The Kickxellomycotina are differentiated from other fungi by the production of septal walls with a lenticular pore, containing a plug of material (Hibbett et al. 2007). This characteristic septal pore has been confirmed from all four orders within the Kickxellomycotina, and has only otherwise been found in the genus Ballocephala and Zygnemomyces within the Entomophthoromycotina (Saikawa 1989, Saikawa et al. 1997). However, no molecular data from these two genera have yet been examined, and the morphology is not conclusive, so the affinity of these genera is uncertain. Members of the Kickxellomycotina produce branched or unbranched septate thalli, sometimes with aseptate regions, such as in the main axis of Pteromaktron. They include arthropod symbionts (Harpellales and Asellariales), haustorial mycoparasites (Dimargaritales), and saprobes (Kickxellales except for Martensella, which is a non-haustorial mycoparasite). Asexual one- or two-spored merosporangia are produced (in Harpellales these merosporangia are referred to as trichospores) as well as zygospores. The sexual spores can vary in shape, being spherical in the Asellariales, Dimargaritales and Kickxellales, biconical (or rarely uniconical) within the Harpellales, and coiled within Orphella (Moss & Young 1978,Valle & Santamaria 2005, Valle & Cafaro 2008).
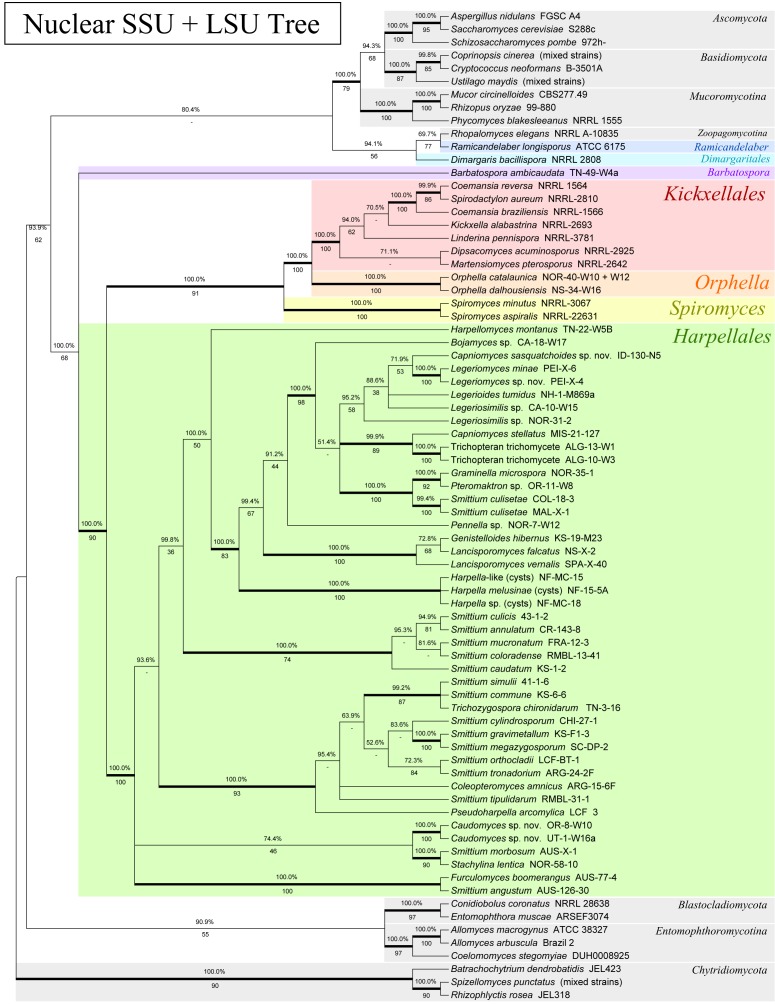
The MCM7 protein tree (Fig. 1) recovered a monophyletic Kickxellomycotina with seven major subclades (BPP: 100.0 %, MLBP: 86/100). These included three of the four known orders; Dimargaritales, Harpellales and Kickxellales, and four genera, Barbatospora, Orphella, Ramicandelaber and Spiromyces, likely to represent new orders (in a subsequent publication). The TSR1 protein tree (Fig. 2) also recovered a monophyletic Kickxellomycotina, with five of the main subclades represented (the Asellariales, Dimargaritales and Orphella have yet to yield sequences), although it was only strongly supported below Ramicandelaber (BPP: 100.0 %, MLBP: 74/100). The 4-gene analysis (Fig. 5) was also able to recover a monophyletic Kickxellomycotina (BPP: 99.8 %, MLBP: 72/100), but the 3-gene analysis (Fig. 4) was not (BPP: 99.9 %, MLBP: 42/100) – it placed Rhopalomyces elegans (Zoopagomycotina) in a clade with Dimargaris and Ramicandelaber. This may be due to long-branch attraction between Dimargaris and Rhopalomyces, as both have highly divergent rDNA sequences.
