Abstract
The basal fungal order Mortierellales constitutes one of the largest orders in the basal lineages. This group consists of one family and six genera. Most species are saprobic soil inhabiting fungi with the ability of diverse biotransformations or the accumulation of unsaturated fatty acids, making them attractive for biotechnological applications. Only few studies exist aiming at the revelation of the evolutionary relationships of this interesting fungal group. This study includes the largest dataset of LSU and ITS sequences for more than 400 specimens containing 63 type or reference strains. Based on a LSU phylogram, fungal groups were defined and evaluated using ITS sequences and morphological features. Traditional morphology-based classification schemes were rejected, because the morphology of the Mortierellales seems to depend on culture conditions, a fact, which makes the identification of synapomorphic characters tedious. This study belongs to the most comprehensive molecular phylogenetic analyses for the Mortierellales up to date and reveals unresolved species and species complexes.
Keywords: internal transcribed spacer, large subunit ribosomal DNA, taxonomic revision, Zygomycetes, Zygomycota
INTRODUCTION
The order Mortierellales – from historical aspects on morphology and systematics to modern approaches in fungal identification
The Mortierellales are a long known, species rich order of the basal fungi. With nearly 100 described species, the Mortierellales is one of the largest basal fungal orders. However, only 13 genera are described in one family, the Mortierellaceae (Kirk et al. 2008, and Species Fungorum January 2013). Out of these genera six are currently accepted with one potential additional genus recently described (Kirk et al. 2008, Jiang et al. 2011, Table 1). The first species of the type genus was described by Coemans (1863) as Mortierella polycephala, originally isolated from a mushroom. The name Mortierella was given in tribute to M. Du Mortier, the president of the Société de Botanique de Belgique (Coemans 1863). Nevertheless, the common life-style of those fungi is as soil inhabiting saprobic organisms on decaying organic matter. Only one species is occasionally described from animal fungal infections (de Hoog et al. 2009). Many mortierellean species possess the ability to produce poly-unsaturated fatty acids or to convert organic compounds, making them highly interesting organisms for biotransformations and other biotechnological applications (Holland 2001, Higashiyama et al. 2002).
Table 1.
Chronological overview of descriptions and name changes for accepted genera in the order Mortierellales Caval.-Sm. 1998 [MB#90555]. The order consists of several genera and one family, the Mortierellaceae A. Fisch. 1892 [MB#81029]. Data based on MycoBank and IndexFungorum (accessed 7 January 2013).
| Year | Genus | Synonyms | Type species | Number of described species | MycoBank no. |
|---|---|---|---|---|---|
| 1863 | Mortierella Coem. | Actinomortierella Chalab. 1968 | |||
| Carnoya Dewèvre 1893 | |||||
| Haplosporangium Thaxt. 1914 | |||||
| Azygozygum Chesters 1933 | |||||
| Naumoviella Novot. 1950 | M. polycephala | 91 | MB#20345 | ||
| 1914 | Dissophora Thaxt. | none | D. decumbens | 3 | MB#20187 |
| 1936 | Modicella Kanouse | none | M. malleola | 2 | MB#20336 |
| 1967 | Aquamortierella Embree & Indoh | none | A. elegans | 1 | MB#20047 |
| 2004 | Gamsiella (R.K. Benj.) Benny & M. Blackw. | none | G. multidivaricata | 1 | MB#28820 |
| 2004 | Lobosporangium M. Blackw. & Benny | Echinosporangium Malloch 1967 | L. transversale | 1 | MB#28819 |
| 2011 | Echinochlamydosporium X.Z. Jiang, X.Y. Liu & Xing Z. Liu | none | E. variabile | 1 | MB#511829 |
MB = Mycobank: http://www.mycobank.org; IndexFungorum: http://www.indexfungorum.org.
