Abstract
The Mucorales (Mucoromycotina) are one of the most ancient groups of fungi comprising ubiquitous, mostly saprotrophic organisms. The first comprehensive molecular studies 11 yr ago revealed the traditional classification scheme, mainly based on morphology, as highly artificial. Since then only single clades have been investigated in detail but a robust classification of the higher levels based on DNA data has not been published yet. Therefore we provide a classification based on a phylogenetic analysis of four molecular markers including the large and the small subunit of the ribosomal DNA, the partial actin gene and the partial gene for the translation elongation factor 1-alpha. The dataset comprises 201 isolates in 103 species and represents about one half of the currently accepted species in this order. Previous family concepts are reviewed and the family structure inferred from the multilocus phylogeny is introduced and discussed. Main differences between the current classification and preceding concepts affects the existing families Lichtheimiaceae and Cunninghamellaceae, as well as the genera Backusella and Lentamyces which recently obtained the status of families along with the Rhizopodaceae comprising Rhizopus, Sporodiniella and Syzygites. Compensatory base change analyses in the Lichtheimiaceae confirmed the lower level classification of Lichtheimia and Rhizomucor while genera such as Circinella or Syncephalastrum completely lacked compensatory base changes.
Keywords: Mucorales, families, phylogeny
INTRODUCTION
The fungal order Mucorales – evolutionary position and characterisation
As a member of the Mucoromycotina, the Mucorales belong to the early diverging, ancient fungi along with the Kickxellomycotina, Zoopagomycotina, Entomophthoromycotina, Mortiellomycotina, Chytridiomycota, Neocallimastigomycota, Blastocladiomycota, and Cryptomycota and Microsporidia with the latter two still highly discussed (Schüßler et al. 2001, James et al. 2006, Hibbett et al. 2007, Hoffmann et al. 2011, Jones et al. 2011a, b, Benny 2012). The Entomophthoromycotina were later elevated to the phylum Entomophthoromycota (Humber 2012).
Mucorales are characterised by a usually abundant, rapidly growing mycelium as well as anamorph structures usually formed in large quantities. The mycelium is typically unseptate or irregularly septate. Anamorphic sporangiospores are produced in multi-spored sporangia, few-spored sporangiola or merosporangia. Chlamydospores, arthrospores and yeast cells are, in most species, rarely formed. Sporangia are characterised by the inclusion of a variously shaped columella. This well-developed columella counts as a synapomorphic character for the Mucorales. Conjugation in homothallic species or between compatible mating types of heterothallic species results in the formation of zygospores. Zygospores often display a specific exospore ornamentation (smooth, rough, warty) and protecting appendages (finger-like, antler-like) born on the supporting cells (suspensors) (Zycha et al. 1969). Some species of the Mucorales exhibit dimorphism, possessing the ability to switch between a filamentous, multi-cellular state to a yeast-like state (Bartnicki-Garcia & Nickerson 1962).
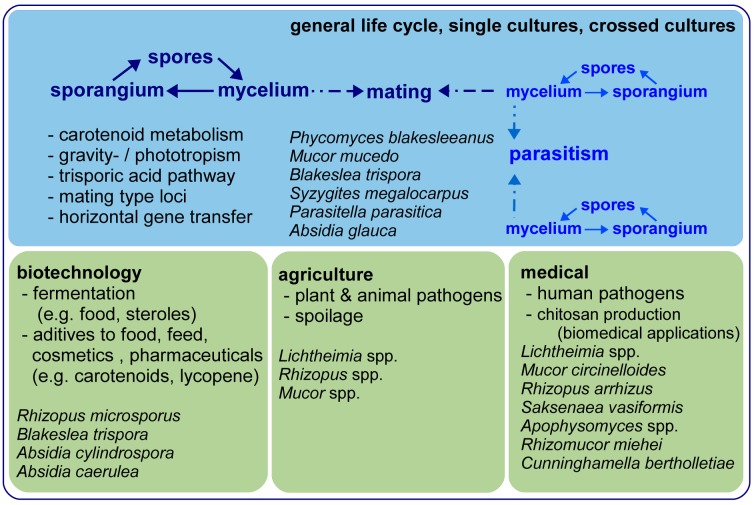
Life styles and applications — Fig. 1
Fig. 1.
General life cycle, important fields of scientific research and main applications of Mucorales. Exemplary and prominent species are given.
Mucoralean fungi are ubiquitous, predominantly saprobic soil organisms on decaying organic material but parasites of plants, fungi and animals also are known. As one of the largest orders in the basal Fungi, the Mucorales is also one of the most studied groups in the early diverging fungi. These studies on mucoralean fungi encompass physiology and biochemistry, as well as taxonomy and systematics, and potential applications in industry. In general, mucoralean fungi reproduce anamorphically via non-motile sporangiospores released from variously shaped sporangia. If not homothallic, a compatible mating partner is needed for the formation of the zygospore, where meiosis occurs. The different sexual modality of either homo- or heterothally in the Mucorales was discovered more than 100 yr ago, with most species found to be heterothallic (Blakeslee 1904). Volatiles are responsible for the formation of sexual reproductive structures (Burgeff 1924). These volatiles were identified as trisporoids, derivatives of beta-carotene (van den Ende 1967, Gooday 1968). The trisporic acid precursors are mutually processed by the compatible mating partners, resulting in the formation of a mature zygospore (Werkman 1976). Although the composition of the compounds is species specific to allow only intra-species matings (Sutter et al. 1989), inter-species zygospores are also described with some impact on systematics (Blakeslee & Cartledge 1927, Stalpers & Schipper 1980). Combining an order-wide trisporoid profiling with the current knowledge on phylogenetic relationships would most likely reveal the ‘languages’ of the different clades and their potentials for interspecific mutual recognition. But currently, only profiles for few species are known: e.g. Phycomyces blakesleeanus (Miller & Sutter 1984) and Blakeslea trispora (Caglioti et al. 1966).
Although a general trisporic acid biosynthesis pathway (Schachtschabel et al. 2005) is widely accepted, the genetic background is resolved only in parts. The synthesis and degradation of beta-carotene is well studied and understood (Almeida & Cerdá-Olmedo 2008, Polaino et al. 2010, Tagua et al. 2012) but most enzymes responsible for trisporic acid production remain undiscovered. So far, only 4-dihydro-methyltrisporate dehydrogenase and 4-dihydrotrisporin dehydrogenase are verified (Czempinski et al. 1996, Wetzel et al. 2009).
Since an interaction of compatible mating types is essential for matured zygospores to be produced, the information for the mating type is probably genetically coded. The appropriate regions were identified first in Phycomyces blakesleeanus (Idnurm et al. 2008) and subsequently discovered in Rhizopus delemar, R. oryzae (Gryganskyi et al. 2010), Mucor circinelloides (Lee et al. 2010) and even in a homothallic species, Syzygites megalocarpus (Idnurm 2011). Although heterothallic strains possess only one gene coding for either plus or minus mating type, the phenomenon of rare switches between mating types (Schipper & Stalpers 1980) is not yet explained.
The importance of zygospores for reproduction and distribution compared to the asexual sporangiospores is still unknown, since germination in the natural habitat could not be observed and germination under laboratory conditions has only been described and illustrated for few species (Michailides & Spotts 1988, Yu & Ko 1997). Nevertheless, zygomycetes are reported from the fossil records. The earliest zygomycotan fossil known, exclusive of the Glomeromycota, may be Jimwhitea circumtecta, possible Endogonaceae, from the middle Triassic (Krings et al. 2012). Many fossil zygomycetes have been found in the Carboniferous and later, including Protoascon missouriensis and others (Taylor et al. 2005, Kar et al. 2010). Calculations of the diverging time of zygomycetes using molecular data suggest an origin of around 600 mya (Berbee & Taylor 2001).
The zygomycetes are known to be useful for a variety of different applications, including food and food additive production and food preservation. Zygomycetes are used as starter cultures for the fermentation of soybean- or rice-based products in Asia, Africa and South America, e.g. beverages, or the well-known tempeh (Henkel 2004, 2005, Hesseltine 1983, 1991, Nout & Kiers 2005, Tamang & Thapa 2006).
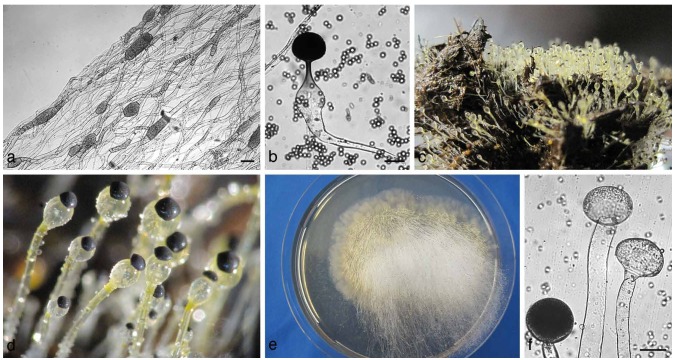
Mucorales also are used for diverse biological transformations (Gładkowski et al. 2004, 2011) as well as the production of additives for food, feed, pharmaceuticals (like lycopene) or various applications of chitosan (reviewed by Shahidi et al. 1999), a cell wall component only known to be produced by Mucorales. Yet, Mucorales also are reported as spoilage agents in stored cereals and other food, especially fruits and vegetables (Martin 1964, Wade & Morris 1982, Ray & Ravi 2005). In addition, some organisms also infect living plants, especially the fruits (e.g., strawberry, yellow summer squash or green beans; Fig. 2c, d) (Dennis 1983). Thus, these fungi play an important role as plant pathogens as well (Shtienberg 1997). Furthermore, some species of the Mucorales are facultative parasites of other fungi. They can be biotrophic or necrotrophic parasites with a few species (Syzygites megalocarpus, Dicranophora fulva, Spinellus fusiger) able to infect the fruit body of agarics (Fig. 2a; Zycha et al. 1969), a feature that is thus far not well studied. However, well studied is the biotrophic fusion parasitism (Fig. 2b) between Absidia glauca and Parasitella parasitica, a model system for studies of horizontal gene transfer and the link between sexual and parasitic interactions (Burgeff 1924, Kellner et al. 1993, Schultze et al. 2005). Trisporic acid and its precursors are also believed to be responsible for recognition of potential hosts for Chaetocladium (another parasite) and Parasitella, which was assumed from an observed mating-type dependent infection (Burgeff 1924, Schultze et al. 2005). Yet, a strict mating-type dependency was rejected as early as 1926 by a mere tendency which, in addition, seems to be restricted to only few species (Satina & Blakeslee 1926). Currently, an order-wide comprehensive survey of host-ranges for all known biotrophic fusion parasites is lacking. A recent investigation revealed an unstudied mycoparasite, Lentamyces parricida, as the most basal with the highest mycoparasitic potential to infect other mucoralean hosts (Hoffmann & Voigt 2009).
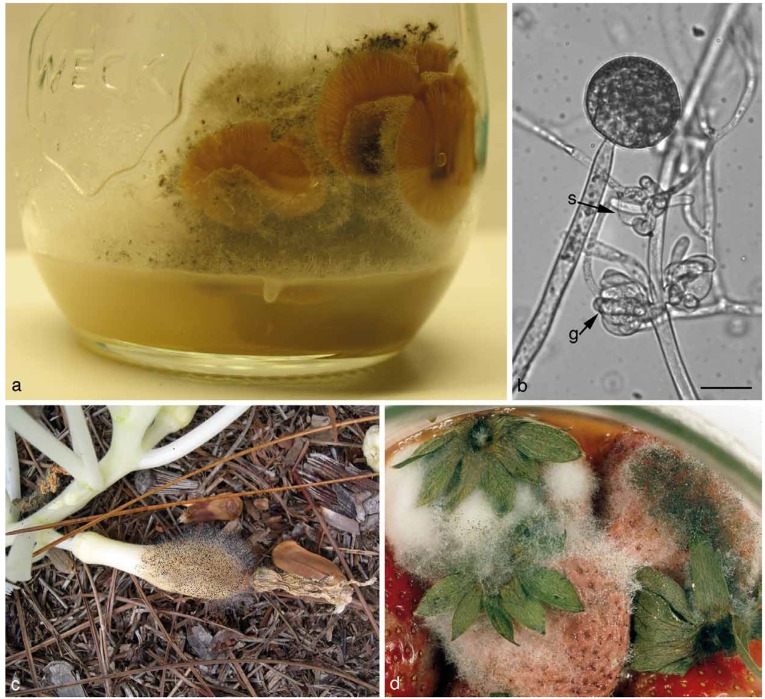
Fig. 2.
a. Syzygites megalocarpus on Pleurotus ostreatus (artificially infected); b. Parasitella parasitica on Mucor circinelloides. Galls (g) and sikyotic cells (s) are marked; c. Choanephora cucurbitarum on yellow summer squash. d. Rhizopus stolonifer on strawberries. — Scale bar = 20 μm.
Mucoralean fungi are also known as human and animal pathogens. Mucor corymbifer (currently Lichtheimia corymbifera) was first reported as causative agent of mycosis in a rabbit (Platauf 1885). In the last decades, the reported number of infections caused by members of the Mucorales (mucormycoses) has constantly increased. This is probably due to a rising awarness, an improved identification by the use of molecular methods, as well as a permanent worldwide increase of risk factors such as immunosupression, malignancies and diabetes (Roden et al. 2005, Skiada et al. 2011).
The symptoms of infections by Mucorales remain unspecific for a long time, making a diagnosis extremely difficult. A fast, proper and effective therapy is required, since these infections can result in death within hours to a few days. Survival rates for mucormycosis are highly dependent on the location of the infection, but they are very low overall at 53 % (Skiada et al. 2011). The large and still increasing numbers of studies pertaining to the susceptibility of Mucorales to known and new fungicides indicate a pressing need for an effective therapy. And with the discovery of species-specific susceptibility profiles, it became obvious, that the causative agents should be identified correctly to species level (e.g. Vitale et al. 2012). To investigate and to understand mucormycoses, their susceptibility and their evolutionary relationships need to be comparatively investigated. Understanding evolutionary relationships will elucidate approaches to improve existing or to invent new applications in industry, agriculture or medicine.
Morphology-based families
Traditionally, Mucorales were classified using their observable characters, for example physiology, biochemistry and, especially, morphology (Table 1). Unfortunately, Mucorales display only a small number of distinguishable morphological characters and only a few of them have proven to be useful for distinction between species, genera and families.
Table 1.
Morphological featured observable in mucoralean fungi.
| Feature/character | Criteria |
|---|---|
| Mycelium | Height, colour, rhizoids, arthrospores, chlamydospores, giant cells |
| Sporangia incl. sporangiophore | Height, origin, branching pattern, size, shape, colour, number of spores, septation, dissolving of the wall, release of spores, response to light |
| Sporangiospores | Shape, size, ornamentation, colour, appendages |
| Zygospores | Homo-/heterothallism, air-borne or submerged, relative placement and size of suspensors, shape, size, colour, ornamentation, appendages |
Nevertheless, in early mucoralean systematics, clustering of morphologically similar species resulted in well-defined genera and families accepted before the implementation of molecular data in phylogenetic reconstruction (Table 2).