All trees presented a branch that contained all members of the orders Harpellales and Kickxellales except for the genus Ramicandelaber. This branch was well supported in both the MCM7 (Fig. 1; BPP: 99.9 %, MLBP: 74/100) and TSR1 (Fig. 2; BPP: 100.0 %, MLBP: 74/100) single-gene analyses, and in both the 3-gene (Fig. 4; BPP: 100.0 %, MLBP: 97/100) and 4-gene (Fig. 5; BPP: 100.0 %, MLBP: 100/100) multi-gene analyses. This strongly suggests that the Harpellales and Kickxellales (except for Ramicandelaber) form a monophyletic group, and Ramicandelaber may not be closely related to the Kickxellales. Within this group, no tree (in which they are present) places the genera Barbatospora or Orphella in a monophyletic clade together with only the Harpellales, and no tree places the genus Spiromyces together with only the Kickxellales. Thus, our suggestion is that these genera may represent distinct evolutionary clades.
Harpellales
Harpellales is a diverse order of symbiotic fungi that live within the guts of aquatic insect larvae or rarely, isopods (White 1999). Along with the Asellariales, they are often referred to as ‘gut fungi’, and can shift between parasitic, commensalistic and mutualistic roles (Lichtwardt et al. 2007). The Harpellales have a unique zygospore, whether biconical or uniconical, that distinguishes them from other orders within the Kickxellomycotina. Most species of Harpellales also produce unispored merosporangia (Moss & Young 1978) for asexual reproduction, referred to as trichospores (noting that Carouxella and Klastostachys spores remain attached to the generative cell, which is dehiscent, similar to the arthrospores of the Asellariales). These spores are specialized for the aquatic environment, with many species having mucilaginous non-motile appendages. Moreover, trichospores are sensitive to the precise condition of the insect gut in which they germinate, and rapidly extrude a sporangiospore when appropriate conditions are detected within the correct host gut. During this extrusion process, a mucilaginous holdfast is excreted which secures the thallus to the gut lining of the host. Some genera of Harpellales (Genistellospora, Harpella and Pennella) are also known to occasionally infest the ovaries of developing black flies, replacing the eggs with ovarian cysts containing spores in the adult black flies (White et al. 2006b). The flying adult then oviposits these cysts among egg masses, allowing for effective dispersal and upstream transmission.
The existing classification of the Harpellales includes two families – the Legeriomycetaceae, which are members that have branched thalli and are usually found in the hindgut, whereas the Harpellaceae are all unbranched and typically found in the midgut of their host (Lichtwardt et al. 2007). However, molecular-based phylogenetic analyses have typically not supported this separation. The most complete phylogenetic analyses of the Harpellales to date were provided by White (2006), White et al. (2006a), and Wang et al. (2013). White (2006) designated a ‘Smittium’ clade consisting of Smittium and a few related genera, and a ‘non-Smittium’ clade for Smittium culisetae and most of the other genera of the Harpellales. Wang et al. (2013) are moving Smittium culisetae to a new genus (we use S. culisetae here, ahead of print). That 2-gene study again found evidence of a Smittium / non-Smittium phylogenetic split and further defined the ‘Smittium allies’ to include Austrosmittium, Coleopteromyces, Furculomyces, Pseudoharpella, Stachylina and Trichozygospora.
Our 3-gene analysis (Fig. 4) provides further evidence of this split, with Coleopteromyces, Furculomyces, Smittium, Stachylina and Trichozygospora all placed together and well supported (BPP: 99.8 %, MLBP: 70/100). Another well-supported clade includes Bojamyces, Capniomyces, Genistelloides, Graminella, Harpella, Lancisporomyces, Legerioides, Legeriomyces, Pennella, Pteromaktron and Smittium culisetae (BPP: 100.0 %, MLBP: 98/100). Harpellomyces and Caudomyces were placed as just outside this group, although only strongly supported by the Bayesian analysis (Harpellomyces: BPP: 100.0 %, MLBP: 58/100). The MCM7 protein analysis alone is not as well-resolved, but still contains all of the same clades, supporting the conclusion that both of these analyses are underlying the correct species tree. This is another indication that family structure will need to be reconsidered, pending improved taxon sampling, to more naturally represent the actual relationships.
The TSR1 analysis of the Harpellales (Fig. 2) does not fully agree with the phylogeny provided by the MCM7 protein or rDNA tree (Fig. 1, 3). Although a monophyletic Harpellales was obtained, the topology within the group is not completely congruent with the other analyses, or analyses using RPB1 and RPB2 (not shown, White et al. unpubl. data). TSR1 presented difficulties from aligning the nucleotide sequences to identifying and removing introns, and finally in aligning the proteins and removing ambiguously aligned regions. Sampled members of the Harpellales seemed to have more introns as well as greater size variation within the protein, compared to related groups. Additional taxon sampling within the Harpellales might help to resolve these issues. The 4-gene tree incorporating the TSR1 protein (Fig. 5) did have the same topology as the one from the 3-gene analysis (Fig. 4).
Fig. 3.