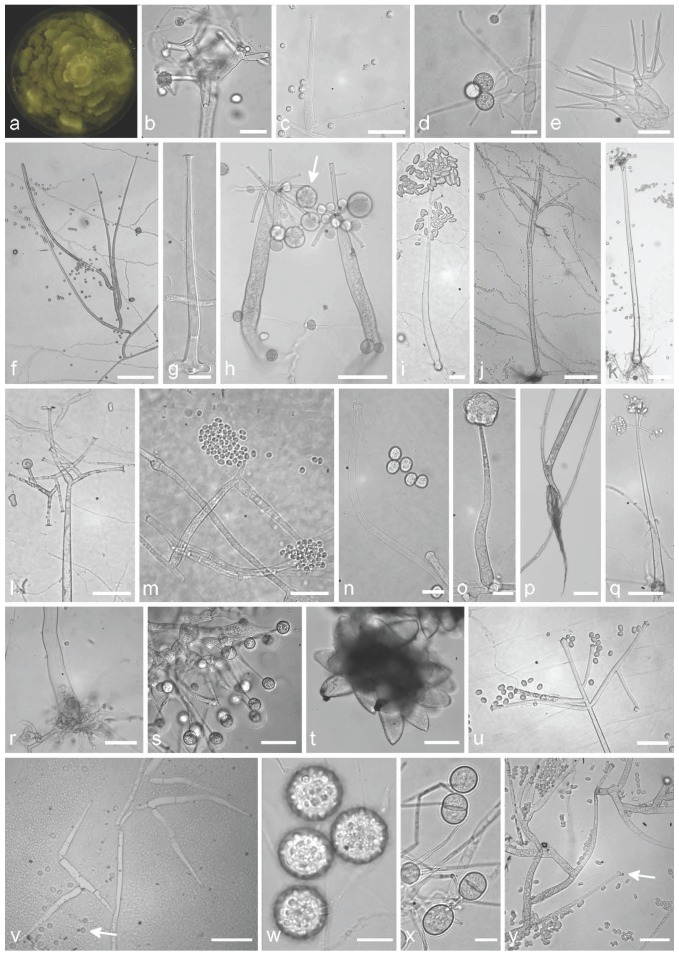
As many basal fungal species, the Mortierellales possess a reduced macro- and micromorphology with only few morphological characters available for differentiation. Examples of micromorphological features are shown in Fig. 1 and 2. Overall appearance of the colonies is the typical zonate, rosette-like growth (Fig. 1a) and the often occurring garlic-like odour. Colonies are in general white to light-grey, young mycelium is coenocytic and septate in aged cultures. Asexual spores are produced in sporangia or sporangiola and are passively released (e.g., Fig. 1h, s). The sporangiophores could be widened at the base (e.g., Fig. 1o) and variously branched (e.g. Fig. 1h, l). A columella is never protruding into the sporangium. Sexual reproductive structures (zygospores, Fig. 2r) are often surrounded by a hyphal sheat. Variously shaped chlamydospores and stylospores are also possible (Fig. 1w, 2l) (Zycha et al. 1969, Gams 1977). Morphological identification based solely on asexual features, leading to the aforementioned traditional classification. Mortierella was furthermore divided into nine sections based on morphology: Actinomortierella, Alpina, Haplosporangium, Hygrophila, Mortierella, Schmuckeri, Simplex, Spinosa and Stylospora (Gams 1977).
Fig. 1.
Typical morphological structures of different isolates of the Mortierellales, which are suitable for species delimitation. a. M. zychae CBS 316.52, macroscopic shape of a growing culture with the typical zonate growth; b. M. hypsicladia CBS 116202, acrotonous branching of a sporangiophore; c. M. epicladia CBS 355.76, sporangiophore and sporangiospores; d. M. zonata CBS 228.35, basitonous branched sporangiophore with sporangioles; e. Gamsiella multidivaricata CBS 227.78, typical branched sporangiophores; f. M. elongata FSU 9721, basitonous branched sporangiophore; g. M. alpina FSU 2698, sporangiophore; h. M. polycephala FSU 867, sporangiospores with sporangia (arrow) and sporangiospores; i. Mortierella cf. wolfii CBS 614.70, sporangiophore with elongated sporangiospores; j. M. parvispora FSU 10759, sporangiophores; k. M. hypsicladia CBS 116202, typical sporangiophore with rhizoid; l. Mortierella cf. wolfii CBS 614.70, acrotonous branching of a sporangiophore; m. Mortierella sp. FSU 10557, sporangiophore and sporangiospores; n. M. paraensis CBS 547.89, tips of a sporangiophore with a pseudocolumella and sporangiospores; o. M. alpina FSU 2698, sporangiophore with unmatured sporangia; p. M. nanthalensis CBS 610.70, typical rhizoid of a sporangiophore; q. M. wolfii CBS 651.93, sporangiospores with unusual remain of the sporangia cover (arrow); r. M. strangulata CBS 455.67, rhizoid of the sporangiophore; s. Gamsiella multidivaricata CBS 227.78, sporangiophores with sporangioles; t. Lobosporangium transversale CBS 357.67, typical sporangia, arranged in clusters, containing numerous spherical sporangiospores; u. M. gamsii FSU 10538, acrotonous branching of a sporangiophore and sporangiospores; v. Dissophora decumbens CBS 592.88, septate sporangiophores along a hypha and sporangiospore (arrow); w. M. polycephala FSU 867, stylospores; x. Gamsiella multidivaricata CBS 227.78, sporangiola containing spores; y. M. kuhlmanii CBS 157.71, branching pattern of the basitonous part of the sporangiophore and elongated sporangiospores, pseudocolumella. — Scale bars: b, c, s–u, x = 30 μm; d, e, i = 20 μm; f, j, k, p = 100 μm; g, n, o, w = 10 μm; h, l, m, q, r, v, y = 50 μm.
