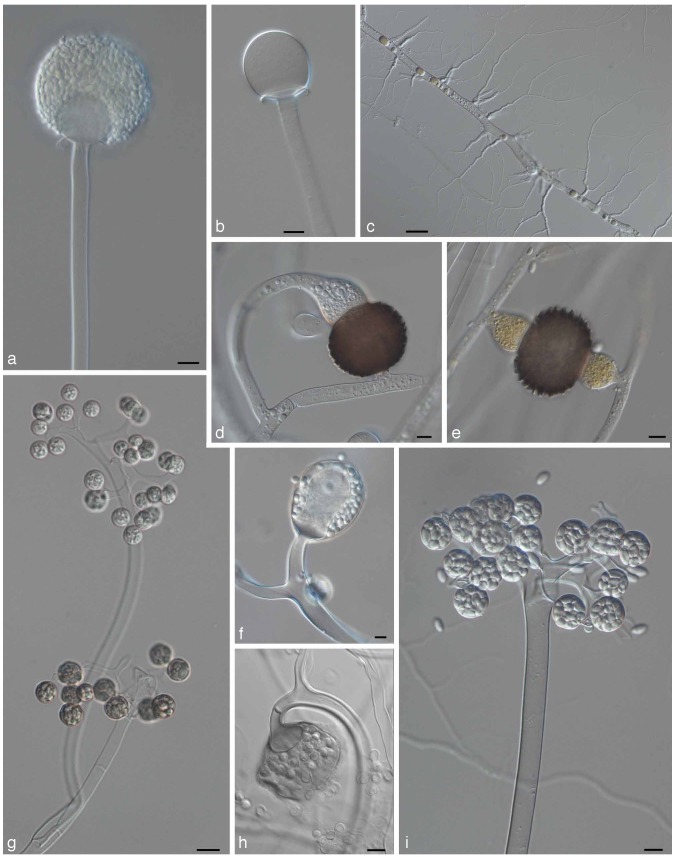
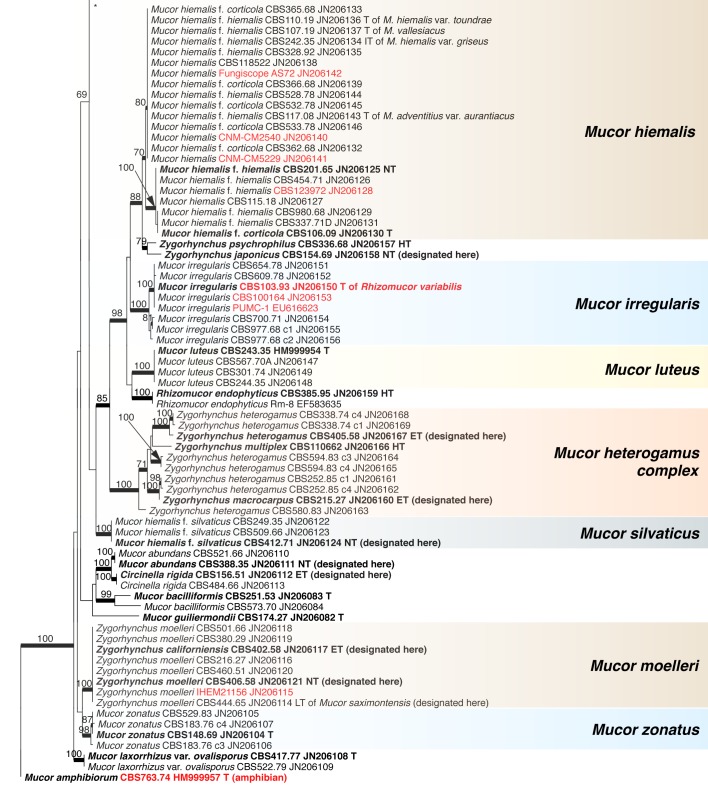
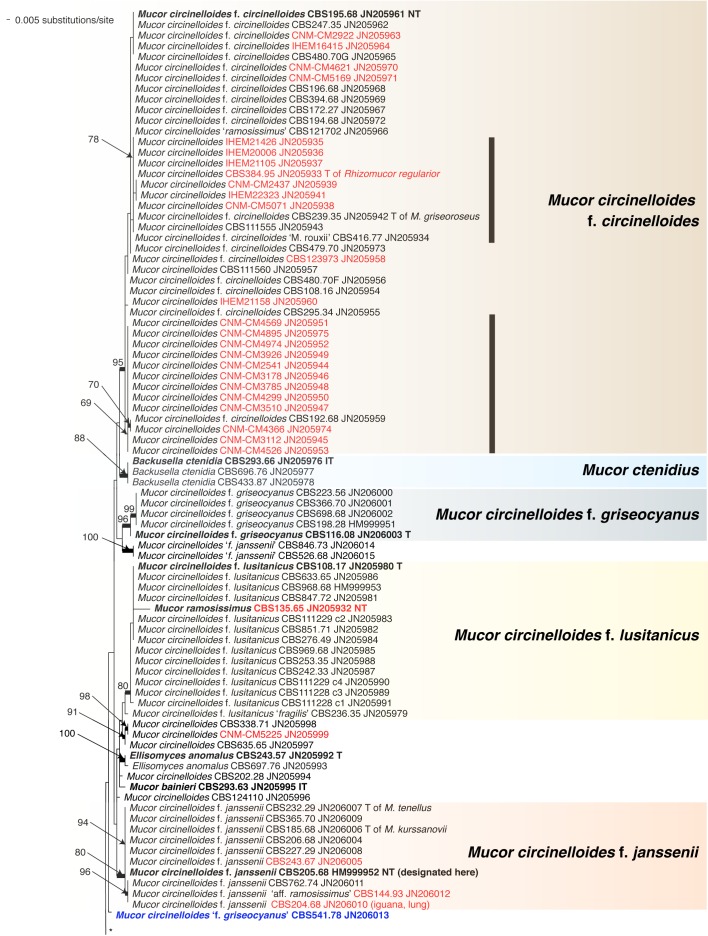
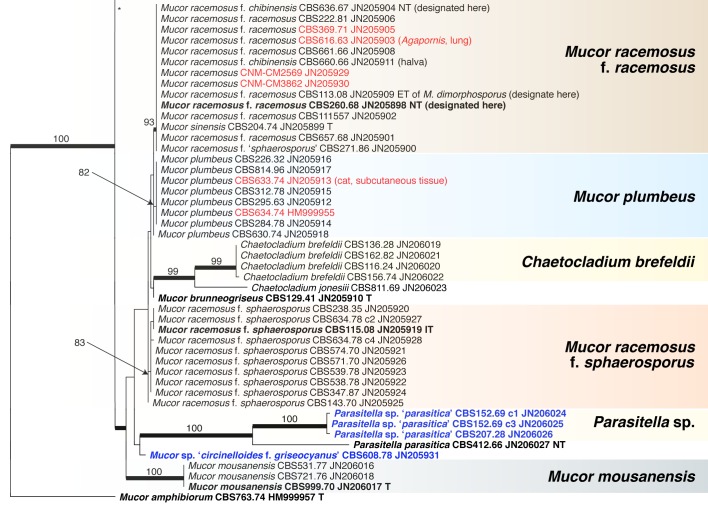
Abstract
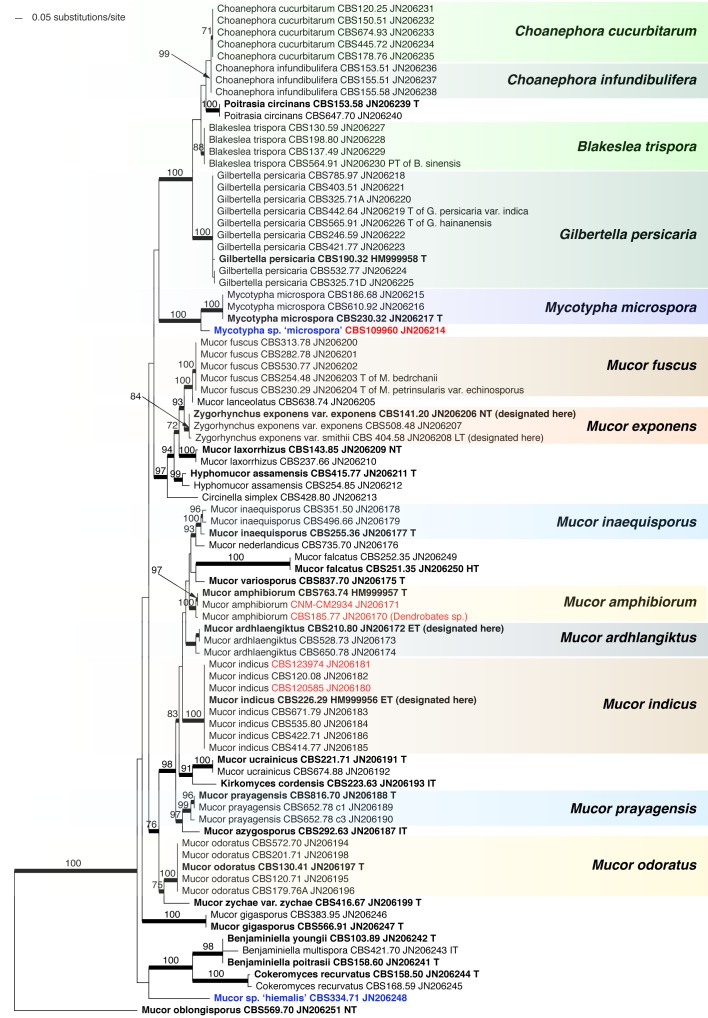
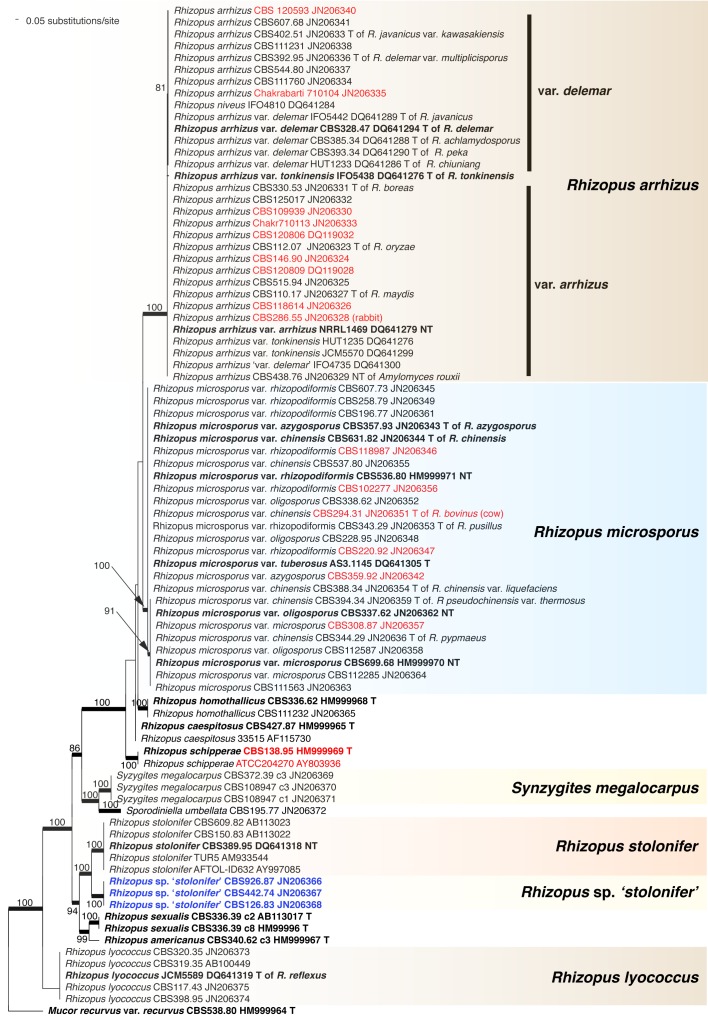
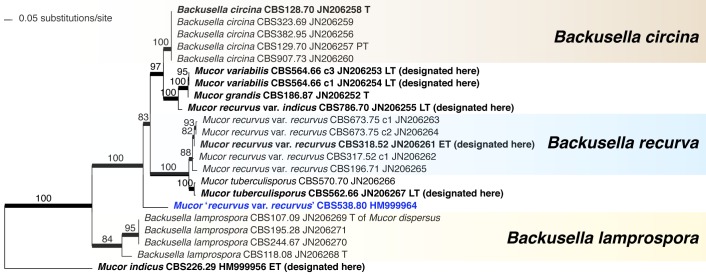
The order Mucorales comprises predominantly fast-growing saprotrophic fungi, some of which are used for the fermentation of foodstuffs but it also includes species known to cause infections in patients with severe immune or metabolic impairments. To inventory biodiversity in Mucorales ITS barcodes of 668 strains in 203 taxa were generated covering more than two thirds of the recognised species. Using the ITS sequences, Molecular Operational Taxonomic Units were defined by a similarity threshold of 99 %. An LSU sequence was generated for each unit as well. Analysis of the LSU sequences revealed that conventional phenotypic classifications of the Mucoraceae are highly artificial. The LSU- and ITS-based trees suggest that characters, such as rhizoids and sporangiola, traditionally used in mucoralean taxonomy are plesiomorphic traits. The ITS region turned out to be an appropriate barcoding marker in Mucorales. It could be sequenced directly in 82 % of the strains and its variability was sufficient to resolve most of the morphospecies. Molecular identification turned out to be problematic only for the species complexes of Mucor circinelloides, M. flavus, M. piriformis and Zygorhynchus moelleri. As many as 12 possibly undescribed species were detected. Intraspecific variability differed widely among mucorealean species ranging from 0 % in Backusella circina to 13.3 % in Cunninghamella echinulata. A high proportion of clinical strains was included for molecular identification. Clinical isolates of Cunninghamella elegans were identified molecularly for the first time. As a result of the phylogenetic analyses several taxonomic and nomenclatural changes became necessary. The genus Backusella was emended to include all species with transitorily recurved sporangiophores. Since this matched molecular data all Mucor species possessing this character were transferred to Backusella. The genus Zygorhynchus was shown to be polyphyletic based on ITS and LSU data. Consequently, Zygorhynchus was abandoned and all species were reclassified in Mucor. Our phylogenetic analyses showed, furthermore, that all non-thermophilic Rhizomucor species belong to Mucor. Accordingly, Rhizomucor endophyticus was transferred to Mucor and Rhizomucor chlamydosporus was synonymised with Mucor indicus. Lecto-, epi- or neotypes were designated for several taxa.
Keywords: Backusella, biodiversity, clinical relevance, DNA barcoding, intraspecific variability, ITS, LSU, Mucor, Mucorales, nomenclature, Rhizomucor, taxonomy, Zygorhynchus
INTRODUCTION
The order Mucorales represents a phylogenetically ancient group of fungi comprising predominantly saprotrophs inhabiting soil, dung and dead plant material, as well as several parasites on plants and on other fungi. Mucoralean strains have been used for centuries in the fermentation of traditional Asian and African food such as tempeh or furu (fermented tofu) (Nout & Aidoo 2010), and they also play a role in the production of several kinds of cheese (Hermet et al. 2012). On the other hand, some members of the Mucorales are responsible for the spoilage of fresh and manufactured food (Pitt & Hocking 2009).
Mucoralean fungi are also known to be involved in human infection. Mucormycoses are still very rare, but their incidence is increasing in hosts with severe immune or metabolic impairment, e.g. due to hemomalignancy, hematopoietic stem cell transplantation or uncontrolled ketoacidotic diabetes mellitus (Skiada et al. 2011). Infections often take a dramatic course and have a high mortality rate. In risk group patients such as those with leukemia or allogenic bone marrow transplant an increase of 8 % and 2 %, respectively, has been noted (Greenberg et al. 2004). In part the clinical strains belong to the same species as the ones used in food fermentation. For example, Mucor circinelloides is used for starter cultures in Asian food (Hesseltine 1983, Nout & Aidoo 2010), but is also able to infect patients with an impaired immune system (e.g. Khan et al. 2009).
Mucorales are among the best represented groups in fungal culture collections. They easily grow in axenic culture and they have been used as model organisms since the late 19th century. A large share of all species described in the order are represented today by living cultures publicly available in fungal reference collections. For example, the Centraalbureau voor Schimmelcultures (www.cbs.knaw.nl) possesses 135 ex-type or authentic strains out of 227 currently accepted species. This is a unique situation, compared e.g. with dermatophytes which were described around the same period (Sabouraud 1910).
Problematic for the nomenclatural stability of the Mucorales is the practice of many early authors to designate a living strain as ‘type’ although this was permitted by the International Code of Botanical Nomenclature. Since 2000 Art. 8.4 of ICBN has allowed deposition of metabolically inactive cultures as types (Greuter et al. 2000). In order to link these original strains to the respective names we designated the vial with the lyophilised strain that was prepared at time of its accession as lectotypes. If the original strain was not lyophilized in the year of its accession we lectotypified the name by the original illustration and designated the original strain as epitype. In the case of Zygorhynchus exponens a neotype was chosen because the original figures were not specific and no other authentic material is known to exist.
Taxonomy of Mucorales has traditionally been based upon microscopic morphology and mating experiments. The classical works of Maria A.A. Schipper (Schipper 1973, 1975, 1976, 1978a, b, 1979, 1984, 1986, 1990, Schipper & Samson 1994, Schipper & Stalpers 1984, 2003) provided model studies and have long remained satisfactory for the identification of major species. A large number of names were synonymised. However, molecular phylogeny has revealed that diversity within and between species is much larger than anticipated, and this has led to a proliferation of the number of taxa recognised. Since the older, morphological synonyms were a priori omitted from most studies, the respective names remained obscure and were not included in nomenclatural comparisons. New names are being introduced today for species that do not match any of the known taxa deposited in GenBank. Verification of the ex-type strains of older, still valid names may prove that some of the new names are later synonyms, and that the historical names have to be re-installed.
DNA barcoding was originally aimed to allow faster and more precise species identification. However, the accuracy of this method strongly depends on completeness of taxon sampling and on taxonomic elaboration (Meyer & Pauley 2005). Since polyphyly was revealed with molecular data in many morphology-based families and genera (O’Donnell et al. 2001, Voigt & Wöstemeyer 2001), several groups have been revised using molecular phylogenetic analyses, e.g. Actinomucor (Zheng & Liu 2005, Khan et al. 2008), Apophysomyces (Álvarez et al. 2010b), Cunninghamella (Liu et al. 2001), Lentamyces (Hoffmann & Voigt 2009), Lichtheimia (Alastruey-Izquierdo et al. 2010), Pilobolus (Foos et al. 2011), Rhizopus (Abe et al. 2006, 2007, 2010, Liu et al. 2007, Gryganskyi 2010), Saksenaea (Álvarez et al. 2010a), Siepmannia (Kwaśna & Nirenberg 2008a, b) and Umbelopsis (Meyer & Gams 2003). However, some genera, e.g. Absidia s.str., Circinella and relatives, or Syncephalastrum have not been revised using molecular data. The largest mucorelean group, Mucor and its relatives, has been investigated only fragmentarily focusing on certain clades (Jacobs & Botha 2008, Budziszewska et al. 2010, Álvarez et al. 2011, Madden et al. 2011, Hermet et al. 2012). Only a few publications (Abe et al. 2007, Alastruey-Izquierdo et al. 2010, Gryganskyi 2010, Hermet et al. 2012) use at least two unlinked molecular markers and apply sufficient strain and taxon sampling to adopt concepts of genealogical concordance phylogenetic species recognition (GCPSR, Taylor et al. 2000) satisfactorily. As a consequence, the criteria of good taxonomy are insufficiently met, and many species in Mucorales are poorly delimited. It was, therefore, the primary aim of the present study to inventory the genetic diversity of Mucorales deposited in the CBS culture collection and to highlight critical groups that need to be studied by a multi-locus approach. Our paper provides DNA barcodes for all ex-type and authentic strains of Mucorales available in the CBS culture collection, and makes these data available by open access as reference for subsequent studies on biodiversity and taxonomy.
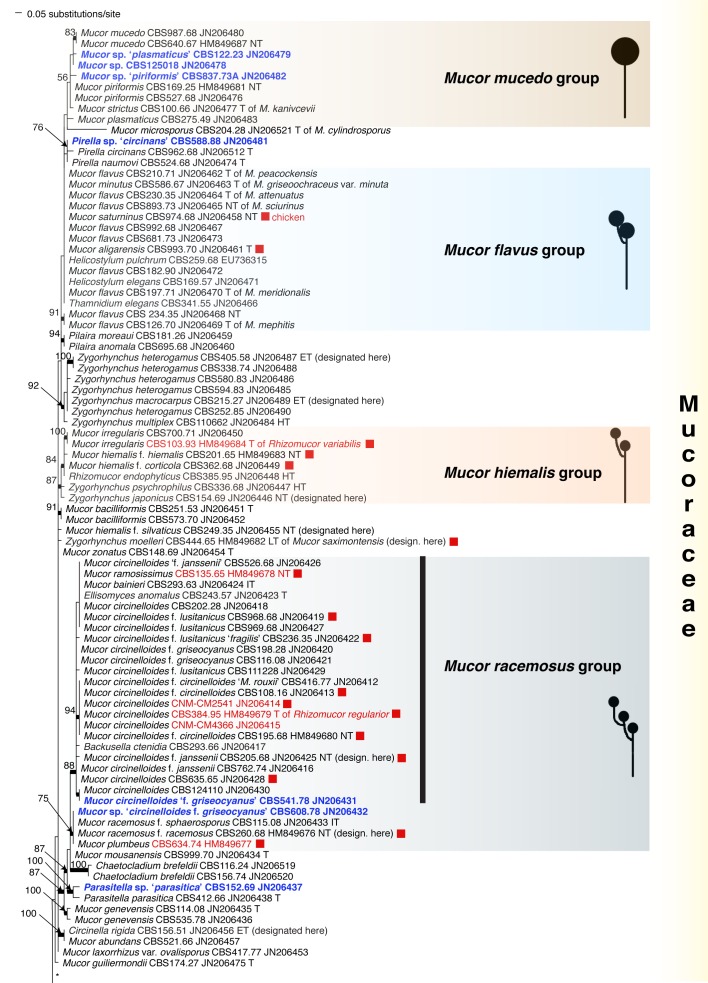
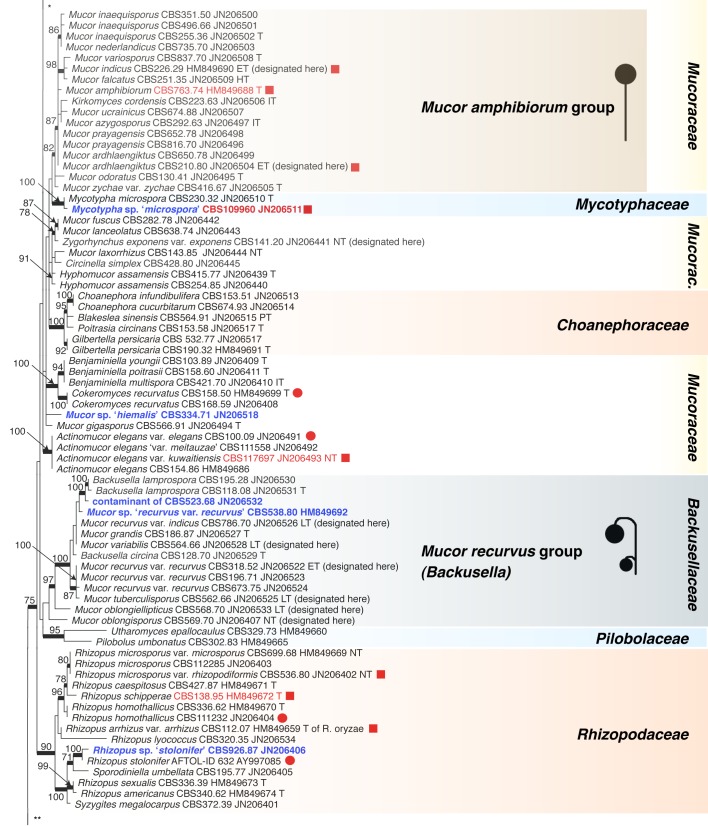
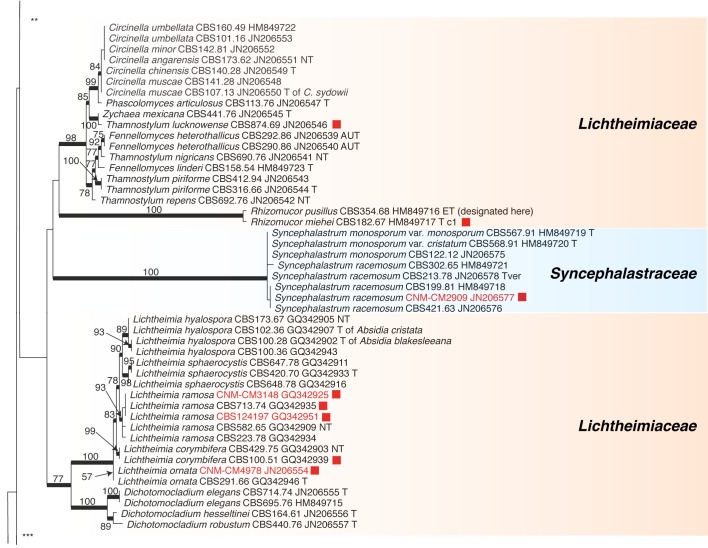
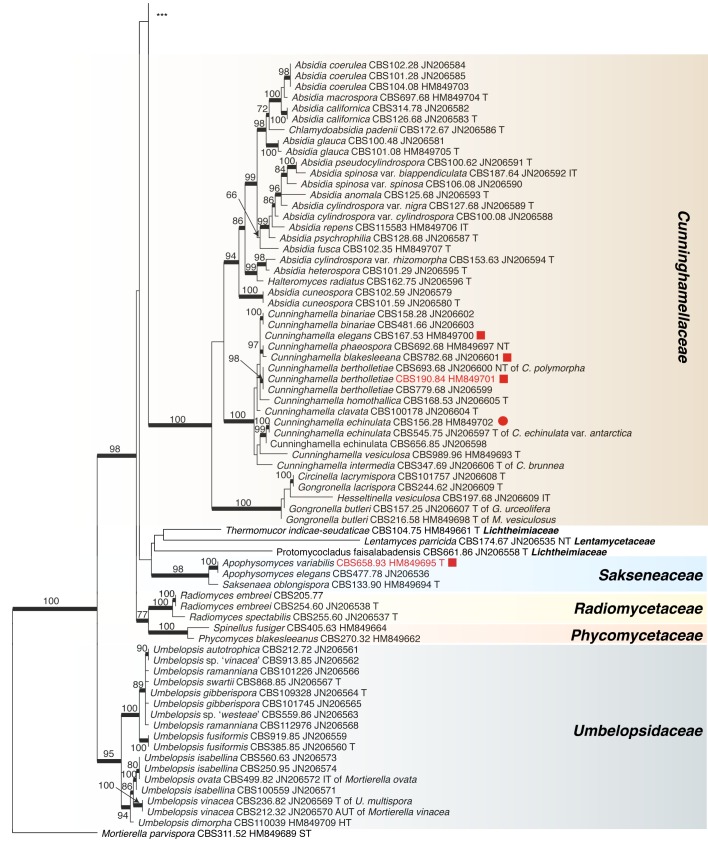
Recently, the Fungal Barcoding Consortium (Schoch et al. 2012) proposed the rDNA internal transcribed spacer (ITS) as a universal DNA barcode marker for fungi. In our study ITS was also applied because of its discriminative power in Mucorales (Meyer & Gams 2003, Kwaśna et al. 2006, Schwarz et al. 2006, Vitale et al. 2012). The ITS region is highly variable between members and is not alignable over the entire order. ITS sequences of some species differ to such an extent that they could not be aligned confidently with their putative sibling species. To establish the phylogenetic position of species and to acquire an overview of the entire order that includes all groups recognized on the basis of molecular data, the D1/D2 region of the large subunit (LSU) rDNA was sequenced from one strain of each Molecular Operational Taxonomic Unit (MOTU). A MOTU is defined by ITS similarities with mutual threshold values of > 99 %. Using this approach a species can be represented by a single or by several MOTUs depending on intraspecific variability.