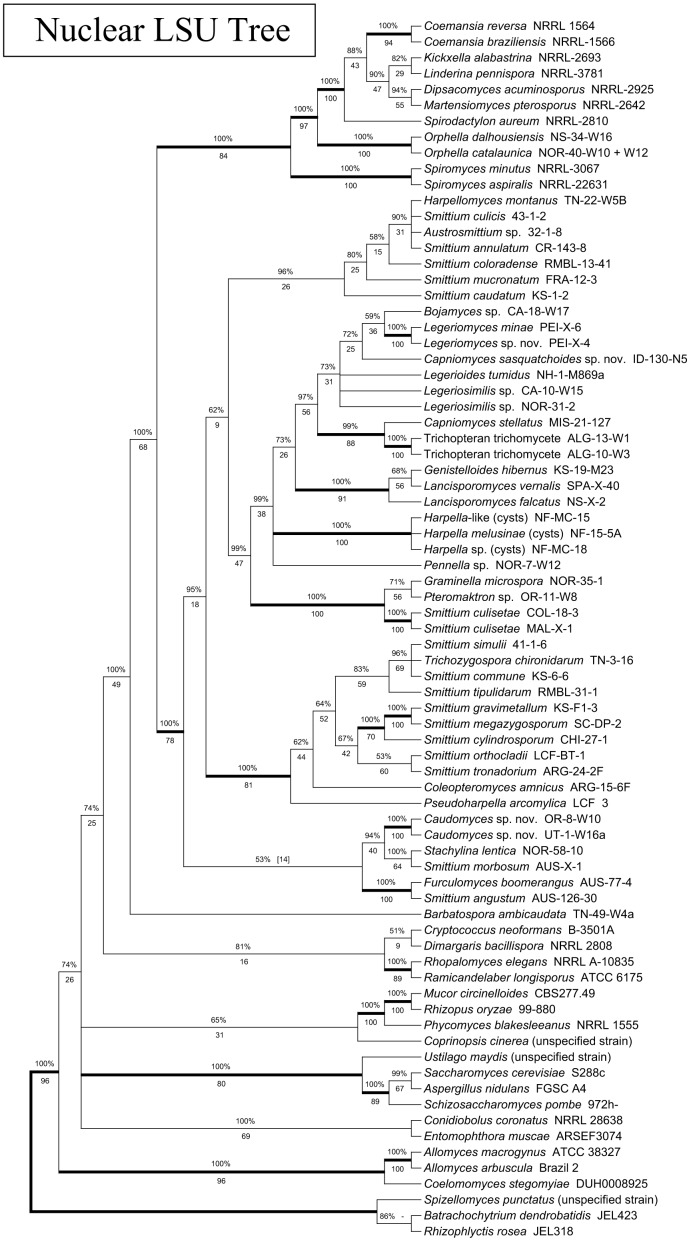
Phylogeny of the Kickxellomycotina based on a concatenated alignment of nuclear small subunit (SSU) and nuclear large subunit (LSU) rDNA. Tree is based on a 50 % majority-rules consensus of 10k trees produced with Bayesian inference (5k used as burn-in). Numbers above branches are Bayesian posterior probabilities. Numbers below branches are maximum-likelihood bootstrap supports produced from 100 bootstrap replicates. Bold branches are highly supported (> 95 % BPP and > .70 MLBP).
Ecological and morphological correlations for these endosymbionts are difficult to place. The ‘non-Smittium’ clade represents a diverse assemblage with variable characteristics, whereas the Smittium clade has much greater morphological similarity. Nearly all members of the Smittium clade have a single appendage as well as a collar left where the trichospore dehisces from the fertile thallus. Trichozygospora is the exception, with its large number of very thin appendages on both the sexual and asexual spores, but is otherwise similar in spore shape and collar presence. Many members of the non-Smittium clade have more than one appendage, and most of them have no collar on the trichospore. A collar is present in Smittium culisetae and Bojamyces, but for both it is flared, and perhaps unlike most species of Smittium. Additionally, phylogenetically related genera Graminella and Pteromaktron have a ball-like or knob-like structure on the appendage near its attachment to the spore. Whether or not this knotted portion of the appendage might represent some remnant of an earlier dehiscent collar or collar-like structures, homologous to the collar of Smittium, is unknown. The Smittium clade is also almost completely restricted to Diptera hosts (except for Coleopteromyces, with one species from aquatic Coleoptera), whereas the non-Smittium clade has members that utilize a diverse group of hosts including not only Diptera, but also Ephemeroptera, Isopoda, Plecoptera and Trichoptera.
Asellariales
The Asellariales represent a much smaller grouping of endosymbiotic fungi, consisting of Asellaria and Baltomyces (within Isopoda) as well as Orchesellaria (in Collembola). The Asellariales produce branched, septate thalli within the hindgut of their host, extending from a specialized holdfast cell with a secreted mucilaginous holdfast (Lichtwardt & Manier 1978). This order is distinguished by the breaking up of the thallus at maturity to produce arthrospores. The general similarity in growth form and life history, along with the similarity between the arthrospores of Asellaria to the asexual reproductive propagules of Carouxella (Harpellales) have been used to suggest that the two orders may be sister taxa (Moss & Young 1978). On the other hand, spherical zygospores have been observed for Asellaria (Valle & Cafaro 2008), unlike the biconical zygospores of the Harpellales. Septal structure has been observed for both Asellaria and Orchesellaria, and is characteristic of the Kickxellomycotina (septa with a lenticular pore and an electron-dense plug), but without the spherical occluding bodies of the Dimargaritales (Moss 1975).
Despite significant effort with all primer combinations listed in this paper (along with some other attempted but unsuccessful primers not provided), we have been unable to amplify and sequence MCM7 or TSR1 for any member of Asellaria. Unpublished RPB1 and RPB2 sequences for Asellaria have been known for some time (Hibbett et al. 2007), and we have successfully amplified additional sequences for these genes as well as the SSU and LSU rDNA (for another manuscript), but even with working genomic samples we were unable to amplify or sequence MCM7. Some bands were visible in the gels, but either would not sequence directly or were deemed incorrect products. Similarly, all attempts to amplify and sequence TSR1 with Asellaria failed to even produce bands. We also attempted to amplify and sequence both of these single-copy genes for Orchesellaria and Baltomyces, but with no success to date.