Table 2.
Morphology based family structure of the Mucorales adopted from Zycha et al. 1969, Hesseltine & Ellis 1973, Benjamin 1979, Benny 1982, von Arx 1982.
| Family | Main characteristics |
|---|---|
| Chaetocladiaceae | Unispored sporangiola formed on fertile vesicles, discoid columella, dichotomous branched fertile hyphae, sterile spines, chlamydospores absent, zygospores rough-walled, suspensors opposed |
| Choanephoraceae | Sporangia and sporangiola, on different sporangiophores, zygospores striate, suspensors apposed or tongs-like |
| Cunninghamellaceae | (Fig. 3a), unispored sporangiola, sporangia absent, zygospores warty, suspensors opposed |
| Gilbertellaceae | Sporangiospores appendaged; zygospores rough-walled, suspensors opposed |
| Mucoraceae | Sporangia present, specialized sporangiola absent, zygospores smooth to warty, variously shaped suspensors: opposed, naked, appendaged, polyphyletic |
| Mycotyphaceae | Sporangiola on pedicels (Fig. 4k) |
| Phycomycetaceae | Sporangiophores large and unbranched, zygospores with coiled tongs-like suspensors and branched appendages |
| Pilobolaceae | Spores are actively liberated, zygospores smooth, suspensors tongs-like or apposed (Fig. 5) |
| Radiomycetaceae | Sporangia absent, sporophores with a primary vesicle bearing secondary ampullae, sporangiola originating from ampullae, zygospores smooth, suspensors apposed, appendaged |
| Saksenaeaceae | Provisionally classified together with Lobosporangium (currently Mortierellomycotina) because of the unusual-shaped sporangia |
| Syncephalastraceae | Merosporangia, zygospores warty, suspensors opposed (Fig. 4g) |
| Thamnidiaceae | Sporangiola present, sporangia absent or apically on the sporangiophores, zygospores warty, suspenors opposed (Fig. 4n, o) |
Delimitations of morphology
Traditional approaches used to classify fungi – fossil records, biochemistry and, especially, morphology (e.g., Paterson & Bridge 1994, Benny 1995, Hawksworth et al. 1995) became less important following the emergence of molecular systematics (White et al. 1990). Applying molecular data to phylogenetic analyses has led to the breakdown of the former phylum Zygomycota, combined by the morphological feature ‘zygospore’ into the subphyla Mucoromycotina, Kickxellomycotina, Zoopagomycotina and Entomophthoromycotina (James et al. 2006, Hibbett et al. 2007).
The family structure of the Mucorales is still rather unstable, but with the discovery of new, potentially phylogenetic informative characters (molecular data) and with the availability of higher resolution microscopy (e.g., fluorescence, SEM, TEM) it becomes feasible to reveal smaller, presumably monophyletic clades.
The most significant changes have affected the Thamnidiaceae, Mucoraceae, Chaetocladiceae and Absidiaceae. The first molecular studies addressing the entire order (O’Donnell et al. 2001, Voigt & Wöstemeyer 2001) showed that species traditionally assigned to Thamnidiaceae and Mucoraceae were scattered over the entire order. A widely accepted classification predominantly based on morphological traits was published by Benny et al. (2001) and is summarised with the molecular studies in Table 3.
Table 3.
Summary of the family structure of the Mucorales based predominantly on morphology (Benny et al. 2001) as well as on combination with molecular data (O’Donnell et al. 2001, Voigt & Wöstemeyer 2001).
| Family | Genera |
|---|---|
| Chaetocladiaceae | Chaetocladium, Dichotomocladium |
| Choanephoraceae | Blakeslea, Choanephora, Poitrasia |
| Cunninghamellaceae | Cunninghamella |
| Gilbertellaceae | Gilbertella |
| Mortierellaceae | Aquamortierella, Dissophora, Echinosporangium, Modicella, Mortierella, Umbelopsis |
| Mucoraceae | Absidia, Actinomucor, Apophysomyces, Chlamydoabsidia, Circinella, Circinomucor, Dicranophora, Gongronella, Halteromyces, Hyphomucor, Micromucor, Mucor, Mycocladus, Parasitella, Protomycocladus, Rhizomucor, Rhizopodopsis, Rhizopus, Spinellus, Sporodiniella, Syzygites, Thermomucor, Zygorhynchus |
| Mycotyphaceae | Benjaminiella, Mycotypha |
| Phycomycetaceae | Phycomyces |
| Pilobolaceae | Pilaira, Pilobolus, Utharomyces |
| Radiomycetaceae | Hesseltinella, Radiomyces |
| Saksenaeaceae | Saksenaea |
| Syncephalstraceae | Syncephalastrum |
| Thamnidiaceae | Backusella, Cokeromyces, Ellisomyces, Fennellomyces, Helicostylum, Kirkomyces, Phascolomyces, Pirella, Thamnidium, Thamnostylum, Zychaea |
Over the following years, several species and genera were studied in more detail, re-evaluated and revised (for a complete list see Walther et al. 2013 in this issue of Persoonia). In the following only studies that influenced family concepts by the dissection of the genus, the exclusion of a genus from a family or the fusion of families are addressed.
Absidia, Lichtheimia and Lentamyces
The genus Absidia was originally defined by its pyriform, apophysate sporangia (Fig. 3b, c, 4h–j). The first phylogenetic analyses (O’Donnell et al. 2001, Voigt & Wöstemeyer 2001) revealed a paraphyletic origin of this genus, a separation was accomplished later. Mesophilic species were retained in the genus Absidia (Cunninghamellaceae), whereas thermotolerant species form a separate phylogenetic clade as genus Lichtheimia (Hoffmann et al. 2007, 2009). In addition to the thermotolerant species separated from Absidia, potential mycoparasitic species were also distinguished in a new genus, Lentamyces (Hoffmann & Voigt 2009). This genus harbours two species, L. parricida and L. zychae. At the same time, two new species were isolated from nature and described as Siepmannia lariceti and S. pineti (Kwaśna & Nirenberg 2008a, b). This genus also was supposed to include both species of Lentamyces. Since molecular data for Siepmannia includes only ITS sequences, with no living material accessible, the relationship between the two genera remains unclear.
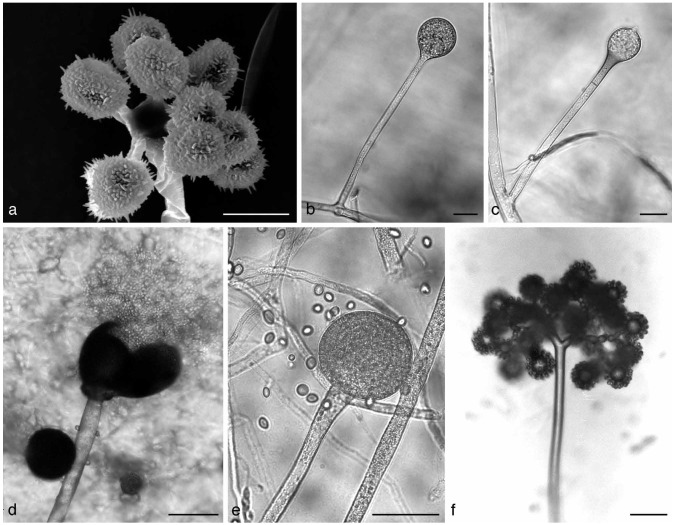
Fig. 3.
a. Cunninghamella sp. Sporangiophore with apical vesicle and sporangiola on stalks; b. apophysate sporangia of Absidia sp.; c. columella of Absidia sp. with typical apical projection and subsporangial septae; d, e. sporangium of Gilbertella persicaria: d. ruptured sporangial wall and released spores; f. branching sporangiophore of Blakeslea trispora with apical vesicles bearing few spored sporangiola. — Scale bars: a = 5 μm; b, c = 20 μm; d–f = 50 μm.
Fig. 4.
a. Umbelopsidaceae. Sporangium and sporangiophore with the highly reduced columella (arrow). — b, c. Lentamycetaceae. b. Warty zygospore, species are homothallic; c. sporangium. — d–f. Dichotomocladium. d. Zygospores; e, f. dichotomous branched sporangiophores. — g. Syncephalastrum racemosum, merosporangia. — h–j. Lichtheimia. h. Columella; i, j. apophysate sporangia. — k. Mycotypha sp., cylindrical vesicle covered with sporangiola. — l, m. Chaetocladium sp., branched fertile head with sporangiola. Branches often terminate in sterile spines. — n, o. Thamnidium elegans. n. Dichotomous branched sporangiophores with sporangiola; o. main multi-spored sporangia. — p, q. Columella and sporangia borne on circinate sporangiophores of Circinella sp. — Scale bars: all = 20 μm.
Choanephora and Gilbertella
Although there are morphological differences in zygosporogenesis in the Gilbertellaceae and the Choanephoraceae, a molecular study combined with ultrastructure supported merging these two families, under the older name, Choanephoraceae (Voigt & Olsson 2008).
Pilaira
Due to morphological similarities, this genus was placed traditionally within the Pilobolaceae together with Pilobolus and Utharomyces. But molecular data (O’Donnell et al. 2001, Voigt & Wöstemeyer 2001) revealed a non-relationship of Pilaira to both other genera, followed by an assignment to the Mucoraceae as published in Index Fungorum. This classification was also suggested on the base of a comprehensive molecular study of the Pilobolaceae (Foos et al. 2011).
Molecular systematics and implications on Mucorales
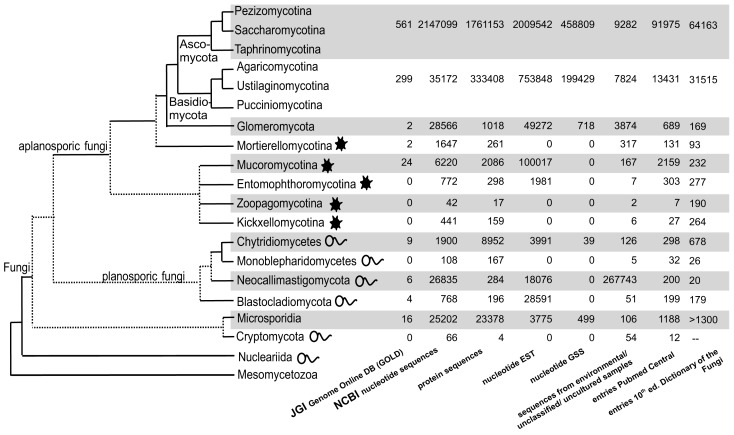
Molecular systematics is rapidly developing. Taxon samplings, possibilities to combine data and the number of applicable analytical tools are constantly increasing. In addition, with the ability to sequence whole genomes at relatively moderately cost combined with appropriate annotation software, computing capability and open access, genome-wide phylogeny comes within reach (Fitzpatrick et al. 2006, Kuramae et al. 2006, Huerta-Cepas et al. 2008). However, as only a few mucoralean fungi are fully sequenced, elucidating the phylogenetic relationships within this order is usually based on single genes or the combination of a few genes. Currently (April 2012), 24 genome/transcriptome projects for Mucorales are listed in the JGI Genome Online Database (GOLD; Fig. 6), but this includes only four different taxa (Mucor circinelloides, Rhizopus oryzae, Rhizopus stolonifer (each one project), and Phycomyces blakesleeanus (21 projects).
Fig. 6.
Schematic fungal tree and important data about the fungal groups. The topology resembles the current understanding of the relationships of the fungal groups according to Hibbett et al. (2007), James et al. (2006) and Schoch et al. (2012) (data retrieved April 2012).
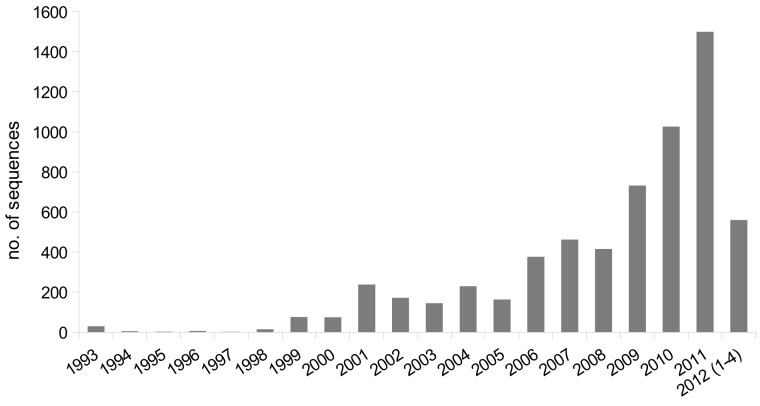
There are currently more than 6 000 sequences of zygomycota deposited in GenBank, approximately one-third of these are protein coding sequences. This is the third largest fraction for basal fungi, but still far behind the derived fungi, the Dikarya (Ascomycota and Basidiomycota; Fig. 6). Molecular data for the Mucorales have been submitted to GenBank since 1993, with a constantly increasing number, reaching more than 1 000 sequences in 2010 and more then 1 400 last year (Fig. 7). Nevertheless, the submitted sequences are restricted to only a few genera and species, with half of the sequences from the two genera Mucor and Rhizopus (Table 4). Around 50 species for Mucor and nine species for Rhizopus are listed in the 10th edition of the Dictionary of the Fungi (Kirk et al. 2008) which is 24 % and 4 %, respectively, of all species accepted in the Mucorales.
Fig. 7.
Chronology of sequences submitted to GenBank since 1993 for the Mucorales (data retrieved April 2012).
Table 4.