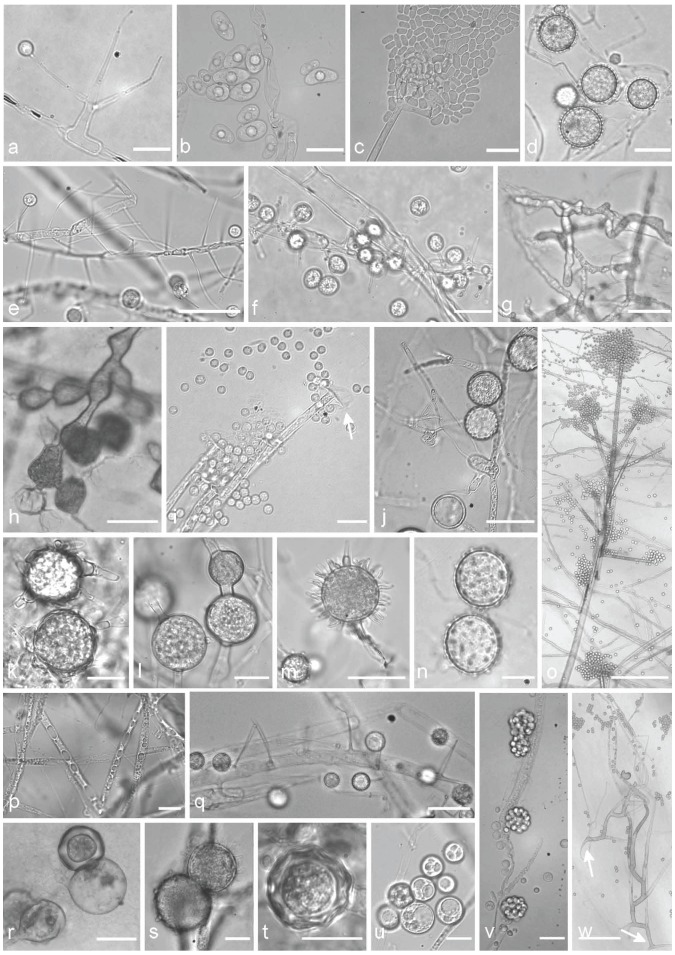
Fig. 2.
Typical morphological structures of different isolates of the Mortierellales, which are suitable for species delimitation. a. M. verticillata CBS 315.52, sporangiophore with a sporangiola; b. M. elongata FSU 9721, elongated sporangiospores containing central oil droplets; c. M. wolfii CBS 651.93, cracked sporangia releasing sporangiospores, on acrotonous branched tip of the sporangiophore; d. M. indohii CBS 720.71, stylospores; e. M. schmuckeri CBS 295.59, sporangiophores alongside a hypha with sporangiola; f. M. claussenii CBS 294.59, sporangiophores along a hypha with sporangiola; g. M. clonocystis CBS 357.76, typical swollen hyphae; h. M. zychae FSU 719, typical swollen hyphae arranged in clusters; i. M. parvispora FSU 10759, tip of a sporangiophore, sporangia leaving a collar (arrow), globose sporangiospores; j. M. lignicola CBS 207.37, sporangiophores, sporangiola (arrow 1), stylospores (arrow 2); k. M. exigua CBS 655.68, chlamydospores with typical outgrowing hyphae; l. M. gemmifera CBS 134.45, chlamydospores; m. M. hypsicladia CBS 116202, stylospores with projections; n. M. polygonia CBS 685.71, stylospores; o. M. nanthalensis CBS 610.70, acrotonous branching part of a sporangiophore; p. M. alpina FSU 2698, oil droplets containing hypha; q. M. camargensis CBS 221.58, sporangiophores along a hypha with sporangiola; r. M. epigama CBS 489.70, zygospores; s. M. echinosphaera CBS 575.75, chlamydospores; t. M. microszygospora CBS 880.97, microzygospore; u. M. camargensis CBS 221.58, oil droplets containing spheric sporangiola; v. Dissophora decumbens CBS 592.88, sporangiophores with sporangia; w. M. paraensis CBS 547.89, two sporangiophores with typical basitonous branchings (arrows mark the basal part). — Scale bars: a, b, i, n, p, r, u = 10 μm; c, j, q = 20 μm; d, e, g, h, m, v = 30 μm; f, k, l, s, t = 15 μm; o = 250 μm; w = 100 μm.