Kickxellales
The Kickxellales are primarily saprobic (except one genus, Martensella, which is mycoparasitic) fungi in the Kickxellomycotina. Saprobic members of this group have been found associated with soil, dung, and insect carcasses (Benny 2005). Members of this order reproduce asexually by means of sporocladia that produce multiple, unispored merosporangia supported upon small basal cells, the pseudophialides, and also sexually through spherical zygospores (Benny 2005). The sporocladia may be either single- or multi-celled (Benny 2005). Most Kickxellales genera release their spores in a droplet of liquid at maturity, referred to by Moss & Young (1978) as ‘slime spores’, with only the genera Spiromyces and Spirodactylon being dry-spored (Benny 2005). Moss & Young (1978) described this slime as possibly being related to a special intracellular structure found in the pseudophialide, referred to as the ‘labyrinthiform organelle’ and possibly homologous to the trichospores appendage produced by the Harpellales. This study also compared the morphology of the reproductive structure of the two groups, describing the structures as having a shared ‘coemansoid’ morphology and suggesting the two groups may be closely related. This relationship has since been supported by several molecular-based phylogenetic studies (O’Donnell et al. 1998, James et al. 2006a, White et al. 2006a). The Kickxellales have a septal structure similar to the Harpellales and Asellariales, with a lenticular septal pore with an electron-dense plug (O’Donnell et al. 1998).
All trees inferred for this study revealed a strongly-supported and monophyletic Kickxellales clade that includes Coemansia, Dipsacomyces, Kickxella, Linderina, Martensiomyces and Spirodactylon (3-gene: BPP: 100.0 %, MLBP: 100/100; 4-gene: BPP: 100.0 %, MLBP: 100/100). This clade never included Ramicandelaber. Spiromyces is included as a strongly-supported sister clade to this group in the four-gene analysis (BPP: 100.0 %, MLBP: 100/100), but for that analysis Orphella was not available. Within the 3-gene analysis (Fig. 4; BPP: 100.0 %, MLBP: 100/100) and the rDNA-based analysis (Fig. 3; BPP: 100.0 %, MLBP: 100/100), Orphella seems to be more closely related to the monophyletic Kickxellales group than Spiromyces. As such, it appears that Ramicandelaber is not part of the Kickxellales, and Spiromyces may not be, unless Orphella (currently, still a member of the Harpellales) is considered to be a member of the Kickxellales as well (see more on this in the Spiromyces and Ramicandelaber sections).
Within the monophyletic Kickxellales, relationships are difficult to resolve. The group consisting of Coemansia braziliensis, Coemansia reversa and Spirodactylon aureum was first shown by O’Donnell et al. (1998) and again by White et al. (2006a). This relationship is further demonstrated by both the MCM7 (Fig. 1; BPP: 98.2 %, MLBP: 75/100) and TSR1 phylogenies (Fig. 2; BPP: 100.0 %, MLBP: 100/100), providing multi-gene support. However, in both the MCM7 (Fig. 1; BPP: 100.0 %, MLBP: 100/100) and TSR1 (Fig. 2; BPP: 100.0 %, MLBP: 100/100) analyses, C. reversa and C. braziliensis are placed together, while in the rDNA analysis (as for the previous published analyses, which also utilised rDNA) C. reversa is placed together with S. aureum, which renders Coemansia polyphyletic (Fig. 3; BPP: 99.9 %, MLBP: 86/100). This may represent a true instance of incomplete lineage sorting within the Kickxellales. Alternately, it may be due to long-branch attraction related to Spirodactylon, which appears to be unusually diverged from the other Kickxellales with regard to rDNA but not MCM7 or TSR1.
Other relationships between the members of the Kickxellales are more difficult to resolve. Both the MCM7 and TSR1 individual gene trees (Fig. 1, 2) are not well resolved within the Kickxellales clade. The 3-gene tree (Fig. 4) is well resolved with regard to internal members of the group; a poorly-supported group consisting of Dipsacomyces and Martensiomyces is the most early-diverging member (BPP: 86.5 %, MLBP: not present), followed by well-supported individual branches containing Linderina (BPP: 100.0 %, MLBP: 78/100) and Kickxella (BPP: 100.0 %, MLBP: 81/100). The 4-gene analysis (Fig. 5), however, does not strongly support these internal branches (possibly due to contrasting signal from TSR1), but does strongly support the relationship between Dipsacomyces and Martensiomyces (BPP: 100.0 %, MLBP: 83/100). This relationship is present, although not as strongly supported, in all four individual-gene analyses (Fig. 1, 2, S2, S3) as well as previously by O’Donnell et al. (1998).
Fig. S2.
Phylogeny of the Kickxellomycotina based on an alignment of nuclear small subunit (SSU) rDNA. Tree is based on a 50 % majority-rules consensus of 10k trees produced with Bayesian inference (5k used as burn-in). The three codon positions were all considered to be on different, unlinked partitions during tree calculation. Numbers above branches are Bayesian posterior probabilities. Numbers below branches are maximum-likelihood bootstrap supports produced from 100 bootstrap replicates. Bold branches are highly supported (> 95 % BPP and > .70 MLBP).