Sequences available at GenBank (April 2012) for mucoralean genera.
| Genus | No. of seq. | Genus | No. of seq. | Genus | No. of seq. |
|---|---|---|---|---|---|
| Absidia | 184 | Gongronella | 31 | Rhizomucor | 200 |
| Actinomucor | 68 | Halteromyces | 5 | Rhizopus | 1928 |
| Ambomucor | 3 | Helicostylum | 20 | Saksenaea | 50 |
| Amylomyces | 163 | Hesseltinella | 4 | Siepmannia | 2 |
| Apophysomyces | 69 | Hyphomucor | 4 | Spinellus | 7 |
| Backusella | 6 | Kirkomyces | 4 | Sporodiniella | 4 |
| Benjaminiella | 12 | Lentamyces | 23 | Syncephalastrum | 64 |
| Blakeslea | 93 | Lichtheimia | 679 | Syzygites | 26 |
| Chaetocladium | 25 | Mucor | 1501 | Thamnidium | 11 |
| Chlamydoabsidia | 6 | Mycotypha | 11 | Thamnostylum | 14 |
| Choanephora | 27 | Parasitella | 17 | Thermomucor | 6 |
| Circinella | 6 | Phascolomyces | 6 | Umbelopsis | 243 |
| Cokeromyces | 16 | Phycomyces | 110 | Utharomyces | 20 |
| Cunninghamella | 141 | Pilaira | 59 | Zychaea | 4 |
| Dichotomocladium | 29 | Pilobolus | 149 | ||
| Dicranophora | 4 | Pirella | 5 | ||
| Ellisomyces | 8 | Poitrasia | 11 | environmental/ | |
| Fennellomyces | 10 | Protomycocladus | 4 | uncultured/ | |
| Gilbertella | 16 | Radiomyces | 8 | unclassified | 108 |
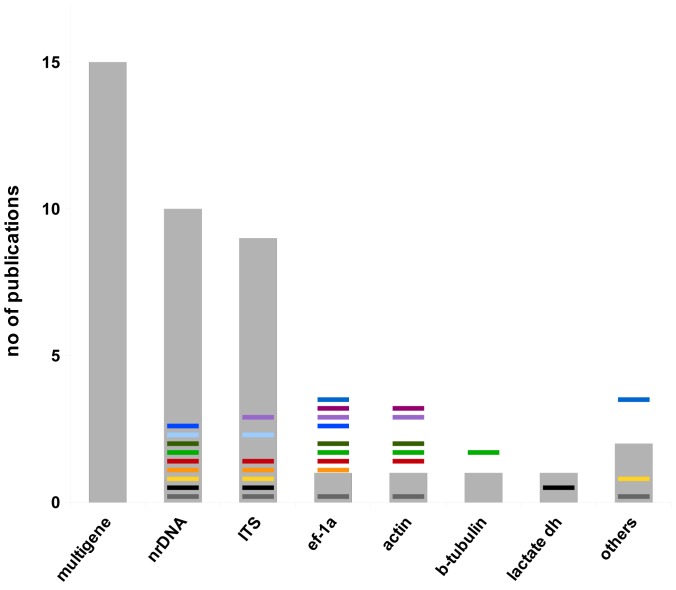
Studies predominately concerned with molecular phylogenetic as-pects of zygomycetes, especially Mucorales, are still relatively rare. Searching NCBI and the ISI Web of Science with ‘zygomycetes or Mucorales AND phylogeny’ resulted only in between 40 and 50 analyses including 15 studies where at least 2 loci were applied (April 2012, Fig. 8).
Fig. 8.
Number of publications predominantly focused on mucoralean phylogeny retrieved from NCBI and ISI Web of Science by searching ‘Zygomycota/ Mucorales AND phylogeny’. Publications are separated by the molecular marker applied for phylogeny. Nearly half of all published studies included more than one molecular marker. Published combinations of molecular markers are indicated by different colours (data retrieved April 2012).
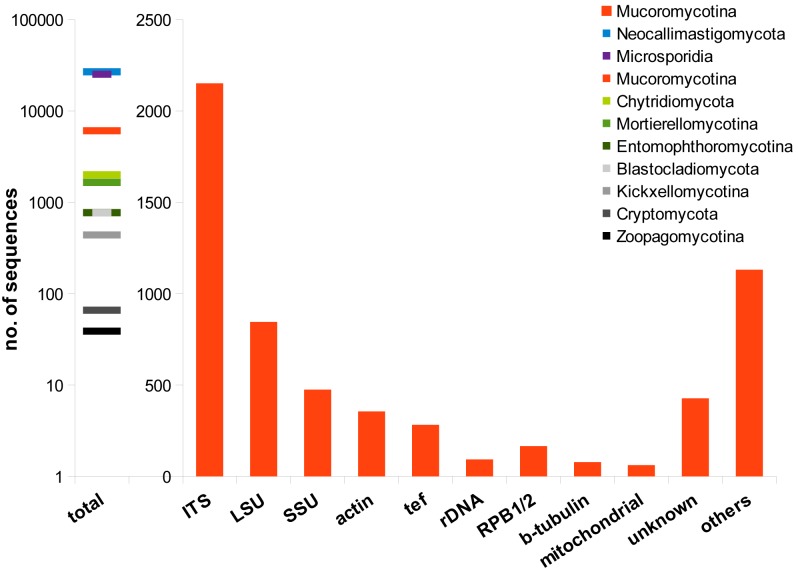
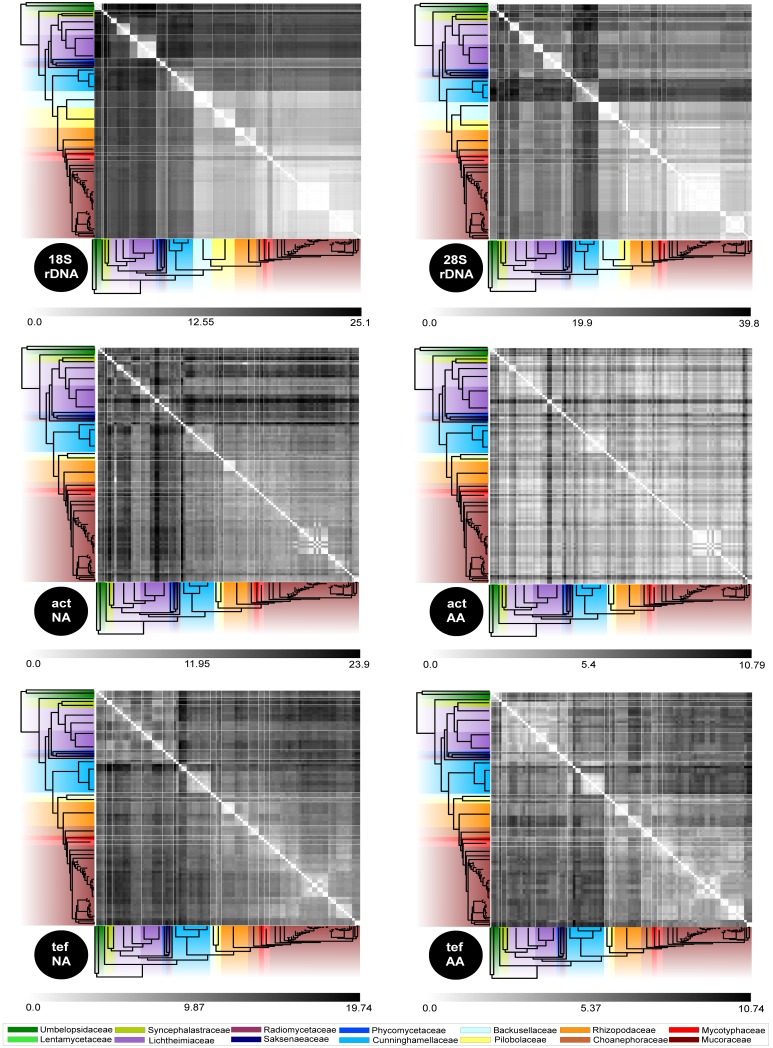
Commonly applied markers for phylogeny are sequences coding for rDNA (especially 18S rDNA for relationship levels of families, orders and above-order as well as ITS1 & 2 for relationships of species and genera). Therefore, the majority of studies are using rDNA sequences for phylogenetic approaches although ITS sequences represent the largest fraction of sequences in GenBank (Fig. 9). Protein coding genes predominantely applied so far are actin and translation elongation factor 1-alpha. Establishment of alternative protein coding markers for the whole order remains difficult. Whereas largest and second-largest subunit of RNA polymerase II (RPB1 & 2), ATPase subunit 6 (ATP6), a DNA replication licensing factor (MCM7), a gene required for rRNA accumulation (TSR1) or cytochrome c oxidase I (COX1) proved to be suitable for other fungal groups mostly belonging to the Basidiomycota and Ascomycota (Matheny et al. 2002, Reeb et al. 2004, Seifert et al. 2007, Schmitt et al. 2009), these genes have not be successfully amplified for a broad range of Mucorales and are still under represented in GenBank (Schoch et al. 2012).
Fig. 9.
Distribution of available sequences in GenBank for the Mucoromycotina. Also the total number of available sequences for all basal fungal linages are given (data retrieved April 2012).
The present study focuses on the family structure of the Mucorales. Family boundaries are inferred from a molecular phylogeny based on four markers and including 201 isolates and all currently accepted genera. Historical approaches and changes in recent years are revised, the support of the families by the current data is discussed and the families are characterised morphologically and ecologically. The resulting changes on the higher level nomenclature of the Mucorales were already briefly introduced by Voigt (2012). In order to ensure that these changes were based on a stable lower level taxonomy the internal transcribed spacer 2 region (ITS2) was analysed for of compensatory base changes (CBCs) as indicators for species boundaries (Müller et al. 2007).
MATERIAL AND METHODS
Strains, DNA isolation, PCR
Strains used for the generation of additional sequences (bold accession numbers in Table 5) were cultivated on 3 % malt extract medium at room temperature. Genomic DNA was extracted as described in Hoffmann et al. (2007). For phylogenetic analyses, sequences of large (LSU) and small (SSU) subunit of ribosomal DNA, ITS (internal transcribed spacer 1 & 2, incl. 5.8 SrDNA), actin (act) and translation elongation factor 1-alpha (tef) were either generated in this study or retrieved from GenBank (www.ncbi.nlm.nih.gov/; Table 5). Primers used for the amplification of LSU were NL1 and NL4 (O’Donnell 1993), NS1 and NS4 for SSU (White et al. 1990), ITS1 and ITS4 for ITS (White et al. 1990), Act1/Act1b and Act4R/Act4Ra for actin (Voigt & Wöstemeyer 2000) and MEF1 and MEF4/UEF4 for Tef (O’Donnell et al. 2001). PCR fragments were purified using the protocol of Vogelstein & Gillespie (1979) and sequenced on an Applied Biosystems 3730xL DNA Analyzer (ABI, Carlsbad) according to the manufacturer’s instructions.
Table 5.
Taxa and sequences used for the phylogenetic analyses. GenBank accession numbers in bold are generated within this study.
| Internal no. | Species | Isolate | 18S rDNA | 28S rDNA | Act | Tef |
|---|---|---|---|---|---|---|
| Ascomycota | ||||||
| KV5 | Archaeorhizomyces finlayi | Ny10 | JF836020 | JF836022 | na | JF836025 |
| P248 | Saccharomyces bayanus | CBS380 | X97777 | AF113892 | na | na |
| Basidiomycota | ||||||
| P249 | Agaricus bisporus | AFTOL448 | AY787216 | AY635775 | na | na |
| Blastocladiomycota | ||||||
| P251 | Blastocladiella emersonii | AFTOL302 | AY635842 | DQ273808 | na | na |
| Chytridiomycota | ||||||
| P250 | Batrachochytrium dendrobatidis | AFTOL21 | AH009052 | NG_027619 | na | na |
| Eccrinales | ||||||
| KV1 | Enterobryus sp. | AY336711 | AY336693 | na | na | |
| KV2 | Enteromyces callianassae | CA12c8 | AY336702 | AY336696 | na | na |
| KV4 | Palavascia patagonica | ARGD1c15 | AY682845 | AY336695 | na | na |
| KV3 | Taeniellopsis sp. | MA5C17 | AY336704 | AY336697 | na | na |
| Endogonales | ||||||
| P011 | Endogone pisiformis | AFTOL539 | DQ322628 | DQ273811 | AB609182 | DQ282618 |
| Entomophthoromycotina | ||||||
| P006 | Basidiobolus ranarum | AFTOL301 | AY635841 | DQ273807 | na | DQ282610 |
| P021 | Batkoa major | ARSEF2936 | EF392559 | EF392401 | na | na |
| P017 | Conidiobolus coronatus | NRRL28638 | NG_017182 | NG_027617 | HM117709 | na |
| P024 | Entomophaga maimaiga | ARSEF1400 | EF392556 | EF392395 | na | na |
| P025 | Entomophthora muscae | ARSEF3074 | NG_017183 | NG_027647 | na | na |
| P026 | Erynia radicans | ATCC60281 | JQ014018 | JN939182 | na | na |
| P027 | Eryniopsis caroliniana | ARSEF640 | EF392552 | EF392387 | na | na |
| P030 | Massospora cicadina | ARSEF374 | EF392548 | EF392377 | na | na |
| P033 | Pandora neoaphidis | ARSEF3240/ARSEF835 | EF392560 | EF392405 | na | na |
| P007 | Schizangiella serpentis | ARSEF203 | AF368523 | EF392428 | na | na |
| P037 | Zoophthora radicans | ARSEF4784/ARSEF6003 | EF392561 | EF392406 | na | na |
| Glomeromycota | ||||||
| P253 | Glomus intraradices | AFTOL845 | DQ322630 | DQ273828 | na | na |
| Kickxellomycotina | ||||||
| P053 | Austrosmittium biforme | 32-1-9/ 32-1-8 | DQ367462 | DQ367494 | na | na |
| P056 | Bojamyces repens | ME-JL-2 | DQ367447 | DQ367478 | na | na |
| P057 | Capniomyces stellatus | mis-21-127 | EF396191 | EF396194 | na | na |
| P088 | Coemansia reversa | NRRL1564 | NG_017186 | NG_027615 | AB609183 | DQ282615 |
| P091 | Dipsacomyces acuminosporus | NRRL2925 | AF007534 | AF031065 | na | na |
| P062 | Furculomyces boomerangus | AFTOL303 | AF007535 | DQ273809 | na | na |
| P065 | Genistelloides hibernus | 2-16-2 | DQ367448 | DQ367479 | na | na |
| P066 | Genistellospora homothallica | VT-3-W14 | DQ367454 | DQ367495 | na | na |
| P048 | Harpella melusinae | NF-15-4b | DQ367514 | DQ367518 | na | na |
| P049 | Harpellomyces sp. | PA-3-1d | EF396192 | EF396195 | na | na |
| P092 | Kickxella alabastrina | NRRL2693 | AF007537 | AF031064 | na | na |
| P093 | Linderina pennispora | NRRL3781 | AF007538 | AF031063 | na | na |
| P095 | Martensiomyces pterosporus | NRRL2642 | AF007539 | AF031066 | na | na |
| P097 | Myconymphaea yatsukahoi | NBRC100467 | AB287984 | AB287998 | na | na |
| P075 | Pennella simulii | NY-5-3/ NF-19-8 | DQ367515 | DQ367502 | na | na |
| P089 | Pinnaticoemansia coronantispora | NBRC100470 | AB287986 | AB288000 | na | na |
| P076 | Plecopteromyces sp. | 37-1-2 | DQ367445 | DQ367476 | na | na |
| P080 | Smittium culisetae | AFTOL29/IAM14394/BL023 | AF007540 | DQ273773 | HM117719 | AB077104 |
| P100 | Spirodactylon aureum | NRRL2810 | AF007541 | AF031068 | na | na |
| P087 | Zygopolaris ephemeridarum | CA-4-W9 | DQ367463 | DQ367508 | na | na |
| Mortierellomycotina | ||||||
| P106 | Dissophora decumbens | NRRL22416 | AF157133 | AF157187 | AJ287155 | AF157247 |
| P107 | Echinosporangium transversale | NRRL3116 | AF113424 | AF113462 | AJ287156 | AF157248 |
| P108 | Gamsiella multidivaricata | NRRL6456 | AF157144 | AF157198 | AJ287168 | AF157260 |
| P111 | Mortierella longicollis | CBS209.32 | JQ040249 | JN940876 | na | na |
| P110 | Mortierella verticillata | CBS374.95 | HQ667482 | JN940872 | na | na |
| KH001 | NRRL6337 | AB016017 | DQ273794 | AJ287170 | AF157262 | |
| Mucoromycotina | ||||||
| P121a | Absidia caerulea | NRRL1315 | AF113405 | AF113443 | AJ287133 | AF157226 |
| P121f | Absidia californica | CBS126.68 | EU736273 | EU736300 | AY944758 | EU736246 |
| P121b | Absidia glauca | CBS101.48 | AF157118 | AF157172 | AJ287135 | X54730 |
| P121e | Absidia macrospora | CBS696.68 | EU736276 | EU736303 | AY944760 | EU736249 |
| P121d | Absidia psychrophilia | CBS128.68 | EU736279 | EU736306 | AY944762 | EU736252 |
| P121 | Absidia repens | NRRL1336 | AF113410 | AF113448 | AJ287136 | AF157228 |
| P121c | Absidia spinosa | ATCC22755 | EU736280 | EU736307 | EU736227 | EU736253 |
| P137 | Actinomucor elegans | NRRL3104/CBS111559 | AF157119 | AF157173 | AJ287137 | AF157229 |
| P190 | Apophysomyces elegans | NRRL22325 | AF113411 | FN554250 | na | na |
| P190a | NRRL28632 | AF113412 | AF113450 | AJ287139 | AF157231 | |
| P140 | Backusella circina | NRRL2446 | AF157121 | AF157175 | AJ287140 | AF157232 |
| kH1 | FSU2455 | JX644458 | JX644491 | na | na | |
| kH9 | FSU10121 | JX644459 | JX644492 | na | na | |
| kH10 | FSU10122 | JX644460 | JX644493 | na | na | |
| kH11 | FSU10123 | JX644461 | JX644494 | na | na | |
| kH12 | FSU10124 | JX644462 | JX644495 | na | na | |
| P169g | Backusella recurva | NRRL3247 | AF157146 | AF157200 | AJ287179 | AF157270 |