Analyses of the ITS region as a single locus can not be used to define species boundaries, but, conversely, hypotheses on species limits can be developed by plotting morphospecies on the ITS trees. Therefore only those taxonomic rearrangements were made that did not require exact knowledge on species limits. These revisions will be discussed, and nomenclatural status of names analysed.
ITS barcodes of 668 strains in 203 taxa (178 species, 19 varieties, 6 forms) were generated for this study covering 78.4 % of the recognized species in Mucorales. Seventy-six percent of the species are represented by ex-type strains or other authentic material. LSU sequences were generated for 43.5 % of the strains. Special attention was paid to the inclusion of a high proportion of clinical strains predominantly provided by the Spanish National Center for Microbiology in Madrid (Spain) and the Postgraduate Institute of Medical Education and Research in Chandigarh (India). The paper focuses on the genera Actinomucor, Mucor and allies, Rhizomucor and Rhizopus because ITS trees have been published for the remaining medically important genera: Apophysomyces (Álvarez et al. 2010b), Cunninghamella (Liu et al. 2001), Lichtheimia (Alastruey-Izquierdo et al. 2010), Saksenaea (Álvarez et al. 2010a) and Syncephalastrum (Vitale et al. 2012).
In main traits we follow the nomenclature of the species provided by Species Fungorum (http://www.speciesfungorum.org) based largely on the 10th edition of the Dictionary of the Fungi. We adopted the family structure by Hoffmann et al. (2013).
MATERIALS AND METHODS
Strains
A total number of 668 mucoralean strains belonging to 178 species, 19 varieties and 6 formae and covering 78.4 % of the currently accepted species was studied. For 22.2 % of the taxa it was possible to include five or more strains per lowest taxonomic level (form, variety or species), 60.1 % of the taxa were represented by 2 or more isolates. Ex-type strains or other authentic material was available for 75.8 % of the studied species. In order to cover intraspecific variability, strains from the most distant localities and from a wide range of substrates were selected. Special attention was paid to the inclusion of clinical isolates. Studied strains originated from the reference collection of the CBS-KNAW Fungal Biodiversity Centre (CBS; Utrecht, The Netherlands), the Instituto de Salud Carlos III National Centre of Microbiology (CNM-CM; Madrid, Spain), the Departments of Medical Microbiology (PGIMER; Chandigarh, India) or the Belgian Co-ordinated Collections of Micro-organisms (IHEM; Brussels, Belgium). The studied strains, source information and GenBank accession numbers are listed in Table 1.
Table 1.
Source information and GenBank accession numbers of the studied strains. Strains marked with an asterisk belong to different genera based on their ITS- or LSU-sequences: * Absidia idahoensis CBS 103.91 belongs to Circinella; ** Circinella lacrymispora CBS 101757 belongs to Gongronella, *** Circinella simplex CBS 428.80 belongs to Mucor. Ex-type strains are designated by: T = ex-type strain, ET = ex-epitype strain, HT = ex-holotype strain, IT = ex-isotype strain, LT = ex-lectotype strain, NT = ex-neotype strain, PT = ex-paratype strain, ST = ex-syntype strain and AUT = authentic material. Type information was checked with original literature only for those taxa that are treated in the taxonomy part; the remaining data were derived from the CBS database.
| Strain number | Species | Species name at the beginning of the study | Status | Country | Source | ITS | LSU |
|---|---|---|---|---|---|---|---|
| CBS 125.68 | Absidia anomala | T | Cuba | soil | JN205815 | JN206593 | |
| CBS 126.68 | Absidia californica | T | USA | dung of rat | JN206583 | ||
| CBS 314.78 | Absidia californica | USA | dung of mouse | JN205816 | JN206582 | ||
| CBS 101.28 | Absidia coerulea | USA | dung of rabbit | JN205818 | JN206585 | ||
| CBS 102.28 | Absidia coerulea | USA | soil | JN205821 | JN206584 | ||
| CBS 104.08 | Absidia coerulea | n.a. | soil | JN205811 | HM849703 | ||
| CBS 628.70B | Absidia coerulea | Denmark | forest soil | JN205812 | |||
| CBS 101.59 | Absidia cuneospora | T | USA | sandy soil | JN206580 | ||
| CBS 102.59 | Absidia cuneospora | USA | clay soil | JN205819 | JN206579 | ||
| CBS 100.08 | Absidia cylindrospora var. cylindrospora | n.a. | n.a. | JN205822 | JN206588 | ||
| CBS 127.68 | Absidia cylindrospora var. nigra | T | USA | soil of pastured hardwood | JN206589 | ||
| CBS 153.63 | Absidia cylindrospora var. rhizomorpha | T | Honduras | rhizosphere of Musa sapientum | JN206594 | ||
| CBS 102.35 | Absidia fusca | T | Germany | soil of pine forest | JN205814 | HM849707 | |
| CBS 346.97 | Absidia fusca | A. cylindrospora var. nigra | Netherlands | myxomycete | JN205817 | ||
| CBS 100.48 | Absidia glauca | Germany | manure, in asparagus field | JN205820 | JN206581 | ||
| CBS 101.08 | Absidia glauca | T | n.a. | n.a. | JN205810 | HM849705 | |
| CBS 101.29 | Absidia heterospora | T | France | soil of pine forest | JN206595 | ||
| CBS 103.91 | Absidia idahoensis* | T | USA | brood chamber of Nomia melanderi | JN205847 | ||
| CBS 697.68 | Absidia macrospora | T | former Czechovakia | soil | HM849704 | ||
| CBS 100.62 | Absidia pseudocylindrospora | T | Tanzania | virgin soil | JN206591 | ||
| CBS 128.68 | Absidia psychrophilia | T | Canada | ambrosia beetle; gland | JN206587 | ||
| CBS 115583 | Absidia repens | IT | UK, England | wallpaper | JN205813 | HM849706 | |
| CBS 187.64 | Absidia spinosa var. biappendiculata | IT | USA | Comandra pallida; leaf | JN206592 | ||
| CBS 106.08 | Absidia spinosa var. spinosa | n.a. | n.a. | JN205809 | JN206590 | ||
| CBS 100.09 | Actinomucor elegans | n.a. | n.a. | JN205827 | JN206491 | ||
| CBS 100.22 | Actinomucor elegans | USA | n.a. | JN205828 | |||
| CBS 111556 | Actinomucor elegans | JN205826 | |||||
| CBS 154.86 | Actinomucor elegans | Egypt | n.a. | JN205829 | HM849686 | ||
| CBS 338.72 | Actinomucor elegans | Nepal | soil | JN205824 | |||
| CBS 117697 | Actinomucor elegans var. kuwaitiensis | A. kuwaitiensis | T | Kuwait | human; wound | JN205823 | JN206493 |
| CBS 111558 | Actinomucor elegans var. meitauzae | A. taiwanensis | n.a. | n.a. | JN205825 | JN206492 | |
| CBS 476.78 | Apophysomyces elegans | T | India | soil of mango orchard | JN206279 | ||
| CBS 477.78 | Apophysomyces elegans | India | soil of grassy site | JN206280 | JN206536 | ||
| CBS 658.93 | Apophysomyces variabilis | A. elegans | T | Netherlands Antilles | human; osteomyelitis | JN206281 | HM849695 |
| CBS 128.70 | Backusella circina | T | USA | soil with lichens | JN206258 | JN206529 | |
| CBS 129.70 | Backusella circina | PT | USA | soil | JN206257 | ||
| CBS 323.69 | Backusella circina | Japan | n.a. | JN206259 | |||
| CBS 382.95 | Backusella circina | China | forest soil | JN206256 | |||
| CBS 907.73 | Backusella circina | Japan | n.a. | JN206260 | |||
| CBS 786.70 | Backusella indica | M. recurvus var. indicus | LT (designated here) | India | n.a. | JN206255 | JN206526 |
| CBS 107.09 | Backusella lamprospora | T of Mucor dispersus | Norway | n.a. | JN206269 | ||
| CBS 118.08 | Backusella lamprospora | T | Switzerland | n.a. | JN206268 | JN206531 | |
| CBS 195.28 | Backusella lamprospora | USA | fallen leaf | JN206271 | JN206530 | ||
| CBS 244.67 | Backusella lamprospora | France | moist wall | JN206270 | |||
| CBS 568.70 | Backusella oblongielliptica | Mucor oblongiellipticus | LT (designated here) | Japan | agaric | JN206278 | JN206533 |
| CBS 569.70 | Backusella oblongispora | Mucor oblongisporus | NT (designated here) | Japan | soil | JN206251 | JN206407 |
| CBS 196.71 | Backusella recurva | M. recurvus var. recurvus | JN206265 | JN206523 | |||
| CBS 317.52 | Backusella recurva | M. recurvus var. recurvus | Macedonia | decaying wood | JN206262 (c1) | ||
| CBS 318.52 | Backusella recurva | M. recurvus var. recurvus | ET (designated here) | USA | Fragaria; diseased root | JN206261 | JN206522 |
| CBS 673.75 | Backusella recurva | M. recurvus var. recurvus | Australia | soil | JN206263 (c1) | JN206524 | |
| JN206264 (c2) | |||||||
| CBS 538.80 | Backusella sp. | M. recurvus var. recurvus | Egypt | Medicago sativa | HM999964 | HM849692 | |
| CBS 562.66 | Backusella tuberculispora | Mucor tuberculisporus | LT (designated here) | India | n.a. | JN206267 | |
| CBS 570.70 | Backusella tuberculispora | Mucor tuberculisporus | Japan | cultivated soil | JN206266 | ||
| CBS 564.66 | Backusella variabilis | Mucor variabilis | LT (designated here) | India | excrements of human | JN206254 (c1) | JN206528 |
| JN206253 (c3) | |||||||
| CBS 421.70 | Benjaminiella multispora | IT | India | humus-rich soil | JN206243 | JN206410 | |
| CBS 158.60 | Benjaminiella poitrasii | T | USA | dung of rat | JN206241 | JN206411 | |
| CBS 103.89 | Benjaminiella youngii | T | Spain | dung of lizard | JN206242 | JN206409 | |
| CBS 130.59 | Blakeslea trispora | Panama | soil | JN206227 | |||
| CBS 137.49 | Blakeslea trispora | Indonesia | Hibiscus rosa-sinensis; flower | JN206229 | |||
| CBS 198.80 | Blakeslea trispora | Sweden | n.a. | JN206228 | |||
| CBS 564.91 | Blakeslea trispora | B. sinensis | PT of B. sinensis | China | soil | JN206230 | JN206515 |
| CBS 116.24 | Chaetocladium brefeldii | n.a. | n.a. | JN206020 | JN206519 | ||
| CBS 136.28 | Chaetocladium brefeldii | n.a. | dung of horse | JN206019 | |||
| CBS 156.74 | Chaetocladium brefeldii | Netherlands | litter | JN206022 | JN206520 | ||
| CBS 162.82 | Chaetocladium brefeldii | Netherlands | dung of rat | JN206021 | |||
| CBS 811.69 | Chaetocladium jonesiii | Norway | meadow soil | JN206023 | |||
| CBS 172.67 | Chlamydoabsidia padenii | T | USA | Pisum sativum; root | JN206294 | JN206586 | |
| CBS 120.25 | Choanephora cucurbitarum | C. infundibulifera f. cucurbitarum | n.a. | n.a. | JN206231 | ||
| CBS 150.51 | Choanephora cucurbitarum | C. infundibulifera f. cucurbitarum | n.a. | n.a. | JN206232 | ||
| CBS 178.76 | Choanephora cucurbitarum | C. infundibulifera f. cucurbitarum | T. of C. heterospora | India | dead insect | JN206235 | |
| CBS 445.72 | Choanephora cucurbitarum | C. infundibulifera f. cucurbitarum | n.a. | n.a. | JN206234 | ||
| CBS 674.93 | Choanephora cucurbitarum | C. infundibulifera f. cucurbitarum | China | n.a. | JN206233 | JN206514 | |
| CBS 153.51 | Choanephora infundibulifera | C. infundibulifera f. infundibulifera | n.a. | n.a. | JN206236 | JN206513 | |
| CBS 155.51 | Choanephora infundibulifera | C. infundibulifera f. infundibulifera | n.a. | n.a. | JN206237 | ||
| CBS 155.58 | Choanephora infundibulifera | C. infundibulifera f. infundibulifera | Japan | n.a. | JN206238 | ||
| CBS 172.62 | Circinella angarensis | USA | dung of rodent | JN205848 | |||
| CBS 173.62 | Circinella angarensis | NT | USA | dung | JN205849 | JN206551 | |
| CBS 140.28 | Circinella chinensis | T | Japan | n.a. | JN205855 | JN206549 | |
| CBS 101757 | Circinella lacrymispora** | T | Argentina | soil | JN206289 | JN206608 | |
| CBS 102.16 | Circinella minor | C. umbellata | n.a. | n.a. | JN205860 | ||
| CBS 142.81 | Circinella minor | Netherlands | dung of mouse | JN205861 | JN206552 | ||
| CBS 143.56 | Circinella minor | n.a. | n.a. | JN205862 | |||
| CBS 107.13 | Circinella muscae | T of C. sydowii | n.a. | gold mine, depth of 600 m | JN205854 | JN206550 | |
| CBS 141.28 | Circinella muscae | n.a. | n.a. | JN205853 | JN206548 | ||
| CBS 159.49 | Circinella muscae | Indonesia | soil | JN205851 | |||
| CBS 342.79 | Circinella muscae | Netherlands | preserved meat | JN205852 | |||
| CBS 720.76A | Circinella muscae | USA | n.a. | JN205850 | |||
| CBS 428.80 | Circinella simplex*** | Colombia | paramo soil under undisturbed vegetation | JN206213 | JN206445 | ||
| CBS 101.16 | Circinella umbellata | n.a. | dung of dog | JN205857 | JN206553 | ||
| CBS 160.49 | Circinella umbellata | Italy | air | JN205858 | HM849722 | ||
| CBS 195.74 | Circinella umbellata | Kuwait | soil | JN205856 | |||
| CBS 837.97 | Circinella umbellata | Germany | dung of mouse | JN205859 | |||
| CBS 158.50 | Cokeromyces recurvatus | T | USA | dung of rabbit | JN206244 | HM849699 | |
| CBS 168.59 | Cokeromyces recurvatus | USA | dung of rat | JN206245 | JN206408 | ||
| CBS 151.80 | Cunninghamella bertholletiae | C. elegans | USA | human; lung (leukemic patient) | JN205875 | ||
| CBS 182.84 | Cunninghamella bertholletiae | USA | human | JN205877 | |||
| CBS 186.84 | Cunninghamella bertholletiae | USA | human; lung (leukemic patient) | JN205876 | |||
| CBS 190.84 | Cunninghamella bertholletiae | USA | human; heart, patient with lymphosarcoma | JN205878 | HM849701 | ||
| CBS 191.84 | Cunninghamella bertholletiae | USA | human; tibia | JN205879 | |||
| CBS 372.95 | Cunninghamella bertholletiae | C. polymorpha | China | forest soil | JN205872 | ||
| CBS 373.95 | Cunninghamella bertholletiae | C. polymorpha | China | rotten log | JN205873 | ||
| CBS 693.68 | Cunninghamella bertholletiae | C. polymorpha | NT of C. polymorpha | former Yugoslavia | soil | JN205871 | JN206600 |
| CBS 779.68 | Cunninghamella bertholletiae | C. polymorpha | n.a. | n.a. | JN205874 | JN206599 | |
| CNM-CM3628 | Cunninghamella bertholletiae | Spain | human; sputum | JN205881 | |||
| CNM-CM3650 | Cunninghamella bertholletiae | Spain | human; skin | JN205880 | |||
| CBS 158.28 | Cunninghamella binariae | C. elegans | n.a. | n.a. | JN205888 | JN206602 | |
| CBS 481.66 | Cunninghamella binariae | C. elegans | Brazil | soil | JN205889 | JN206603 | |
| CBS 133.27 | Cunninghamella blakesleeana | T | Switzerland | n.a. | JN205865 | ||
| CBS 177.36 | Cunninghamella blakesleeana | n.a. | n.a. | JN205868 | |||
| CBS 224.64 | Cunninghamella blakesleeana | Canada | Linum issitatissimum | JN205870 | |||
| CBS 433.84 | Cunninghamella blakesleeana | Kuwait | n.a. | JN205867 | |||
| CBS 720.85 | Cunninghamella blakesleeana | C. elegans | Croatia | decaying fruit | JN205866 | ||
| CBS 782.68 | Cunninghamella blakesleeana | n.a. | n.a. | JN205869 | JN206601 | ||
| CBS 100178 | Cunninghamella clavata | T | n.a. | n.a. | JN205890 | JN206604 | |
| CBS 362.95 | Cunninghamella clavata | China | soil | JN205891 | |||
| CBS 156.28 | Cunninghamella echinulata | NT | n.a. | n.a. | JN205895 | HM849702 | |
| CBS 656.85 | Cunninghamella echinulata | Egypt | soil | JN205896 | JN206598 | ||
| CBS 766.68 | Cunninghamella echinulata | n.a. | n.a. | JN205894 | |||
| CBS 545.75 | Cunninghamella echinulata var. antarctica | C. antarctica | T | Chile | soil | JN205893 | JN206597 |
| CBS 167.53 | Cunninghamella elegans | Canada | Linum usitatissimum; seed | JN205882 | HM849700 | ||
| CBS 318.78 | Cunninghamella elegans | C. phaeospora | Turkey | soil under shrub vegetation | JN205886 | ||
| CBS 773.68 | Cunninghamella elegans | former Czechoslovakia | n.a. | JN205887 | |||
| CBS 781.68 | Cunninghamella elegans | C. phaeospora | n.a. | n.a. | JN205885 | ||
| CNM-CM5114 | Cunninghamella elegans | Spain | human; lung, biopsy sample | JN205884 | |||
| CBS 168.53 | Cunninghamella homothallica | T | Japan | soil | JN205863 | JN206605 | |
| CBS 347.69 | Cunninghamella intermedia | C. phaeospora | T of C. brunnea | India | n.a. | JN205892 | JN206606 |
| CBS 692.68 | Cunninghamella phaeospora | NT | Indonesia | soil | JN205864 | HM849697 | |
| CBS 989.96 | Cunninghamella vesiculosa | T | India | soil of Shorea robusta forest | JN205897 | HM849693 | |
| CBS 695.76 | Dichotomocladium elegans | USA | dung of rodent | JN205840 | HM849715 | ||
| CBS 714.74 | Dichotomocladium elegans | T | USA | dung of mouse | JN205839 | JN206555 | |
| CBS 164.61 | Dichotomocladium hesseltinei | T | India | soil of a cultivated field | JN205841 | JN206556 | |
| CBS 439.76 | Dichotomocladium robustum | USA | dung of mouse | JN205842 | |||
| CBS 440.76 | Dichotomocladium robustum | T | USA | dung of mouse | JN205843 | JN206557 | |
| CBS 243.57 | Ellisomyces anomalus | T | USA | dung of lizard | JN205992 | JN206423 | |
| CBS 697.76 | Ellisomyces anomalus | USA | dung of mouse | JN205993 | |||
| CBS 290.86 | Fennellomyces heterothallicus | AUT | India | dung of house lizard | JN205844 | JN206540 | |
| CBS 292.86 | Fennellomyces heterothallicus | AUT | India | dung of shrew | JN206539 | ||
| CBS 158.54 | Fennellomyces linderi | T | USA | poplin | JN205846 | HM849723 | |
| CBS 190.32 | Gilbertella persicaria | G. persicaria var. persicaria | T | USA | Prunus persica; fruit | HM999958 | HM849691 |
| CBS 246.59 | Gilbertella persicaria | G. persicaria var. persicaria | USA | trickling filter plant | JN206222 | ||
| CBS 325.71A | Gilbertella persicaria | G. persicaria var. persicaria | Indonesia | Saccharum officinarum; leaf | JN206220 | ||
| CBS 325.71D | Gilbertella persicaria | G. persicaria var. persicaria | Indonesia | wood | JN206225 | ||
| CBS 403.51 | Gilbertella persicaria | Rhizopus stolonifer | Japan | n.a. | JN206221 | ||
| CBS 421.77 | Gilbertella persicaria | G. persicaria var. persicaria | Pakistan | soil | JN206223 | ||
| CBS 442.64 | Gilbertella persicaria | G. persicaria var. indica | T of G. persicaria var. indica | India | n.a. | JN206219 | |
| CBS 532.77 | Gilbertella persicaria | G. persicaria var. persicaria | India | dung of mouse | JN206517 | ||
| CBS 565.91 | Gilbertella persicaria | G. hainanensis | T of G. hainanensis | China | dung of swine | JN206226 | |
| CBS 785.97 | Gilbertella persicaria | Mucor thermophilus | n.a. | n.a. | JN206218 | ||
| CBS 102.44 | Gongronella butleri | Italy | n.a. | JN206284 | |||
| CBS 157.25 | Gongronella butleri | T of G. urceolifera | Indonesia | Cocos nucifera; root | JN206607 | ||
| CBS 179.28 | Gongronella butleri | n.a. | n.a. | JN206286 | |||
| CBS 216.58 | Gongronella butleri | T of Mucor vesiculosus | UK | garden soil | JN206285 | HM849698 | |
| CBS 415.67 | Gongronella butleri | Brazil | soil | JN206288 | |||
| CBS 969.73 | Gongronella butleri | Chile | volcanic ash soil | JN206287 | |||
| CBS 244.62 | Gongronella lacrispora | T | USA | dooryard soil | JN206609 | ||
| CBS 162.75 | Halteromyces radiatus | T | Australia | mud from mangrove forest, contaminated with effluent | JN206290 | JN206596 | |
| CBS 223.63 | Helicostylum cordense | IT | India | forest soil | JN206193 | JN206506 | |
| CBS 169.57 | Helicostylum elegans | UK | dead isopod (woodlouse) | JN206471 | |||
| CBS 107.23 | Helicostylum pulchrum | n.a. | n.a. | JN206053 | |||
| CBS 258.59 | Helicostylum pulchrum | T of H. venustellum | UK, England | isopod, on the underside of a disturbed pine stump | JN206054 | ||
| CBS 259.68 | Helicostylum pulchrum | Germany | air-dried raw sausage | JN206052 | |||
| CBS 639.69 | Helicostylum pulchrum | USA | spoiled beef crackling | JN206055 | |||
| CBS 197.68 | Hesseltinella vesiculosa | IT | Brazil | rice-field soil | JN206610 | ||
| CBS 254.85 | Hyphomucor assamensis | Malaysia | Burmannia | JN206212 | JN206440 | ||
| CBS 415.77 | Hyphomucor assamensis | T | India | n.a. | JN206211 | JN206439 | |
| CBS 174.67 | Lentamyces parricida | NT | UK | grassland soil, surface layer | JN206293 | JN206535 | |
| CBS 100.17 | Lichtheimia corymbifera | Mycocladus corymbifer | n.a. | n.a. | GQ342885 | GQ342942 | |
| CBS 100.51 | Lichtheimia corymbifera | Mycocladus corymbifer | n.a. | n.a. | GQ342886 | GQ342939 | |
| CBS 102.48 | Lichtheimia corymbifera | Mycocladus corymbifer | India | mouldy shoe | GQ342888 | GQ342910 | |
| CBS 115811 | Lichtheimia corymbifera | Mycocladus corymbifer | Germany | indoor air | GQ342887 | GQ342932 | |
| CBS 429.75 | Lichtheimia corymbifera | Mycocladus corymbifer | NT | Afghanistan | soil | GQ342878 | GQ342903 |
| CBS 100.31 | Lichtheimia corymbifera | Mycocladus corymbifer | n.a. | aborted cow | GQ342879 | GQ342914 | |
| CBS 101040 | Lichtheimia corymbifera | Mycocladus corymbifer | France | human; keratomycosis | GQ342882 | GQ342918 | |
| CBS 109940 | Lichtheimia corymbifera | Mycocladus corymbifer | Norway | human; finger tissue | GQ342881 | GQ342917 | |
| CBS 120580 | Lichtheimia corymbifera | Mycocladus corymbifer | France | human; lung | GQ342884 | GQ342919 | |
| CBS 120581 | Lichtheimia corymbifera | Mycocladus corymbifer | France | human; bronchia | GQ342883 | GQ342948 | |
| CBS 120805 | Lichtheimia corymbifera | Mycocladus corymbifer | France | human; bone | GQ342880 | GQ342915 | |
| CBS 519.71 | Lichtheimia corymbifera | Absidia griseola | T of Absidia griseola | Japan | n.a. | GQ342889 | GQ342904 |
| CBS 100.28 | Lichtheimia hyalospora | Mycocladus blakesleeanus; Absidia blakesleeana | T of Absidia blakesleeana | USA | Bertholletia excelsa; nut | GQ342896 | GQ342902 |
| CBS 100.36 | Lichtheimia hyalospora | Mycocladus blakesleeanus; Absidia blakesleeana | n.a. | n.a. | GQ342898; GQ342897 | GQ342943 | |
| CBS 102.36 | Lichtheimia hyalospora | Mycocladus blakesleeanus; Absidia blakesleeana | T of Absidia cristata | Ghana | Manihot esculenta; stem | GQ342895 | GQ342907 |
| CBS 518.71 | Lichtheimia hyalospora | Mycocladus blakesleeanus var. atrosporus; Absidia blakesleeana var. atrospora | T of Absidia blakesleeana var. atrospora | Japan | n.a. | GQ342894 | GQ342944 |
| CBS 173.67 | Lichtheimia hyalospora | Mycocladus hyalosporus; Absidia hyalospora | NT | Philippines | fermented food taosi | GQ342893 | GQ342905 |
| CBS 291.66 | Lichtheimia ornata | Mycocladus corymbifer | T of Absidia ornata | India | dung of bird | GQ342891 | GQ342946 |
| CNM-CM4978 | Lichtheimia ornata | Spain | human; wound | GQ342892 | JN206554 | ||
| CBS 958.68 | Lichtheimia ornata | Mycocladus corymbifer | n.a. | n.a. | GQ342890 | GQ342936 | |