Fig. S3.
Phylogeny of the Kickxellomycotina based on an alignment of nuclear large subunit (LSU) rDNA. Tree is based on a 50 % majority-rules consensus of 10k trees produced with Bayesian inference (5k used as burn-in). The three codon positions were all considered to be on different, unlinked partitions during tree calculation. Numbers above branches are Bayesian posterior probabilities. Numbers below branches are maximum-likelihood bootstrap supports produced from 100 bootstrap replicates. Bold branches are highly supported (> 95 % BPP and > .70 MLBP).
Dimargaritales
Dimargaritales is an unusual group of Kickxellomycotina. Mycoparasites of Mucorales and Ascomycota, they have several morphological and life history features that differentiate them from other Kickxellomycotina. While they retain the diagnostic lenticular septal cavity with an electron-dense plug (Jeffries & Young 1979, Brain et al. 1982), the plug has globose bodies to either side of the septum in the Dimargaritales (Benjamin 1959), which can be dissolved in 2–3 % KOH, unlike the septal plugs of the Kickxellales. Other unique features include bispored merosporangia (all other Kickxellomycotina are unispored) and the presence of haustoria.
We attempted to amplify and sequence MCM7 and TSR1 for our single representative of this order, Dimargaris bacillosporus (see Table 2). We were able to successfully sequence MCM7. Unfortunately, for TSR1, our sequence appears to be that of a mucoralean contaminant. Dimargaris bacillosporus is often grown in co-culture with its host, Cokeromyces recurvatus. The phylogenetic position of the Dimargaris within the TSR1 tree suggests strongly that our sequence is that of the host fungus. Our MCM7 sequence does not appear to show any affinity to the Mucorales, and thus be genuine.
The MCM7 analysis reveals a monophyletic Kickxellomycotina that includes Dimargaris (BPP: 100.0 %, MLBP: 86/100) as part of an early-diverging group that also contains Ramicandelaber, although the connection between Ramicandelaber and Dimargaritales was only supported by the Bayesian analysis (BPP: 97.5 %, MLBP: 59/100). Multi-gene analysis was less clear because the three-gene analysis placed Dimargaris in an unsupported group with Rhopalomyces (BPP: 66.7 %, MLBP: not present). The notion that Dimargaritales is one of the most early-diverging members of the Kickxellomycotina is evocative. Several features of Dimargaritales bear close resemblance to members of the Zoopagomycotina, particularly Piptocephalis and Syncephalis that are mucoralean mycoparasites (Benny 2005). Beyond the lifestyle, these genera also have multispored merosporangia and appear to have a similar growth form. It may be that the Kickxellomycotina either descend from within the Zoopagomycotina or form a sister clade to it. Molecular analyses thus far, including this one, have been unable to fully resolve the phylogenetic position of the Zoopagomycotina, and point to the need for further study.
We suggest that the MCM7 gene will be particularly useful for Dimargaritales due to the consistent sequence length and reliable alignment. Dimargaritales have demonstrated extremely diverged and variable rDNA sequences that make them difficult to align and result in long-branch attraction artifacts (White et al. 2006a). MCM7 does not suffer from this problem and trees have relatively consistent branch-lengths, at least as demonstrated by our Dimargaris representative.
Distinct lineages: Barbatospora, Orphella, Ramicandelaber and Spiromyces
Several genera of Harpellales and Kickxellales have consistently not clustered with their respective orders. These unique genera (lineages) are examined.
Barbatospora has not been reported since the type B. ambicaudata was described from blackflies in the Great Smoky Mountains National Park, USA (White et al. 2006c). Although the general growth form of Barbatospora resembles the Harpellales, with a branched, septate thallus and a secreted holdfast, it also presents unique morphological features. These include a ‘cap-like’ structure at the terminal end of the trichospores, which typically falls away at maturity, to reveal a set of appendages or appendage-like structures on either end of the asexual spore. However, much about the morphology of this species is not known, including the presence and form of zygospores, the septal wall structure, and the method of spore extrusion and germination. Barbatospora was placed, on morphological grounds, in Harpellales within the family Legeriomycetaceae.
Phylogenetically, Barbatospora consistently places within a branch that includes the Harpellales, the Kickxellales, Orphella and Spiromyces. This placement is well-supported in the MCM7 (Fig. 2; BPP: 99.9 %, MLBP: 74/100), TSR1 (Fig. 2; BPP: 100.0 %, MLBP: 74/100), 3-gene (Fig. 4; BPP: 100.0 %, MLBP: 97/100), and 4-gene (Fig. 5; BPP: 100.0 %, MLBP: 100/100) analyses, and is present (but not completely supported) in the rDNA (Fig. 3; BPP: 100.0 %, MLBP: 68/100) analysis as well. Within this group, the Harpellales, Kickxellales, Orphella and Spiromyces are together on a strongly-supported branch within the TSR1 (Fig. 2; BPP: 100.0 %, MLBP: 75/100), rDNA (Fig. 3; BPP: 100.0 %, MLBP: 90/100), 3-gene (Fig. 4; BPP: 99.9 %, MLBP: 91/100), and 4-gene analyses (Fig. 5; BPP: 100.0 %, MLBP: 95/100), and a branch that is not strongly supported within the MCM7 (Fig. 1; BPP: 86.4 %, MLBP: 48/100) analysis. The position of Barbatospora, which is one of the most consistent and well supported evolutionary hypotheses provided by this study, may suggest that the species is an ‘offshoot’ from an ancestral clade that split to form the Kickxellales and Harpellales. Thus, Barbatospora might offer valuable insights into the early evolution of this group.