| kH5 | FSU10115 | JX644463 | JX644496 | na | na | |
| kH6 | FSU10116 | JX644464 | JX644497 | na | na | |
| kH7 | FSU10117 | JX644465 | JX644498 | na | na | |
| kH8 | FSU10118 | JX644466 | JX644499 | na | na | |
| P143 | Benjaminiella poitrasii | NRRL2845 | AF157123 | AF157177 | AJ287142 | AF157234 |
| P114 | Blakeslea trispora | CBS130.59 | AF157124 | AF157178 | AJ287143 | AF157235 |
| P146 | Chaetocladium brefeldii | CBS136.28 | EU736284 | EU736311 | EU736230 | EU736257 |
| P146a | NRRL1349 | AF157125 | AF157179 | AJ287144 | AF157236 | |
| P146b | Chaetocladium jonesii | NRRL2343 | AF157126 | AF157180 | AJ287145 | AF157237 |
| P123 | Chlamydoabsidia padenii | NRRL2977 | AF113415 | AF113453 | AJ287146 | AF157238 |
| P115 | Choanephora infundibulifera | CBS150.51/NRRL2744 | AF157127 | AF157181 | AJ287147 | AF157239 |
| P151a | Circinella sp. | NRRL13768 | JX644467 | JX644500 | JX644524 | JX644574 |
| P151b | NRRL13768 | JX644468 | JX644501 | JX644525 | JX644575 | |
| P151 | Circinella umbellata | NRRL1351 | AF157128 | AF157182 | AJ287148 | AF157240 |
| P154 | Cokeromyces recurvatus | AFTOL627 | AY635843 | DQ273812 | na | na |
| P154a | NRRL2243 | AF113416 | AF113454 | AJ287150 | AF157242 | |
| P124a | Cunninghamella bainierii | FSU319/NRRL1375 | EU736286 | EU736313 | EU736232 | EU736259 |
| P124b | Cunninghamella bertholletiae | NRRL6436 | AF113421 | AF113459 | AJ287151 | AF157243 |
| P124 | Cunninghamella echinulata | NRRL1382/CBS156.28 | AF157130 | AF157184 | AJ287152 | AF157244 |
| P194 | Dichotomocladium elegans | NRRL6236 | AF157131 | AF157185 | AJ287153 | AF157245 |
| P194a | NRRL2664 | JQ775463 | JQ775492 | EU826394 | EU826399 | |
| P194e | Dichotomocladium floridanum | FSU8694 | JQ775462 | JQ775491 | JX644526 | JX644576 |
| P194b | Dichotomocladium hesseltinei | NRRL5912 | JQ775464 | JQ775493 | JX644527 | JX644577 |
| P194c | Dichotomocladium robustum | NRRL6234 | JQ775465 | JQ775494 | JX644528 | JX644578 |
| P194d | NRRL6235 | JQ775466 | JQ775495 | JX644529 | na | |
| P194f | Dichotomocladium sphaerosporum | FSU8696 | JQ775469 | JQ775498 | na | JX644579 |
| P194g | FSU8697 | JQ775467 | JQ775496 | JX644530 | JX644580 | |
| P194h | Dichotomocladium sphaerosporum 2 | FSU8698 | JQ775468 | JQ775497 | JX644531 | JX644581 |
| P194i | Dichotomocladium sphaerosporum 3 | FSU8698 | JX644469 | JX644502 | JX644532 | JX644582 |
| P156 | Dicranophora fulva | NRRL22204 | AF157132 | AF157186 | AJ287154 | AF157246 |
| P157 | Ellisomyces anomalus | NRRL2749 | AF157134 | AF157188 | AJ287157 | AF157249 |
| P195 | Fennellomyces linderi | NRRL2342 | AF157135 | AF157189 | AJ287158 | AF157250 |
| P119 | Gilbertella persicaria | NRRL2357/CBS442.64 | AF157136 | AF157190 | AJ287159 | AF157251 |
| P125 | Gongronella butleri | NRRL1340/ATCC8989 | AF157137 | AF157191 | AJ287160 | AF157252 |
| P126 | Halteromyces radiatus | NRRL6197 | AF157138 | AF157192 | AJ287161 | AF157253 |
| P160 | Helicostylum elegans | NRRL2568/CBS258.59 | AF157139 | AF157193 | AJ287162 | AF157254 |
| P160c | Helicostylum pulchrum | CBS639.69 | EU736289 | EU736316 | EU736235 | EU736262 |
| P160b | CBS259.68 | EU736288 | EU736315 | EU736234 | EU736261 | |
| P127 | Hesseltinella vesiculosa | CBS197.68 | AF157140 | AF157194 | AJ287163 | AF157255 |
| P162 | Hyphomucor assamensis | NRRL22324 | AF157141 | AF157195 | AJ287164 | AF157256 |
| P164 | Kirkomyces cordensis | NRRL22618 | AF157142 | AF157196 | AJ287165 | AF157257 |
| P160a | CBS223.63 | EU736287 | EU736314 | EU736233 | EU736260 | |
| P216a | Lentamyces zychae | CBS104.35 | EU736282 | EU736309 | EU736228 | EU736255 |
| P134 | Lichtheimia corymbifera | CBS429.75 | JQ014052 | GQ342903 | GQ342831 | FJ719483 |
| P134a | NRRL2982 | AF113407 | FJ719429 | AJ287134 | AF157227 | |
| P134b | Lichtheimia hyalospora | NRRL1304 | AF157117 | AF157171 | AJ287132 | AF157225 |
| P134d | NRRL2916 | EU826360 | EU826368 | EF030531 | JX644583 | |
| P134c | Lichtheimia ramosa | FSU6197 | JX644470 | JX644503 | JX644533 | JX644584 |
| P169a | Mucor amphibiorum | NRRL28633 | AF113426 | AF113466 | AJ287172 | AF157263 |
| P152 | Mucor circinelloides | NRRL22652 | AF157129 | AF157183 | AJ287149 | AF157241 |
| P169 | Mucor circinelloides f. circinelloides | CBS195.68/FSU6169 | EU484248 | FN650667 | na | na |
| P169h | CBS416.77 | EU736294 | EU736321 | EU736240 | EU736267 | |
| P169b | Mucor circinelloides f. lusitanicus | NRRL3631 | AF113427 | AF113467 | AJ287173 | AF157264 |
| P141 | Mucor ctenidius | NRRL6238 | AF157122 | AF157176 | AJ287141 | AF157233 |
| kH2 | FSU10112 | JX644471 | JX644504 | na | na | |
| kH3 | FSU10113 | JX644472 | JX644505 | na | na | |
| kH4 | FSU10114 | JX644473 | JX644506 | na | na | |
| P169c | Mucor hiemalis | NRRL3624 | AF113428 | AF113468 | AJ287174 | AF157265 |
| P169d | Mucor indicus | NRRL28634 | AF113429 | AF113469 | AJ287175 | AF157266 |
| P135c | Mucor irregularis | NRRL28773 | AF113435 | AF113476 | AJ287193 | AF157284 |
| P180 | Mucor moelleri | FSU779/FSU514 | EU736298 | EU736325 | EU736244 | EU736271 |
| P180b | FSU531 | EU736297 | EU736324 | EU736243 | EU736270 | |
| P168 | Mucor mucedo | CBS144.24 | X89434 | AF113470 | AJ287176 | AF157267 |
| P169i | Mucor plumbeus | FSU283 | EU736295 | EU736322 | EU736241 | EU736268 |
| P169j | FSU289 | EU736296 | EU736323 | EU736242 | EU736269 | |
| P169e | Mucor racemosus | NRRL3640 | AF113430 | AF113471 | AJ287177 | AF157268 |
| P169f | Mucor ramosissimus | NRRL3042 | AF113431 | AF113472 | AJ287178 | AF157269 |
| P182a | Mycotypha africana | NRRL2978 | AF157147 | AF157201 | AJ287180 | AF157271 |
| P182 | Mycotypha microspora | NRRL1572/F169 | AF157148 | AF157202 | AJ287181 | AF157272 |
| P170 | Parasitella parasitica | NRRL1461/CBS412.66/NRRL2501 | HQ845295 | HQ845307 | AJ287182 | HQ845318 |
| P197 | Phascolomyces articulosus | NRRL2880 | AF157150 | AF157204 | AJ287183 | AF157274 |
| P197a | CBS113.76 | JX644474 | JX644507 | JX644534 | na | |
| P183 | Phycomyces blakesleeanus | NRRL1555 | NG_017190 | NG_027559 | genome1 | DQ282620 |
| kH20 | Pilaira sp. | FSU2463 | JX644475 | JX644508 | na | na |
| P171 | Pilaira anomala | NRRL2526 | AF157152 | AF157206 | AJ287185 | AF157276 |
| kH19 | FSU774 | JX644476 | JX644509 | JX644535 | JX644585 | |
| kH22 | NRRL2526 | AF157152 | na | AJ287185 | AF157276 | |
| P171a | Pilaira caucasica | NRRL6282 | JX644477 | JX644510 | JX644536 | JX644586 |
| kH14 | FSU10081 | JX644478 | JX644511 | JX644537 | na | |
| kH16 | FSU10083 | JX644479 | JX644512 | JX644538 | na | |
| kH17 | FSU10084 | JX644480 | JX644513 | JX644539 | na | |
| kH18 | FSU10085 | JX644481 | JX644514 | JX644540 | na | |
| KH21 | FSU6229 | EU826363 | EU826369 | EU826376 | EU826385 | |
| kH13 | Pilaira sp. | FSU10080 | JX644482 | JX644515 | JX644541 | na |
| kH15 | . | FSU10082 | JX644483 | JX644516 | JX644542 | na |
| kH25 | Pilobolus crystallinus | FSU6210 | JX644484 | JX644517 | na | na |
| kH28 | Pilobolus longipes | IUE563 | EU595654 | na | na | na |
| KH29 | IUE409 | DQ211054 | na | na | na | |
| KH30 | IUE340 | DQ211053 | na | na | na | |
| KH31 | Pilobolus roridus | IUE415 | EU595649 | na | na | na |
| kH23 | Pilobolus sp. | DSM1343 | JX644485 | JX644518 | na | JX644587 |
| P186 | Pilobolus umbonatus | NRRL6349 | AF157153 | AF157207 | AJ287186 | AF157277 |
| kH24 | CBS302.83 | JX644486 | JX644519 | na | na | |
| kH26 | UAMH7297 | DQ211050 | na | na | na | |
| kH27 | NRRL6349 | AF157153 | na | na | na | |
| KH32 | UAMH7298 | DQ211051 | na | na | na | |
| P172 | Pirella circinans | NRRL2402/Kh-BI-O | AF157154 | AF157208 | AJ287187 | AF157278 |
| P120 | Poitrasia cicinans | CBS153.58 | AF157155 | AF157209 | AJ287188 | AF157279 |
| P198 | Protomycocladus faisalabadensis | NRRL22826 | AF157156 | AF157210 | AJ287189 | AF157280 |
| P198a | CBS661.86 | JX644487 | JX644520 | na | na | |
| P191 | Radiomyces spectabilis | NRRL2753 | AF157157 | AF157211 | AJ287190 | AF157281 |
| P135a | Rhizomucor miehei | NRRL28774 | AF113432 | AF113473 | AJ287191 | AF157282 |
| P135d | CBS182.67 | JX644488 | JX644521 | JX644543 | na | |
| P135 | Rhizomucor pusillus | NRRL3695 | HQ845298 | HQ845310 | na | HQ845321 |
| P135b | NRRL2543 | AF113433 | AF113474 | AJ287192 | AF157283 | |
| P135e | CBS354.68 | JX644489 | JX644522 | na | HQ845320 | |
| P175 | Rhizopus arrhizus | CBS112.07 | AB250164 | AB250187 | AB281499 | AB281528 |
| P205 | CBS438.76 | AB250171 | AB250194 | na | na | |
| P205a | NRRL3139 | AF157120 | AF157174 | AJ287138 | AF157230 | |
| P176a | Rhizopus microsporus var. azygosporus | NRRL28627 | AF113436 | AF113477 | AJ287194 | AF157285 |
| P176 | Rhizopus microsporus var. microsporus | CBS699.68 | AB250155 | JN939137 | AB512247 | AB512270 |
| P176b | NRRL28775 | AF113438 | AF113479 | AJ287195 | AF157286 | |
| P176c | Rhizopus microsporus var. oligosporus | NRRL2710 | AF157158 | AF157212 | AJ287197 | AF157288 |
| P176d | Rhizopus microsporus var. rhizopodiformis | NRRL28630 | AF113439 | AF113480 | AJ287196 | AF157287 |
| P176e | Rhizopus stolonifer | NRRL1477 | AF113441 | AF113482 | AJ287199 | AF157290 |
| P193 | Saksenaea vasiformis | NRRL2443 | AF113442 | AF113483 | AJ287200 | AF157291 |
| P184 | Spinellus fusiger | NRRL22323 | AF157159 | AF157213 | AJ287201 | AF157292 |
| P213 | Sporodiniella umbellata | NRRL20824 | AF157160 | AF157214 | AJ287202 | AF157293 |
| P199 | Syncephalastrum monosporum | NRRL54019/NRRL22812 | AF157161 | AF157215 | AJ287203 | AF157294 |
| P199b | S. monosporum var. pluriproliferum | CBS569.91 | JX644490 | JX644523 | na | JX644588 |
| P199a | Syncephalastrum racemosum | NRRL2496 | X89437 | AF113484 | AJ287204 | AF157295 |
| P215 | Syzygites megalocarpus | NRRL6288/xsd08121 | AF157162 | AF157216 | AJ287205 | AF157296 |
| P178 | Thamnidium elegans | NRRL2467/CBS341.55 | AF157163 | AF157217 | AJ287206 | AF157297 |
| P200 | Thamnostylum piriforme | NRRL6240 | AF157164 | AF157218 | AJ287207 | AF157298 |
| P136 | Thermomucor indicae-seudaticae | NRRL6429 | AF157165 | AF157219 | AJ287208 | AF157299 |
| P202a | Umbelopsis isabellina | NRRL1757 | AF157166 | AF157220 | AJ287209 | AF157300 |
| P202c | Umbelopsis nana | NRRL22420 | AF157167 | AF157221 | AJ287210 | AF157301 |
| P202b | Umbelopsis ramanniana | NRRL5844 | X89435 | AF113463 | AJ287166 | AF157258 |
| P202 | Umbelopsis sp. | FSU10157 | JQ014049 | JN939141 | na | na |
| P189 | Utharomyces epallocaulus | NRRL3168 | AF157168 | AF157222 | AJ287211 | AF157302 |
| P201 | Zychaea mexicana | NRRL6237 | AF157169 | AF157223 | AJ287212 | AF157303 |
| P180a | Zygorhynchus heterogamus | NRRL1489 | AF157170 | AF157224 | AJ287213 | AF157304 |
| Neocallimastigomycota | ||||||
| P252 | Neocallimastix sp. | AFTOL638 | DQ322625 | DQ273822 | na | na |
| Zoopagomycotina | ||||||
| P230 | Kuzuhaea moniliformis | NRRL13723 | AB016010 | DQ273796 | na | na |
| P234 | Piptocephalis corymbifera | ATCC12665 | AB016023 | AY546690 | na | DQ282619 |
na = not available; 1 = estExt_Genewise1Plus.C_200172.