| CBS 100.24 | Lichtheimia ramosa | Mycocladus corymbifer | n.a. | n.a. | GQ342876 | GQ342941 | |
| CBS 100.49 | Lichtheimia ramosa | Mycocladus corymbifer | Indonesia | dung of cow | GQ342858 | GQ342940 | |
| CBS 100.55 | Lichtheimia ramosa | Mycocladus corymbifer | n.a. | n.a. | GQ342851 | GQ342938 | |
| CBS 101.51 | Lichtheimia ramosa | Mycocladus corymbifer | Netherlands | Guinea pig; lung | GQ342859 | GQ342945 | |
| CBS 101.55 | Lichtheimia ramosa | Mycocladus corymbifer | Switzerland | human; cornea | GQ342865 | GQ342947 | |
| CBS 103.35 | Lichtheimia ramosa | Mycocladus corymbifer | T of Absidia gracilis | n.a. | Musa sapientum; fruit | GQ342847 | GQ342908 |
| CBS 124198 | Lichtheimia ramosa | Mycocladus corymbifer | Netherlands | culture contaminant | GQ342848 | GQ342906 | |
| CBS 223.78 | Lichtheimia ramosa | Mycocladus corymbifer | n.a. | cocoa soil | GQ342877 | GQ342934 | |
| CBS 271.65 | Lichtheimia ramosa | Mycocladus corymbifer | n.a. | n.a. | GQ342875 | GQ342937 | |
| CBS 582.65 | Lichtheimia ramosa | Mycocladus corymbifer | NT | Ghana | Theobroma cacao; seed | GQ342874 | GQ342909 |
| CBS 649.78 | Lichtheimia ramosa | Mycocladus corymbifer | India | cultivated field soil | GQ342849 | GQ342912 | |
| CBS 713.74 | Lichtheimia ramosa | Mycocladus corymbifer | n.a. | n.a. | GQ342856 | GQ342935 | |
| CNM-CM2166 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; sputum | GQ342863 | GQ342926 | |
| CNM-CM3590 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human | GQ342869 | GQ342924 | |
| CNM-CM4119 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; skin | GQ342862 | GQ342923 | |
| CNM-CM4228 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; skin | GQ342861 | GQ342922 | |
| CNM-CM4253 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; skin | GQ342860 | GQ342921 | |
| CNM-CM4261 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; lung | GQ342854 | GQ342953 | |
| CNM-CM4337 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; skin | GQ342852 | GQ342920 | |
| CNM-CM4427 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; bronchoaspirate | GQ342853 | GQ342931 | |
| CNM-CM4537 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; skin | GQ342873 | GQ342930 | |
| CNM-CM4849 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; skin | GQ342855; GQ342868 | GQ342929 | |
| CNM-CM5111 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; sputum | GQ342871 | GQ342928 | |
| CNM-CM5171 | Lichtheimia ramosa | Mycocladus corymbifer | Belgium | human | GQ342864 | GQ342927 | |
| AS 3.4808 | Lichtheimia ramosa | Mycocladus corymbifer | T of Absidia idahoensis var. thermophila | China | soil | GQ342867 | GQ342955 |
| CBS 112528 | Lichtheimia ramosa | Mycocladus corymbifer | Germany | human; wound, double infection with Candida albicans | GQ342850 | GQ342913 | |
| CBS 124197 | Lichtheimia ramosa | Mycocladus corymbifer | Greece | human; abscess of the flank | GQ342870 | GQ342951 | |
| CBS 269.65 | Lichtheimia ramosa | Mycocladus corymbifer | n.a. | n.a. | GQ342857 | GQ342949 | |
| CNM-CM1638 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; gastric juice | GQ342866 | GQ342954 | |
| CNM-CM3148 | Lichtheimia ramosa | Mycocladus corymbifer | Spain | human; corneal exudate | GQ342872 | GQ342925 | |
| CBS 420.70 | Lichtheimia sphaerocystis | Mycocladus aff. blakesleeanus | India | n.a. | GQ342900 | GQ342933 | |
| CBS 647.78 | Lichtheimia sphaerocystis | Mycocladus aff. blakesleeanus | India | dung of mouse | GQ342899 | GQ342911 | |
| CBS 648.78 | Lichtheimia sphaerocystis | Mycocladus aff. blakesleeanus | India | soil | GQ342901 | GQ342916 | |
| CBS 388.35 | Mucor abundans | M. hiemalis f. silvaticus | NT (designated here) | Germany | forest soil | JN206111 | |
| CBS 521.66 | Mucor abundans | Russia | forest soil | JN206110 | JN206457 | ||
| CBS 244.58 | Mucor aligarensis | UK | human; ear | JN206057 | |||
| CBS 993.70 | Mucor aligarensis | T | India | soil | JN206056 | JN206461 | |
| CBS 185.77 | Mucor amphibiorum | Central America | diseased Dendrobates sp. | JN206170 | |||
| CBS 763.74 | Mucor amphibiorum | T | Germany | amphibian | HM999957 | HM849688 | |
| CNM-CM2934 | Mucor amphibiorum | Germany | human | JN206171 | |||
| CBS 210.80 | Mucor ardhlaengiktus | ET (designated here) | India | garden soil | JN206172 | JN206504 | |
| CBS 528.73 | Mucor ardhlaengiktus | Mozambique | Gossypium; seed | JN206173 | |||
| CBS 650.78 | Mucor ardhlaengiktus | M. aff. variisporus | India | dung of lizard | JN206174 | JN206499 | |
| CBS 292.63 | Mucor azygosporus | IT | USA | dung of lizard | JN206187 | JN206497 | |
| CBS 251.53 | Mucor bacilliformis | T | USA | soil | JN206083 | JN206451 | |
| CBS 573.70 | Mucor bacilliformis | Japan | agaric | JN206084 | JN206452 | ||
| CBS 293.63 | Mucor bainieri | IT | India | forest soil | JN205995 | JN206424 | |
| CBS 129.41 | Mucor brunneogriseus | M. plumbeus | T | Netherlands | aphid | JN205910 | |
| CBS 124110 | Mucor circinelloides (non of the described formae) | Hawaii island, USA | Morinda citrifolia; fermented fruit juice | JN205996 | JN206430 | ||
| CBS 202.28 | Mucor circinelloides (non of the described formae) | M. circinelloides f. janssenii | n.a. | n.a. | JN205994 | JN206418 | |
| CBS 338.71 | Mucor circinelloides (non of the described formae) | M. hiemalis f. hiemalis | Turkey | n.a. | JN205998 | ||
| CBS 526.68 | Mucor circinelloides (non of the described formae) | M. circinelloides f. janssenii | Armenia | soil | JN206015 | JN206426 | |
| CBS 635.65 | Mucor circinelloides (non of the described formae) | M. hiemalis f. hiemalis | UK, England | faeces of diseased Apis mellifera | JN205997 | JN206428 | |
| CBS 846.73 | Mucor circinelloides (non of the described formae) | M. flavus | Chile | soil | JN206014 | ||
| CNM-CM5225 | Mucor circinelloides (non of the described formae) | Austria | human | JN205999 | |||
| CBS 108.16 | Mucor circinelloides f. circinelloides | Japan | n.a. | JN205954 | JN206413 | ||
| CBS 111555 | Mucor circinelloides f. circinelloides | M. hiemalis | China | sufu starter | JN205943 | ||
| CBS 111560 | Mucor circinelloides f. circinelloides | M. racemosus f. racemosus | Vietnam | sufu, chao | JN205957 | ||
| CBS 121702 | Mucor circinelloides f. circinelloides | M. ramosissimus | Spain | commercial honey | JN205966 | ||
| CBS 123973 | Mucor circinelloides f. circinelloides | Germany | human; thigh necrosis after trauma | JN205958 | |||
| CBS 172.27 | Mucor circinelloides f. circinelloides | n.a. | Artocarpus; fruit | JN205967 | |||
| CBS 192.68 | Mucor circinelloides f. circinelloides | Netherlands | dung of pig | JN205959 | |||
| CBS 194.68 | Mucor circinelloides f. circinelloides | South Africa | n.a. | JN205972 | |||
| CBS 195.68 | Mucor circinelloides f. circinelloides | NT | Netherlands | air | JN205961 | HM849680 | |
| CBS 196.68 | Mucor circinelloides f. circinelloides | Turkey | Triticum aestivum | JN205968 | |||
| CBS 239.35 | Mucor circinelloides f. circinelloides | T of M. griseoroseus | Germany | soil | JN205942 | ||
| CBS 247.35 | Mucor circinelloides f. circinelloides | Germany | air | JN205962 | |||
| CBS 295.34 | Mucor circinelloides f. circinelloides | Ukraine | n.a. | JN205955 | |||
| CBS 384.95 | Mucor circinelloides f. circinelloides | Rhizomucor variabilis var. regularior | T of Rhizomucor variabilis var. regularior | China | human; face | JN205933 | HM849679 |
| CBS 394.68 | Mucor circinelloides f. circinelloides | Netherlands | thawing beef meat | JN205969 | |||
| CBS 416.77 | Mucor circinelloides f. circinelloides | M. rouxii | n.a. | fermenting rice, component of ‘Chinese yeast’ | JN205934 | JN206412 | |
| CBS 479.70 | Mucor circinelloides f. circinelloides | Finland | soil | JN205973 | |||
| CBS 480.70F | Mucor circinelloides f. circinelloides | India | garden soil | JN205956 | |||
| CBS 480.70G | Mucor circinelloides f. circinelloides | n.a. | n.a. | JN205965 | |||
| CNM-CM2437 | Mucor circinelloides f. circinelloides | Spain | human; nail | JN205939 | |||
| CNM-CM2541 | Mucor circinelloides f. circinelloides | Spain | human; nail | JN205944 | JN206414 | ||
| CNM-CM2922 | Mucor circinelloides f. circinelloides | Spain | human; wound exudate | JN205963 | |||
| CNM-CM3112 | Mucor circinelloides f. circinelloides | Spain | human; peritoneal dialysis | JN205945 | |||
| CNM-CM3178 | Mucor circinelloides f. circinelloides | Spain | human; urine | JN205946 | |||
| CNM-CM3510 | Mucor circinelloides f. circinelloides | Spain | human; reservoir | JN205947 | |||
| CNM-CM3785 | Mucor circinelloides f. circinelloides | Spain | human; catheter (low respiratory tract infection) | JN205948 | |||
| CNM-CM3926 | Mucor circinelloides f. circinelloides | Spain | human; arm wound (traumatism with arm amputation) | JN205949 | |||
| CNM-CM4299 | Mucor circinelloides f. circinelloides | Spain | human; skin (surgical wound) | JN205950 | |||
| CNM-CM4366 | Mucor circinelloides f. circinelloides | Spain | human; wound exudate (polytraumatism, esplenomectomy) | JN205974 | JN206415 | ||
| CNM-CM4526 | Mucor circinelloides f. circinelloides | Spain, Canaries | human; otic exudate | JN205953 | |||
| CNM-CM4569 | Mucor circinelloides f. circinelloides | Spain | human; bronchioaspirate (acute lymphoblastic leukemia B cells) | JN205951 | |||
| CNM-CM4621 | Mucor circinelloides f. circinelloides | Spain | human; skin of arm (burned patient) | JN205970 | |||
| CNM-CM4895 | Mucor circinelloides f. circinelloides | Spain | human; exudate (traumatism) | JN205975 | |||
| CNM-CM4974 | Mucor circinelloides f. circinelloides | Spain | human; wound skin | JN205952 | |||
| CNM-CM5071 | Mucor circinelloides f. circinelloides | Spain | human; bronchioaspirate | JN205938 | |||
| CNM-CM5169 | Mucor circinelloides f. circinelloides | Belgium | human | JN205971 | |||
| IHEM 16415 | Mucor circinelloides f. circinelloides | Italy | human; skin of hand, trauma | JN205964 | |||
| IHEM 20006 | Mucor circinelloides f. circinelloides | Belgium | human; wound | JN205936 | |||
| IHEM 21105 | Mucor circinelloides f. circinelloides | France | human | JN205937 | |||
| IHEM 21158 | Mucor circinelloides f. circinelloides | Belgium | human; wound (burned patient) | JN205960 | |||
| IHEM 21426 | Mucor circinelloides f. circinelloides | Belgium | human (burned patient) | JN205935 | |||
| IHEM 22323 | Mucor circinelloides f. circinelloides | Belgium | human | JN205941 | |||
| CBS 116.08 | Mucor circinelloides f. griseocyanus | T | Norway | soil | JN206003 | JN206421 | |
| CBS 198.28 | Mucor circinelloides f. griseocyanus | n.a. | n.a. | HM999951 | JN206420 | ||
| CBS 223.56 | Mucor circinelloides f. griseocyanus | Netherlands | n.a. | JN206000 | |||
| CBS 366.70 | Mucor circinelloides f. griseocyanus | Netherlands | canned strawberries | JN206001 | |||
| CBS 698.68 | Mucor circinelloides f. griseocyanus | South Africa | Zea mays | JN206002 | |||
| CBS 144.93 | Mucor circinelloides f. janssenii | M. aff. ramosissimus | USA? | human; thigh | JN206012 | ||
| CBS 185.68 | Mucor circinelloides f. janssenii | T of M. kurssanovii | Russia | grassland soil | JN206006 | ||
| CBS 204.68 | Mucor circinelloides f. janssenii | n.a. | iguana; lung | JN206010 | |||
| CBS 205.68 | Mucor circinelloides f. janssenii | NT (designated here) | South Africa | forest soil | HM999952 | JN206425 | |
| CBS 206.68 | Mucor circinelloides f. janssenii | India | n.a. | JN206004 | |||
| CBS 227.29 | Mucor circinelloides f. janssenii | n.a. | n.a. | JN206008 | |||
| CBS 232.29 | Mucor circinelloides f. janssenii | T of M. tenellus | France | n.a. | JN206007 | ||
| CBS 243.67 | Mucor circinelloides f. janssenii | South Africa | human | JN206005 | |||
| CBS 365.70 | Mucor circinelloides f. janssenii | n.a. | dung of Dixippus morosus | JN206009 | |||
| CBS 762.74 | Mucor circinelloides f. janssenii | Netherlands | milk powder | JN206011 | JN206416 | ||
| CBS 108.17 | Mucor circinelloides f. lusitanicus | T | n.a. | n.a. | JN205980 | ||
| CBS 111228 | Mucor circinelloides f. lusitanicus | M. racemosus f. racemosus | China | sufu starter, from sufu factory | JN205991 (c1) | JN206429 | |
| JN205989 (c3) | |||||||
| CBS 111229 | Mucor circinelloides f. lusitanicus | M. indicus | China | sufu starter, from sufu factory | JN205983 (c2) | ||
| JN205990 (c4) | |||||||
| CBS 236.35 | Mucor circinelloides f. lusitanicus | M. fragilis | Germany | Tremella | JN205979 | JN206422 | |
| CBS 242.33 | Mucor circinelloides f. lusitanicus | n.a. | n.a. | JN205987 | |||
| CBS 253.35 | Mucor circinelloides f. lusitanicus | USA | Zea mays; grain | JN205988 | |||
| CBS 276.49 | Mucor circinelloides f. lusitanicus | n.a. | n.a. | JN205984 | |||
| CBS 633.65 | Mucor circinelloides f. lusitanicus | South Afrika | Zea mays | JN205986 | |||
| CBS 847.72 | Mucor circinelloides f. lusitanicus | Portugal | n.a. | JN205981 | |||
| CBS 851.71 | Mucor circinelloides f. lusitanicus | n.a. | n.a. | JN205982 | |||
| CBS 968.68 | Mucor circinelloides f. lusitanicus | Mexico | n.a. | HM999953 | JN206419 | ||
| CBS 969.68 | Mucor circinelloides f. lusitanicus | Russia | forest soil | JN205985 | JN206427 | ||
| CBS 293.66 | Mucor ctenidius | Backusella ctenidia | USA | desert soil | JN205976 | JN206417 | |
| CBS 433.87 | Mucor ctenidius | Backusella ctenidia | Kenya | dead plant material | JN205978 | ||
| CBS 696.76 | Mucor ctenidius | Backusella ctenidia | USA | dung of pack rat | JN205977 | ||
| CBS 156.51 | Mucor durus | Circinella rigida | ET (designated here) | UK, England | soil | JN206112 | JN206456 |
| CBS 484.66 | Mucor durus | Circinella rigida | Ukraine | soil | JN206113 | ||
| CBS 385.95 | Mucor endophyticus | Rhizomucor endophyticus | HT | China | Triticum aestivum; leaves | JN206159 | JN206448 |
| CBS 141.20 | Mucor exponens | Zygorhynchus exponens var. exponens | NT (designated here) | Germany | n.a. | JN206206 | JN206441 |
| CBS 404.58 | Mucor exponens | Zygorhynchus exponens var. smithii | LT of Z. exponens var. smithii (designated here) | UK, England | ploughed field soil | JN206208 | |
| CBS 508.48 | Mucor exponens | Zygorhynchus exponens var exponens | Germany | n.a. | JN206207 | ||
| CBS 251.35 | Mucor falcatus | HT | Germany | honeycomb | JN206250 | JN206509 | |
| CBS 252.35 | Mucor falcatus | Germany | dung of rabbit | JN206249 | |||
| CBS 126.70 | Mucor flavus | T of M. mephitis | USA | dung of mouse | JN206049 | JN206469 | |
| CBS 182.90 | Mucor flavus | Czech Republic | floor of room in cave, used for speleotherapy | JN206065 | JN206472 | ||
| CBS 197.71 | Mucor flavus | T of M. meridionalis | Ukraine | dung of wood mouse | JN206066 | JN206470 | |
| CBS 210.71 | Mucor flavus | T of M. peacockensis | India | dung of peacock | JN206050 | JN206462 | |
| CBS 230.35 | Mucor flavus | T of M. attenuatus | Germany | dung of roe | JN206061 | JN206464 | |
| CBS 234.35 | Mucor flavus | NT | Germany | n.a. | JN206051 | JN206468 | |
| CBS 664.67 | Mucor flavus | Finland | forest litter | JN206064 | |||
| CBS 681.73 | Mucor flavus | Germany | wheat field soil | JN206069 (c2) | JN206473 | ||
| JN206070 (c3) | |||||||
| CBS 893.73 | Mucor flavus | NT of M. sciurinus | Russia | forest soil | JN206062 (c1) | JN206465 | |
| JN206063 (c2) | |||||||
| CBS 992.68 | Mucor flavus | Antarctica | coastal gravel flat covered by Bryum argenteum | JN206067 (c1) | JN206467 | ||
| JN206068 (c3) | |||||||
| CBS 230.29 | Mucor fuscus | T of M. petrinsularis var. echinosporus | France | n.a. | JN206204 | ||
| CBS 254.48 | Mucor fuscus | T of M. bedrchanii | Germany | n.a. | JN206203 | ||
| CBS 282.78 | Mucor fuscus | France | cheese | JN206201 | JN206442 | ||
| CBS 313.78 | Mucor fuscus | France | cheese | JN206200 | |||
| CBS 530.77 | Mucor fuscus | India | dung of mouse | JN206202 | |||
| CBS 336.68 | Mucor fusiformis | Zygorhynchus psychrophilus | HT | Finland | Picea abies; brown needle | JN206157 | JN206447 |
| CBS 114.08 | Mucor genevensis | T | Switzerland | soil | JN206041 | JN206435 | |
| CBS 404.71 | Mucor genevensis | Ukraine | dung of field-mouse | JN206042 (c1) | |||
| JN206043 (c2) | |||||||
| CBS 535.78 | Mucor genevensis | USA | n.a. | JN206045 (c1) | |||
| JN206046 (c3) | JN206436 | ||||||
| JN206047 (c4) | |||||||
| CBS 564.75 | Mucor genevensis | Netherlands | Malus sylvestris; fruit | JN206044 (c3) | |||
| CBS 383.95 | Mucor gigasporus | China | soil | JN206246 | |||
| CBS 566.91 | Mucor gigasporus | T | China | soil | JN206247 | JN206494 | |
| CBS 186.87 | Mucor grandis | T | India | dung of mouse | JN206252 | JN206527 | |
| CBS 174.27 | Mucor guiliermondii | T | Russia | dung of Periplaneta americana | JN206082 | JN206475 | |
| CBS 252.85 | Mucor heterogamus | Zygorhynchus heterogamus | Netherlands | cattle feed | JN206161 (c1) | JN206490 | |
| JN206162 (c4) | |||||||
| CBS 338.74 | Mucor heterogamus | Zygorhynchus heterogamus | Sweden | sediment in drain pipe | JN206169 (c1) | JN206488 | |
| JN206168 (c4) | |||||||
| CBS 405.58 | Mucor heterogamus | Zygorhynchus heterogamus | ET (designated here) | n.a. | n.a. | JN206167 | JN206487 |
| CBS 580.83 | Mucor heterogamus | Zygorhynchus heterogamus | Netherlands | sandy soil in potato field | JN206163 | JN206486 | |
| CBS 594.83 | Mucor heterogamus | Zygorhynchus heterogamus | Colombia | soil | JN206164 (c3) | JN206485 | |
| JN206165 (c4) | |||||||
| CBS 115.18 | Mucor hiemalis | M. hygrophilus | n.a. | n.a. | JN206127 | ||
| CBS 118522 | Mucor hiemalis | Denmark | agricultural soil | JN206138 | |||
| CNM-CM2540 | Mucor hiemalis | Spain | human; nail | JN206140 | |||
| CNM-CM5229 | Mucor hiemalis | Spain | human; nail | JN206141 | |||
| Fungiscope AS72 | Mucor hiemalis | Austria | human | JN206142 | |||
| CBS 106.09 | Mucor hiemalis f. corticola | T | Norway | n.a. | JN206130 | ||
| CBS 362.68 | Mucor hiemalis f. corticola | Russia | forest soil | JN206132 | JN206449 | ||
| CBS 365.68 | Mucor hiemalis f. corticola | Austria | soil | JN206133 | |||
| CBS 366.68 | Mucor hiemalis f. corticola | Austria | soil | JN206139 | |||
| CBS 532.78 | Mucor hiemalis f. corticola | Netherlands | soil | JN206145 | |||
| CBS 533.78 | Mucor hiemalis f. corticola | Netherlands | greenhouse soil under Lycopersicon esculentum | JN206146 | |||
| CBS 107.19 | Mucor hiemalis f. hiemalis | T of M. vallesiacus | Switzerland | n.a. | JN206137 | ||
| CBS 110.19 | Mucor hiemalis f. hiemalis | T of M. hiemalis var. toundrae | Switzerland | n.a. | JN206136 | ||
| CBS 117.08 | Mucor hiemalis f. hiemalis | T of M. adventitius var. aurantiacus | Switzerland | n.a. | JN206143 | ||
| CBS 123972 | Mucor hiemalis f. hiemalis | Germany | human; sputum | JN206128 | |||
| CBS 201.65 | Mucor hiemalis f. hiemalis | NT | USA | n.a. | JN206125 | HM849683 | |
| CBS 242.35 | Mucor hiemalis f. hiemalis | IT of M. hiemalis var. griseus | Germany | forest soil | JN206134 | ||
| CBS 328.92 | Mucor hiemalis f. hiemalis | Italy | lake sediment, submerged | JN206135 | |||
| CBS 337.71D | Mucor hiemalis f. hiemalis | Norway | soil | JN206131 | |||
| CBS 454.71 | Mucor hiemalis f. hiemalis | n.a. | dung of rabbit | JN206126 | |||
| CBS 528.78 | Mucor hiemalis f. hiemalis | Netherlands | agaric | JN206144 | |||
| CBS 980.68 | Mucor hiemalis f. hiemalis | Netherlands | Coccinella | JN206129 | |||
| CBS 255.36 | Mucor inaequisporus | T | Ghana | Spondias mombin; fruit | JN206177 | JN206502 | |
| CBS 351.50 | Mucor inaequisporus | Indonesia | Musa sapientum; fruit | JN206178 | JN206500 | ||
| CBS 496.66 | Mucor inaequisporus | Japan | Diospyros kaki; immature fruit | JN206179 | JN206501 | ||
| CBS 120.08 | Mucor indicus | n.a. | n.a. | JN206182 | |||
| CBS 120585 | Mucor indicus | India | human; muscle | JN206180 | |||
| CBS 123974 | Mucor indicus | Germany | human; gastrointestinal infection | JN206181 | |||
| CBS 226.29 | Mucor indicus | ET (designated here) | Switzerland | n.a. | HM999956 | HM849690 | |
| CBS 414.77 | Mucor indicus | Ascidiophora sp. | India | dung of berber goat | JN206185 | ||
| CBS 422.71 | Mucor indicus | Indonesia | Dioscorea; tuber | JN206186 | |||
| CBS 535.80 | Mucor indicus | South Africa | sorghum malt | JN206184 | |||
| CBS 671.79 | Mucor indicus | Indonesia | n.a. | JN206183 | |||
| CBS 100164 | Mucor irregularis | M. hiemalis f. luteus | China | human; nasolabial infection | JN206153 | ||
| CBS 103.93 | Mucor irregularis | Rhizomucor variabilis | T of Rhizomucor variabilis | China | human; hand | JN206150 | HM849684 |
| CBS 609.78 | Mucor irregularis | M. hiemalis f. hiemalis | Nigeria | garden soil | JN206152 | ||
| CBS 654.78 | Mucor irregularis | M. aff. variosporus | India | owl pellet | JN206151 | ||
| CBS 700.71 | Mucor irregularis | M. hiemalis f. luteus | T | India | soil | JN206154 | JN206450 |
| CBS 977.68 | Mucor irregularis | M. hiemalis f. hiemalis | India | n.a. | JN206155 (c1) | ||
| JN206156 (c2) | |||||||
| CBS 154.69 | Mucor japonicus | Zygorhynchus japonicus | NT (designated here) | Russia | forest soil | JN206158 | JN206446 |
| CBS 638.74 | Mucor lanceolatus | M. fuscus | France | cheese | JN206205 | JN206443 | |
| CBS 143.85 | Mucor laxorrhizus | NT | UK | lake mud | JN206209 | JN206444 | |
| CBS 237.66 | Mucor laxorrhizus | Ukraine | peat | JN206210 | |||
| CBS 243.35 | Mucor luteus | M. hiemalis f. luteus | T | Germany | n.a. | HM999954 | HM849685 |
| CBS 244.35 | Mucor luteus | M. hiemalis f. luteus | Germany | n.a. | JN206148 | ||
| CBS 301.74 | Mucor luteus | M. hiemalis f. luteus | Germany | n.a. | JN206149 | ||
| CBS 567.70A | Mucor luteus | M. hiemalis f. luteus | Japan | agaric | JN206147 | ||
| CBS 215.27 | Mucor megalocarpus | Zygorhynchus macrocarpus | ET (designated here) | France | n.a. | JN206160 | JN206489 |