Orphella, also currently a member of the Harpellales, has unusual morphological features for that order (see review by Valle & Santamaria 2005). Orphella is unique among gut fungi in releasing both trichospores and zygospores as multi-celled dissemination units, and in having allantoid to coiled asexual spores and (to some extent) coiled zygospores (Valle & Santamaria 2005). At maturity, both spore forms extend, attached to the thallus, beyond the anus of the host. Valle & Santamaria (2005) reported that Orphella has a characteristic ‘coemansoid’ growth form, which pointed to a relationship with the Kickxellales. This relationship was also suggested by molecular-based studies (James et al. 2006a, White et al. 2006a), where Orphella is clustered with the Kickxellales. Aside from the unusual spore features, its morphology resembles the Harpellales, with an extruded mucilaginous holdfast, a specialized holdfast cell, and a branched, septate thallus.
Again, for Orphella, we were able to sequence MCM7 but not TSR1. The MCM7 analysis does not offer any additional insight into the relationship between Orphella and the other taxa within the Kickxellomycotina, beyond suggesting that Orphella is separate from the other Harpellales. The 3-gene analysis (Fig. 4; BPP: 100.0 %, MLBP: 100/100 both above and below the branch containing Orphella) supports the phylogeny demonstrated by previous studies (James et al. 2006a, White et al. 2006a), noting the possible disproportionate phylogenetic signal from rDNA.
Spiromyces is currently a member of the Kickxellales, although previous phylogenetic analyses have placed it apart from that order. It is separated from the Kickxellales by Orphella (White et al. 2006a), but sometimes it appears ancestral to both the Kickxellales and Harpellales (James et al. 2006a). Morphologically, Spiromyces is an unusual member of the Kickxellales because rather than pseudophialides, it produces merosporangia from enlarged sections of the sporangiophore, similar to the collar regions of the generative cells of Harpellales (Moss & Young 1978). It is also one of the few Kickxellales that is dry-spored at maturity. Spiromyces species are saprobic and usually associated with dung. We were able to amplify and sequence both MCM7 and TSR1 for Spiromyces, but neither single-gene tree is able to place it reliably (Fig. 1, 2). Within the 3-gene tree (Fig. 4), Spiromyces is placed as a sister clade to a group consisting of the Kickxellales (except Ramicandelaber) and Orphella (BPP: 100.0 %, MLBP: 88/100). Within the 4-gene tree (Fig. 5), Spiromyces is with the Kickxellales (except Ramicandelaber) as the earliest-diverging member (but recall Orphella is not available for this tree) (BPP: 100.0 %, MLBP: 100/100).
Ramicandelaber is another genus within the Kickxellales that may not belong with the order. This genus has an unusual growth form for the Kickxellales. It forms both stolons and rhizoids and in R. brevisporus, may form supporting branches (Benny 2005). It is also unusual how, in age, Ramicandelaber sporocladia broaden and become covered with more pseudophialides, which become subspherical (Ogawa et al. 2001). Previous molecular studies have placed Ramicandelaber apart from the Harpellales and Kickxellales (Ogawa et al. 2005, White et al. 2006a). On the MCM7 tree (Fig. 1), Ramicandelaber is placed within the Kickxellomycotina on an early-diverging branch along with Dimargaris (note however that this branch was only supported by the Bayesian analysis; BPP: 97.5 %, MLBP: 59/100). Within the TSR1 analysis (Fig. 2), it was placed as an unsupported branch as the earliest-diverging member of the Kickxellomycotina (recall that the Dimargaritales sample was not placed correctly on this tree due to amplification of the fungal host of Dimargaris; BPP: 85 %, MLBP: 40/100). Within all five analyses (Fig. 1–5) Ramicandelaber is placed outside of a well-supported clade that contains all other members of the Kickxellales and the Harpellales (except, in the case of the rDNA analysis, Barbatospora). These results suggest that the rDNA-based placement of Ramicandelaber outside of the Kickxellales is likely an accurate one, and that this genus may well represent a unique, early-diverging lineage of the Kickxellomycotina.
CONCLUSIONS
The comparison between the rDNA-based and MCM7-based phylogenies suggest that MCM7 is likely to be a valuable gene for phylogenetic inference within the Kickxellomycotina, although it does not seem to have the same degree of resolving power that it does within Ascomycota (Schmitt et al. 2009, Raja et al. 2011). While it is unlikely to be sufficient to resolve complex relationships on its own, the relative ease of amplification and sequencing (for a single-copy, protein-coding gene) and the high degree of resolving power make it a valuable addition to rDNA-based or multi-gene studies. In addition, MCM7 seems to not be plagued with the long-branch attraction problems demonstrated by the rDNA of the Dimargaritales, and to a lesser extent, Ramicandelaber, making it an excellent alternative to consider for accurate phylogenetic inference.