Sequence alignment, phylogeny, distance matrices, CBC
Multiple sequence alignments were generated using MAFFT v. 6.901b (server: mafft.cbrc.jp/alignment/server/) or v. 6.822 as implemented at the CIPRES portal (//www.phylo.org/; Miller et al. 2010). Alignments comprised 201 taxa and 1 586 characters for 18S rDNA, 358 characters for 28S rDNA, 807 characters for actin and 1 092 characters for translation elongation factor 1-alpha. Phylogenetic trees were calculated using RAxML v. 7.3.0 and MrBayes v. 3.1.2 from the CIPRES portal under the default settings with the following adjustments: RAxML was run choosing rapid bootstrapping (GTRCAT) and GTRGAMMA for final tree inference with 1 000 bootstrap iterations. Bayesian inference was run setting the number of substitution types to 6 (GTR), with among-site rate variation set to invgamma. Analysis was run with four chains each in two runs for 5 million generations. 5 001 trees were sampled, and 2 501 trees were analysed discarding the first 50 % of the samples as burnin. Bootstrapping was done with 1 000 iterations. Dataset was partitioned for both analyses. Alignments and phylogenetic trees are deposited in TreeBase2 under TB2:S13469. Distances were calculated using distMat from the EMBOSS suite (Rice et al. 2000; http://emboss.sourceforge.net/) with the alignments as input. Distances are expressed as substitutions per 100 bases or amino acids. CBC analyses were done as described previously (Pawłowska et al. In press).
RESULTS AND DISCUSSION
Species recognition is an essential step to higher level classification. Yet, morphology and/or mating behaviour played a major role in traditional fungal species concepts. Depending on the experience of the mycologist and on experimental conditions, morphology and mating behaviour could profoundly vary, and today, both methods were shown to be unsuitable to define mucoralean species if they are not combined with DNA data. Additional concepts have been surveyed and evaluated for fungi (Mayden 1997) with the genealogical concordance phylogenetic species recognition (GCPSR, Taylor et al. 2000) being the most likely one to recognize natural species. Phylogenetic species recognition (PSR) already revealed more species within originally identified species using morphological or biological species recognition (e.g., Hibbett et al. 1995, Taylor et al. 1999). The underlying problems of interbreeding and geographic/allopatric speciation were extensively discussed by Taylor et al. (2000). Following the discovery of phylogenetic species, additional biological and morphological characters were revealed that supported those species (reviewed by Taylor et al. 2000).
In Mucorales, the application of GCPSR resulted in the detection of several new species (Álvarez et al. 2010a, b, Alastruey-Izquierdo et al. 2010, Hermet et al. 2012) but on the other hand several taxa were synonymized based on comparisons of ITS sequences (Abe et al. 2006, Álvarez et al. 2010a, Walther et al. 2013).
In contrast to the naturally existing species there are no concepts for the recognition of higher or lower taxonomic levels. Traditionally, certain morphological features (Table 2) that were regarded as synapomorphies were used to define families (Zycha et al. 1969, Hesseltine & Ellis 1973, Benjamin 1979, Benny 1982, von Arx 1982). Later they were adapted based on results of molecular phylogeny. Undoubtedly higher taxa should represent monophyletic groups but the taxonomic rank that a group deserves remains a subjective decision. Genetic distances are helpful in this decision but they cannot be translated directly into higher level taxonomy because of dramatic difference in the phylogenetic age in fungal groups.
Even though studies implementing molecular data are still very rare for Mucorales compared to other fungal groups, the number of sequences submitted to GenBank is constantly increasing (Fig. 7). Yet, sequences deposited are predominantly sequences of the rDNA cluster, (Fig. 9). Protein coding sequences are still under represented. This may be due to the lack of appropriate primers which are able to work over a broad range of isolates and an often encountered problem of direct sequencing of the amplificates (Schoch et al. 2012) and the frequent presence of paralogs (Alastruey-Izquierdo et al. 2010). Studies which do apply this kind of molecular data and which are predominantly focused on mucoralean phylogeny count far below 100 if searching ISI Web of Science and NCBI. Furthermore, most of these studies using only one marker for the analyses (Fig. 8). If different sequences are combined in an analysis, it is often rDNA and the genomically linked ITS region, but also rDNA combined with protein coding genes (Fig. 8).
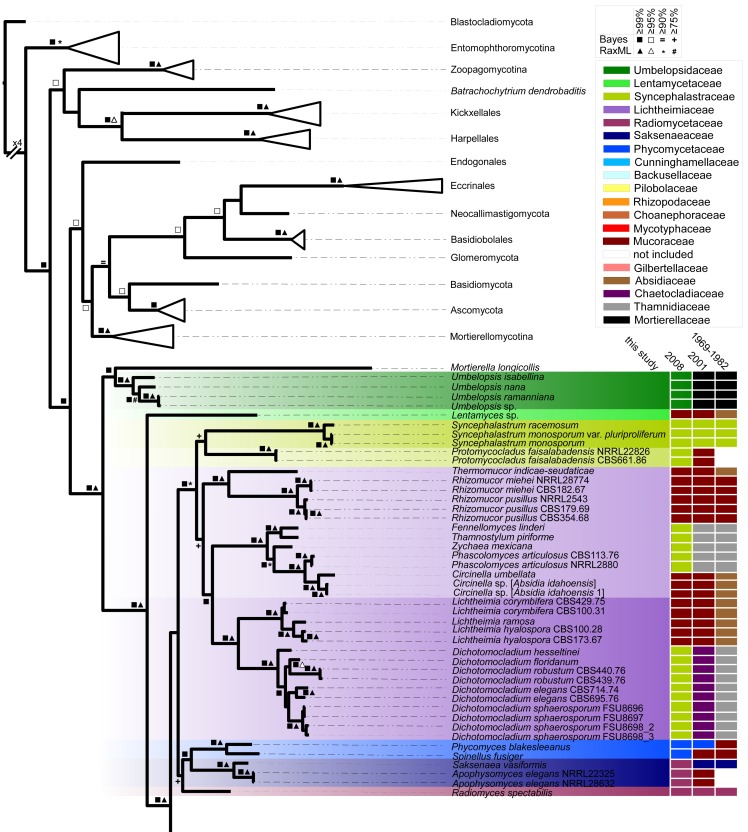
The phylogenetic analysis in Fig. 10 consists of combined sequences coding for LSU, SSU, actin and translation elongation factor. At least one member of all accepted genera is included with a total of 201 isolates, 151 belonging to the Mucorales, and 103 unique species representing around half of all described species in the order. Species were included if at least two loci were present in the alignment.
Fig. 10.
Bayesian analysis of combined sequences coding for actin, translation elongation factor 1-alpha, 18S rDNA and 28S rDNA. Bootstrap values and posterior probabilities are given for branches supported with equal or higher than 75 % in maximum likelihood (RAxML) and Bayesian analysis (see legend within figure for explanation of the symbols). Strain numbers are given in parts to distinguish different isolates (compare with Table 5). Furthermore, a rough outline about the historical family structures and changes are given on the right site including benchmark studies since 1969 (Zycha et al. 1969, Hesseltine & Ellis 1973, Benjamin 1979, Benny 1982, von Arx 1982, Benny et al. 2001, Voigt & Wöstemeyer 2001, O’Donnell et al. 2001, Kirk et al. 2008). Families accepted here, are colour coded over the whole tree branches.
A distance matrix was calculated for each locus. The order-wide distance analyses were based solely on the isolates in the illustrated tree (Fig. 10). Species-specific variations for each locus were not considered. The inclusion and analyses of all available sequences from GenBank would constitute a separate research project that goes beyond the scope of this study.
As expected, distance matrices derived from protein coding genes vary less if based on amino acids instead of nucleic acids. Based on the underlying data, amino acid sequences of actin are more conserved within the Mucorales with relatively similar distances over the whole order versus the situation for the translation elongation factor. When comparing all distance matrices, three major groups can be distinguished (Fig. 11):
Fig. 11.
Distance matrices for all applied loci based on nucleic acid and amino acid sequences. The range of distances is given for each locus. Families are coded according to Fig. 10.
-
i) Low to moderate distances for the most derived clade of the ‘Mucorineae’ including the Mucoraceae, Mycotyphaceae, Choanephoraceae, Pilobolaceae, Rhizopodaceae and Backusellaceae. All matrices show the lowest distances for Mucoraceae (incl. Mycotyphaceae).
All other groups and clades in the tree show no low distance values to any other group. Shortest distances exist only with-in each group whereas distances to all other groups are more or less similar. Clades included here are:
ii) the Cunninghamellaceae. Within this family, the shortest distances are between the genera Absidia, Halteromyces, Chlamydoabsidia and Cunninghamella (except for translation elongation factor, where distances between Gongronella/Hesseltinella and Absidia/Halteromyces/Chlamydoabsidia are shorter than to the embedded Cunninghamella.
-
iii) Lichtheimiaceae/Syncephalastraceae/Lentamycetaceae/Umbelopsidiaceae/Radiomycetaceae/Phycomyceteaceae.
High distance values for the more ancient clades of the phy-logenetic tree result from the different evolutionary times of origin which gives the more basal groups more time to evolve separately.
In the following, clades of the phylogenetic tree will be discussed including proposed/necessary changes in nomenclature or family delimitation.
A) Well-established and supported clades:
Aa) Umbelopsidaceae W. Gams & W. Mey.
Species of this family were thought to belong most likely to the Mortierellales, rather than to the Mucorales, mainly because of the highly reduced columella (nearly ‘acolumellatae’, Fig. 4a) and non-mucoralean colony morphology. The colony mycelium is very dense and velvety as opposed to floccose. And unlike the colonies formed by species of Mortierella, those of Umbelopsis are reddish, brownish or ochraceous and lack a typical garlic-like odour. This distinction and a relationship to Mucorales are surveyed in detail by Meyer & Gams (2003, including a detailed description of the family). With those slight morphological differences compared to all other mucoralean fungi, this group is currently regarded as the most basal in this order. The family is a monogeneric group with a clade support (CS, bootstrap support from the Likelihood analysis and Posterior Probabilities from the Bayesian analysis) of at least 99 %, and a clear distinction from the core Mucorales (CS ≥ 99 %) (Fig. 10).
Ab) Phycomycetaceae Arx
This clade (CS ≥ 99 %) includes only two genera with different life styles. Species of Phycomyces are saprobic, whereas those of Spinellus are facultatively parasitic on the basidioma of Agaricomycotina (Fig. 2a). Species of Phycomyces are model organisms for studies of phototropism and geotropism as well as carotenoide synthesis, carotenoide degradation and zygosporogenesis.
Ac) Pilobolaceae Corda
The Pilobolaceae is one of the few families recognized from the pre-genomics era with one taxonomic change. The genus Pilaira (Fig. 5e, f), thought to be a member of the family due to morphological characteristics, was placed in the Mucoraceae and is related most closely to Helicostylum, Thamnidium, Pirella and Mucor mucedo.
Fig. 5.
Pilobolaceae. a. Substrate mycelium with trophocysts; b. sporangium of Utharomyces epallocaulus with subsporangial swelling; c. colony morphology of Pilobolus sp. on horse dung. Sporangia are phototrophic; d. sporangiophores with subsporangial swelling and the black sporangium. Light is focused through the swelling towards carotenoids at the base of the vesicle, the ocellus (orange colour); e. colony morphology of Pilaira sp. Sporangiophores are also light sensitive; f. sporangium and columellae of Pilaira sp. — Scale bar = 50 μm.