| CBS 204.28 | Mucor microsporus | T of M. cylindrosporus | France | n.a. | JN206272 | JN206521 | |
| CBS 245.35 | Mucor microsporus | Austria | soil | JN206273 | |||
| CBS 586.67 | Mucor minutus | T of M. griseoochraceus var. minuta | India | n.a. | JN206048 | JN206463 | |
| CBS 216.27 | Mucor moelleri | Zygorhynchus moelleri | France | n.a. | JN206116 | ||
| CBS 380.29 | Mucor moelleri | Zygorhynchus moelleri | Netherlands | wood mixed with soil | JN206119 | ||
| CBS 406.58 | Mucor moelleri | Zygorhynchus moelleri | NT (designated here) | USA | soil | JN206121 | |
| CBS 444.65 | Mucor moelleri | Zygorhynchus moelleri | LT of M. saximontensis (designated here) | USA | soil | JN206114 | HM849682 |
| CBS 460.51 | Mucor moelleri | Zygorhynchus moelleri | UK, England | culture contaminant | JN206120 | ||
| CBS 501.66 | Mucor moelleri | Zygorhynchus moelleri | Austria | soil | JN206118 | ||
| IHEM 21156 | Mucor moelleri | Zygorhynchus moelleri | France | human | JN206115 | ||
| CBS 402.58 | Mucor moelleri f. californiensis | Zygorhynchus californiensis | ET (designated here) | USA | soil | JN206117 | |
| CBS 531.77 | Mucor mousanensis | India | dung of mouse | JN206016 | |||
| CBS 721.76 | Mucor mousanensis | Taiwan | dung of Rattus norvegensis | JN206018 | |||
| CBS 999.70 | Mucor mousanensis | T | India | dung of mouse | JN206017 | JN206434 | |
| CBS 228.29 | Mucor mucedo | T of M. murorum | Russia | n.a. | JN206088 | ||
| CBS 296.35 | Mucor mucedo | Germany | dung of rabbit | JN206090 | |||
| CBS 525.68 | Mucor mucedo | Armenia | dung of field-mouse | JN206087 | |||
| CBS 542.66 | Mucor mucedo | Ukraine | water | JN206086 | |||
| CBS 640.67 | Mucor mucedo | NT | Netherlands | cow; nose effluent | JN206085 | HM849687 | |
| CBS 834.73 | Mucor mucedo | n.a. | dung of dog | JN206093 (c1) | |||
| JN206091 (c2) | |||||||
| CBS 836.73 | Mucor mucedo | M. piriformis | Germany | wheat field soil | JN206092 (c1) | ||
| CBS 987.68 | Mucor mucedo | Netherlands | dung of rabbit | JN206089 | JN206480 | ||
| CBS 110662 | Mucor multiplex | Zygorhynchus multiplex | HT | China | paddy field soil | JN206166 | JN206484 |
| CBS 735.70 | Mucor nederlandicus | n.a. | n.a. | JN206176 | JN206503 | ||
| CBS 120.71 | Mucor odoratus | USA | n.a. | JN206195 | |||
| CBS 130.41 | Mucor odoratus | T | Denmark | laboratory air | JN206197 | JN206495 | |
| CBS 179.76A | Mucor odoratus | Germany | dung of cow | JN206196 | |||
| CBS 201.71 | Mucor odoratus | Netherlands | dung of horse | JN206198 | |||
| CBS 572.70 | Mucor odoratus | Japan | soil | JN206194 | |||
| CBS 417.77 | Mucor parviseptatus | M. laxorrhizus var. ovalisporus | T | Australia | sandy heath soil | JN206108 | JN206453 |
| CBS 522.79 | Mucor parviseptatus | M. laxorrhizus var. ovalisporus | Austria | soil | JN206109 | ||
| CBS 111230 | Mucor piriformis | n.a. | n.a. | JN206030 (c1) | |||
| CBS 169.25 | Mucor piriformis | NT | n.a. | Pyrus communis; decaying fruit | JN206028 | HM849681 | |
| CBS 175.27 | Mucor piriformis | France | n.a. | JN206029 | |||
| CBS 256.85 | Mucor piriformis | USA | soil and decaying pear fruit | JN206031 | |||
| CBS 527.68 | Mucor piriformis | Romania | river water | JN206034 | JN206476 | ||
| CBS 528.68 | Mucor piriformis | Russia | ant hill | JN206032 | |||
| CBS 177.46 | Mucor plasmaticus | UK, England | dung of rabbit | JN206076 (c1) | |||
| JN206077 (c4) | |||||||
| CBS 275.49 | Mucor plasmaticus | Netherlands | dung of mouse | JN206078 (c2) | JN206483 | ||
| JN206079 (c4) | |||||||
| CBS 402.73 | Mucor plasmaticus | Russia | dung of forest-mouse | JN206081 (c1) | |||
| JN206080 (c4) | |||||||
| CBS 226.32 | Mucor plumbeus | Canada | forest soil | JN205916 | |||
| CBS 284.78 | Mucor plumbeus | Netherlands | lettuce | JN205914 | |||
| CBS 295.63 | Mucor plumbeus | Zaire | leaf litter | JN205912 | |||
| CBS 312.78 | Mucor plumbeus | Netherlands | horse meat | JN205915 | |||
| CBS 630.74 | Mucor plumbeus | France | cheese | JN205918 | |||
| CBS 633.74 | Mucor plumbeus | Switzerland | cat; subcutaneous tissue | JN205913 | |||
| CBS 634.74 | Mucor plumbeus | Germany | human; biopsy material | HM999955 | HM849677 | ||
| CBS 814.96 | Mucor plumbeus | Sweden | wheat bran | JN205917 | |||
| CBS 652.78 | Mucor prayagensis | M. aff. variisporus | India | dung of shrew | JN206189 (c1) | JN206498 | |
| JN206190 (c3) | |||||||
| CBS 816.70 | Mucor prayagensis | T | India | n.a. | JN206188 | JN206496 | |
| CBS 111557 | Mucor racemosus f. racemosus | M. circinelloides f. circinelloides | China | sufu starter | JN205902 | ||
| CBS 113.08 | Mucor racemosus f. racemosus | M. racemosus f. sphaerosporus | IT of M. dimorphosporus | Switzerland | n.a. | JN205909 | |
| CBS 204.74 | Mucor racemosus f. racemosus | M. sinensis | T of M. sinensis | China | soya cheese | JN205899 | |
| CBS 222.81 | Mucor racemosus f. racemosus | Netherlands | Juglans regia; nut | JN205906 | |||
| CBS 260.68 | Mucor racemosus f. racemosus | NT (designated here) | Switzerland | n.a. | JN205898 | HM849676 | |
| CBS 271.86 | Mucor racemosus f. racemosus | M. racemosus f. sphaerosporus | Spain, Tenerife | dung | JN205900 | ||
| CBS 369.71 | Mucor racemosus f. racemosus | Finland | human; skin | JN205905 | |||
| CBS 616.63 | Mucor racemosus f. racemosus | n.a. | Agapornis; lung | JN205903 | |||
| CBS 636.67 | Mucor racemosus f. racemosus | M. racemosus f. chibinensis | NT (designated here) of M. racemosus f. chibinensis | Russia | grassland soil | JN205904 | |
| CBS 657.68 | Mucor racemosus f. racemosus | Belgium | contaminated cheese | JN205901 | |||
| CBS 660.66 | Mucor racemosus f. racemosus | M. racemosus f. chibinensis | Ukraine | halva | JN205911 | ||
| CBS 661.66 | Mucor racemosus f. racemosus | Austria | soil | JN205908 | |||
| CNM-CM2569 | Mucor racemosus f. racemosus | Spain | human; nail | JN205929 | |||
| CNM-CM3862 | Mucor racemosus f. racemosus | Spain | human | JN205930 | |||
| CBS 115.08 | Mucor racemosus f. sphaerosporus | IT | Norway | n.a. | JN205919 | JN206433 | |
| CBS 143.70 | Mucor racemosus f. sphaerosporus | n.a. | storage rot of Cucurbita maxima | JN205925 | |||
| CBS 238.35 | Mucor racemosus f. sphaerosporus | Germany | wood | JN205920 | |||
| CBS 347.87 | Mucor racemosus f. sphaerosporus | USA | soil and litter | JN205924 | |||
| CBS 538.78 | Mucor racemosus f. sphaerosporus | Germany | sausage | JN205922 | |||
| CBS 539.78 | Mucor racemosus f. sphaerosporus | France | cheese | JN205923 | |||
| CBS 571.70 | Mucor racemosus f. sphaerosporus | M. plasmaticus | Japan | dung of horse | JN205926 | ||
| CBS 574.70 | Mucor racemosus f. sphaerosporus | Japan | steamed sweet potato | JN205921 | |||
| CBS 634.78 | Mucor racemosus f. sphaerosporus | M. racemosus f. racemosus | France | cheese | JN205927 (c2) | ||
| JN205928 (c4) | |||||||
| CBS 135.65 | Mucor ramosissimus | NT | Uruguay | human; nasal lesion | JN205932 | HM849678 | |
| CBS 598.78 | Mucor saturninus | Netherlands | fruit body of Hygrophoropsis aurantiaca | JN206074 | |||
| CBS 599.78 | Mucor saturninus | Netherlands | chicken leg | JN206073 (c3) | |||
| JN206075 (c4) | |||||||
| CBS 974.68 | Mucor saturninus | NT | Netherlands | soil | JN206072 | JN206458 | |
| CBS 249.35 | Mucor silvaticus | M. hiemalis f. silvaticus | Germany | soil of Picea forest | JN206122 | JN206455 | |
| CBS 412.71 | Mucor silvaticus | M. hiemalis f. silvaticus | NT (designated here) | Denmark | forest soil | JN206124 | |
| CBS 509.66 | Mucor silvaticus | M. hiemalis f. silvaticus | Germany | forest soil | JN206123 | ||
| CBS 122.23 | Mucor sp. | M. plasmaticus | Germany | n.a. | JN206040 | JN206479 | |
| CBS 125018 | Mucor sp. | France | strawberries | JN206038 (c1) | JN206478 | ||
| JN206039 (c2) | |||||||
| CBS 334.71 | Mucor sp. | M. hiemalis f. hiemalis | Benin | tropical vegetable | JN206248 | JN206518 | |
| CBS 541.78 | Mucor sp. | M. circinelloides f. griseocyanus | South Africa | Zea mays | JN206013 | JN206431 | |
| CBS 608.78 | Mucor sp. | M. circinelloides f. griseocyanus | USA | gymnosperm litter | JN205931 | JN206432 | |
| CBS 837.73A | Mucor sp. | M. piriformis | Netherlands | Ribes rubrum | JN206033 | JN206482 | |
| CBS 100.66 | Mucor strictus | T of M. kanivcevii | Ukraine | grassland soil | JN206035 | JN206477 | |
| CBS 368.71A | Mucor strictus | Norway | soil of dried up bog | JN206036 | |||
| CBS 576.66 | Mucor strictus | NT | Austria | soil at lake shore | JN206037 | ||
| CBS 221.71 | Mucor ucrainicus | T | Ukraine | dung of mouse | JN206191 | ||
| CBS 674.88 | Mucor ucrainicus | Germany | soil of litter layer | JN206192 | JN206507 | ||
| CBS 837.70 | Mucor variisporus | T | India | n.a. | JN206175 | JN206508 | |
| CBS 148.69 | Mucor zonatus | T | Germany | soil | JN206104 | JN206454 | |
| CBS 183.76 | Mucor zonatus | Sweden | forest soil under Picea abies | JN206106 (c3) | |||
| JN206107 (c4) | |||||||
| CBS 529.83 | Mucor zonatus | Sweden | mould-infected Pinus wood | JN206105 | |||
| CBS 416.67 | Mucor zychae var. zychae | T | India | manured soil | JN206199 | ||
| CBS 186.68 | Mycotypha microspora | Germany | decaying wood | JN206215 | |||
| CBS 230.32 | Mycotypha microspora | T | Netherlands | Citrus aurantium; peel, contaminant | JN206217 | JN206510 | |
| CBS 610.92 | Mycotypha microspora | Germany | washroom in hospital | JN206216 | |||
| CBS 109960 | Mycotypha sp. | M. microspora | Thailand | human; pus of wound | JN206214 | JN206511 | |
| CBS 412.66 | Parasitella parasitica | T | USA | Paeonia | JN206027 | JN206438 | |
| CBS 152.69 | Parasitella sp. | P. parasitica | n.a. | n.a. | JN206024 (c1) | JN206437 | |
| JN206025 (c3) | |||||||
| CBS 207.28 | Parasitella sp. | P. parasitica | USA | n.a. | JN206026 | ||
| CBS 113.76 | Phascolomyces articulosus | T | Panama | dung of bat | JN206547 | ||
| CBS 112.20 | Phycomyces blakesleeanus | P. blakesleeanus var. piloboloides | T | Germany | n.a. | JN206303 | |
| CBS 188.27 | Phycomyces blakesleeanus | USA | n.a. | JN206304 | |||
| CBS 269.32 | Phycomyces blakesleeanus | P. blakesleeanus var. piloboloides | Germany | n.a. | JN206309 | ||
| CBS 270.32 | Phycomyces blakesleeanus | Germany | n.a. | JN206305 | HM849662 | ||
| CBS 282.35 | Phycomyces blakesleeanus | Netherlands | n.a. | JN206306 | |||
| CBS 284.35 | Phycomyces blakesleeanus | Germany | n.a. | JN206308 | |||
| CBS 286.35 | Phycomyces blakesleeanus | P. blakesleeanus var. piloboloides | Germany | n.a. | JN206307 | ||
| CBS 131.23 | Pilaira anomala | n.a. | n.a. | JN206097 | |||
| CBS 396.71C | Pilaira anomala | UK, Scotland | dung of rabbit | JN206099 | |||
| CBS 424.70 | Pilaira anomala | UK | dung | JN206101 | |||
| CBS 695.68 | Pilaira anomala | Netherlands | dung | JN206098 | JN206460 | ||
| CBS 699.71 | Pilaira anomala | Germany | dung of cow | JN206100 | |||
| CBS 181.26 | Pilaira moreaui | France | n.a. | JN206094 | JN206459 | ||
| CBS 411.67 | Pilaira moreaui | Backusella lamprospora | India | n.a. | JN206095 | ||
| CBS 496.71 | Pilaira moreaui | Germany | dung of cow | JN206096 | |||
| CBS 523.68 | Pilaira moreaui var. caucasica | P. caucasica | T | Armenia | dung of mouse | JN206299 | JN206532 |
| CBS 302.83 | Pilobolus umbonatus | Netherlands | dung of deer | JN206274 | HM849665 | ||
| CBS 425.50 | Pilobolus umbonatus | Germany | n.a. | JN206275 | |||
| CBS 962.68 | Pirella circinans | T | India | soil | JN206102 | JN206512 | |
| CBS 524.68 | Pirella naumovi | T | Armenia | dung of mouse | JN206103 | JN206474 | |
| CBS 588.88 | Pirella sp. | P. circinans | USA | beneath Joshua trees | JN206071 | JN206481 | |
| CBS 153.58 | Poitrasia circinans | T | Trinidad and Tobago | soil | JN206239 | JN206516 | |
| CBS 647.70 | Poitrasia circinans | Sri Lanka | soil | JN206240 | |||
| CBS 661.86 | Protomycocladus faisalabadensis | India | dung of rodent | JN206558 | |||
| CBS 205.77 | Radiomyces embreei | USA | dung of lizard (?) | JN206291 | HM849663 | ||
| CBS 206.77 | Radiomyces embreei | USA | dung of pack rat | JN206292 | |||
| CBS 254.60 | Radiomyces embreei | T | USA | dung of mouse | JN206538 | ||
| CBS 255.60 | Radiomyces spectabilis | T | USA | dung of lizard | JN206537 | ||
| CBS 182.67 | Rhizomucor miehei | T | USA | retting Parthenium argentatum | HM999959 (c1) | HM849717 | |
| HM999960 (c2) | |||||||
| HM999961 (c4) | |||||||
| CBS 209.77A | Rhizomucor miehei | USA | soil | JN206322 (c1) | |||
| JN206321 (c2) | |||||||
| CBS 360.92 | Rhizomucor miehei | Australia | human; kidney and liver (leukemic patient) | JN206318 | |||
| CBS 429.70 | Rhizomucor miehei | UK, England | Hordeum; stored grains | JN206319 (c1) | |||
| JN206320 (c4) | |||||||
| CBS 179.69 | Rhizomucor pusillus | R. tauricus | T of R. tauricus | Ukraine | forest soil | JN206310 | |
| CBS 219.31 | Rhizomucor pusillus | n.a. | pig; kidney | JN206311 | |||
| CBS 354.68 | Rhizomucor pusillus | ET (designated here) | Netherlands | corn meal | JN206312 | HM849716 | |
| CBS 425.78 | Rhizomucor pusillus | USA | soil | HM999962 | |||
| CNM-CM2752 | Rhizomucor pusillus | Spain | human; sputum | JN206313 | |||
| CNM-CM2935 | Rhizomucor pusillus | n.a. | n.a. | JN206314 | |||
| CNM-CM2974 | Rhizomucor pusillus | Spain | human; peritoneal fluid | JN206315 | |||
| CNM-CM4727 | Rhizomucor pusillus | Spain | human; biopsy sample | JN206316 | |||
| CNM-CM5124 | Rhizomucor pusillus | Spain | human; lung, biopsy sample | JN206317 | |||
| CBS 340.62 | Rhizopus americanus | R. sexualis var. americanus | T | USA | air | HM999967 (c3) | HM849674 |
| CBS 109939 | Rhizopus arrhizus var. arrhizus | R. oryzae | Canada | human; dermal lesions of back | JN206330 | ||
| CBS 110.17 | Rhizopus arrhizus var. arrhizus | R. oryzae | T of R. maydis | Switzerland | n.a. | JN206327 | |
| CBS 112.07 | Rhizopus arrhizus var. arrhizus | R. oryzae | T of R. oryzae | Netherlands | n.a. | JN206323 | HM849659 |
| CBS 118614 | Rhizopus arrhizus var. arrhizus | R. oryzae | Turkey | human; palate | JN206326 | ||
| CBS 125017 | Rhizopus arrhizus var. arrhizus | R. oryzae | Greece | human; sinus (leukemic patient) | JN206332 | ||
| CBS 146.90 | Rhizopus arrhizus var. arrhizus | R. oryzae | Netherlands | human; palatum molle | JN206324 | ||
| CBS 286.55 | Rhizopus arrhizus var. arrhizus | R. oryzae | n.a. | rabbit brain | JN206328 | ||
| CBS 330.53 | Rhizopus arrhizus var. arrhizus | R. oryzae | T of R. boreas | Japan | n.a. | JN206331 | |
| CBS 438.76 | Rhizopus arrhizus var. arrhizus | R. oryzae | NT of Amylomyces rouxii | Thailand | Look Pang (sweet fermented glutenous rice) | JN206329 | |
| CBS 515.94 | Rhizopus arrhizus var. arrhizus | R. oryzae | Singapore | tempeh | JN206325 | ||
| Chakrabarti710113 | Rhizopus arrhizus var. arrhizus | India | human | JN206333 | |||
| CBS 111231 | Rhizopus arrhizus var. delemar | R. oryzae | n.a. | n.a. | JN206338 | ||
| CBS 111760 | Rhizopus arrhizus var. delemar | R. oryzae | Vietnam | starch-based rice wine starters | JN206334 | ||
| CBS 120593 | Rhizopus arrhizus var. delemar | R. oryzae | France | human; lung | JN206340 | ||
| CBS 392.95 | Rhizopus arrhizus var. delemar | R. oryzae | T of R. delemar var. multiplicisporus | n.a. | air | JN206336 | |
| CBS 402.51 | Rhizopus arrhizus var. delemar | R. oryzae | T of R. javanicus var. kawasakiensis | Japan | n.a. | JN206339 | |
| CBS 544.80 | Rhizopus arrhizus var. delemar | R. oryzae | South Africa | sorghum malt | JN206337 | ||
| CBS 607.68 | Rhizopus arrhizus var. delemar | R. oryzae | n.a. | spaghetti | JN206341 | ||
| Chakrabarti710104 | Rhizopus arrhizus var. delemar | India | human | JN206335 | |||
| CBS 427.87 | Rhizopus caespitosus | T | India | n.a. | HM999965 | HM849671 | |
| CBS 111232 | Rhizopus homothallicus | India | n.a. | JN206365 | JN206404 | ||
| CBS 336.62 | Rhizopus homothallicus | T | Guatemala | tropical desert soil | HM999968 | HM849670 | |
| CBS 117.43 | Rhizopus lyococcus | R. stolonifer var. reflexus | Netherlands | Hordeum vulgare; grain | JN206375 | ||
| CBS 320.35 | Rhizopus lyococcus | R. stolonifer var. reflexus | n.a. | n.a. | JN206373 | JN206534 | |
| CBS 398.95 | Rhizopus lyococcus | n.a. | n.a. | JN206374 | |||
| CBS 111563 | Rhizopus microsporus | Vietnam | sufu starter: rice wine tablet | JN206363 | |||
| CBS 357.93 | Rhizopus microsporus var. azygosporus | T | Indonesia | tempeh | JN206343 | HM849666 | |
| CBS 359.92 | Rhizopus microsporus var. azygosporus | Australia | human; liver of premature infant, necrotising enterocolitis | JN206342 | |||
| CBS 294.31 | Rhizopus microsporus var. chinensis | T of R. bovinus | France | cow foetus | JN206351 | ||
| CBS 344.29 | Rhizopus microsporus var. chinensis | T of R. pypmaeus | Russia | n.a. | JN206360 | ||
| CBS 388.34 | Rhizopus microsporus var. chinensis | T of R. chinensis var. liquefaciens | Japan | n.a. | JN206354 | ||
| CBS 394.34 | Rhizopus microsporus var. chinensis | T of R. pseudochinensis var. thermosus | Japan | n.a. | JN206359 | ||
| CBS 537.80 | Rhizopus microsporus var. chinensis | South Africa | sorghum malt | JN206355 | |||
| CBS 631.82 | Rhizopus microsporus var. chinensis | T | China | bread | JN206344 | HM849668 | |
| CBS 112285 | Rhizopus microsporus var. microsporus | Mozambique | maize; ground nuts | JN206364 | JN206403 | ||
| CBS 308.87 | Rhizopus microsporus var. microsporus | Australia | human; hand, necrotic tissue | JN206357 | |||
| CBS 699.68 | Rhizopus microsporus var. microsporus | NT | Ukraine | soil | HM999970 | HM849669 | |
| CBS 112587 | Rhizopus microsporus var. oligosporus | Indonesia | tempeh | JN206358 | |||
| CBS 228.95 | Rhizopus microsporus var. oligosporus | Indonesia | tempeh | JN206348 | |||
| CBS 337.62 | Rhizopus microsporus var. oligosporus | NT | Indonesia | probably from tempeh | JN206362 | ||
| CBS 338.62 | Rhizopus microsporus var. oligosporus | Indonesia | tempeh fermentation | JN206352 | |||
| CBS 102277 | Rhizopus microsporus var. rhizopodiformis | n.a. | human; rhinocerebral infection | JN206356 | |||
| CBS 118987 | Rhizopus microsporus var. rhizopodiformis | France | human; cutaneous lesion | JN206346 | |||
| CBS 196.77 | Rhizopus microsporus var. rhizopodiformis | USA | herbal tea from Borbonia cordata | JN206361 | |||
| CBS 220.92 | Rhizopus microsporus var. rhizopodiformis | Netherlands | human; lung | JN206347 | |||
| CBS 258.79 | Rhizopus microsporus var. rhizopodiformis | Sweden | dust in saw mill | JN206349 | |||
| CBS 343.29 | Rhizopus microsporus var. rhizopodiformis | T of R. pusillus | Russia | n.a. | JN206353 | ||
| CBS 536.80 | Rhizopus microsporus var. rhizopodiformis | NT | South Africa | sorghum malt | HM999971 | JN206402 | |
| CBS 607.73 | Rhizopus microsporus var. rhizopodiformis | former Yugoslavia | stored cereals | JN206345 | HM849667 | ||
| CBS 113206 | Rhizopus microsporus var. tuberosus | China | koji in brewery | JN206350 | |||
| CBS 138.95 | Rhizopus schipperae | T | USA | human; bronchial wash of patient with myeloma | HM999969 | HM849672 | |
| CBS 336.39 | Rhizopus sexualis | T | UK | Fragaria; decaying fruit | HM999966 | HM849673 | |
| CBS 126.83 | Rhizopus sp. | R. stolonifer | n.a. | ragi | JN206368 | ||
| CBS 442.74 | Rhizopus sp. | R. stolonifer | Netherlands | coffee-ground | JN206367 | ||
| CBS 926.87 | Rhizopus sp. | R. stolonifer | Spain, Tenerife | Spartocytisus supranubius; stem | JN206366 | JN206406 | |
| CBS 133.90 | Saksenaea oblongispora | S. vasiformis | Brazil | rain forest soil | JN206282 (c1) | HM849694 | |
| JN206283 (c3) | |||||||
| CBS 405.63 | Spinellus fusiger | n.a. | n.a. | JN206298 | HM849664 | ||
| CBS 515.75 | Spinellus fusiger | Netherlands | fruit bodies of Mycena galericulata | JN206297 | |||
| CBS 633.80 | Spinellus fusiger | Germany | fruit body of Mycena cf. leptocephala | JN206295 | |||
| CBS 894.73 | Spinellus fusiger | Switzerland | fruit body of Mycena pura | JN206296 | |||
| CBS 195.77 | Sporodiniella umbellata | Ecuador | Umbonia | JN206372 | JN206405 | ||
| CBS 122.12 | Syncephalastrum monosporum | S. racemosum | Switzerland | n.a. | HM999977 (c1) | JN206575 | |
| HM999976 (c2) | |||||||
| CBS 568.91 | Syncephalastrum monosporum var. | T | China | soil | HM999975 | HM849720 | |
| cristatum | |||||||
| CBS 567.91 | Syncephalastrum monosporum var. | T | China | soil | HM999974 | HM849719 | |
| monosporum | |||||||
| CBS 120811 | Syncephalastrum racemosum | France | human; skin | HM999979 (c1) | |||
| HM999980 (c4) | |||||||
| CBS 199.81 | Syncephalastrum racemosum | Kuwait | tidal mud-flat soil | HM999972 | HM849718 | ||
| CBS 213.78 | Syncephalastrum racemosum | T of S. verruculosum | India | air | HM999978 | JN206578 | |
| CBS 302.65 | Syncephalastrum racemosum | Brazil | soil | HM999981 (c2) | HM849721 | ||
| HM999984 (c4) | |||||||
| CBS 370.49 | Syncephalastrum racemosum | Indonesia | air | HM999983 | |||
| CBS 421.63 | Syncephalastrum racemosum | Zaire | humose soil under Linum usitatissimum | HM999973 | JN206576 | ||
| CBS 440.59 | Syncephalastrum racemosum | USA | soil | HM999982 | |||
| CBS 441.59 | Syncephalastrum racemosum | USA | dung of coyote | HM999985 | |||
| CNM-CM2909 | Syncephalastrum racemosum | Spain | environmental | JN206577 | |||
| CBS 108947 | Syzygites megalocarpus | Netherlands | Amanita rubescens; decaying fruit body | JN206371 (c1) | |||
| JN206370 (c3) | |||||||
| CBS 372.39 | Syzygites megalocarpus | n.a. | n.a. | JN206369 (c3) | JN206401 | ||
| CBS 341.55 | Thamnidium elegans | USA | n.a. | JN206060 | JN206466 | ||
| CBS 411.52 | Thamnidium elegans | Poland | dung of bat | JN206058 | |||
| CBS 641.69 | Thamnidium elegans | USA | n.a. | JN206059 | |||