Judging from the proposed total number of fungi with 1.5 million species and the current number of described and registered species with 75 000 (Hawksworth 2001) it seems likely that also for the order Mortierellales an unknown percentage of undescribed species may exists, a fact which might influence phylogenetic analyses. Yet, a recent study challenged previous estimations of the potential number of undescribed fungal species and proposed that, at least for Mortierella, nearly all species are most likely described already (Nagy et al. 2011). Based on this knowledge, phylogenetic analyses including sequences of an extensive amount of type and reference strains could reveal the natural evolutionary relationships.
Nevertheless, the phylogenetic position of the Mortierellales is controversial discussed. They are either placed within the subphylum Mucoromycotina (Hibbett et al. 2007) or elevated to an own subphylum, the Mortierellomycotina (Hoffmann et al. 2011). Furthermore, relationships within this order are also poorly understood and were extensively analysed only in few studies until now (Nagy et al. 2011, Petkovits et al. 2011). Our study contributes to the effort to elucidate natural phylogenetic relationships based on one of the largest datasets assembled so far. This study concerns the extension of previous datasets and facilitates an approach to molecular identification of the Mortierellales. We surveyed the diversity of the Mortierellales including a re-evaluation of the morphology based classifications. This study based on the broad sampling of specimens which are maintained at the fungal culture collections CBS (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands) and JMRC (Jena Microbial Resource Collection, Jena, Germany).
MATERIALS AND METHODS
Taxon sampling, culture conditions and light microscopic investigations
For this study, a total of 421 isolates were obtained from the Centraalbureau voor Schimmelcultures (CBS, Utrecht, The Netherlands) and the Jena Microbial Resource Collection (JMRC, Jena, Germany) (Table 2). Strains were cultivated on malt-extract medium (3 % malt extract, 0.5 % yeast extract) for DNA isolation and on oatmeal agar (OA, 3 %), soil extract agar (Gams 1969) or synthetic nutrient deficient agar (SNA, Nirenberg 1981) for morphological examinations. Cultivation was done at 20–37 °C for 7–20 days depending on the requirements of the fungus to sporulate. The light microscopical examinations shown in Fig. 1 and 2 were performed with an Axiophot (Zeiss, Germany). The best method to observe micro-scopic features is to grow cultures directly on cover slips.
Table 2.
Strains used in this study.
| Original name | Strain numbers | Microscopic identification | Type status | Locality | Substrate | Accession no. ITS | Accession no. LSU |
|---|---|---|---|---|---|---|---|
| Dissophora decumbens | CBS301.87, FSU9780 | D. decumbens | Kingston, Rhode Island | ground-up litter of Quercus-Acer wood-land, incubated at 0°C for two months | JX976001 | HQ667354.1 | |
| CBS592.88, FSU801 | D. decumbens | Rhode Island | ground-up Quercus and Acer leaves, incubated at 0°C for 21 months | HQ630276.1 | HQ667355.1 | ||
| Dissophora ornata | CBS347.77, FSU9782 | – | Holotype of Mortierella ornata | Cordillera Central, Cauca en Huila, Parque Nacional del Puracé, Colombia | soil, in mountain forest under Wein-mannia, Clusia etc., alt. 3100 m | HQ630278.1 | HQ667357.1 |
| CBS348.77, FSU9783 | – | Holotype of Mortierella ornata | Cordillera Central, Cauca en Huila, Parque Nacional del Puracé, Colombia | soil, in mountain forest under Wein-mannia, Clusia etc., alt. 3100 m | JX976036 | HQ667356.1 | |
| Gamsiella multidivaricata | CBS227.78, FSU9784 | G. multidivaricata | Isotype of Mortierella multidivaricata | Moskva, Sokolniki Park, Russia | decaying stump | JX975871 | HQ667355.1 |