Main characteristics of this family are the formation of trophocysts (Fig. 5a), the mode of spore release and the growth on dung of herbivores and rodents (Fig. 5c, d). Both included genera possess a vesicle/swelling below the sporangium, which functions in Pilobolus during active discharge of the sporangium (Page 1964, Zycha et al. 1969). In Utharomyces (Fig. 5b) spores are passively released. Pilobolus is especially difficult to cultivate on artificial media over several generations, resulting in changes in morphology and eventually in death of the culture. Based on analyses of molecular data, only the size and shape of the sporangiospores is retained as of relevance in species delimitation since this feature is the only one that correlates with molecular phylogenies (Foos et al. 2011).
Ad) Choanephoraceae J. Schröt.
This clade (CS ≥ 99 %) includes species producing only sporangia (Poitrasia, Gilbertella; Fig. 3d, e), or also sporangiola (Choanephora, Blakeslea; Fig. 3f). Sporangia and sporangiola are produced on separate sporangiophores. The wall of the sporangium is persistent. At maturity the wall ruptures at preformed sutures to release sporangiospores with hyaline, hair-like polar appendages representing a synapomorphy of this family. The species are saprobes or fruit and vegetable inhabiting parasites, sometimes occurring as major post-harvest pathogens in tropical and subtropical regions (Fig. 2c). The newly introduced subfamilies Gilbertelloideae (MycoBank IF550022) and Choanephoroideae (MycoBank IF550021) are distinguished by the characters of the zygospore, e.g. suspensors opposed or apposed, zygosporangium ornamented or smooth (Voigt 2012).
Ae) Cunninghamellaceae Naumov ex R.K. Benj.
Although this clade is highly supported (CS ≥ 99 %), it is one family that should be studied in more detail. While two recent studies dealt with the genus Cunninghamella and incorporated the largest number of isolates studied so far, the sister genera lack such a profound study. The authors evaluated all available information ranging from morphology to growth temperatures, mating experiments and molecular data (Liu et al. 2001, Zheng & Chen 2001). Currently, only Absidia and Cunninghamella are well sampled; Gongronella, and especially Hesseltinella, Halteromyces and Chlamydoabsidia, definitely need more isolates to study. Since Chlamydoabsidia is always nested within Absidia, its status as a distinct genus should be evaluated; this might also be extended to Halteromyces. The distances between sequences are very high in this family representing one of the highest variabilities when compared to other clades (Fig. 11).
Af) Lentamycetaceae K. Voigt & P.M. Kirk — MycoBank IF550009
Since the first analyses including species of the genus Lentamyces (formerly Absidia) it was obvious, that these species should be separated. And since there are no other species of the Mucorales closely related to this genus, a separate family is introduced (Voigt 2012). Species of the Lentamycetaceae (Fig. 4b, c) are homothallic and mycoparasitic, although the mycoparasitic potential of L. zychae was lost during cultivation (Zycha et al. 1969). Kwaśna & Nirenberg (2008a, b) introduced the genus Siepmannia that included the two Lentamyces species besides the new species S. pineti and S. lariceti. A correct classification of these taxa is still unclear because only ITS sequences and no living material are available from S. pineti and S. lariceti. A resampling of strains of Siepmannia is necessary to perform multilocus studies and to determine their mycoparasitic potential.
Ag) Backusellaceae K. Voigt & P.M. Kirk — MycoBank IF550011
Species included here originally were placed in the Mucoraceae or Thamnidiaceae. Like other described families once included in the Mucoraceae (e.g. Pilobolaceae, Choanephoraceae), this clade should also be distinguished from the Mucoraceae. The monogeneric Backusellaceae are characterised by transitorily recurved sporangiophores and the tendency to produce sporangiola in addition to the sporangia. Several Mucor species owning these characters were transferred to Backusella. Clade support for the Backusellaceae is ≥ 99 % (Fig. 10) and it contains three species: Backusella lamprospora, B. circina, B. recurva. The members of the Backusellaceae seem to be saprotrophs found e.g. in soil, on wood and fallen leaves (Walther et al. 2013).
Ah) Rhizopodaceae K. Voigt & P.M. Kirk — MycoBank IF550010
Like the Backusellaceae, the Rhizopodaceae forms a well-supported clade, distinct from the Mucoraceae (CS ≥ 99 %). Within this clade, a trifurcation is observed (each with a CS ≥ 99 %), with one Rhizopus microsporus-clade containing predominantely thermotolerant fungi (growth up to 45 °C), a sub-thermotolerant R. arrhizus-group (37–40 °C) and a meso-philic group containing R. stolonifer, Sporodiniella, and Syzygites. This was already observed applying morphology and growth temperatures (Schipper & Stalpers 1984), establishing a classification accepted as standard for many decades. The application of molecular data and biochemistrical features (e.g. production of lactic acids) supported those three major clades, but revealed also new/cryptic species (Abe et al. 2006, 2007). The implementation of GCPSR, including different genetic markers, resulted in the publication of a new, reliable Rhizopus classification (Abe et al. 2010). Yet, the final clustering in the Rhizopodaceae (Fig. 10) remains unresolved, because some species (R. caespitosus, R. homothallicus, R. lyococcus, R. schip-perae, R. sexualis) were not included because of missing data. But the thermotolerant species R. caespitosus, R. homothallicus and R. schipperae, seem to be closely related to the R. microsporus clade (rDNA analysis, Abe et al. 2006). In this study, R. sexualis (mesophilic) is related to R. stolonifer and R. lyococcus (mesophilic) appears as a very basal species (Abe et al. 2006). All species of the Rhizopodaceae are reported to be pathogenic to other organisms. Whereas Syzygites is a parasite of Dikarya (Kovacs & Sundberg 1999), Sporodiniella is a parasite of insect larvae (Evans & Samson 1977, Chien & Huang 1997), and species of Rhizopus are pathogens of plants and opportunists of animals, including humans.
Ai) Radiomycetaceae Hesselt. & J.J. Ellis & Saksenaeaceae Hesselt. & J.J. Ellis
The Radiomycetaceae contains only one genus with three species (Benny & Benjamin 1991). Radiomyces is coprophilous and pathogenic to mice (experimental infections, Kitz et al. 1980). The unispored or multispored sporangia are produced on pedicels, which originate from a vesicle. The Saksenaeaceae contain two genera, Saksenaea and Apophysomyces are saprobic in soil and compost. Some species are also known to infect animals and humans (Álvarez et al. 2010a, b).
B) Moderately supported clades:
Ba) Mycotyphaceae Benny & R.K. Benj.
The Mycotyphaceae currently contains only one genus (Benny & Benjamin 1976). Although the inclusion of adjacent species is proposed (Voigt 2012), the results of molecular phylogenetics are still controversial (Fig. 10). Furthermore, CS is ≥ 99 % for Mycotyphaceae, but strong support for the separation from Mucoraceae is only given for Bayesian analysis (CS ≥ 90 %). Although molecular distances (Fig. 11) of Mycotypha are similar to those of the Mucoraceae, the Mycotyphaceae is maintained as the sister family to Mucoraceae also because of the exceptional sporangiophores bearing terminal, elongate, cylindrical vesicles (Fig. 4k). The unispored sporangiola are of two types, an inner layer that consists of globose spores and an outer layer of spores that are either obovoid or more or less cylindrical.
Bb) Lichtheimiaceae Kerst. Hoffm., G. Walther & K. Voigt & Syncephalastraceae Naumov ex R.K. Benj.
Species of the genus Absidia growing well at elevated temperatures were transferred to the genus Lichtheimia based on both molecular and physiological data (Hoffmann et al. 2007, 2009). Lichtheimia has appeared as a well-supported sister taxon to Dichotomocladium in many phylogenetic analysis (e.g. O’Donnell et al. 2001, White et al. 2006) requiring an emendation of the Lichtheimiaceae. Dichotomocladium has been included in the Chaetocladiaceae (Benny & Benjamin 1993) based on morphological structures such as sterile spines, unispored sporangiola and branched, tree-like sporangiophores (Fig. 4d–f, l, m). Molecular data, however, revealed that these morphological features are of no phylogenetic significance. A shared feature of Lichtheimia and Dichotomocladium is their tolerance of higher temperatures. Species of Lichtheimia are consistently able to grow at and above 37 °C (Hoffmann et al. 2007), the species of Dichotomocladium tolerate 35 °C and some species, namely D. hesseltinei, D. floridanum and D. robustum are even able to grow at 37 °C (unpubl. data). The subfamilies Lichtheimioideae (MycoBank IF550086) and Dichotomocladioideae (MycoBank IF550087) are proposed for the Lichtheimiaceae (Voigt 2012). Based on a smaller set of sequences a third subclade within the Lichtheimiaceae was suggested: namely the Rhizomucoroideae (MycoBank IF550085) (Voigt 2012) but this classification could not be verified in this study.
Syncephalastrum (Syncephalastraceae) is the only genus in the Mucorales producing sporangiola with the spores arranged in a linear series (merosporangia, Fig. 4g). Whether other genera (e.g. Protomycocladus) should be included in this family needs to be studied in more detail because of the low phylogenetic branch support (Fig. 10), leaving Syncephalastrum the only genus in this family. The final position of Protomycocladus could not be resolved unquestionable due to low branch support in this study as well as other publications (e.g. O’Donnell et al. 2001, Voigt & Wöstemeyer 2001, White et al. 2006, Walther et al. 2013).
Closely related to the families, Lichtheimiaceae and the Syncephalastraceae, are three additional clades: i) Protomycocladus faisalabadensis; ii) Rhizomucor/Thermomucor; iii) Fennellomyces/Circinella/Thamnostylum/Zychaea/Phascolomyces. Clades i) and ii) include thermotolerant species with growth temperature maxima at 45 °C for Protomycocladus (Schipper & Samson 1994), and thermophilic species with growth temperature maxima at 55–57 °C for Rhizomucor (de Hoog et al. 2000) or above 60 °C for Thermomucor (Subrahamanyam et al. 1977). Clade iii) contains species that are predominantly mesophilic, not growing at elevated temperatures. Furthermore, this clade is characterised by circinate (strong or less pronounced) elements in the sporangiophores (Fig. 4p, q).
For a reliable placement of clades i–iii, in relation to the Lichtheimiaceae and Syncephalastraceae, additional data are needed, since the relationships of the former clades are not significantly supported in any published analyses. Therefore, these clades gain the status incertae sedis till their relationships could be solved unambiguously.
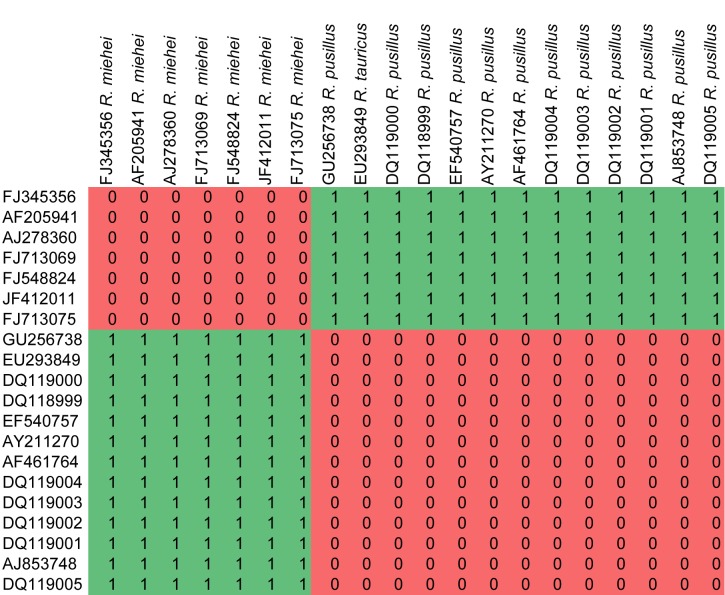
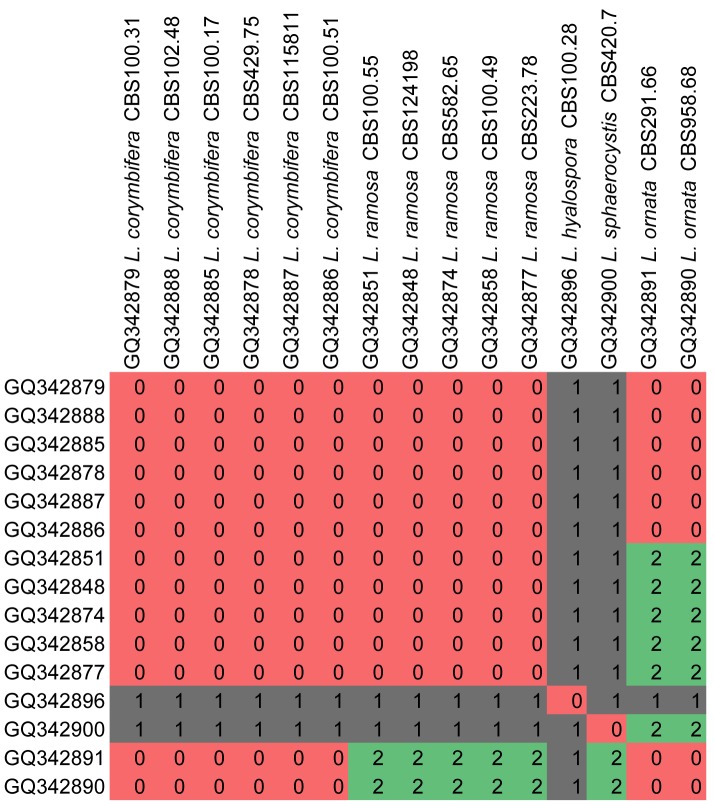
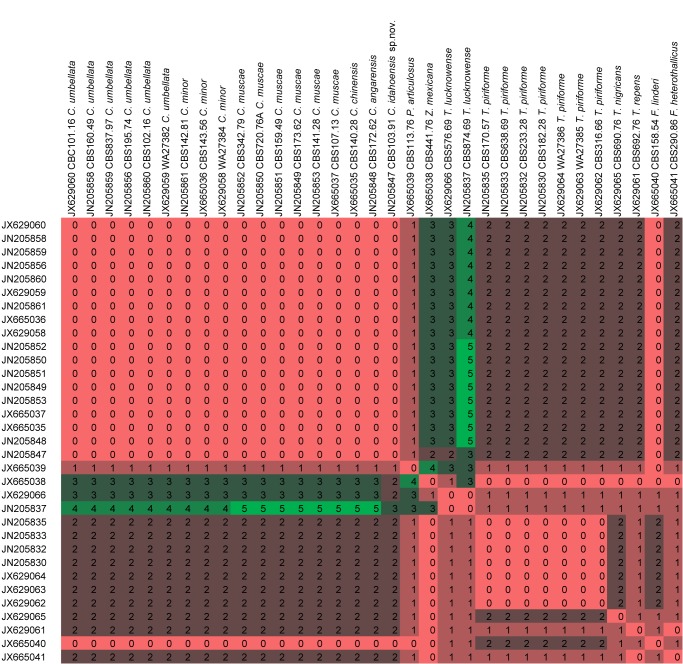
In order to test the taxonomic stability in the newly delimitated Lichtheimiaceae, ITS2 sequences of all isolates were searched for compensatory base changes (CBC) as indicators for species boundaries. A comprehensive study on CBC suggests that with a reliability of 93.11 % one CBC is present in two specimens belonging to two different species. But the lack of CBCs does not indicate that two specimens do belong to the same species (Müller et al. 2007). Applying CBC analyses to several clades within the Lichtheimiaceae/Syncephalastraceae, CBC is widely concordant with species concepts in Rhizomucor (Fig. 12), Lichtheimia (Fig. 13, except L. corymbifera and L. ornata), Dichotomocladium (Fig. 14), Zychaea and Thamnostylum (Fig. 15).