| CBS 874.69 | Thamnostylum lucknowense | USA | dung of pack rat | JN205837 | JN206546 | ||
| CBS 690.76 | Thamnostylum nigricans | NT | Mexico | dung of lizard | JN205838 | JN206541 | |
| CBS 170.57 | Thamnostylum piriforme | USA | dung of rat | JN205835 | |||
| CBS 182.28 | Thamnostylum piriforme | USA | Bertholletia excelsa | JN205830 | HM849724 | ||
| CBS 233.28 | Thamnostylum piriforme | USA | dung of rat | JN205832 | |||
| CBS 316.66 | Thamnostylum piriforme | T | France | n.a. | JN205836 | JN206544 | |
| CBS 412.94 | Thamnostylum piriforme | Cuba | dung of Capromys sp. | JN205834 | JN206543 | ||
| CBS 480.69 | Thamnostylum piriforme | Canada | soil under Thuja occidentalis | JN205831 | |||
| CBS 638.69 | Thamnostylum piriforme | Brazil | dung of pig | JN205833 | |||
| CBS 692.76 | Thamnostylum repens | NT | USA | dung of mouse | JN206542 | ||
| CBS 104.75 | Thermomucor indicae seudaticae | T | India | municipal compost | JN206300 | HM849661 | |
| CBS 446.78 | Thermomucor indicae seudaticae | Nigeria | Zea mays | JN206302 | |||
| CBS 447.78 | Thermomucor indicae seudaticae | Germany | compost | JN206301 | |||
| CBS 603.68 | Umbelopsis angularis | HT | Netherlands | soil | JN206380 | HM849710 | |
| CBS 212.72 | Umbelopsis autotrophica | Sweden | forest soil | JN206561 | |||
| CBS 110039 | Umbelopsis dimorpha | HT | New Zealand | soil of basaltic parent material | JN206387 | HM849709 | |
| CBS 117350 | Umbelopsis dimorpha | USA | soil of native decidious forest | JN206388 | |||
| CBS 385.85 | Umbelopsis fusiformis | T | Australia | soil of forest of Eucalyptus regnans | JN206386 | JN206560 | |
| CBS 919.85 | Umbelopsis fusiformis | Australia | soil | JN206559 | |||
| CBS 101745 | Umbelopsis gibberispora | Japan | Pinus luchuensis; decaying needle | JN206565 | |||
| CBS 109328 | Umbelopsis gibberispora | T | Japan | Fagus crenata; leaf litter | JN206384 | JN206564 | |
| CBS 100559 | Umbelopsis isabellina | USA | soil | JN206396 | JN206571 | ||
| CBS 167.80 | Umbelopsis isabellina | Colombia | fruit body of Panellus pusillus | JN206398 | |||
| CBS 250.95 | Umbelopsis isabellina | Netherlands | Varanus comodoensis; granulomatous ulcer | JN206399 | JN206574 | ||
| CBS 309.93 | Umbelopsis isabellina | UK | soil | JN206397 | |||
| CBS 560.63 | Umbelopsis isabellina | Germany | soil of Larix forest | JN206400 | JN206573 | ||
| CBS 150.81 | Umbelopsis nana | T of U. versiformis | USA | root | JN206389 | ||
| CBS 309.52 | Umbelopsis nana | Belgium | forest soil | JN206390 | |||
| CBS 373.67 | Umbelopsis nana | Georgia | forest soil | JN206394 | |||
| CBS 444.68 | Umbelopsis nana | USA | pine stump wood | JN206392 | |||
| CBS 669.83 | Umbelopsis nana | Canada | washed mycorrhizal root | JN206393 | |||
| CBS 858.68 | Umbelopsis nana | T of Mortierella alba | Poland | forest soil | JN206391 | ||
| CBS 499.82 | Umbelopsis ovata | IT | Australia | Isopogon ceratophyllus; rhizoplane | JN206395 | JN206572 | |
| CBS 101226 | Umbelopsis ramanniana | USA | soil, wood scraps and other debris | JN206383 | JN206566 | ||
| CBS 112976 | Umbelopsis ramanniana | New Zealand | litter of ? Carex sp. | JN206382 | JN206568 | ||
| CBS 366.95 | Umbelopsis ramanniana | China | forest soil | JN206385 | |||
| CBS 913.85 | Umbelopsis sp. | U. vinacea | Germany | Picea abies; root | JN206376 | JN206562 | |
| CBS 914.85 | Umbelopsis sp. | U. angularis | Germany | root | JN206377 | ||
| CBS 559.86 | Umbelopsis sp. | U. westeae | Australia | soil | JN206381 | JN206563 | |
| CBS 868.85 | Umbelopsis swartii | T | Australia | soil under Eucalyptus regnans | JN206378 | JN206567 | |
| CBS 212.32 | Umbelopsis vinacea | AUT | Australia | sandy loam | JN206570 | ||
| CBS 236.82 | Umbelopsis vinacea | T of U. multispora | Japan | Fragaria; root | JN206569 | ||
| CBS 870.85 | Umbelopsis westeae | T | Australia | soil of acid heathland | JN206379 | ||
| CBS 329.73 | Utharomyces epallocaulus | India | soil | JN206276 | HM849660 | ||
| CBS 342.73 | Utharomyces epallocaulus | Mexico | dung of rat | JN206277 | |||
| CBS 441.76 | Zychaea mexicana | T | Mexico | dung of mouse | JN205845 | JN206545 |
DNA extraction, PCR amplification, cloning and sequencing
Genomic DNA was extracted in most cases from 2d-old cultures grown on malt extract agar (MEA 5 %, Oxoid, Badhoevedorp, The Netherlands) following the protocol given by Möller et al. (1992) with diverse modifications described in detail by Alastruey-Izquierdo et al. (2010). The primer pair V9G (de Hoog & Gerrits van den Ende 1998) and LR3 (Vilgalys & Hester 1990) was used to amplify a DNA segment consisting of the complete ITS region and the D1/D2 region of the LSU. The PCR reaction mixture (25 μl) contained 0.4 μM of each primer, 0.185 mM of each deoxynucleoside triphosphate (GC Biotech, Alphen aan den Rijn, The Netherlands), 10× NH4 BioTaq Reaction buffer (GC Biotech), 1.5 mM MgCl2, 0.8 U BioTaq DNA polymerase (GC Biotech), and about 20 ng DNA. PCR reactions were conducted on a Thermal cycler 2720 (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands) as follows: one initial cycle at 94 °C for 5 min, followed by 35 cycles of 1 min at 94 °C, 1 min at 53 °C and 2 min at 72 °C, and one final cycle of 7 min at 72 °C. PCR products were analysed in 1 % agarose gels. Direct cycle-sequencing was performed for both strands of the PCR products using the Big dye sequencing kit (Applied Biosystems) and the primer set ITS1 and ITS4 (White et al. 1990) for the complete ITS region, and NL1 and LR3 for the D1/D2 region of the LSU (O’Donnell 1993). Cycle-sequencing products were analysed on an ABI 3730XL automatic sequencer (Applied Biosystems).
In cases where direct sequencing failed or double bands were detected in the agarose gels PCR products were ligated into pGEM-T Easy Vector (Promega, Leiden, The Netherlands) and cloned in E. coli JM109 competent cells (Promega) following the manufacturer’s instructions. Colony-PCRs were performed using the primer pair M13f (5’-GTAAAACGACGGCCAGT-3’) and M13r (5’-GGAAACAGCTATGACCATG-3’).
Sequence analyses
Consensus sequences were constructed by means of the SeqMan program v. 7.2.2 (DNASTAR, Lasergene) and deposited in GenBank. A total of 592 ITS sequences (GenBank accession numbers JN205809 to JN206400) and 210 LSU sequences (GenBank accession numbers JN206401 to JN206610) were newly generated for this study. Ninety-two ITS sequences and 113 LSU sequences of strains included in this study were published in previous papers (Alastruey-Izquierdo et al. 2010, Vitale et al. 2012).
The highly diverse ITS sequences were divided in subsets of alignable sequences for phylogenetic analyses. Additionally, a set of LSU sequences was generated representing every MOTU detected in the ITS alignments. The MOTUs were defined by a ≥ 99 % similarity threshold. Similarity values based on the uncorrected distances were calculated with BioNumerics (v. 0.4.61) software (Applied Maths, Sint-Martens-Latem, Belgium).
ITS and LSU sequences were aligned using the server version of the MAFFT program (www.ebi.ac.uk/Tools/mafft) and manually corrected using the program Se-Al v. 2.0a11 (Rambaut 2002; http://tree.bio.ed.ac.uk/software/seal/). All alignments are lodged in TreeBASE (www.treebase.org/). Mortierella parvispora, a member of the Mortierellales, was selected as outgroup of the LSU tree because the Mortierellales is the sibling order of the Mucorales (e.g. O’Donnell et al. 2001, Voigt & Wöstemeyer 2001). The ITS trees were rooted by the nearest neighbours of the respective group in the LSU tree. The maximum likelihood algorithm with the server version of RAxML-VI-HPC v. 7.0.0 (Stamatakis et al. 2008) as implemented on the Cipres portal was used to estimate phylogenetic relationships. The robustness of all phylogenetic trees was assessed by bootstrap analyses with 1 000 replicates. ITS trees were only calculated for Actinomucor, Mucor (4 trees), Rhizomucor and Rhizopus, but all of the taxa are included in the LSU tree.
Morphology
Strains with a position in the phylogenetic tree that was in conflict with their original classification were verified microscopically. These strains were cultivated on MEA at 24 °C. After 2 and 7 d colonies were observed using a stereomicroscope (SMZ 1500; Nikon) and slides for microscopic examination were made. Fungal material was mounted in lactic acid with cotton blue (2 mg cotton blue/mL lactic acid) and in lactic acid only and examined using a Nikon eclipse 80i microscope (Nikon, Amstelveen, The Netherlands). Measurements were performed using the software NIS-Elements D 3.0 (Nikon). For a few strains mounts from slide cultures were photographed using a Nikon 05SM digital camera.
RESULTS AND DISCUSSION
Performance of ITS and LSU sequencing
Direct sequencing was possible in 82 % of the strains for the ITS and in all strains for the LSU region. ITS sequences of strains that failed direct sequencing could be obtained by cloning in most taxa, while cloning did not solve the problem in the group of Rhizopus stolonifer; this was probably due to extended poly-A- and poly-T-regions. In the majority of cloned strains ITS copies differed only by a single basepair. However, in Syncephalastrum we found ITS sequences of two types forming two clades in the ITS tree (illustrated in Vitale et al. 2012). Divergent copies of ITS have been found in Fusarium (O’Donnell & Cigelnik 1997) and Laetiporus (Lindner & Banik 2011). The proportion of strains that had to be cloned was especially high in Absidia, Umbelopsis and Syncephalastrum.
Phylogenetic relationships inferred from LSU data
The family structure of the Mucorales revealed by the LSU cladogram in main traits corresponds with that given by Hoffmann et al. (2013), although the Lichtheimiaceae and the Mucoraceae do not form clades (Fig. 1), probably because our analysis is based on a single locus. Some remaining taxonomic inconsistencies between phenotypic and sequence-based classifications were found, particularly in the upper part of the LSU cladogram (Fig. 1). The Mycotyphaceae and the Choanephoraceae are nested within the Mucoraceae and these families together with the Backusellaceae, the Pilobolaceae and the Rhizopodaceae compose a well-supported clade (Mucorineae clade; bootstrap 75 %). The Mucorineae clade is dominated by the polyphyletic genus Mucor (Fig. 2a), which comprises many groups that are intermingled by other sporangia-forming genera such as Circinella p.p. (Fig. 2h), Parasitella, Pilaira, Pirella (Fig. 2f), Rhizomucor p.p. (Fig. 2b), and Zygorhynchus (Fig. 2d), sporangia- and sporangiola-forming genera such as Backusella (Fig. 8) and Thamnidium (Fig. 2i), as well as exclusively sporangiola-forming genera such as Chaetocladium, Ellisomyces (Fig. 2g), Helicostylum and Kirkomyces. Based on the LSU (Fig. 1) and ITS trees (Fig. 3 to 6), the genera Backusella, Circinella, Mucor, Rhizomucor and Zygorhynchus appear to be polyphyletic. The lower part of the LSU tree shows a more resolved taxonomy. The majority of genera here are clearly defined and appear to be monophyletic based on our sampling. Exceptions were the paraphyletic genus Absidia that includes Halteromyces and Chlamydoabsidia, and the polyphyletic genera Circinella and Rhizomucor that are addressed in more detail below.
Fig. 1.
RAxML phylogram of the Mucorales based on the D1/D2 region of the LSU. Each LSU sequence covers for a MOTU in ITS defined by a similarity threshold of 99 %. Branches with bootstrap values of 75 % or higher are printed in bold. Morphological groups according to sporangiophore branching and diameter of the sporangium: Mucor mucedo group, M. flavus group, M. hiemalis group, M. racemosus group, M. amphibiorum group, M. recurvus group. Black bar highlights the Mucor circinelloides complex. Ex-type strains are designated by: T = ex-type strain, ET = ex-epitype strain, HT = ex-holotype strain, IT = ex-isotype strain, LT = ex-lectotype strain, NT = ex-neotype strain, PT = ex-paratype strain, ST = ex-syntype strain, AUT = authentic material, c = clone, clinical strains are highlighted by red strain and GenBank accession numbers, strains representing MOTUs that comprise clinical strains are marked by red squares. Clinically relevant species are indicated by a red circle if no ITS sequence of a clinical isolate was available for the assignment to a MOTU. Potentially undescribed taxa are indicated by bold blue font.
Fig. 2.
Morphological diversity of Mucoraceae. a. CBS 243.35 Mucor luteus, sporangiophore and sporangium with collar, columella and sporangiospores; b. CBS 385.95 Rhizomucor endophyticus (recombined into Mucor endophyticus in this paper), sporangiophore with columella and collar; c. CBS 243.35 Mucor luteus, rhizoids formed on glass slides; d. CBS 110662 Zygorhynchus multiplex (recombined into Mucor multiplex in this paper), zygospore; e. CBS 385.95 Rhizomucor endophyticus (recombined into Mucor endophyticus in this paper), zygospore; f. CBS 588.88 Pirella circinans, lateral sporangium; g. CBS 243.57 Ellisomyces anomalus, multispored sporangiola; h. Circinella rigida (recombined into Mucor durus in this paper), circinate sporangiophore branch with apophysate sporangium; i. CBS 341.55 Thamnidium elegans, multispored sporangiola. — Scale bars = 10 μm, except c = 100 μm.
Fig. 8.
Morphology of the genus Backusella. a, b. CBS 128.70 Backusella circina, a. unispored sporangiola; b. transitorily recurved sporangiophore; c. CBS 318.52 Mucor recurvus var. recurvus (Backusella recurva here), transitorily recurved sporangiophore; d. CBS 564.66 Mucor variabilis (Backusella variabilis here), transitorily recurved sporangiophore. — Scale bar: a = 10 μm, b–d = 50 μm.
Fig. 3.
RAxML phylogram of the Mucor mucedo group, M. flavus group, M. hiemalis group and related taxa based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, ET = ex-epitype strain, HT = ex-holotype strain, IT = ex-isotype strain, LT = ex-lectotype strain, NT = ex-neotype strain. Ex-type strains of currently accepted taxa are printed in bold. Clones are specified by a ‘c’ followed by the clone number. Clinical strains are highlighted by red strain and GenBank accession numbers, non-human hosts are given in brackets. Potentially undescribed taxa are indicated by bold blue font. Morphological identifications are given in quotation marks.
Although the polyphyly of Mucor is indisputable, only few clear lineages within Mucor are recognisable. Some of these lineages show a characteristic combination of sporangium size and the branching of the tall sporangiophores: in the Mucor mucedo group (Fig. 1), consisting, in addition to the name-giving species M. mucedo, of M. piriformis, M. plasmaticus and M. strictus, all form unbranched tall sporangiophores and large sporangia often exceeding 200 μm diam. The morphology of Mucor strictus is strongly temperature-dependent. The species develops the typical group morphology only at low temperatures (Schipper 1975: 24). The Mucor flavus group, comprising M. aligarensis, M. minutus and M. saturninus in addition to M. flavus consists of species developing sympodially branched tall sporangiophores and medium-sized sporangia that are over 80 μm but not exceeding 200 μm diam. A clade including M. hiemalis, M. irregularis, M. luteus, Rhizomucor endophyticus and Zygorhynchus psychrophilus, the Mucor hiemalis group, is characterised by poorly sympodially branched tall sporangiophores and small sporangia not exceeding 80 μm diam. The Mucor racemosus group contains M. circinelloides with the formae circinelloides, griseocyanus, lusitanicus and janssenii, Mucor bainieri, M. plumbeus, M. racemosus, M. ramosissimus and Backusella ctenidia. The characteristics of this group are repeatedly sympodially branched sporangiophores and small sporangia not exceeding 80 μm diam. Species of the Mucor amphibiorum group, namely M. amphibiorum, M. ardhlaengiktus, M. azygosporus, M. falcatus, M. inaequisporus, M. indicus, M. nederlandicus, M. odoratus, M. prayagensis, M. ucrainicus, M. variosporus and M. zychae form − with the exception of M. indicus and M. falcatus − usually unbranched tall sporangiophores that bear, unlike species of the M. mucedo group, small- to medium-sized sporangia (maximum diameter between 70 and 175 μm). The Mucor recurvus group comprises Mucor and Backusella species with transitorily recurved sporangiophores and is discussed below in more detail. All of these groups were recognised, to a large extent, by Schipper (1973, 1975, 1976, 1978a).
Phenotypic characters with restricted taxonomic relevance
Genera preferably are defined as monophyletic. This may be in conflict with phenotypic definitions when single characters are concerned. For example, sporangiola-forming taxa such as Backusella, Chaetocladium, Ellisomyces, Helicostylum, Kirkomyces and Thamnidium, appear sporadically among Mucor species (Fig. 1, 3, 5, 6) and only the clade consisting of the sporangiola-forming genera Benjaminiella and Cokeromyces (Fig. 1) possess phylogenetic support. Hence we consider the potential to develop sporangiola in addition to or instead of sporangia as a plesiomorphic character in the Mucoraceae that is genetically fixed but not revealed in all species. Consequently, the presence of sporangiola should not be used to define new genera, as was done recently in the newly described genus Isomucor (de Souza et al. 2012) with sporangiola as the only distinctive trait. A classical generic criterion is the presence or absence of rhizoids. Our finding of rhizoids in Mucor species (Fig. 2c), the genus originally separated from Rhizomucor by the absence of rhizoids, and the reclassification of Rhizomucor species such as R. regularior, R. variabilis (Álvarez et al. 2010a), R. chlamydosporus and R. endophyticus (this paper) into Mucor suggests that the potential to form rhizoids might be plesiomorphic in Mucorales as well. Homo- and heterothallic species cluster randomly in our trees demonstrating the reduced taxonomic value of this trait. Circinate sporangiophores branches (Fig. 2h), considered to be a characteristic of the genus Circinella, evolved at least three times.
Fig. 5.
RAxML phylogram of the Mucor racemosus group and related taxa based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, ET = ex-epitype strain, IT = ex-isotype strain, NT = ex-neotype strain. Ex-type strains of currently accepted taxa are printed in bold. Clones are specified by a ‘c’ followed by the clone number. Clinical strains are highlighted by red strain and GenBank accession numbers, non-human hosts are given in brackets. Potentially undescribed taxa are indicated by bold blue font. Morphological identifications are given in quotation marks. Black bars indicate groups within Mucor circinelloides f. circinelloides including a large number of clinical strains.
Fig. 6.
RAxML phylogram of the Mucor amphibiorum group and related taxa based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, ET = ex-epitype strain, HT = ex-holotype strain, IT = ex-isotype strain, LT = ex-lectotype strain, NT = ex-neotype strain, PT = ex-paratype strain. Ex-type strains of currently accepted taxa are printed in bold. Clones are specified by a ‘c’ followed by the clone number. Clinical strains are highlighted by red strain and GenBank accession numbers, non-human hosts are given in brackets. Potentially undescribed taxa are indicated by bold blue font. Morphological identifications are given in quotation marks.
Species identification by DNA barcoding
In our study the ITS region turned out to be an appropriate DNA barcoding marker in Mucorales because of its power to discriminate the currently accepted species, including predominantly morphologically defined species (morphospecies), and also all species in Mucor (Hermet et al. 2012), Lichtheimia (Alastruey-Izquierdo et al. 2010) and Rhizopus (Abe et al. 2007) recognized by GCPSR studies. The statement of Schwarz et al. (2006) that ITS is able to discriminate most but not all clinically relevant members of Mucorales was due to the fact that the authors treated R. azygosporus as a separate species, while now it is classified as a variety of R. microsporus (Zheng et al. 2007). Indeed the taxonomy of the Mucoraceae is unsatisfactory according to current molecular characteristics and consequently identification by DNA barcoding is reliable only for species that form clearly delimited clades in the ITS trees. In Rhizomucor (Fig. 11) and Rhizopus (Fig. 10), all species can be identified by their ITS sequence, and even two varieties of Rp. arrhizus proved to be distinguishable (see sections on Rhizomucor or Rhizopus, respectively). ITS data also separate a surprisingly high number of species of Mucor and its allies as monophyletic groups (Fig. 2–5), allowing reliable species identification despite unclear generic boundaries. Problematic for molecular identification are the species complexes of Mucor circinelloides, M. flavus, M. piriformis and Zygorhynchus moelleri. The complex of Mucor circinelloides constitutes a supported clade (bootstrap support 88 %) in the LSU phylogram (Fig. 1) and comprises all formae of M. circinelloides but also other species such as M. bainieri and M. ramosissimus, Ellisomyces anomalus and Backusella ctenidia that cannot be separated by ITS and LSU data from M. circinelloides (Fig. 5). All formae of M. circinelloides described by Schipper (1976) can be discriminated by their ITS sequences, but there are several strains in the complex with a M. circinelloides morphology that do not group in any of these formae (see sections Mucor and Backusella).