As we have pursued the use of TSR1 over a shorter time frame, its potential utility is more difficult to ascertain. While the large-scale phylogenetic resolution of TSR1 appears to be quite good, difficulties in identifying and removing introns and incongruities between the TSR1 tree and the rDNA tree make it uncertain at those levels, and specifically how trustworthy it may be within the Harpellales. Additional studies with it for more representatives of the Harpellales for TSR1 should make it easier to reliably remove introns and align amino acid sequences. To date, TSR1 appears to be more difficult to amplify and sequence compared to MCM7, although in our laboratory we have found it to be far easier to work with than RPB1 or RPB2 (White et al. unpubl. data). When TSR1 amplifies, it is specific to the correct gene and to Fungi, and the variability could be an asset within groups of closely related species.
In addition to their utility within the Kickxellomycotina, general congruence with accepted phylogenetic studies across the broader fungal tree and successful amplification within several early-diverging lineages suggests that MCM7 and TSR1 can potentially be used by those studying other groups of early-diverging fungi. In particular, they are also likely to be useful within the Entomophthoromycotina and Zoopagomycotina, groups that traditionally have proven difficult to culture (for some taxa at least) and to place. While the gene regions require some manual adjustment (intron removal, translation, and removal of poorly aligned regions) to be used, this is true of the majority of phylogenetically-informative genes, including well-accepted ones (such as RPB1 and RPB2), when used over wide taxonomic ranges. We are poised to consider them for our revisionary efforts on the gut fungi, within the Kickxellomycotina, and hope they will be considered by others exploring the earliest branches of the fungal tree of life.
Acknowledgments
Financial support from NSF REVSYS Awards DEB-0918182 (to MMW) and DEB-0918169 (to collaborator R.W. Lichtwardt, University of Kansas) are gratefully acknowledged for this and ongoing studies toward a molecular-based reclassification of the Harpellales and Asellariales. MMW received funding for some sequences from a Martin-Baker Award from the Mycological Society of America. We would especially like to thank all who have contributed samples to our efforts, without which they would have never been able to proceed. In particular, we would like to thank A. Gryganskyi (and the laboratory of Rytas Vilgalys) as well R. Humber. Additionally, G. Benny, M.J. Cafaro, L.C. Ferrington Jr., D.B. Strongman, L.G. Valle provided sample vouchers and support. T. James provided kind research support during earlier (AFTOL) training sessions to MMW, and more recently as well. Thanks to Paula Clarke for proofreading final versions of the manuscript. Comments from committee members I. Robertson, J. Smith and S. Novak were incorporated at the proof stage. Finally, we would like to thank Dr. Robert W. Lichtwardt, without whose tireless efforts on the Harpellales and other trichomycetes, especially with his extended efforts on the web-based trichomycete resources, this lineage of studies have never been possible and as poised for continuity.
REFERENCES
- Abascal F, Zardoya R, Posada D.2005. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics: 21, 9: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Aguileta G, Marthey S, Chiapello H, Lebrun M-H, Rodolphe F, et al. 2008. Assessing the performance of single-copy genes for recovering robust phylogenies. Systematic Biology 57: 613–627 [DOI] [PubMed] [Google Scholar]
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F.2004. Parallel metropolis-coupled markov chain monte carlo for Bayesian phylogenetic inference. Bioinformatics 20: 407–415 [DOI] [PubMed] [Google Scholar]
- Benjamin RK.1959. The merosporangiferous Mucorales. Aliso 4: 321–433 [Google Scholar]
- Benny GL.2005. Zygomycetes. Published on the internet at http://www.zygomycetes.org [Google Scholar]
- Brain APR, Jeffries P, Young TWK.1982. Ultrastructure of septa in Tieghemiomyces californicus. Mycologia 74: 173–181 [Google Scholar]
- Drummond A, Strimmer K.2001. PAL: An object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics 17: 662–663 [DOI] [PubMed] [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M, Bruns TD.1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118 [DOI] [PubMed] [Google Scholar]
- Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK.2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. Rna-a Publication of the Rna Society 7: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hermet A, Méheust D, Mounier J, Barbier G, Jany J-L.2012. Molecular systematics in the genus Mucor with special regards to species encountered in cheese. Fungal Biology 116: 692–705 [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509–547 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. 2006a. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: 818–822 [DOI] [PubMed] [Google Scholar]
- James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, et al. 2006b. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98: 860–871 [DOI] [PubMed] [Google Scholar]
- Jeffries P, Young TWK.1979. Ultrastructure of septa in Dimargaris cristalligena R.K. Benjamin. Journal of General Microbiology 111: 303–311 [Google Scholar]
- Kearsey SE, Labib K.1998. MCM proteins: evolution, properties, and role in DNA replication. Biochimica et Biophysica Acta 1398: 113–136 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.2008. An improved general amino acid replacement matrix. Molecular Biology and Evolution 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lichtwardt RW, Cafaro MR, White MM.2007. Taxonomy and co-evolution of Trichomycetes (gut-inhabiting fungi) and their Chironomidae (Diptera) hosts. Published at http://www.nhm.ku.edu/fungi/ [Google Scholar]