Fig. 12.
CBC analyses of ITS2 sequences from the genus Rhizomucor. Numbers of detected CBCs are given.
Fig. 13.
CBC analyses of ITS2 sequences from the genus Lichtheimia. Numbers of detected CBCs are given.
Fig. 14.
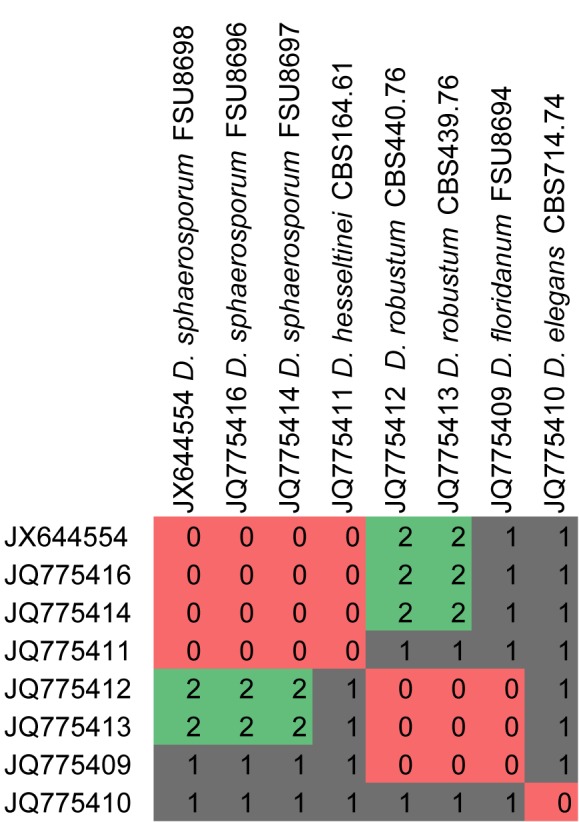
CBC analyses of ITS2 sequences from the genus Dichotomocladium. Numbers of detected CBCs are given.
Fig. 15.
CBC analyses of ITS2 sequences from the clade Circinella / Phascolomyces / Zychaea / Fennellomyces / Thamnostylum. Numbers of detected CBCs are given.
There are few species in the analyses which could not be clearly separated from others, which is due to the lack of CBCs (e.g. Dichotomolcadium hesseltinei and D. floridanum, Fennellomyces heterothallicus, Thamnostylum repens). However, no CBCs at all were detected in the genera Syncephalastrum and Circinella showing that CBC analyses cannot be used generally as a tool for species recognition in Mucorales. CBC analyses between different genera remains difficult if not impossible (especially in the ancient clades of the Mucorales) due to highly diverse ITS2 sequences and thus secondary structure. If differing too much, no comparison of the secondary structure is possible, which results in no detectable CBCs. CBC analyses are in parts suitable for distinguishing species that are highly similar in their morphology (e.g. Lichtheimia ramosa and L. corymbifera) and could assist in supporting molecular phylogenies.
Bc) Mucoraceae Dumort.
The Mucoraceae is undoubtly the largest family and presumably the most derived in the Mucorales (Fig. 10). Traditionally all species lacking features for classification within any other family where assigned to the Mucoraceae making the family polyphyletic. This study has circumscribed a monophyletic Mucoraceae with highly diverse features that characterise different species and genera. All species are saprobes except Dicranophora, Parasitella and Chaetocladium which are facultative mycoparasites (Dicranophora on Agaricomycetes, Parasitella and Chaetocladium on Mucorales). A few species are also described as opportunistic pathogens causing deep and systemic mycoses. Species are either homothallic or heterothallic, the zygospores form a warty to smooth zygosporangial wall with naked (without appendages) opposed suspensors. Sporangia are borne on branched or unbranched, sometimes phototrophic sporangiophores, sporangiola are rare and the sporangia are ± lageniform, ± apophysate and columellate.
SUMMARY AND CONCLUDING REMARKS
Traditional classification in Mucorales was done, as in all Eumycetes, mainly by using morphological characters. Already eleven years ago large deficiencies in the morphology-based system were revealed by molecular data. The distinctly extended dataset of the current study gives now a clearer picture of the family structure in the Mucorales. Our phylogeny based on four markers and contains 14 clades that we interpret as families: 1) Umbelopsidaceae; 2) the newly erected monogeneric Lentamyetaceae; 3) Syncephalastraceae presumably including Protomycocladus; 4) Lichtheimiaceae containing Lichtheimia and Dichotomocladium; 5) Phycomycetaceae; 6) Saksenaeaceae; 7) Radiomycetaceae; 8) Cunninghamellaceae inclusively Absidia s.str.; 9) the newly erected monogeneric Backusellaceae; 10) Pilobolaceae; 11) the newly erected Rhizopodaceae including the genera Rhizopus, Sporodiniella and Syzygites; 12) Choanephoraceae; 13) Mycotyphaceae; and 14) Mucoraceae. Most of these family clades were well supported. Only the delimitation between the Mucoraceae and the Mycotyphaceae as well as the Lichtheimiaceae and the Syncephalastraceae could not be defined doubtlessly, few subclades are classified as incertae sedis. The Mucoraceae, Mycotyphaceae and Cunninghamellaceae involve several taxonomic deficiencies and a detailed study of the phylogenetic relationships in these families is needed.
Acknowledgments
KH and KV thank Dr. H. Vogel and D. Schnabelrauch from the MPI for Chemical Ecology Jena, Germany for their support in sequencing. Financial support was partially provided by the Polish Ministry of Science and Higher Education (MNiSW), grant no. NN303_548839 to JP and MW. We wish to thank the reviewers for critically reviewing and valuable comments on the manuscript.
Footnotes
Note:
New taxa in Voigt 2012 were validated in Kirk 2012 and Kirk & Voigt 2012.
REFERENCES
- Abe A, Asano K, Sone T.2010. A molecular phylogeny-based taxonomy of the genus Rhizopus. Bioscience, Biotechnology and Biochemistry 74: 1325–1331 [DOI] [PubMed] [Google Scholar]
- Abe A, Oda Y, Asano K, Sone T.2006. The molecular phylogeny of the genus Rhizopus based on rDNA sequences. Bioscience, Biotechnology and Biochemistry 70: 2387–2393 [DOI] [PubMed] [Google Scholar]
- Abe A, Oda Y, Asano K, Sone T.2007. Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia 99: 714–722 [DOI] [PubMed] [Google Scholar]
- Alastruey-Izquierdo A, Hoffmann K, Hoog GS de, Rodriquez-Tudela JL, Voigt K, et al. 2010. Species recognition and clinical relevance of the zygomycetous genus Lichtheimia syn. Absidia pro parte, Mycocladus. Journal of Clinical Microbiology 48: 2154–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida ER, Cerdá-Olmedo E.2008. Gene expression in the regulation of carotene biosynthesis in Phycomyces. Current Genetics 53: 129–137 [DOI] [PubMed] [Google Scholar]
- Álvarez E, Garcia-Hermoso D, Sutton DA, Cano JF, Stchigel AM, et al. 2010a. Molecular phylogeny and proposal of two new species of the emerging pathogenic fungus Saksenaea. Journal of Clinical Microbiology 48: 4410–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez E, Stchigel AM, Cano J, Sutton DA, Fothergill AW, et al. 2010b. Molecular phylogenetic diversity of the emerging mucoralean fungus Apophysomyces: Proposal of three new species. Revista Iberoamericana de Micología 27: 80–89 [DOI] [PubMed] [Google Scholar]
- Arx JA von.1982. ‘1984. ’. On Mucoraceae s.str. and other families of the Mucorales. Sydowia 35: 10–26 [Google Scholar]
- Bartnicki-Garcia S, Nickerson WJ.1962. Induction of yeast-like development in Mucor by carbon dioxide. Journal of Bacteriology 84: 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin RK.1979. Zygomycetes and their spores. In: Kendrick B. (ed), The whole fungus: the sexual-asexual synthesis. Vol. 2: 573–616 National Museums of Canada, Ottawa, Canada [Google Scholar]
- Benny GL.1982. Zygomycetes. In: Parker SP. (ed), Synopsis and classification of living organisms. Vol. I: 184–195 McGraw-Hill Book Company, Inc., New York [Google Scholar]
- Benny GL.1995. Classical morphology in zygomycete taxonomy. Canadian Journal of Botany 73Suppl. 1: 725–730 [Google Scholar]
- Benny GL.2012. Current systematics of the zygomycotan fungi with a brief discussion of their biology. In: Misra JK, Tewari JP, Deshmukh SK. (eds), Systematics and evolution of fungi. Progress in mycological research: 55–105 Science Publishers, Enfield, New Hampshire, USA [Google Scholar]
- Benny GL, Benjamin RK.1976. Observations on Thamnidiaceae (Mucorales). II. Chaetocladium, Cokeromyces, Mycotypha, and Phascolomyces. Aliso 8: 391–424 [Google Scholar]
- Benny GL, Benjamin RK.1991. ‘1992. ’. The Radiomycetaceae (Mycorales; Zygomycetes). III. A new species of Radiomyces, and cladistic analysis and taxonomy of the family; with a discussion of the evolutionary ordinal relationships in Zygomycotina. Mycologia 83: 713–735 [Google Scholar]
- Benny GL, Benjamin RK.1993. Observations on Thamnidiaceae (Mucorales). VI. Two new species of Dichotomocladium and the zygospores of D. hesseltinei (Chaetocladiaceae). Mycologia 85: 660–671 [Google Scholar]
- Benny GL, Humber RA, Morton JB.2001. Zygomycota: Zygomycetes. In: McLaughlin DJ, McLaughlin EG, Lemke PA. (eds), The mycota. Vol. VIIA Systematics and evolution: 113–146 Springer-Verlag, Berlin, Germany [Google Scholar]
- Berbee ML, Taylor JW.2001. Fungal molecular evolution: gene trees and geologic time. In: McLaughlin DJ, McLaughlin EG, Lemke PA. (eds), The mycota. Vol. VII Part B. Systematics and evolution: 229–245 Springer Verlag, Berlin, Germany [Google Scholar]
- Blakeslee AF.1904. Sexual reproduction in the Mucorineae. Proceedings of the National Academy of Arts and Sciences 40: 205–319 [Google Scholar]
- Blakeslee AF, Cartledge JL.1927. Sexual dimorphism in Mucorales II. Interspecific reactions. Botanical Gazette 84: 51–57 [Google Scholar]
- Burgeff H.1924. Untersuchungen über Sexualität und Parasitismus bei Mucorineen I. Botanische Abhandlungen 4: 1–135 [Google Scholar]
- Caglioti L, Cainelli G, Camerino B, Mondelli R, Prieto A, et al. 1966. The structure of trisporic-C acid. Tetrahedron 22, Suppl. 7: 175–1875927157 [Google Scholar]
- Chien C-Y, Huang B-C.1997. First record of the occurrence of Sporodiniella umbellata Mucorales in Taiwan. Mycoscience 38: 343–346 [Google Scholar]
- Czempinski K, Kruft V, Wöstemeyer J, Burmester A.1996. 4-Dihydromethyltrisporate dehydrogenase from Mucor mucedo, an enzyme of the sexual hormone pathway: purification, and cloning of the corresponding gene. Microbiology 142: 2647–2654 [DOI] [PubMed] [Google Scholar]
- Dennis C.1983. Soft fruits. In: Dennis C. (ed), Post-harvest pathology of fruits and vegetables: 23–42 Academic Press, London [Google Scholar]
- Ende H van den.1967. Sexual factor of the Mucorales. Nature 215: 211–212 [DOI] [PubMed] [Google Scholar]
- Evans HC, Samson RA.1977. Sporodiniella umbellata, an entomogenous fungus of the Mucorales from cocoa farms in Ecuador. Canadian Journal of Botany 55: 2981–2984 [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G.2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evolutionary Biology 6: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foos KM, May NL, Beach DL, Pomper M, Sheehan KB, Ruch DG.2011. Phylogeny of Pilobolaceae. Mycologia 103: 36–44 [DOI] [PubMed] [Google Scholar]
- Gładkowski W, Grabarczyk M, Konopka M, Wawrzeńczyk C.2004. Lactones 20. Biohydroxylation of saturated bicyclic gamma-lactones with the substituted cyclohexane system. Journal of Molecular Catalysis B: Enzymatic 29: 13–7 [Google Scholar]
- Gładkowski W, Mazur M, Białońska A, Wawrzeńczyk C.2011. Lactones 35 [1]. Metabolism of iodolactones with cyclohexane ring in Absidia cylindrospora culture. Enzyme and Microbial Technology 48: 326–333 [DOI] [PubMed] [Google Scholar]
- Gooday GW.1968. Hormonal control of sexual reproduction in Mucor mucedo. New Phytologist 67: 815–821 [Google Scholar]
- Gryganskyi AP, Lee SC, Litvintseva AP, Smith ME, Bonito G, et al. 2010. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PloS ONE 5: e15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Kirk PM, Sutton BC, Pegler DN.1995. Ainsworth and Bisby’s dictionary of the fungi, 8th ed International Mycological Institute, Egham, UK [Google Scholar]
- Henkel TW.2004. Manufacturing procedures and microbiological aspects of Parakari, a novel fermented beverage of the Wapisiana Amerindians of Guyana. Economic Botany 58: 25–37 [Google Scholar]
- Henkel TW.2005. Parakari, an indigenous fermented beverage using amylolytic Rhizopus in Guyana. Mycologia 97: 1–11 [DOI] [PubMed] [Google Scholar]
- Hermet A, Méheust D, Mounier J, Barbier G, Jany JL.2012. Molecular systematics in the genus Mucor with special regards to species encountered in cheese. Fungal Biology 116: 692–705 [DOI] [PubMed] [Google Scholar]
- Hesseltine CW.1983. Microbiology of oriental fermented food. Annual Review of Microbiology 37: 575–601 [DOI] [PubMed] [Google Scholar]
- Hesseltine CW.1991. Zygomycetes in food fermentations. Mycologist 5: 162–169 [Google Scholar]
- Hesseltine CW, Ellis JJ.1973. Mucorales. In: Ainsworth GC, Sparrow FK, Sussman AS. (eds), The fungi. Vol. 4b: 187–217 Academic Press, New York [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509–547 [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Fukumasa-Nakai Y, Tsuneda A, Donoghue MJ.1995. Phylogenetic diversity in shiitake inferred from nuclear ribosomal DNA sequences. Mycologia 87: 618–638 [Google Scholar]
- Hoffmann K, Discher S, Voigt K.2007. Revision of the genus Absidia Mucorales, Zygomycetes based on physiological, phylogenetic and morphological characters: Thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycological Research 111: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Voigt K.2009. Absidia parricida plays a predominant role in biotrophic fusion parasitism among mucoralean fungi Zygomycetes: reclassi-fication as Lentamyces parricida gen. nov., comb. nov. Plant Biology 11: 537–554 [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Voigt K, Kirk PM.2011. Mortierellomycotina subphyl. nov., based on multi-gene genealogies. Mycotaxon 115: 353–363 [Google Scholar]
- Hoffmann K, Walther G, Voigt K.2009. Mycocladus vs. Lichtheimia: a correction Lichtheimiaceae fam. nov., Mucorales, Mucoromycotina. Mycological Research 113: 277–278 [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MJ.2000. Atlas of clinical fungi, 2nd ed Utrecht, Reus [Google Scholar]