Fig. 11.

RAxML phylogram of the genus Rhizomucor based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, LT = ex-lectotype strain, ET = ex-epitype strain. The identifying ex-type strain of a clade is printed in bold. Clones are specified by a ‘c’ followed by the clone number. Clinical strains are highlighted by red strain and GenBank accession numbers.
Fig. 10.
RAxML phylogram of the genus Rhizopus based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, NT = ex-neotype strain. Ex-type strains of currently accepted taxa are printed in bold. Clones are specified by a ‘c’ followed by the clone number. Clinical strains are highlighted by red strain and GenBank accession numbers, non-human hosts are given in brackets. Potentially undescribed taxa are indicated by bold blue font. Morphological identifications are given in quotation marks.
Criteria for taxonomic changes based on ITS and LSU data
In general, multilocus studies are needed to establish boundaries between taxa forming sister clades in single-locus trees. However, in this stage of research we consider ITS and LSU data sufficient to justify taxonomic changes in the following cases: 1) synonymy if the ex-type strains have identical ITS sequences (in case the ITS sequences are identical but large morphological differences have been reported we maintain varieties or formae); 2) attribution of the species rank to varieties or formae if they appear distant to the type variety or forma in ITS and LSU trees; 3) reclassifications based on polyphyly shown by ITS and LSU data (e.g. Zygorhynchus); and 4) extension of a genus based on ITS and LSU analyses and morphological synapomorphies (e.g. Backusella).
Species concepts: conflicts between grouping in the ITS trees and mating results
The broad species concepts in Mucor maintained by Schipper (1973, 1975, 1976, 1978a), based on positive matings, are partly in conflict with the grouping in our ITS trees. Species in the sense of Schipper, e.g. Mucor circinelloides, M. hiemalis (including M. luteus) and M. flavus are not monophyletic. Their clades include other species or even other genera showing that the simple presence of zygospores is not a sufficient marker for conspecifity. However, a detailed study of their size, shape, colour and number may allow to differentiate between inter- and intraspecific zygospores, as shown for Mucor irregularis (Schell et al. 2011) and Lichtheimia (Alastruey-Izquierdo et al. 2010). Consequently, the study of zygospores should not be devaluated by our results but is ideally combined with multi-locus studies to recognize species.
Detection of undescribed species by DNA barcoding
Twelve MOTUs are revealed by ITS sequencing that deviate considerably from species included in this study and might represent undescribed taxa (indicated by bold blue font in all trees). The species to which they had previously been assigned, on the basis of morphological features, are given in quotation marks. In order to establish whether these MOTUs deserve species rank we have initiated multilocus-analyses and detailed morphological studies, which will be published in subsequent papers. Regardless of the question if these MOTUs represent undescribed species, our results suggest that the used phenotypic criteria for species recognition in the Mucorales underestimated existing diversity.
The ITS sequences of numerous strains of Absidia and Umbelopsis could not be obtained by direct sequencing and not all of them could be cloned. Our ITS dataset is therefore incomplete for these genera.
ITS barcoding detected cryptic species, but conversely revealed growth-reduced mutants of existing taxa that had been maintained incorrectly as separate species. Examples include Mucor sinensis, which is synonymised with M. racemosus f. racemosus below, alleged Mucor ramosissimus strains that proved to belong to M. circinelloides f. circinelloides or f. janssenii, and Rhizomucor tauricus which is a synonym of Rm. pusillus.
Intraspecific variability
Intraspecific variability differs widely among mucorealean species and genera. The following dissimilarity values were obtained for the ITS region of species represented by at least 5 strains: Backusella circina (0 %), Choanephora cucurbitarum (0.5 %), Circinella muscae (1.6 %), Cunninghamella bertholletiae (1.0 %), C. blakesleeana (0.9 %), C. echinulata (13.3 %), Gilbertella persicaria (1.2 %), Gongronella butleri (1.1 %), Lichtheimia corymbifera (2.0 %), L. hyalospora (6.3 %), L. ramosa (7.6 %), Mucor circinelloides (5.3 %), M. fuscus (0.2 %), M. hiemalis (4.1 %), M. indicus (0.9 %), M. irregularis (2.6 %), M. mucedo (3.5 %), M. odoratus (0.3 %), M. piriformis (0.6 %), M. plumbeus (0.2 %), M. racemosus (2.1 %), Phycomyces blakesleeanus (0 %), Pilaira anomala (0 %), Rhizomucor pusillus (0.2 %), Rhizopus arrhizus incl. var. delemar (1.2 %), R. lyococcus (0.7 %), R. microsporus (2.8 %), Thamnostylum piriforme (1.6 %), Umbelopsis nana (0 %) and Zygorhynchus moelleri (0 %). Complete identity might be the result of undersampling. High dissimilarity values of morphologically defined species such as Cunninghamella echinulata could be caused by the inclusion of more than one species. However, the taxonomically well-elaborated genus Lichtheimia also contains species with more than 5 % dissimilarity suggesting that intraspecific variability can be comparatively high in Mucorales.
In agreement with Nilsson et al. (2006) we did not find a unifying threshold for intraspecific variability, a result that should caution against formal ITS-based species delimitation. On the other hand, the ≥ 99 % identity threshold given by Balajee et al. (2009) for comparative ITS sequence identification using GenBank in Mucorales is not covering the intraspecific variability of several clinically important species. Our data suggest that a distinct identity threshold has to be defined for every species for a reliable ITS-based identification. GCPSR studies will be required beforehand to define species boundaries in species with conflicting morphological and ITS data especially in those that are part of a species complex.
Clinically relevant taxa
Clinical strains are highlighted by red strain numbers and GenBank accession numbers in all figures. Strains representing a clinically relevant MOTU are marked either by a red square (assignment to the respective MOTU by ITS comparison) or by a red circle (assignment to the respective MOTU by morphology) in the LSU tree (Fig. 1). The following genera contain species that are involved in human opportunistic infections: Actinomucor, Apophysomyces, Cokeromyces, Cunninghamella, Lichtheimia, Mucor, Rhizomucor, Rhizopus, Saksenaea, Syncephalastrum, and possibly Mycotypha, Thamnostylum (Xess et al. 2012) and Zygorhynchus, the latter genus being synonymised with Mucor below. The recently described clinically relevant species of Apophysomyces, viz. A. ossiformis and A. trapeziformis (Álvarez et al. 2010b) and of Saksenaea, viz. S. erythrospora as well as the pathogenic S. vasiformis (Álvarez et al. 2010a) were not included in our study.
No clinical strains of the following species were included in the present study, but clinical relevance of the species, resp. strains originating from clinical samples were reported by other authors: Cokeromyces recurvatus (e.g. Ryan et al. 2011), Cunninghamella blakesleeana (García Rodríguez et al. 2012), C. echinulata (Lemmert et al. 2002), Mucor ardhlaengiktus (as M. ellipsoideus), M. circinelloides f. lusitanicus (as M. lusitanicus) (Álvarez et al. 2011), Rhizopus homothallicus (Chakrabarti et al. 2010), Rhizopus stolonifer (de Hoog et al. 2000) and Thamnostylum lucknowense (Xess et al. 2012). A clinical strain treated as Mucor fragilis by Álvarez et al. (2011) is assigned to M. circinelloides below.
The involvement of Zygorhynchus moelleri and a possibly undescribed Mycotypha species in human mucormycoses has not been proven: strain IHEM 21156 of Zygorhynchus moelleri was isolated by J.P. Bouchara at the University Hospital Angers (France) in 2004 but the exact source is unknown, and Mycotypha strain CBS 109960 was isolated by N. Poonwan from pus of a wound in the RMSC Pitsunalok (Thailand) in 2002. There is also no case report on Mucor plumbeus but the source of isolation of strain CBS 634.74 (human biopsy material) and CBS 633.74 (subcutaneous tissue of a cat) suggest a pathogenic potential of this species. Mucor aligariensis was isolated in 1958 from human and M. saturninus in 1978 from chicken, but no proven case reports have been published. Based on their maximum growth temperatures (< 30 °C, Schipper 1978a) the involvement of these species in infections is questionable.
The clinical strain CNM-CM 5114 (JN205884) isolated in 2008 from a lung biopsy specimen in Barcelona (Spain) was diagnosed as Cunninghamella elegans by a 99.2 % similarity of the ITS region with the ex-neotype strain of this species CBS 160.28 (AF254928). This is the first time that the involvement of C. elegans in human infection has been documented molecularly.
The recent detection of species with clinical relevance, such as Cunninghamella echinulata (Lemmert et al. 2002), Lichtheimia ornata (Alastruey-Izquierdo et al. 2010), Mucor ardhlaengiktus (as M. ellipsoideus, Álvarez et al. 2011) or Rhizopus homothallicus (Chakrabarti et al. 2010) shows that basically all mucorealean taxa, including species regarded as strictly environmental, should be included in the database for molecular identification of clinical strains. Molecular identification by BLAST searches of GenBank may lead to wrong conclusions because of incomplete sampling, inconsistent nomenclature and a high percentage of misidentified taxa (Bridge et al. 2003, Nilsson et al. 2006, Bidartondo et al. 2008, Lian et al. 2011).
The following taxa have been reported from proven case studies (de Hoog et al. 2000, Gomes et al. 2011): Actinomucor elegans, Apophysomyces variabilis, A. trapeziformis (Weddle et al. 2012), Cokeromyces recurvatus, Cunninghamella bertholletiae, C. elegans, Lichtheimia corymbifera, L. ramosa, Mucor amphibiorum, M. ardhlaengiktus (Sugui et al. 2011), M. circinelloides (f. circinelloides, f. janssenii, f. lusitanicus), M. hiemalis, M. irregularis (syn. Rhizomucor variabilis), M. luteus (syn. M. hiemalis f. luteus), M. indicus, M. racemosus f. racemosus, M. ramosissimus, Rhizopus microsporus, Rp. arrhizus, Rp. homothallicus, Rp. schipperae, Rp. stolonifer, Rhizomucor miehei, Rm. pusillus, Saksenaea vasiformis, S. erythrospora (Hospenthal et al. 2011) and Syncephalastrum racemosum.
The forma circinelloides of Mucor circinelloides contains the highest number of clinical isolates in Mucor and forms a well-supported clade (bootstrap 95 %). Two groups within forma circinelloides (highlighted by black bars in Fig. 5) contain a large number of clinical strains, which might be explained to high frequency in the human environment, or to an increased virulence. Interestingly, the forma griseocyanus, being the only forma of Mucor circinelloides that is not able to grow at 37 °C, does not comprise any clinical isolate. In Mucor racemosus we found a similar picture: only the forma racemosus appears to be involved in human infections, while the forma sphaerosporus contains strains only isolated from food and environmental samples. The maximum growth temperature of forma sphaerosporus is distinctly lower (28 °C > Tmax < 25 °C) than that of the clinically relevant forma racemosus (37 °C > Tmax < 30 °C) (Schipper 1976).
Many thermotolerant or thermophilic species (Schipper 1973, 1975, 1976, 1978a, b, 1979, 1984, 1986, 1990, Schipper & Stalpers 1984, Schipper & Samson 1994, de Hoog et al. 2000, Zheng & Chen 2001, Hoffmann et al. 2007, Alastruey-Izquierdo et al. 2010, Álvarez et al. 2010a, b) in the Mucorales are opportunistic pathogens of vertebrates, but there is no direct correlation between thermotolerance and pathogenicity. Several taxa including species of Backusella, Cunninghamella and Mucor, as well as Protomycocladus faisalabadensis and Thermomucor indicae-seudaticae are thermotolerant or even thermophilic, but have never been reported to cause infections.
TAXONOMY
Mucor
Mucor mucedo group, M. flavus group, M. hiemalis group and related taxa
In our ITS tree (Fig. 3) Mucor flavus is divided in several small clades that are intermingled with M. aligarensis, Helicostylum pulchrum and Thamnidium elegans. Several ex-type strains of species that were synonymised by Schipper (1975) with M. flavus based on positive matings and morphological similarity are placed in other clades than the neotype strain CBS 234.35. Their correct taxonomic status needs to be assessed by more detailed studies.
Only two of the four formae established by Schipper (1973) for Mucor hiemalis, viz. f. hiemalis and f. corticola, constitute a clade in the ITS tree (Fig. 3). This clade is divided into two subclades both composed of strains morphologically assigned to f. hiemalis and f. corticola. However, the ex-type strains of both formae are located in the same subclade (Fig. 3). For that reason we consider the small differences in the shape of the spores between the formae as taxonomically insignificant. Strains of Mucor hiemalis f. silvaticus are not part of the well-supported Mucor hiemalis clade, not in the ITS (Fig. 3) nor in the LSU tree (Fig. 1). Therefore we treat this taxon as a discrete species. The same applies to Mucor hiemalis f. luteus that recently was reclassified as Mucor luteus (Budziszewska et al. 2010).
No authentic material is known to exist of Mucor abundans. The protologue (Povah 1917) describes M. abundans as a species with slightly branched sporangiophores, sporangia with a diameter below 100 μm, subglobose to pyriform columellae and small globose to short ellipsoidal sporangiospores. The strains CBS 388.35 (Fig. 4) and CBS 521.66, the latter deposited as M. abundans in the CBS collection, match these microscopic characters including the typical size and shape of the sporangiospores. They only differ by features that may have changed during prolonged cultivation such as the colour of the colony or the colour of the young sporangia that are hyaline with a slight yellow tinge in the studied strains instead of yellowish. Schipper (1973) noted the similarity of both strains with M. abundans but she considered them too similar to M. hiemalis f. corticola to recognise a separate species. Our ITS tree (Fig. 3) however, supports the separate position of these strains and therefore we here designate CBS 388.35 (preserved in a metabolically inactive state by lyophilization, batch nr. 472) as the neotype of Mucor abundans.
Fig. 4.
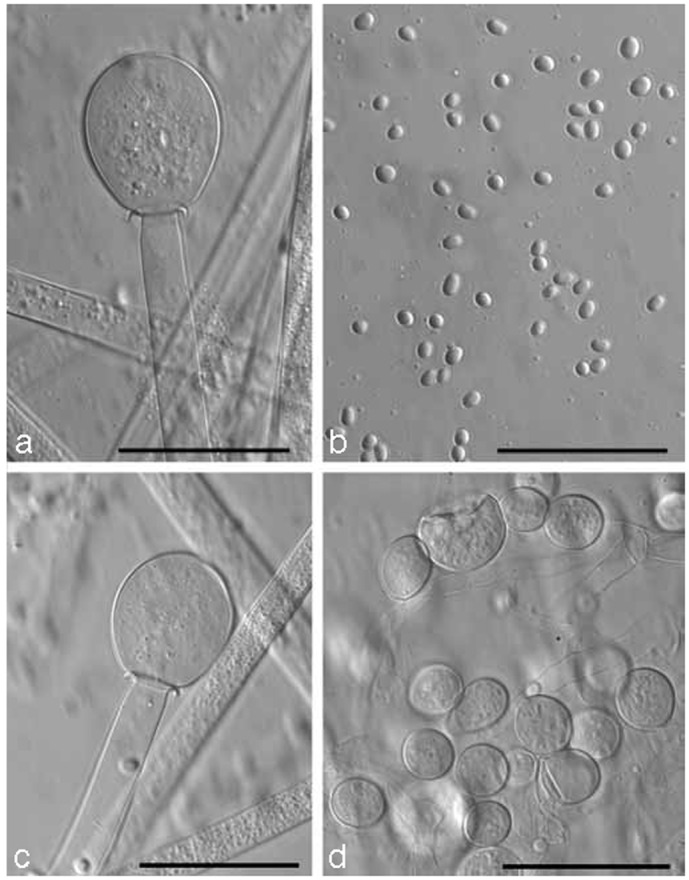
CBS 388.35 Mucor abundans. a, c. Sporangiophore with columella and collar; b. sporangiospores; d. chlamydospores. — Scale bars = 50 μm.
On MEA at room temperature strain CBS 388.35 shows the following features: colonies expanding, cottony, at first white later depending on the intensity of sporulation grey beige or pale grey; reverse uncoloured; sporangiophores slightly sympodially branched, up to 15 μm diam; sporangiophore branches straight; sporangia dark greyish, young hyaline or with a slight yellow tinge, small, up to 70 μm diam; columellae subglobose, ellipsoidal or slightly pyriform (as illustrated by Povah 1917) (Fig. 4a, c), up to 39 by 46 μm in size, often purplish grey; collars distinct; sporangiospores hyaline, smooth, subglobose (3.5–6 μm diam), short ellipsoidal to short cylindrical (4.5–7 by 3.5–5.5 μm) (Fig. 4b); chlamydospores globose, ellipsoidal or pyriform, intercalary, terminally and laterally formed mainly in the submerged mycelium (Fig. 4d); zygospores absent.
Mucor racemosus group and related taxa
Based on our analyses (Fig. 1, 5) Mucor circinelloides represents a species complex, which also includes other Mucor species and sporangiola-forming taxa. The backbone of the M. circinelloides part of the ITS tree is poorly resolved, hampering a decision on the rank of the taxa included. Based on positive matings, Schipper (1976) reduced four species related to Mucor circinelloides to formae, namely f. circinelloides, f. griseocyanus, f. janssenii and f. lusitanicus. Recently Álvarez et al. (2011) proposed species status for f. lusitanicus without considering the positive mating results obtained by Schipper (1976). Strains morphologically assigned to the various formae of M. circinelloides form well-supported clades in the ITS tree (Fig. 5), but several additional strains that morphologically belong to M. circinelloides are located outside these clades. The strains CBS 338.71 and CBS 635.65, for example, are placed basally to the forma lusitanicus clade in the ITS tree (Fig. 5) and develop predominantly globose columellae characteristic of forma lusitanicus, but occasionally they develop obovoid columellae typical of forma circinelloides. Schipper’s (1976) mating results, the presence of intermediate strains and the absence of compensatory base changes (CBC) between f. circinelloides, f. lusitanicus, and f. janssenii (Pawłowska et al. In press) lead us to regard M. circinelloides as a single species that consists of several still interbreeding lineages which result in a high intraspecific ITS variability of 5.3 %.
Some confusion exists because of misapplied names for important model strains. Strain CBS 416.77, deposited in the CBS collection by S. Bartnicki-García as Mucor rouxii, belongs to Mucor circinelloides according to its ITS sequence, a fact that has been noted by several authors (Abe et al. 2006, Schwarz et al. 2006, Liu et al. 2007). Ellis et al. (1976) proposed NRRL 5866 = CBS 438.76 as the neotype for Amylomyces rouxii (Calmette 1892) and found it to be conspecific with that of Rhizopus arrhizus. Wehmer (1900) incorrectly believed that the strains he isolated represented Calmette’s Amylomyces rouxii and proposed the name Mucor rouxii (Calmette) Wehmer for them. However, he probably studied strains of Mucor indicus (Schipper 1978a: 10). Consequently Mucor rouxii sensu Wehmer is a synonym of Mucor indicus, whereas Mucor rouxii (Calmette) Wehmer is Rhizopus arrhizus. Strains of M. indicus are very similar morphologically to Mucor circinelloides at some stages in the life cycle (Schipper 1978a: 10), but the molecular differences between the two species are unambiguous.
Mucor ramosissimus is another member of the Mucor circinelloides complex. Its ITS sequence clusters in the well-supported clade of Mucor circinelloides f. lusitanicus. However, the sequence differs considerably (19 out of 585 basepairs, 3.2 % dissimilarity) from the remaining sequences of M. circinelloides f. lusitanicus. This is expressed by a long branch in Fig. 5. We therefore retain the species rank for this taxon and await studies on other loci. Other strains that had been identified morphologically as M. ramosissimus are positioned distantly in different groups: CBS 144.93 clusters in the Mucor circinelloides f. janssenii clade and CBS 121702 in the M. circinelloides f. circinelloides clade. These strains differ by slow growth and possibly represent growth-reduced mutants of these formae.
CBS 236.35, the only strain listed as Mucor fragilis in the CBS database, is positioned basally in the Mucor circinelloides f. lusitanicus clade. As far as we are aware no type material exists for M. fragilis. Bainier’s (1884) original description assigns the fungus to the Mucor hiemalis group because the zygospores are black and bear characteristically roughened stellate spines. Schipper (1976) treated Mucor fragilis as a doubtful species because strain CBS 236.35, received as Mucor fragilis from Zycha in 1935, deviated from Bainier’s description. In agreement with Schipper (1976) we are not using this name and reidentify strain CBS 236.35 as Mucor circinelloides f. lusitanicus.
Mucor circinelloides f. janssenii splits in two groups in the ITS tree (Fig. 5): a first group containing the ex-type strain, and a second group consisting of CBS 144.93, CBS 204.68 and CBS 762.74 at 0.7 % distance. Considering the high degree of sequence diversity in Mucorales, expressed for example in an ITS sequence dissimilarity of 2.6 % between Mucor circinelloides f. janssenii and f. lusitanicus, these groups within jansenii are being treated as taxonomically insignificant. The ITS sequence of the ex-type strain of the recently described Mucor velutinosus (FN650646; Álvarez et al. 2011) is identical to that of CBS 762.74 of f. janssenii (data not shown). According to their ITS sequences (data not shown) isolates ATCC1209b (HM754254) and UIC-1 (HQ154609) of M. circinelloides forming an unknown group in the phylogenetic analyses of Li et al. (2011) also belong to f. janssenii but type material of this form was not included in the study of Li et al. (2011).
The ITS sequence deposited in GenBank (EF203698) for the newly described species Mucor renisporus (Jacobs & Botha 2008) is identical to that of CBS 480.70F (data not shown) of Mucor circinelloides f. circinelloides. However, according to the description given by Jacobs & Botha (2008) the taxa differ significantly in the sizes of sporangia, columellae and sporangiospores. A microscopical study of the ex-type strain of Mucor renisporus, and repeated ITS sequencing are necessary to verify conspecifity.
The morphological distinction of f. racemosus and f. sphaerosporus of M. racemosus is ambiguous while they can be clearly differentiated based on ITS data. Some strains of f. racemosus such as CBS 271.86 and CBS 113.08 produce a high proportion of spherical sporangiospores.
Mucor racemosus f. chibinensis is grouped with M. racemosus f. racemosus and represents a synonym of the latter. To our knowledge no type material is available for f. chibinensis. Schipper (1976) concluded that CBS 636.67 and CBS 660.66 strains matched the protologue of Mucor chibinensis (Neophytova 1955) and proposed the name Mucor racemosus f. chibinensis. Here we designate CBS 636.67 (preserved in a metabolically inactive state by lyophilization, batch nr. 768) as neotype of Mucor racemosus f. chibinensis because it matches the description of the basionym M. chibinensis and it is well described and illustrated (Schipper 1976); in addition the strain was isolated in Russia, corresponding which the geographic origin of the type.
Mucor sinensis is considered to be conspecific with M. racemosus f. racemosus because it groups with the ex-neotype CBS 260.68 of the latter in the ITS tree. It appears to represent a morphological variant or a growth-reduced mutant rather than a separate species.
Mucor amphibiorum group and related taxa
The ITS sequence of the ex-type strain of the recently described Mucor ellipsoideus (FN650647, Álvarez et al. 2011) is identical to that of CBS 210.80, the ex-type strain of Mucor ardhlaengiktus, except of an A missing at the 3’ terminus probably due to low sequence quality (data not shown). The characteristic azygospores described in M. ardhlaengiktus (Mehrotra & Mehrotra 1978) appear to be a variable feature that was absent from the strain studied by Álvarez et al. (2011) and from strain CBS 650.78. Mucor ardhlaengiktus is the older name (Mehrotra & Mehrotra 1978) and we therefore consider M. ellipsoideus as a synonym. The varieties of Mucor laxorrhizus, var. laxorrhizus and var. ovalisporus, appear distantly positioned in the ITS (Fig. 6) and LSU (Fig. 1) trees and as a result we recognize them as separate species.
Mucor ardhlaengiktus B.S. Mehrotra & B.M. Mehrotra, Sydowia 31: 94. 1979 [1978]. — MycoBank MB317921
= Mucor ellipsoideus E. Álvarez, Stchigel, Cano, D.A. Sutton & Guarro, in Álvarez et al., Med. Mycol. 49: 67. 2011.
Lectotype. Fig. 1 (Mehrotra & Mehrotra, Sydowia 31: 95. 1979 [1978]), designated here.
Epitype. CBS 210.80 (preserved in a metabolically inactive state by lyophilization, batch nr. 968), designated here.
Specimens examined. CBS 210.80, CBS 528.73, CBS 650.78.
Mucor circinelloides f. janssenii (Lendn.) Schipper, Stud. Mycol. 12: 13. 1976. — MycoBank MB348491
Basionym. Mucor janssenii Lendn. (as ‘janseni’), Bull. Herb. Boissier 2, Sér. 7: 251. 1908.
≡ Mucor griseocyanus Hagem f. janssenii (Lendn.) Schipper, Antonie van Leeuwenhoek 36: 486. 1970.
≡ Circinomucor janssenii (Lendn.) Arx, Sydowia 35: 18.1982.
= Mucor tenellus Y. Ling, Rev. Gén. Bot. 42: 736. 1930.
≡ Circinella tenella (Y. Ling) Zycha, Krypt.-Fl. Brandenburg (Leipzig) 6a: 99. 1935.
= Mucor stagnalis Novot., Notul. Syst. Inst. Cryptog. Horti Bot. Petropol. 6: 158. 1950.
= Mucor kurssanovii Milko & Beliakova, Mikrobiologija 36: 118. 1967.