- Lichtwardt RW, Manier JF.1978. Validation of the Harpellales and Asellariales. Mycotaxon 7: 441–442 [Google Scholar]
- Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, et al. 2009. Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Molecular Biology and Evolution 26: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR.2011. Mesquite: a modular system for evolutionary analysis. v 2.75 (http://mesquiteproject.org). [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D.1982. Cold-sensitive cell-division-cycle mutants of yeast: Isolation, properties, and pseudoreversion studies. Genetics 100: 547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern I, Powlowski J, Ishmael N, Darmond C, Marqueteau S, et al. 2012. A molecular phylogeny of thermophilic fungi. Fungal Biology 116: 489–502 [DOI] [PubMed] [Google Scholar]
- Moss ST.1975. Septal structure in the trichomycetes with special reference to Astreptonema gammari (Eccrinales). Transactions of the British Mycological Society 65: 11 5–124 [Google Scholar]
- Moss ST, Young TWK.1978. Phyletic considerations of the Harpellales and Asellariales (Trichomycetes, Zygomycotina) and the Kickxellales (Zygomycetes, Zygomycotina). Mycologia 70: 944–963 [Google Scholar]
- O’Donnell K.1993. Fusarium and its near relatives. In: Reynolds DR, Taylor JW. (eds), The fungal holomorph: mitotic, Meiotic and pleomorphic speciation in fungal systematics: 225–233, CAB International, UK [Google Scholar]
- O’Donnell K, Cigelnik E, Benny GL.1998. Phylogenetic relationships among the Harpellales and Kickxellales. Mycologia 90: 624–639 [Google Scholar]
- Ogawa Y, Hayashi S, Degawa Y, Yaguchi Y.2001. Ramicandelaber, a new genus of the Kickxellales, Zygomycetes. Mycoscience 42: 193–199 [Google Scholar]
- Ogawa Y, Kurihara Y, Suda A, Kusama-Eguchi K, Watanabe K, Tokumasu S.2005. Taxonomic position of the genus Ramicandelaber, Kickxellales, inferred from 18S rDNA. Nippon Kingakukai Kaiho 46: 13–17 [Google Scholar]
- Posada D.2008. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Raja H, Schoch CL, Hustad V, Shearer C, Miller A.2011. Testing the phylogenetic utility of MCM7 in the Ascomycota. MycoKeys 1: 63–94 [Google Scholar]
- Ronquist F, Huelsenbeck JP.2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Saikawa M.1989. Ultrastructure of the septum in Ballocephala verrucospora (Entomophthorales, Zygomycetes). Canadian Journal of Botany 67: 2484–2488 [Google Scholar]
- Saikawa M, Oguchi M, Ruiz RFC.1997. Electron microscopy of two nematode-destroying fungi, Meristacrum asterospermum and Zygnemomyces echinulatus (Meristacraceae, Entomophthorales). Canadian Journal of Botany 75: 762–768 [Google Scholar]
- Schmitt I, Crespo A, Divakar P, Fankhauser J, Herman-Sackett E, et al. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto S, Rochon D, Long J, Dee J, Berbee M.2011. A multigene phylogeny of Olpidium and its implications for early fungal evolution. BMC Evolutionary Biology 11: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Glazko GV, Nei M.2002. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proceedings of the National Academy of Sciences 99: 16138–16143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, Saikawa M, Watanabe MM, Sugiyama J.2004. Molecular phylogeny of Zygomycota based on EF-1α and RPB1 sequences: limitations and utility of alternative markers to rDNA. Molecular Phylogenetics and Evolution 30: 438–449 [DOI] [PubMed] [Google Scholar]
- Valle LG, Cafaro MJ.2008. First report of Zygospores in Asellariales and new species from the Caribbean. Mycologia 100: 122–131 [DOI] [PubMed] [Google Scholar]
- Valle LG, Santamaria S.2005. Zygospores as evidence of sexual reproduction in the genus Orphella. Mycologia 97: 1335–1347 [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M.1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.2012. Ribosomal RNA gene-based and multigene phylogenies of Smittium (Harpellales) and allies – toward unraveling relationships among early diverging fungi. MS thesis Boise State University, USA. [Google Scholar]
- Wang Y, Tretter ED, Lichtwardt RW, White MM.2013. Overview of 75 years of Smittium research, establishing a new genus for Smittium culisetae, and prospects for future revisions of the ‘Smittium’ clade. Mycologia 105: 90–111 [DOI] [PubMed] [Google Scholar]
- White MM.1999. Legerioides, a new genus of Harpellales in isopods and other Trichomycetes from New England, USA. Mycologia 91: 1021–1030 [Google Scholar]
- White MM.2006. Evolutionary implications of a rRNA-based phylogeny of Harpellales. Mycological Research 110: 1011–1024 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee J, Taylor J.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR Protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, USA [Google Scholar]
- White MM, James TY, O’ Donnell K, Cafaro MJ, Tanabe Y, Sugiyama J.2006a. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia 98: 872–884 [DOI] [PubMed] [Google Scholar]
- White MM, Lichtwardt RW, Colbo MH.2006b. Confirmation and identification of parasitic stages of obligate endobionts (Harpellales) in blackflies (Simuliidae) by means of rRNA sequence data. Mycological Research 110: 1070–1079 [DOI] [PubMed] [Google Scholar]
- White MM, Siri A, Lichtwardt RW.2006c. Trichomycete insect symbionts in Great Smoky Mountains National Park and vicinity. Mycologia 98: 333–352 [DOI] [PubMed] [Google Scholar]
- Wilgenbusch JC, Warren DL, Swofford DL.2004. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. http://ceb.csit.fsu.edu/awty [DOI] [PubMed] [Google Scholar]
- Zwickl DJ.2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, The University of Texas at Austin. [Google Scholar]