- Huerta-Cepas J, Bueno A, Dopazo J, Gabaldón T.2008. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Research (database issue): D491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humber RA.2012. Entomophthoromycota: a new phylum and reclassification of entomophthoroid fungi. Mycotaxon 120: 477–492 [Google Scholar]
- Idnurm A.2011. Sex determination in the first-described sexual fungus. Eukaryotic Cell 10: 1485–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Walton FJ, Floyd A, Heitman J.2008. Identification of the sex genes in an early diverged fungus. Nature 451: 193–196 [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. 2006. Recon-structing the early evolution of fungi using a six-gene phylogeny. Nature 443: 818–822 [DOI] [PubMed] [Google Scholar]
- Jones MD, Forn I, Gadelha C, Egan MJ, Bass D, et al. 2011a. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 4747350: 200-3 [DOI] [PubMed] [Google Scholar]
- Jones MD, Richards TA, Hawksworth DL, Bass D.2011b. Validation and justification of the phylum name Cryptomycota phyl. nov. IMA Fungus 2: 173–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar R, Mandaokar BD, Kar RK.2010. Fungal taxa from the Miocene sediments of Mizoram, northeast India. Review of Palaeobotany and Palynology 158: 240–249 [Google Scholar]
- Kellner M, Burmester A, Wöstemeyer A, Wöstemeyer J.1993. Transfer of genetic information from the mycoparasite Parasitella parasitica to its host Absidia glauca. Current Genetics 23: 334–337 [DOI] [PubMed] [Google Scholar]
- Kirk PM.2012. Nomenclatural novelties. Index Fungorum 11: 1 [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA.2008. Ainsworth & Bisby’s Dictionary of the fungi. 10th edn CAB International, Wallingford, UK [Google Scholar]
- Kirk PM, Voigt K.2012. Nomenclatural novelties. Index Fungorum 13: 1 [Google Scholar]
- Kitz DJ, Embree RW, Cazin Jr J.1980. Radiomyces a genus in the Mucorales pathogenic for mice. Sabouaudia 18: 115–121 [DOI] [PubMed] [Google Scholar]
- Kovacs RL, Sundberg WJ.1999. Syzygites megalocarpus Mucorales, Zygo-mycetes in Illinois. Transactions of the Illinois Academy of Science 92: 181–190 [Google Scholar]
- Krings M, Taylor TN, Dotzler N, Persichini G.2012. Fossil fungi with suggested affinities to the Endogonaceae from the Middle Triassic of Antarctia. Mycologia 104: 835–844 [DOI] [PubMed] [Google Scholar]
- Kuramae EE, Robert V, Snel B, Weib M, Boekhout T.2006. Phylogenomics reveal a robust fungal tree of life. FEMS Yeast Research 6: 1213–1220 [DOI] [PubMed] [Google Scholar]
- Kwaśna H, Nirenberg HI.2008a. Siepmannia, a new genus in the Mucoraceae. Mycologia 100: 259–274 [DOI] [PubMed] [Google Scholar]
- Kwaśna H, Nirenberg HI.2008b. Validation of the genus Siepmannia Mucoraceae and its four species. Polish Botanical Journal 53: 187–188 [Google Scholar]
- Lee SC, Ni M, Li W, Shertz C, Heitman J.2010. The evolution of sex: a perspective from the fungal kingdom. Microbiology and Molecular Biology Reviews 74: 298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Huang H, Zheng RY.2001. Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon 80: 77–95 [Google Scholar]
- Martin WJ.1964. Effectiveness of fungicides in reducing soft rot in washed, cured sweet potatoes. Plant Disease Reporter 48: 606–607 [Google Scholar]
- Matheny PB, Liu YJJ, Ammirati JF, Hall BD.2002. Using RPB1 sequences to improve phylogenetic inference among mushrooms Inocybe, Agaricales. American Journal of Botany 89: 688–698 [DOI] [PubMed] [Google Scholar]
- Mayden RL.1997. A hierarchy of species concepts: The denouement in the saga of the species problem. In: Claridge MF, Dawah HA, Wilson MR. (eds), Species: The units of biodiversity: 381–424 Chapman & Hall, London [Google Scholar]
- Meyer W, Gams W.2003. Delimitation of Umbelopsis (Mucorales, Umbelopsidaceae fam. nov.) based on ITS sequence and RFLP data. Mycological Research 107: 339–350 [DOI] [PubMed] [Google Scholar]
- Michailides TJ, Spotts RA.1988. Germination of zygospores of Mucor piri-formis on the life history of Mucor piriformis. Mycologia 80: 837–844 [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010: 1–8, New Orleans, LA [Google Scholar]
- Miller ML, Sutter RP.1984. Methyltrisporate E. A sex pheromone in Phycomyces blakesleeanus. The Journal of Biological Chemistry 259: 6420–6422 [PubMed] [Google Scholar]
- Müller T, Philippi N, Dandekar T, Schultz J, Wolf M.2007. Distinguishing species. RNA 13: 1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nout MJR, Kiers JL.2005. Tempe fermentation, innovation and functionality: update into the third millennium. Journal of Applied Microbiology 98: 789–805 [DOI] [PubMed] [Google Scholar]
- O’Donnell K.1993. Fusarium and its near relatives. In: Reynolds DR, Taylor JW. (eds), The fungal holomorph: Mitotic, Meiotic and Pleomorphic speciation in fungal systematics: 225–233 CAB International, Wallingford, UK [Google Scholar]
- O’Donnell K, Lutzoni FM, Ward TJ, Benny GL.2001. Evolutionary relationships among mucoralean fungi Zygomycota: Evidence for family polyphyly on a large scale. Mycologia 932: 286–296 [Google Scholar]
- Page RM.1964. Sporangium discharge in Pilobolus: a photographic study. Science 146: 925–927 [DOI] [PubMed] [Google Scholar]
- Paterson RRM, Bridge PD.1994. Biochemical techniques for filamentous fungi. CAB International, Wallingford, UK [Google Scholar]
- Pawłowska J, Walther G, Wilk M, Hoog GS de, Wrzosek M. In press. The use of compensatory base changes analysis of ITS2 as a tool in phylogeny of Mucorales illustrated by ‘Mucor circinelloides’ group. Organisms Diversity & Evolution. [Google Scholar]
- Platauf AP.1885. Mycosis mucorina. Virchows Archives 102: 543–564 [Google Scholar]
- Polaino S, Herrador MM, Cerdá-Olmedo E, Barrero AF.2010. Splitting of beta-carotene in the sexual interaction of Phycomyces. Organic and Biomolecular Chemistry 8: 4229–4231 [DOI] [PubMed] [Google Scholar]
- Ray RC, Ravi V.2005. Post harvest spoilage of sweetpotato in tropics and control measures. Critical Reviews in Food Science and Nutrition 45: 623–644 [DOI] [PubMed] [Google Scholar]
- Reeb V, Lutzoni F, Roux C.2004. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes Pezizomycotina, fungi with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32: 1036–1060 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genetics 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Roden MM, Zautis TE, Buchanan WL, Knudsen TA, Sarkisova TA, et al. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clinical Infectious Diseases 41: 634–653 [DOI] [PubMed] [Google Scholar]
- Satina S, Blakeslee AF.1926. The mucor Parasitella parasitica in relation to sex. Proceedings of the National Academy of Sciences 12: 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtschabel D, Schimek C, Wöstemeyer J, Boland W.2005. Biological activity of trisporoids and trisporoid analogues in Mucor mucedo. Phytochemistry 66: 1358–1365 [DOI] [PubMed] [Google Scholar]
- Schipper MAA, Samson RA.1994. Miscellaneous notes on Mucoraceae. Mycotaxon 50: 475–491 [Google Scholar]
- Schipper MAA, Stalpers JA.1980. Various aspects of the mating system in Mucorales. Persoonia 11: 53–63 [Google Scholar]
- Schipper MAA, Stalpers JA.1984. Revision of the genus Rhizopus. Studies in Mycology 25: 1–34 [Google Scholar]
- Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, et al. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences USA 109: 6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze K, Schimek C, Wöstemeyer J, Burmester A.2005. Sexuality and para-sitism share common regulatory pathways in the fungus Parasitella parasitica. Gene 348: 33–44 [DOI] [PubMed] [Google Scholar]
- Schüßler A, Schwarzott D, Walker C.2001. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycological Research 105: 1413–1421 [Google Scholar]
- Seifert KA, Samson RA, Dewaard JR, Houbraken J, Lévesque CA, et al. 2007. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proceedings of the National Academy of Sciences USA 104: 3901–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidi F, Arachchi JKV, Jeon Y-J.1999. Food applications of chitin and chitosan. Trends in Food Science & Technology 10: 73–51 [Google Scholar]
- Shtienberg D.1997. Rhizopus head rot of confectionery sunflower: effects on yield quantity and quality and implications for disease management. Phytopathology 87: 1226–1232 [DOI] [PubMed] [Google Scholar]
- Skiada A, Pagano L, Groll A, Zimmerli S, Dupont B, et al. 2011. Zygomycosis in Europe: analysis of 230 cases accrued by the registry of the European Confederation of Medical Mycology ECMM Working Group on Zygomycosis between 2005 and 2007. Clinical Microbiology and Infection 17: 1859–1867 [DOI] [PubMed] [Google Scholar]
- Stalpers JA, Schipper MAA.1980. Comparison of zygospore ornamentation in intra- and interspecific matings in some related species of Mucor and Backusella. Persoonia 11: 39–52 [Google Scholar]
- Subrahamanyam A, Mehrotra BS, Thirumalacher MJ.1977. Thermomucor – a new genus of Mucorales. Georgia Journal of Science 35: 1–4 [Google Scholar]
- Sutter RP, Dadok J, Bothner-By AA, Smith RR, Mishra PK.1989. Cultures of separated mating types of Blakeslea trispora make D and E forms of trisporic acids. Biochemistry 28: 4060–4066 [DOI] [PubMed] [Google Scholar]
- Tagua VG, Medina HR, Martín-Domínguez R, Eslava AP, Corrochano LM, et al. 2012. A gene for carotene cleavage required for pheromone biosynthesis and carotene regulation in the fungus Phycomyces blakesleeanus. Fungal Genetics and Biology 49: 398–404 [DOI] [PubMed] [Google Scholar]
- Tamang JP, Thapa S.2006. Fermentation dynamics during production of bhaati jaanr, a traditional fermented rice beverage of the eastern Himalayas. Food Biotechnology 20: 251–261 [Google Scholar]
- Taylor JW, Geiser DM, Burt A, Koufopanou V.1999. The evolutionary biology and population genetics underlying strain-typing. Clinical Microbiology Reviews 12: 126–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32 [DOI] [PubMed] [Google Scholar]
- Taylor TN, Krings M, Klavins SD, Taylor EL.2005. Protoascon missouriensis, a complex fossil microfungus revisited. Mycologia 97: 725–729 [DOI] [PubMed] [Google Scholar]
- Vitale RG, Hoog GS de, Schwarz P, Dannaoui E, Deng S, et al. 2012. Antifungal susceptibility and phylogeny of opportunistic members of the order Mucorales. Journal of Clinical Microbiology 50: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Gillespie D.1979. Preparative and analytical purification of DNA from agarose. Proceedings of the National Academy of Sciences USA 76: 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K.2012. Zygomycota. In: Frey W. (ed), Syllabus of plant families, 13th ed: 130–156 Borntraeger, Stuttgart, Germany [Google Scholar]
- Voigt K, Olsson L.2008. Molecular phylogenetic and scanning electron microscopical analyses places the Choanephoraceae and the Gilbertellaceae in a monophyletic group within the Mucorales (Zygomycetes, Fungi). Acta Biologica Hungarica 59: 365–383 [DOI] [PubMed] [Google Scholar]
- Voigt K, Wöstemeyer J.2000. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiological Research 155: 179–195 [DOI] [PubMed] [Google Scholar]
- Voigt K, Wöstemeyer J.2001. Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1α genes. Gene 270: 113–120 [DOI] [PubMed] [Google Scholar]
- Wade NL, Morris SC.1982. Causes and control of cantaloupe postharvest wastage in Australia. Plant Disease 66: 549–552 [Google Scholar]
- Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela JL, et al. 2013. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30: 11–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werkman BA.1976. Localization and partial characterization of a sex-specific enzyme in homothallic and heterothallic mucorales. Archives of Microbiology 109: 209–213 [Google Scholar]
- Wetzel J, Scheibner O, Burmester A, Schimek C, Wöstemeyer J.2009. 4-dihydrotrisporin dehydrogenase, an enzyme of the sex hormone pathway of Mucor mucedo: purification, cloning of the corresponding gene, and developmental expression. Eukaryotic Cell 8: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MM, James TY, O’Donnell K, Cafaro MJ, Tanabe Y, Sugiyama J.2006. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia 98: 872–884 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, San Diego [Google Scholar]
- Yu MQ, Ko WH.1997. Factors affecting germination and mode of germination of zygospores of Choanephora cucurbitarum. Journal of Phytopathology 145: 357–361 [Google Scholar]
- Zheng RY, Chen GQ.2001. A monograph of Cunninghamella. Mycotaxon 80: 1–75 [Google Scholar]
- Zycha H, Siepmann R, Linnemann G.1969. Mucorales. Eine Beschreibung aller Gattungen und Arten dieser Pilzgruppe. Cramer, Lehre [Google Scholar]