= Mucor velutinosus E. Álvarez, Stchigel, Cano, D.A. Sutton & Guarro, in Álvarez et al., Med. Mycol. 49: 64. 2011.
Neotype. CBS 205.68 (preserved in a metabolically inactive state by lyophilization, batch nr. 605), designated here.
Specimens examined. CBS 144.93, CBS 185.68, CBS 204.68, CBS 205.68, CBS 206.68, CBS 227.29, CBS 232.29, CBS 243.67, CBS 365.70, CBS 762.74.
Notes — Schipper (1970) described Mucor griseocyanus f. janssenii based on strain CBS 205.68 because no authentic material of M. janssenii existed. However, she did not designate CBS 205.68 as the neotype though it was considered as such in the following years. Here we designate strain CBS 205.68 (in a lyophilized state) as neotype of M. janssenii because it fits the description of this species morphologically and it is well described (Schipper 1970).
Mucor parviseptatus G. Walther & de Hoog, nom. nov. — MycoBank MB800447
≡ Mucor laxorrhizus Y. Ling var. ovalisporus Schipper, Stud. Mycol. 31: 154. 1989, non Mucor ovalisporus (G. Sm.) Pidopl. & Milko, Atlas Mukor. Grib. (Kiev): 61. 1971.
Etymology. The epithet parviseptatus refers to the few septa that are formed unconnected with branching of the sporangiophores.
Specimens examined. CBS 417.77 ex-type strain of Mucor laxorrhizus var. ovalisporus, CBS 522.79.
Mucor racemosus Fresen. f. racemosus Beitr. Mykol. 1: 12. 1850. — MycoBank MB427116
≡ Circinomucor racemosus (Fresen.) Arx, Sydowia 35: 18. 1982.
= Mucor racemosus f. brunneus Morini, Malpighia 10: 88. 1896.
= Mucor dimorphosporus Lendn., Mat. Fl. Crypt. Suisse 3, 1: 93. 1908.
= Mucor christianensis Hagem, Ann. Mycol. 8: 268. 1910.
= Mucor racemosus var. christianensis (Hagem) Naumov, Opred. Mukor. (Mucorales): 46. 1935.
= Mucor varians Povah, Bull. Torrey Bot. Club 44: 297. 1917.
= Mucor pispekii Naumov, Encycl. Mycol. 9: 47. 1939.
= Mucor chibinensis Neophyt., Notul. Syst. Inst. Cryptog. Horti Bot. Petropol. 10: 160. 1955.
= Mucor racemosus f. chibinensis (Neophyt.) Schipper, Stud. Mycol. 12: 24. 1976.
= Mucor oudemansii VÁňová, Česka Mykol. 45: 25. 1991.
= Mucor sinensis Milko & Beliakova, in Pidopl. & Milko, Atlas Mukor. Grib. (Kiev): 53. 1971.
Neotype. CBS 260.68 (preserved in a metabolically inactive state by lyo-philization, batch nr. 87.1018), designated here.
Specimens examined. CBS 113.08 ex-lectotype strain of Mucor dimorphosporus (lectotype: Fig. 34 (Lendner, Mat. Fl. Crypt. Suisse 3, 1: 93. 1908), designated here; epitype: CBS 113.08, preserved in a metabolically inactive state by lyophilization, batch nr. 479, designated here), CBS 204.74 ex-type strain of Mucor sinensis, CBS 222.81, CBS 260.68, CBS 271.86, CBS 369.71, CBS 616.63, CBS 636.67 ex-neotype strain of Mucor racemosus f. chibinensis, CBS 657.68, CBS 660.66, CBS 661.66, CBS 111557, CNM-CM 2569, CNM-CM 3862.
Notes — Strain CBS 260.68 was used by Schipper (1970) for the description of Mucor racemosus f. racemosus but not designated as neotype. Here we designate strain CBS 260.68 (in a lyophilized state) as neotype of this species because it is well characterized (Schipper 1970) and because its morphology fully complies with the original description.
Mucor silvaticus Hagem, Skr. Vidensk.-Selsk. Christiana, Math.-Naturvidensk. Kl. 7: 31. 1908. — MycoBank MB182519
≡ Mucor hiemalis f. silvaticus (Hagem) Schipper, Stud. Mycol. 4: 31. 1973.
Neotype. CBS 412.71 (preserved in a metabolically inactive state by lyophilization, batch nr. 853), designated here.
Specimens examined. CBS 249.35, CBS 412.71, CBS 509.66.
Notes — To our knowledge, authentic material of this species has been lost. Schipper (1973) studied five strains that matched the description of Hagem (CBS 249.35, CBS 250.35, CBS 508.66, CBS 509.66 and CBS 412.71). Schipper’s description and drawings are based on CBS 412.71. It was isolated in Denmark, while the remaining strains originated in Germany. Here we designate CBS 412.71 as the neotype of Mucor silvaticus, because it was well described and illustrated as M. hiemalis f. silvaticus in Schipper (1973). Compared to the strains available in international fungal culture collections, it was isolated closest to the locality of the type, Norway.
Mucor recurvus group and Backusella
The LSU phylogram (Fig. 1) resolved a clade (Mucor recurvus group) consisting of Mucor grandis, M. oblongiellipticus, M. oblongisporus, M. recurvus, M. tuberculisporus, M. variabilis, two potentially undescribed species as well as Backusella circina and B. lamprospora that exclusively unifies taxa with transitorily recurved sporangiophores (Fig. 8). In this clade the sporangiophores are curved during maturation of the sporangium and become upright afterwards. Schipper (1978a) recognised the Mucor portion of this group on morphological grounds. The type species of Backusella, B. circina (Ellis & Hesseltine 1969: 865) (Fig. 8a, b), as well as B. lamprospora (Benny & Benjamin 1975: 320) also have been described to form transitorily recurved sporangiophores.
The genus Backusella differs from Mucor only by the formation of sporangiola in addition to sporangia. However, sporangiola, though in low frequency, have also been described in Mucor recurvus var. indicus and in M. tuberculisporus (Schipper 1978a). In our opinion the clade in the LSU phylogram including Mucor and Backusella species represents a natural group characterised by transitorily recurved sporangiophores. Consequently, we transfer all Mucor species belonging to that clade to the genus Backusella. The two varieties of Mucor recurvus (var. recurvus and var. indicus) are located in different supported subclades of our ITS tree (Fig. 7) and likely represent separate species.
Fig. 7.
RAxML phylogram of the Mucor recurvus group and Backusella based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, ET = ex-epitype strain, LT = ex-lectotype strain, PT = ex-paratype strain. Ex-type strains of currently accepted taxa are printed in bold. Clones are specified by a ‘c’ followed by the clone number. Potentially undescribed taxa are indicated by bold blue font. Morphological identifications are given in quotation marks.
Sequence diversity is high in the emended genus Backusella. ITS sequences of M. oblongisporus CBS 569.70, M. oblongiellipticus CBS 568.70, and a contaminant strain of CBS 523.68 deviate significantly from the remaining members of the group and cannot be aligned with confidence. However, their LSU sequences and the formation of transitorily recurved sporangiophores clearly assign these taxa to the emended genus Backusella.
Backusella ctenidia is positioned inside the Mucor circinelloides complex (Fig. 1, 5) and does not belong in Backusella. For that reason we propose transferring it to Mucor.
We propose the following emendation for Backusella:
Backusella Ellis & Hesseltine emend. Walther et al.
Type species. Backusella circina J.J. Ellis & Hesselt.
Sporophores arising directly from the substrate mycelium, simple or sympodially branched, recurved when young, erect at maturity (transitorily recurved), smooth or roughened, producing terminal sporangia and in some species few to many lateral, pedicellate sporangiola. Terminal sporangia columellate, multispored, globose to subglobose, size ranging from 90 to 375 μm diam, nonapophysate, wall encrusted, deliquescent; columellae subglobose, ellipsoidal, slightly pyriform or conical, smooth. Collars small or consisting of needle-shaped spines. Sporangiolar pedicels straight, curved, or recurved, simple or branched, smooth or encrusted. Sporangiola columellate, multi- or unispored; wall verrucose or spinulose or both, persistent. Sporangiospores of sporangia and multispored sporangia identical, large, subglobose, ellipsoidal or irregularly polyhedral, smooth. Columellae, hyphae and sporangiospores in some species with yellowish or brownish content. Zygospores globose to subglobose; wall dark, opaque or translucent, ornamented with conical or rounded projections; suspensors opposed, smooth or roughened, equal or unequal.
Notes — The emended genus Backusella includes the following species: Backusella circina, B. grandis, B. indica, B. lamprospora, B. oblongielliptica, B. oblongispora, B. recurva, B. tuberculispora and B. variabilis.
Backusella grandis (Schipper & Samson) G. Walther & de Hoog, comb. nov. — MycoBank MB800453
Basionym. Mucor grandis Schipper & Samson, Mycotaxon 50: 479. 1994.
Specimen examined. CBS 186.87 ex-type strain of Mucor grandis.
Notes — The ITS sequence of Mucor grandis differs by only 6 basepairs from that of M. variabilis, while different clones of M. variabilis vary at four positions. The small sequence differences suggest conspecifity but the diameter of the sporangia varies significantly between the species. More detailed taxonomic studies are needed to clarify species limits.
Backusella indica (Baijal & B.S. Mehrotra) G. Walther & de Hoog, comb. nov. — MycoBank MB800449
Basionym. Mucor recurvus var. indicus Baijal & B.S. Mehrotra, Sydowia 19: 207. 1965.
Lectotype. CBS 786.70 (preserved in a metabolically inactive state by lyophilization, batch nr. 344), designated here.
Specimen examined. CBS 786.70.
Backusella oblongielliptica (H. Nagan., Hirahara & Seshita ex Pidopl. & Milko) G. Walther & de Hoog, comb. nov. — MycoBank MB800451
Basionym. Mucor oblongiellipticus H. Nagan., Hirahara & Seshita, Essays Stud. Fac. Hiroshima Jogakuin College 18: 167. 1969, nom. inval., Art. 36.1
≡ Mucor oblongiellipticus H. Nagan., Hirahara & Seshita ex Pidopl. & Milko, Atlas Mukor. Grib. (Kiev): 81. 1971.
Lectotype. CBS 568.70 (preserved in a metabolically inactive state by lyophilization, batch nr. 113), designated here.
Specimen examined. CBS 568.70.
Backusella oblongispora (Naumov) G. Walther & de Hoog, comb. nov. — MycoBank MB800452
Basionym. Mucor oblongisporus Naumov, Mater. Mykol. Fitopatol. Rossii 1(4): 12. 1915.
Neotype. CBS 569.70 (preserved in a metabolically inactive state by lyophilization, batch nr. 55), designated here.
Specimen examined. CBS 569.70.
Backusella recurva (E.E. Butler) G. Walther & de Hoog, comb. nov. — MycoBank MB800448; Fig. 8c
Basionym. Mucor recurvus E.E. Butler, Mycologia 44: 561. 1952.
Lectotype. Fig. 1 (Butler, Mycologia 44: 562. 1952).
Epitype. CBS 318.52 (preserved in a metabolically inactive state by lyophilization, batch nr. 717), designated here.
Specimens examined. CBS 196.71, CBS 317.52, CBS 318.52, CBS 673.75.
Backusella tuberculispora (Schipper) G. Walther & de Hoog, comb. nov. — MycoBank MB800450
Basionym. Mucor tuberculisporus Schipper, Stud. Mycol. 17: 23. 1978.
Lectotype. CBS 562.66 (preserved in a metabolically inactive state by lyophilization, batch nr. 88.1007), designated here.
Specimens examined. CBS 562.66, CBS 570.70.
Backusella variabilis (A.K. Sarbhoy) G. Walther & de Hoog, comb. nov. — MycoBank MB800454; Fig. 8d
Basionym. Mucor variabilis A.K. Sarbhoy, Trans. Brit. Mycol. Soc. 48: 559. 1965.
Lectotype. CBS 564.66 (preserved in a metabolically inactive state by lyophilization, batch nr. 22), designated here.
Specimen examined. CBS 564.66.
Mucor ctenidius (Durrell & M. Fleming) G. Walther & de Hoog, comb. nov. — MycoBank MB800455
Basionym. Thamnidium ctenidium Durrell & M. Fleming, Mycologia 58: 797. 1966.
≡ Backusella ctenidia (Durrell & M. Fleming) Pidopl. & Milko, Atlas Mukor. Grib. (Kiev): 85. 1971, ex Benny & R.K. Benj., Aliso 8: 325. 1975.
Specimens examined. CBS 293.66 ex-isotype strain of Thamnidium ctenidium, CBS 433.87, CBS 696.76.
Pilaira
Recently, Zheng & Liu (2009a) studied the genus Pilaira morphologically and reclassified P. caucasica as a variety of P. moreaui. The variety differed from var. moreaui in the size of sporangiophores and sporangiospores (Zheng & Liu 2009a). We found identical ITS sequences for both varieties supporting conspecifity (Fig. 3). We retain both varieties despite of identical ITS sequences because of the clear morphological distinction.
Zygorhynchus
Phenotypically Zygorhynchus and Mucor differ in the following features. First, species of Zygorhynchus are exclusively homothallic, while the majority of Mucor species is heterothallic (Watanabe 1994). Second, the suspensors of the zygospores are unequal in Zygorhynchus and equal in Mucor. Third, the two suspensors originate from the same hypha in Zygorhynchus, the ‘Zygorhynchus pattern’, while they arise from different hyphae in Mucor, the ‘Mucor pattern’ (Hesseltine et al. 1959, Schipper 1986). However, these differences are gradual (Schipper 1986). Zygorhynchus exponens may develop equal but Mucor plumbeus more or less unequal suspensors (Schipper 1986). In Zygorhynchus exponens (Hesseltine et al. 1959), Z. japonicus (Schipper 1986) and Z. moelleri (Green 1927) zygospores are also produced between different hyphae.
Based on our LSU (Fig. 1) and ITS (Fig. 3, 6) data, Zygorhynchus is polyphyletic. Our analyses indicate that unequal suspensors and the Zygorhynchus pattern of zygospore production do not represent synapomorphies in the genus Zygorhynchus, but appear to be convergent characters within Mucor. Therefore we recombine all Zygorhynchus species in Mucor.
The ex-type strains of Zygorhynchus moelleri and Z. californiensis have identical ITS sequences suggesting conspecifity. However, Z. californiensis has regularly globose spores, while the spores of Z. moelleri are oblong to ovoidal in shape, 2.0–3.3 × 3.0–6.5 μm (Hesseltine et al. 1959). For that reason we propose reclassifying Z. californiensis as a forma of Z. moelleri.
The two varieties described in Zygorhynchus exponens, var. exponens and var. smithii differ by only a single basepair in their ITS sequences. Also small morphological differences such as the lighter and browner sporangia and columellae in var. smithii, do not justify the maintenance of a separate variety and consequently we consider both varieties as synonymous.
Mucor exponens (Burgeff) G. Walther & de Hoog, comb. nov. — MycoBank MB800461
Basionym. Zygorhynchus exponens Burgeff, Bot. Abh. 4: 34. 1924.
= Zygorhynchus exponens Burgeff var. smithii Hesselt., C.R. Benj. & B.S. Mehrotra, Mycologia 51: 179. 1959.
Neotype. CBS 141.20 (preserved in a metabolically inactive state by lyophilization, batch nr. 563), designated here.
Specimens examined. CBS 141.20, CBS 404.58 ex-lectotype strain of Zygorhynchus exponens var. smithii (lectotype: CBS 404.58, preserved in a metabolically inactive state by lyophilization, batch nr. 40, designated here), CBS 508.48.
Mucor fusiformis G. Walther & de Hoog, nom. nov. — MycoBank MB800459
≡ Zygorhynchus psychrophilus Schipper & Hintikka, Antonie van Leeuwenhoek 35: 29. 1969, non Mucor psychrophilus Milko, in Pidopl. & Milko, Atlas Mukor. Grib. (Kiev): 73. 1971.
Etymology. The epithet refers to the shape of the sporangiospores.
Specimens examined. CBS 336.68 ex-type strain of Zygorhynchus psychrophilus.
Mucor heterogamus Vuill., Bull. Séanc. Soc. Sci. Nancy 8: 50. 1887. — MycoBank MB249261
≡ Zygorhynchus heterogamus (Vuill.) Vuill., Bull. Trimestriel. Soc. Mycol. France 19: 117. 1903.
Lectotype. Pl. II, f. 27-48 (Vuill., Bull. Séanc. Soc. Sci. Nancy 8. 1887), designated here.
Epitype. CBS 405.58 (preserved in a metabolically inactive state by lyophilization, batch nr. 658), designated here.
Specimens examined. CBS 252.85, CBS 338.74, CBS 405.58, CBS 580.83, CBS 594.83.
Notes — The original material of this species consists of slides labelled as “Mucor heterogamus P.V. Zygospores Mis de pain 17-3-86” (Hesseltine et al. 1959). Hesseltine et al. (1959) studied five strains: NRRL 1489, NRRL 1490, NRRL 1491, NRRL 1616 (= CBS 405.58) and a fresh isolate without an NRRL number designated as ‘No. 1957’ and compared these strains with the original material. The authors found remarkable intraspecific variation in colony appearance, but micromorphologically Vuillemin’s material was almost identical with their living cultures except for some differences in lengths of the zygospore projections. Therefore they considered the type material and their strains as conspecific. Here we designate CBS 405.58 (NRRL 1616, preserved in a lyophilized state) verified by Hesseltine et al. (1959) as epitype of Mucor heterogamus.
Isolates of M. heterogamus vary considerably in their ITS sequences (maximum dissimilarity of 10 %) and might represent a complex of several species. Isolates that were morphologically assigned to M. heterogamus form a well-supported group with Z. multiplex and Z. macrocarpus, but at distances to the designated ex-epitype strain of 10.8 % and 7.7 %, respectively. The precise definition of species boundaries awaits detailed multilocus DNA sequence-based analyses.
Mucor japonicus (Komin.) G. Walther & de Hoog, comb. nov. — MycoBank MB800458
Basionym. Zygorhynchus japonicus Komin., Mykol. Zentbl. 5: 3. 1915 (1914).
Neotype. CBS 154.69 (preserved in a metabolically inactive state by lyophilization, batch nr. 409), designated here.
Specimen examined. CBS 154.69.
Notes — The authentic strain of Zygorhynchus japonicus studied by Kominami (1915) has been lost (Schipper 1986). Strain CBS 154.69 (preserved in a lyophilized state) is selected as neotype of Z. japonicus because it resembles the original strain and it is well described and illustrated (Schipper 1986).
Mucor megalocarpus G. Walther & de Hoog, nom. nov. — MycoBank MB800456
≡ Zygorhynchus macrocarpus Y. Ling, Rev. Gén. Bot. 42: 150. 1930, non Mucor macrocarpus Corda, Icon. Fungorum 2: 21. 1838.
Lectotype. Fig. 1 (Ling, Rev. Gén. Bot. 42: 152. 1930), designated here.
Epitype. CBS 215.27 (preserved in a metabolically inactive state by lyophilization, batch nr. 748), designated here.
Specimen examined. CBS 215.27.
Mucor moelleri (Vuill.) Lendn. f. moelleri, Mat. Fl. Crypt. Suisse 3, 1: 72. 1908.
Basionym. Zygorhynchus moelleri Vuill., Bull. Trimestriel Soc. Mycol. France 19: 117. 1903.
= Zygorhynchus vuilleminii Namysl., Ann. Mycol. 8: 154. 1910.
= Zygorhynchus vuilleminii race agamus Namysl., Bull. Int. Acad. Sci. Cracovie, Cl. Sci. Math., Ser. B, Sci. Nat. 6: 479. 1911.
= Zygorhynchus dangeardii Moreau, Bull. Soc. Bot. France 59: 717. 1912.
= Mucor saximontensis Rall, Mycologia 57: 874. 1965.
Neotype. CBS 406.58 (preserved in a metabolically inactive state by lyophilization, batch nr. 656), designated here.
Specimens examined. CBS 216.27, CBS 380.29, CBS 406.58, CBS 444.65 ex-lectotype strain of Mucor saximontensis (lectotype: CBS 444.65, preserved in a metabolically inactive state by lyophilization, batch nr. 803, designated here), CBS 460.51, CBS 501.66, IHEM 21156.
Notes — No authentic material of this species is known to be preserved. Hesseltine et al. (1959) reported NRRL 2660 (= CBS 406.58) as the type of Z. moelleri but the strain studied by Vuillemin was isolated in Eberswalde (Germany) while NRRL 2660 originated from soil in Wisconsin (USA).
Mucor moelleri f. californiensis (Hesselt., C.R. Benj. & B.S. Mehrotra) G. Walther & de Hoog, comb. nov. — MycoBank MB800460
Basionym. Zygorhynchus californiensis Hesselt., C.R. Benj. & B.S. Mehrotra, Mycologia 51: 185. 1959.
Lectotype. Fig. 8–10 (Hesseltine, Benjamin & Mehrotra, Mycologia 51: 176. 1959), designated here.
Epitype. CBS 402.58 (preserved in a metabolically inactive state by lyophilization, batch nr. 90.0055), designated here.
Specimen examined. CBS 402.58.
Mucor multiplex (R.Y. Zheng) G. Walther & de Hoog, comb. nov. — MycoBank MB800457
Basionym. Zygorhynchus multiplex R.Y. Zheng, Mycotaxon 84: 370. 2002.
Specimen examined. CBS 110662 ex-type strain of Zygorhynchus multiplex.
Actinomucor
Currently there are three varieties in Actinomucor: A. elegans var. elegans, var. meitauzae (syn. A. taiwanensis, Zheng & Liu 2005) and var. kuwaitiensis (Khan et al. 2008). Characters distinguishing the varieties are shape, size and ornamentation of the sporangiospores (Zheng & Liu 2005, Khan et al. 2008). The var. meitauzae and var. kuwaitiensis show reduced growth on Czapek’s agar. In contrast to earlier reports (Jong & Yuan 1985), the maximum growth temperature does not discriminate the varieties (Zheng & Liu 2005, Khan et al. 2008). However, the relationships deduced from our ITS data (Fig. 9) contradict current taxonomic concepts. Strains with the characteristics of var. meitauzae are scattered over nearly all parts of the tree, and only a part of the strains belonging to var. elegans is included in a well-supported clade around the ex-type strain of var. elegans. A detailed taxonomic revision is required.
Fig. 9.
RAxML phylogram of the genus Actinomucor based on the ITS region. Branches with bootstrap values of 75 % or higher are printed in bold. Ex-type strains are designated by: T = ex-type strain, NT = ex-neotype strain. Ex-type strains are printed in bold. Clinical strains are highlighted by red strain and GenBank accession numbers.
Rhizopus
Based on our phylogenetic trees the genus Rhizopus is para-phyletic because Sporodiniella umbellata and Syzygites megalocarpus cluster among Rhizopus species. All currently accepted Rhizopus species are well recognizable in the ITS tree. However, three strains of Rhizopus stolonifer, CBS 126.83, CBS 442.74 and CBS 926.87, exhibit widely deviating ITS sequences, forming a separate group that may represent a new species. In agreement with our results, Vágvölgyi et al. (2004) found strains morphologically assigned to R. stolonifer with strongly deviating randomly amplified polymorphic DNA (RAPD) patterns and consequently the authors suspected an undescribed variety or even species. The varieties arrhizus and delemar of Rhizopus arrhizus are also recognized in the ITS tree, in accordance with Abe et al. (2007) and Gryganskyi (2010) who treated them as separate species. Strains identified morphologically as R. arrhizus var. tonkinensis by Zheng et al. (2007) do not form a separate cluster but are distributed in var. arrhizus and var. delemar clades. However, by using short tandem repeat motives of IGS rDNA sequences Liu et al. (2008) were able to characterize all three varieties of R. arrhizus.
The morphological varieties described in Rhizopus microsporus are not supported genetically; a single, supported clade includes strains representing the varieties microsporus, chinensis and oligosporus. ITS sequences of the remaining R. microsporus strains that had been assigned morphologically to the varieties azygosporus, chinensis, oligosporus, rhizopodiformis and tuberosus are all identical. The ITS identities imply in agreement with Liu et al. (2008), Abe et al. (2010) and Dolatabadi et al. (In press) that enlarged size and indistinct ornamentation of sporangiospores have no genetic basis in R. microsporus.
Circinella
The genus Circinella was erected by van Tieghem & le Monnier (1873) in order to accommodate strains differing from Mucor by circinate sporangiophore branches that terminate in globose sporangia with persistent walls (Hesseltine & Fennell 1955). Based on our LSU tree (Fig. 1) Circinella is polyphyletic, resolved in a well-supported group around the type species C. umbellata within the Lichtheimiaceae, and two separate species, C. simplex and C. rigida, positioned distantly within different clades of Mucor. As a consequence, we propose assigning C. rigida to Mucor. No type material is known to have been preserved of C. simplex. We studied five strains of this species but we only obtained good sequence data for CBS 428.80. More detailed taxonomic studies on the numerous strains of this species that are available in public collections are necessary to test its monophyly and to select a neotype. Excluding the unrelated species C. rigida and C. simplex, the genus Circinella is restricted to species that develop sporangiophores either with sterile spines or umbels with circinate branches.
The ex-type strain of Circinella lacrymispora clusters with the ex-type strain of Gongronella lacrispora in the Gongronella clade of the LSU phylogram (Fig. 1). Based on this finding Circinella lacrymispora should be reclassified in Gongronella. The LSU sequences of C. lacrymispora and G. lacrispora differ only in 3 basepairs and conspecifity cannot be excluded. We defer recombination until we obtain the ITS sequences of both taxa and until we perform a detailed morphological study.
Mucor durus G. Walther & de Hoog, nom. nov. — MycoBank MB800462
≡ Circinella rigida G. Sm., Trans. Brit. Mycol. Soc. 34: 19. 1951, non Mucor rigidus Léger, Rech. Struct. Mucor (Thèse, Paris): 71. 1895.
Lectotype. Pl. 2, Fig. 7–8 (Smith, Trans. Brit. Mycol. Soc. 34: 17–22. 1951), designated here.
Epitype. CBS 156.51 (preserved in a metabolically inactive state by lyophilization, batch nr. 389), designated here.
Etymology. Named after the rigid wall of the sporangium.
Specimens examined. CBS 156.51, CBS 484.66.
Notes — The species differs markedly from other Mucor species by the extremely rigid sporangial walls, the often curved branches of the sporangiophores, the common formation of subsporangial septa and the frequent presence of distinct apophyses (Fig. 2h).
Rhizomucor
All non-thermophilic Rhizomucor species, namely Rm. chlamydosporus, Rm. endophyticus, Rm. regularior and Rm. variabilis belong to Mucor based on our LSU tree (Fig. 1). Rhizomucor regularior and Rm. variabilis have recently been reclassified: Rm. variabilis has been renamed as Mucor irregularis, whereas Rm. regularior has been synonymised with M. circinelloides (Álvarez et al. 2011). Our ITS data (Fig. 3) indicate that Rhizomucor endophyticus represents a discrete species closely related to Mucor luteus. The ITS sequence of the ex-type strain of Rm. chlamydosporus (GenBank EF583634) is identical to that of Mucor indicus (data not shown); the morphological description of the species (Zheng & Liu 2009b) fully matches with that of M. indicus.
Mucor hiemalis and M. luteus develop distinct rhizoids when they grow over glass slides (Fig. 2c). These findings demonstrate that not only Rhizomucor, but also Mucor species have the ability to produce rhizoids, at least under certain conditions. Consequently, this feature should not be used as sole criterion for the distinction of Mucor and Rhizomucor.
After removal of the above species, Rhizomucor with its type species Rhizomucor parasiticus (which is a synonym of Rm. pusillus) is monophyletic. The genus is restricted to thermophilic species with predominantly subglobose spores, as was recognized previously by Schipper (1978b) applying only phenotypic characters. Currently four thermophilic Rhizomucor species are accepted: Rm. miehei, Rm. nainitalensis, Rm. pakistanicus and Rm. pusillus. Rhizomucor tauricus is considered to be conspecific with Rm. pusillus because their ITS sequences are identical (Fig. 11). Analysis of its carbon source utilization, isoenzyme patterns and PCR-coupled RFLP of the ITS suggested that Rm. tauricus represented a heterothallic mutant strain of Rm. pusillus (Vágvölgyi et al. 1999).
Rhizomucor nainitalensis forms sporangiospores of different shapes, varying from subglobose to irregularly shaped (Joshi 1982). The sporangiospores of Rhizomucor pakistanicus are globose or ovoidal (Mirza et al. 1979). The following description of Rhizomucor is slightly modified from Schipper (1978b: 53):
Rhizomucor Lucet & Costantin (1900)
Thermophilic; sporangiophores originating from aerial mycelium, either from short aerial hyphae or from distinct stolons, both with simple or weakly branched rhizoids; sporangiophores branched, each branch bearing a multispored terminal sporangium; sporangia borne in an upright position, globose, dark (coloured), distinctly columellate, non-apophysate; sporangiospores consistently or partly subglobose; zygospores globose, covered with blunt projections, and formed in the aerial mycelium between non-ornamented, isogamous opposed suspensors.
Mucor endophyticus (R.Y. Zheng & H. Jiang) J. Pawłowska & G. Walther, comb. nov. — MycoBank MB800463
Basionym. Rhizomucor endophyticus R.Y. Zheng & H. Jiang, Mycotaxon 56: 456. 1995.
Specimens examined. CBS 385.95 ex-type strain of Rhizomucor endophyticus.
Mucor indicus Lendn., Bull. Soc. Bot. Genève, Ser. 2, 21: 258. 1930. — MycoBank MB267842
≡ Zygorhynchus indicus (Lendn.) Arx, Sydowia 35: 16. 1982.
= Rhizomucor chlamydosporus R.Y. Zheng, X.Y. Liu & R.Y. Li, Sydowia 61: 142. 2009.
Lectotype. Fig. 1–3 (Lendner, Bull. Soc. Bot. Genève, Ser. 2, 21: 258–260. 1930), designated here.
Epitype. CBS 226.29 (preserved in a metabolically inactive state by lyophilization, batch nr. 679), designated here.
Specimens examined. CBS 120.08, CBS 226.29, CBS 414.77, CBS 422.71, CBS 535.80, CBS 671.79, CBS 120585, CBS 123974.
Rhizomucor pusillus (Lindt) Schipper, Stud. Mycol. 17: 54. 1978. — MycoBank MB322484
Basionym. Mucor pusillus Lindt, Arch. Exp. Path. Pharmacol. 21: 272. 1886.
= Mucor septatus Bezold, Schimmelmyc. Menschl. Ohres: 97. 1889.
≡ Rhizomucor septatus (Bezold) Lucet & Costantin, Archs Parasitol. 4: 362. 1901.
= Mucor parasiticus Lucet & Costatin, Compt. Rend. Hebd. Séances Acad. Sci. 129: 1033. 1899.
≡ Rhizomucor parasiticus (Lucet & Costantin) Lucet & Costantin, Rev. Gén. Bot. 12: 81. 1900.
≡ Rhizopus parasiticus (Lucet & Costantin) Lendn., Mat. Fl. Crypt. Suisse 3: 115. 1908.
= Mucor buntingii Lendn., Bull. Soc. Bot. Genève, Ser. 2, 21: 260. 1930.
= Mucor tauricus Milko & Schkur., Novosti Sist. Nizsh. Rast. 7: 139. 1970.
≡ Rhizomucor tauricus (Milko & Schkur.) Schipper, Stud. Mycol. 17: 62. 1978.
≡ Rhizomucor pusillus var. tauricus (Milko & Schkur.) R.Y. Zheng, X.Y. Liu & R.Y. Li, Sydowia 61: 144. 2009.
Lectotype. Pl. II.III, Fig. 1–6 (Lindt, Arch. Exp. Path. Pharmacol. 21: 269–298. 1886), designated here.
Epitype. CBS 354.68 (preserved in a metabolically inactive state by lyophilization, batch nr. 85.0901), designated here.
Specimens examined. CBS 179.69 ex-lectotype strain of Rhizomucor tauricus (lectotype: CBS 179.69, preserved in a metabolically inactive state by lyophilization, batch nr. 87.3168, designated here), CBS 219.31, CBS 354.68, CBS 425.78, CBS 120586, CBS 120587, CNM-CM 2752, CNM-CM 2935, CNM-CM 2974, CNM-CM 4727, CNM-CM 5124.
Umbelopsis
Our ITS dataset is incomplete for Umbelopsis because of the high proportion of strains that needed to be cloned. For that reason, we refrain from taxonomic changes. The ex-type strains of Umbelopsis dimorpha and Umbelopsis nana possess identical ITS sequences and it is likely that these species are conspecific. The ITS sequences of the ex-type strains of U. swartii and U. westeae, as well as those of U. gibberosa and U. ramanniana are very similar necessitating a critical revision of Umbelopsis taxonomy.
Acknowledgments
We are grateful to Willem van Boekel for excellent technical assistance. We thank Maria Vehreschild of Fungiscope for providing several clinical isolates. We also express our thanks to Kerstin Voigt, Paul Kirk and Scott Redhead for critically reading of the manuscript and useful advice. The reviewers Gerald Benny and Kerry O’Donnell are thanked for their helpful corrections and suggestions.
REFERENCES
- Abe A, Asano K, Sone T.2010. A molecular phylogeny-based taxonomy of the genus Rhizopus. Bioscience, Biotechnology and Biochemistry 74: 1325–1331 [DOI] [PubMed] [Google Scholar]
- Abe A, Oda Y, Asano K, Sone T.2006. The molecular phylogeny of the genus Rhizopus based on rDNA sequences. Bioscience, Biotechnology and Biochemistry 70: 2387–2393 [DOI] [PubMed] [Google Scholar]
- Abe A, Oda Y, Asano K, Sone T.2007. Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia 99: 714–722 [DOI] [PubMed] [Google Scholar]
- Alastruey-Izquierdo A, Hoffmann K, Hoog GS de, Rodriguez-Tudela JL, Voigt K, et al. 2010. Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Mycocladus, Absidia pp.). Journal of Clinical Microbiology 48: 2154–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez E, Cano J, Stchigel AM, Sutton DA, Fothergill AW, et al. 2011. Two new species of Mucor from clinical samples. Medical Mycology 49: 62–72 [DOI] [PubMed] [Google Scholar]
- Álvarez E, Garcia-Hermoso D, Sutton DA, Cano JF, Stchigel AM, et al. 2010a. Molecular phylogeny and proposal of two new species of the emerging pathogenic fungus Saksenaea. Journal of Clinical Microbiology 48: 4410–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez E, Stchigel AM, Cano J, Sutton DA, Fothergill AW, et al. 2010b. Molecular phylogenetic diversity of the emerging mucoralean fungus Apophysomyces: Proposal of three new species. Revista Iberoamericana de Micología 27: 80–89 [DOI] [PubMed] [Google Scholar]
- Bainier M.1884. Nouvelles observations sur les zygospores des Mucorinées. Annales des Sciences Naturelles, Botanique 19: 200–214 [Google Scholar]
- Balajee SA, Borman AM, Brandt ME, Cano J, Cuenca-Estrella M, et al. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: Where are we and where should we go from here? Journal of Clinical Microbiology 47: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benny GL, Benjamin RK.1975. Observations on Thamnidiaceae (Mucorales). New taxa, new combination, and notes on selected species. Aliso 8: 301–351 [Google Scholar]
- Bidartondo MI, Bruns T, Blackwell M, Edwards I, Taylor AFS, et al. 2008. Preserving accuracy in GenBank. Science 21: 1616. [DOI] [PubMed] [Google Scholar]
- Bridge PD, Roberts PJ, Spooner BM, Pancha G.2003. On the unreliability of published DNA sequences. New Phytologist 160: 43–48 [DOI] [PubMed] [Google Scholar]
- Budziszewska J, Piątkowska J, Wrzosek M.2010. Taxonomic position of Mucor hiemalis f. luteus. Mycotaxon 111: 75–85 [Google Scholar]
- Calmette LCA.1892. Contribution à l’étude des ferments de l’amidon; la levûre chinoise. Annales de l’Institut Pasteur 6: 604–620 [Google Scholar]
- Chakrabarti A, Marak RSK, Shivaprakash MR, Gupta S, Garg R, et al. 2010. Cavitary pulmonary zygomycosis caused by Rhizopus homothallicus. Journal of Clinical Microbiology 48: 1965–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatabadi S, Walther G, Hoog GS de. In press. Diversity and delimitation of Rhizopus microsporus. Fungal diversity. [Google Scholar]
- Ellis JJ, Hesseltine CW.1969. Two new members of the Mucorales. Mycologia 61: 863–872 [Google Scholar]
- Ellis JJ, Rhodes LJ, Hesseltine CW.1976. The genus Amylomyces. Mycologia 68: 131–143 [Google Scholar]
- Foos KM, May NL, Beach DL, Pomper M, Sheehan KB, Ruch DG.2011. Phylogeny of Pilobolaceae. Mycologia 103: 36–44 [DOI] [PubMed] [Google Scholar]
- García Rodríguez J, Quiles I, Humala K, Monzon A, Cuenca-Estrella M.2012. Isolation of Cunninghamella blakesleeana in an immunodepressed patient. Mycoses 55: 463–465 [DOI] [PubMed] [Google Scholar]
- Gomes MZR, Lewis RE, Kontoyiannis DP.2011. Mucormycosis caused by unusual Mucormycetes, non-Rhizopus, -Mucor, and -Lichtheimia species. Clinical Microbiology Reviews 24: 411–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E.1927. The life-history of Zygorhynchus moelleri Vuill. Annals of Botany 41: 419–435 [Google Scholar]
- Greenberg RN, Scott LJ, Vaughn HH, Ribes JA.2004. Zygomycosis (mucormycosis): emerging clinical importance and new treatments. Current Opinion in Infectious Diseases 17: 517–525 [DOI] [PubMed] [Google Scholar]
- Greuter W, McNeill J, Barrie FR, Burdet H-M, Demoulin V, et al. (eds) 2000. International Code of Botanical Nomenclature (St Louis Code). Regnum Vegetabile 138. Koeltz Scientific Books, Königstein [Google Scholar]
- Gryganskyi AP, Lee SC, Litvintseva AP, Smith ME, Bonito G, et al. 2010. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLoS ONE 5: e15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermet A, Méheust D, Mounier J, Barbier G, Jany JL.2012. Molecular systematics within the genus Mucor with special regard to species encountered in cheese. Fungal Biology 116: 692–705 [DOI] [PubMed] [Google Scholar]
- Hesseltine CW.1983. Microbiology of oriental fermented foods. Annual Review of Microbiology 37: 575–606 [DOI] [PubMed] [Google Scholar]
- Hesseltine CW, Benjamin CR, Mehrotra BS.1959. The genus Zygorhynchus. Mycologia 51: 173–194 [Google Scholar]
- Hesseltine CW, Fennel DI.1955. The genus Circinella. Mycologia 47: 193–212 [Google Scholar]
- Hoffmann K, Discher S, Voigt K.2007. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycological Research 111: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Pawłowska J, Walther G, Wrzosek M, Hoog GS de, et al. 2013. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30: 57–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Voigt K.2009. Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biology 11: 537–554 [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG.1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41: 183–189 [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Gené F, Figueras MJ.2000. Atlas of clinical fungi, 2nd ed Reus, Utrecht [Google Scholar]
- Hospenthal DR, Chung KK, Lairet K, Thompson EH, Guarro J, et al. 2011. Saksenaea erythrospora infection following combat trauma. Journal of Clinical Microbiology 49: 3707–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs K, Botha A.2008. Mucor renisporus sp. nov., a new coprophilous species from Southern Africa. Fungal Diversity 29: 27–35 [Google Scholar]
- Jong SC, Yuan GF.1985. Actinomucor taiwanensis sp. nov., for manufacture of fermented soybean food. Mycotaxon 23: 261–264 [Google Scholar]
- Joshi MC.1982. A new species of Rhizomucor from India. Sydowia 35: 100–103 [Google Scholar]
- Khan ZU, Ahmad S, Brazda A, Chandy R.2009. Mucor circinelloides as a cause of invasive maxillofacial zygomycosis: an emerging dimorphic pathogen with reduced susceptibility to posaconazole. Journal of Clinical Microbiology 47: 1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan ZU, Ahmad S, Mokaddas E, Chandy R, Cano J, Guarro J.2008. Actinomucor elegans var. kuwaitiensis isolated from the wound of a diabetic patient. Antonie van Leeuwenhoek 94: 343–352 [DOI] [PubMed] [Google Scholar]
- Kominami K.1915. Zygorhynchus japonicus, une nouvelle Mucorinée hétérogame, isolée du sol du Japon. Mycologisches Centralblatt 5: 1–4 [Google Scholar]
- Kwaśna H, Nirenberg HI.2008a. Siepmannia, a new genus in the Mucoraceae. Mycologia 100: 259–274 [DOI] [PubMed] [Google Scholar]
- Kwaśna H, Nirenberg HI.2008b. Validation of the genus Siepmannia (Mucoraceae) and its four species. Polish Botanical Journal 53: 187–188 [Google Scholar]
- Kwaśna H, Ward E, Bateman GL.2006. Phylogenetic relationships among Zygomycetes from soil based on ITS1/2 rDNA sequences. Mycological Research 110: 501–510 [DOI] [PubMed] [Google Scholar]
- Lemmert K, Losert H, Rickerts V, Just-Nübling G, Sander A, et al. 2002. Identification of Cunninghamella spec. by molecular methods. Mycoses 45, Suppl. 1: 31–36 [DOI] [PubMed] [Google Scholar]
- Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, et al. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathogens 7: e1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Lackner M, Hoog GS de, Gerrits van de Ende AHG, Priha O, et al. 2011. Assessment of identity of filamentous fungi colonizing from water-damaged building materials. Sydowia 63: 49–66 [Google Scholar]
- Lindner DL, Banik MT.2011. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia 103: 731–740 [DOI] [PubMed] [Google Scholar]
- Liu XY, Huang H, Zheng RY.2001. Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon 80: 77–95 [Google Scholar]
- Liu XY, Huang H, Zheng RY.2007. Molecular phylogenetic relationships within Rhizopus based on combined analyses of ITS rDNA and pyrG gene sequences. Sydowia 59: 235–253 [Google Scholar]
- Liu XY, Huang H, Zheng RY.2008. Delimitation of Rhizopus varieties based on IGS rDNA. Sydowia 60: 93–112 [Google Scholar]
- Lucet A, Costantin J.1900. Rhizomucor parasiticus, espèce pathogéne de l’homme. Revue Générale Botanique 12: 81–98 [Google Scholar]
- Madden AA, Stchigel AM, Guarro J, Sutton DA, Starks PT.2011. Mucor nidicola sp. nov., a novel fungal species isolated from an invasive paper wasp nest. International Journal of Systematic and Evolutionary Microbiology10.1099/ijs.0.033050–0. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Mehrotra BS, Mehrotra BM.1978. Another azygosporic species of Mucor from India. Sydowia 31: 94–96 [Google Scholar]
- Meyer CP, Paulay G.2005. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biology 3: e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W, Gams W.2003. Delimitation of Umbelopsis (Mucorales, Umbelopsidaceae fam. nov.) based on ITS sequence and RFLP data. Mycological Research 107: 339–350 [DOI] [PubMed] [Google Scholar]
- Mirza JH, Khan SM, Begum S, Shagufta S.1979. Mucorales of Pakistan. University of Agriculture, Faisalabad, Pakistan [Google Scholar]
- Möller EM, Bahnweg G, Sandermann H, Geiger HH.1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Research 22: 6115–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytova VK.1955. Novye vidy gribov iz sfagnovogo torfãnika. (Fungorum species novae e palude sphagnosa.) Notulae Systematicae E Sectione Cryptogamica Instituti Botanici 10: 159–162 [Google Scholar]
- Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson K-H, Kõljalg U.2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nout MJR, Aidoo KE.2010. Asian fungal fermented food. In: Hofrichter M.(ed), Industrial applications. The Mycota, Vol. 10, ed 2: 30–58 Springer, Berlin, Heidelberg [Google Scholar]
- O’Donnell K.1993. Fusarium and its near relatives. In: Reynolds DR, Taylor JW. (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233 CAB International, Wallingford, UK [Google Scholar]
- O’Donnell K, Cigelnik E.1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Lutzoni FM, Ward TJ, Benny GL.2001. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia 93: 286–296 [Google Scholar]
- Pawłowska J, Walther G, Wilk M, Hoog GS de, Wrzosek M. In press. The use of compensatory base changes analysis of ITS2 as a tool in phylogeny of Mucorales illustrated by ‘Mucor circinelloides’ group. Organisms Diversity & Evolution. [Google Scholar]
- Pitt JI, Hocking AD.2009. Fungi and food spoilage. 3rd ed, Springer, New York [Google Scholar]
- Povah AHW.1917. A critical study of certain species of Mucor. Bulletin of the Torrey Botanical Club 44: 287–310 [Google Scholar]
- Rambaut A.2002. Se–Al. http://tree.bio.ed.ac.uk/software/seal/ [Google Scholar]
- Ryan LJ, Ferrieri P, Powell RD, Paddock CD, Zaki SR, Pambuccian SE.2011. Fatal Cokeromyces recurvatus pneumonia: report of a case highlighting the potential for histopathologic misdiagnosis as Coccidioides. International Journal of Surgical Pathology 19: 373–376 [DOI] [PubMed] [Google Scholar]
- Sabouraud R.1910. Maladies du cuir chevelu. III. – Les maladies cryptogamiques. Les teignes. Masson & Cie, Paris [Google Scholar]
- Schell WA, O’Donnell K, Alspaugh JA.2011. Heterothallic mating in Mucor irregularis and first isolate of the species outside of Asia. Medical Mycology 49: 714–723 [DOI] [PubMed] [Google Scholar]
- Schipper MAA.1970. Two species of Mucor with oval- and spherical-spored strains. Antonie van Leeuwenhoek 36: 475–488 [DOI] [PubMed] [Google Scholar]
- Schipper MAA.1973. A study on variability in Mucor hiemalis and related species. Studies in Mycology 4: 1–40 [Google Scholar]
- Schipper MAA.1975. On Mucor mucedo, Mucor flavus and related species. Studies in Mycology 10: 1–33 [Google Scholar]
- Schipper MAA.1976. On Mucor circinelloides, Mucor racemosus and related species. Studies in Mycology 12: 1–40 [Google Scholar]
- Schipper MAA.1978a. On certain species of Mucor with a key to all accepted species. Studies in Mycology 17: 1–52 [Google Scholar]
- Schipper MAA.1978b. On the genera Rhizomucor and Parasitella. Studies in Mycology 17: 53–71 [Google Scholar]
- Schipper MAA.1979. Thermomucor (Mucorales). Antonie van Leeuwenhoek 45: 275–280 [DOI] [PubMed] [Google Scholar]
- Schipper MAA.1984. A revision of the genus Rhizopus. 1. The Rhizopus stolonifer-group and Rhizopus oryzae. Studies in Mycology 25: 1–19 [Google Scholar]
- Schipper MAA.1986. Notes on Zygorhynchus species. Persoonia 13: 97–105 [Google Scholar]
- Schipper MAA.1990. Notes on Mucorales – 1. Observations on Absidia. Persoonia 14: 133–148 [Google Scholar]
- Schipper MAA, Samson RA.1994. Miscellaneous notes on Mucoraceae. Mycotaxon 50: 475–491 [Google Scholar]
- Schipper MAA, Stalpers JA.1984. A revision of the genus Rhizopus. II. The Rhizopus microsporus-group. Studies in Mycology 25: 20–34 [Google Scholar]
- Schipper MAA, Stalpers JA.2003. Zygomycetes: the order Mucorales. In: Howard DH. (ed), Pathogenic fungi in human and animals: 67–125 Marcel Dekker, Inc., USA [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. 2012. The nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences USA 109: 6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz P, Bretagne S, Gantier JC, Garcia-Hermoso D, Lortholary O, et al. 2006. Molecular identification of zygomycetes from culture and experimentally infected tissue. Journal of Clinical Microbiology 44: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiada A, Pagano L, Groll A, Zimmerli S, Dupont B, et al. 2011. Zygomycosis in Europe: analysis of 230 cases accrued by the registry of the European Confederation of Medical Mycology (ECMM) Working Group on Zygomycosis between 2005 and 2007. Clinical Microbiology and Infection 17: 1859–1867 [DOI] [PubMed] [Google Scholar]
- de Souza JI, Pires-Zottarelli CLA, dos Santos JF, Costa JP, Harakava R.2012. Isomucor (Mucoromycotina): a new genus from a Cerrado reserve in state of São Paulo, Brazil. Mycologia 104: 232–241 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.2008. A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology 75: 758–771 [DOI] [PubMed] [Google Scholar]
- Sugui JA, Christensen JA, Bennett JE, Zelazny AM, Kwon-Chung KJ.2011. Hematogenously disseminated skin disease cured by Mucor velutinosus in a patient with acute myeloid leukemia. Journal of Clinical Microbiology 49: 2728–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32 [DOI] [PubMed] [Google Scholar]
- Tieghem P van, Monnier G le.1873. Recherches sur les Mucorinées. Annales des Sciences Naturelles, Botanique 17: 261–399 [Google Scholar]
- Vágvölgyi C, Heinrich H, Ács K, Papp T.2004. Genetic variability in the species Rhizopus stolonifer, assessed by random amplified polymorphic DNA analysis. Antonie van Leeuwenhoek 86: 181–188 [DOI] [PubMed] [Google Scholar]
- Vágvölgyi C, Vastag M, Ács K, Papp T.1999. Rhizomucor tauricus: a questionable species of the genus. Mycological Research 103: 1318–1322 [Google Scholar]
- Vilgalys R, Hester M.1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale RG, Hoog GS de, Schwarz P, Dannaoui E, Deng S, et al. 2012. Antifungal susceptibility and phylogeny of opportunistic members of Mucorales. Journal of Clinical Microbiology 50: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Wöstemeyer J.2001. Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1α genes. Gene 270: 113–120 [DOI] [PubMed] [Google Scholar]
- Watanabe T.1994. Two new species of homothallic Mucor in Japan. Mycologia 86: 691–695 [Google Scholar]
- Weddle G, Gandy K, Bratcher D, Pahud B, Jackson MA.2012. Apophysomyces trapeziformis infection associated with a tornado related injury. The Pediatric Infectious Disease Journal 31: 640–642 [DOI] [PubMed] [Google Scholar]
- Wehmer C.1900. Studien über technische Pilze. VII. Die ‘Chinesische Hefe’ und der sogenannte Amylomyces (= Mucor Rouxii). Centralblatt für Bakteriologie, Parasitenkunde und Infektionskrankheiten, Abt. II, 6: 353–365 [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, Inc., New York, USA [Google Scholar]
- Xess I, Mohapatra S, Shivaprakash MR, Chakrabarti A, Benny GL, et al. 2012. Evidence implicating Thamnostylum lucknowense as an etiological agent of rhino-orbital mucormycosis. Journal of Clinical Microbiology 50: 1491–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng RY, Chen GQ.2001. A monograph of Cunninghamella. Mycotaxon 80: 1–75 [Google Scholar]
- Zheng RY, Chen GQ, Huang H, Liu XY.2007. A monograph of Rhizopus. Sydowia 59: 273–372 [Google Scholar]
- Zheng RY, Liu XY.2005. Actinomucor elegans var. meitauzae, the correct name for A. taiwanensis and Mucor meitauzae (Mucorales, Zygomycota). Nova Hedwigia 80: 419–432 [Google Scholar]
- Zheng RY, Liu XY.2009a. Taxa of Pilaira (Mucorales, Zygomycota) from China. Nova Hedwigia 88: 255–267 [Google Scholar]
- Zheng RY, Liu XY.2009b. More Rhizomucor causing human mucormycosis from China: R. chlamydosporus sp. nov. Sydowia 61: 135–147 [Google Scholar]