| Lobosporangium transversale | CBS357.67, FSU9785 | – | Type of Echinosporangium transversale | Nevada, Virginia City | soil | – | HQ667404.1 |
| Mortierella acrotona | CBS386.71, FSU9788 | – | Type of Mortierella acrotona | Jaipur, Rambagh Palace Hotel, Rajasthan | soil | JX975921 | HQ667405.1 |
| Mortierella alliacea | CBS106.78 | – | France | gymnosperm litter | JX976019 | KC018349 | |
| CBS894.68 | – | Tirol, Obergurgl, Austria | alpine raw humus soil | JX975990 | JX976148 | ||
| Mortierella alpina | CBS110518 | – | South Africa | soil, dry sandy highveld grassland | JX975906 | – | |
| CBS210.32, FSU9789 | M. alpina | Authentic strain of Mortierella renispora | Victoria | sandy loam soil | JX975853 | HQ667421.1 | |
| CBS219.35 | – | – | JX976018 | KC018359 | |||
| CBS250.53 | – | – | JX975955 | KC018184 | |||
| CBS384.71C | – | Jaipur, Rambagh Palace Hotel, Rajasthan | soil | JX976098 | JX976154 | ||
| CBS387.71 | – | Gran Canaria, Spain | soil, under Pinus canariensis | JX976038 | KC018378 | ||
| CBS396.91 | – | Washington | air bladder of juvenile fish | JX975994 | KC018375 | ||
| CBS529.72 | – | North Carolina | pasture soil | JX976124 | KC018320 | ||
| CBS585.81 | M. kuhlmanii | Netherlands | agricultural soil | JX976132 | JX976152 | ||
| CBS608.70 | – | Netherlands | agricultural soil | JX976046 | KC018438 | ||
| CBS696.70 | M. cystojenkinii | Wageningen, Mansholtlaan, Netherlands | agricultural soil | JX975947 | KC018328 | ||
| FSU2698 | M. alpina | Argentinia | JX976004 | KC018272 | |||
| FSU6524 | M. alpina | Geisenheim, Germany | JX976045 | KC018273 | |||
| Mortierella ambigua | CBS373.96 | – | Fukiagehama, Kagoshima, Japan | soil of salt marsh | JX976062 | JX976147 | |
| CBS450.88 | – | – | JX976067 | KC018411 | |||
| CBS457.66 | – | Armenia | soil | JX976041 | KC018398 | ||
| CBS474.96 | – | Ootomi, Iriomotejima Island, Okinawa, Japan | calcareous soil in ditch | JX976056 | KC018416 | ||
| CBS521.80 | – | Delhi, India | dung | JX976120 | KC018423 | ||
| Mortierella amoeboidea | CBS889.72, FSU9790 | M. alpina | Type of Mortierella amoeboidea | Teutoburger Wald, Beller Holz, Germany | JX976073 | HQ667422.1 | |
| Mortierella angusta | CBS293.61, FSU9791 | M. angusta | Neotype of Mortierella polycephala var. angusta | Chesh., Delamere Forest, England | podzol soil, pH up to 2.8 | JX976061 | HQ667358.1 |
| Mortierella antarctica | CBS194.89 | – | Northern Foothills, Northern Victoria Land, Antarctica | soil | JX976087 | KC018345 | |
| CBS195.89 | – | Northern Victoria Land, Edmonson Point, Antarctica | soil | JX975843 | – | ||
| CBS196.89 | – | Northern Victoria Land, Cape King, Antarctica | soil | JX976059 | – | ||
| CBS609.70, FSU9792 | – | Type of Mortierella antarctica | near Hallett Station, Antarctica | soil, rock crevice near glacier | JX975907 | HQ667503.1 | |
| Mortierella armillariicola | CBS105.78 | – | Putten, Schovenhorst, Netherlands | JX976100 | KC018432 | ||
| CBS914.73, FSU9793 | – | Type of Mortierella armillariicola | Baarn, Groeneveld, Netherlands | attacked by Dipodascus armillariae | JX975924 | HQ667446.1 | |
| Mortierella bainieri | CBS220.35 | – | former West-Germany | JX975901 | KC018324 | ||
| CBS272.71 | M. kuhlmanii | South Carolina | soil under Pinus taeda | JX975964 | JX976155 | ||
| CBS273.71 | M. kuhlmanii | South Carolina | soil under Pinus taeda | JX975920 | KC018355 | ||
| CBS442.68 | – | Georgia | soil from pine forest | JX975864 | KC018331 | ||
| CBS508.81 | – | Getzbach near Eupen, Belgium | JX975844 | KC018393 | |||
| CBS552.80 | – | Eifel, Hundsbachtal near Gerolstein, Germany | JX975850 | JX976174 | |||
| Mortierella basiparvispora | CBS517.72 | – | Valdivia, Cordillera Pelada, Chile | soil, under Fitzroya cupressoides | JX976048 | JX976167 | |
| Mortierella beljakovae | CBS102878 | – | Toronto High Park, Ontario | infrabuccal pellet of Camponotus pennsylvanicus (carpenter ant) on Pinus | JX976090 | KC018350 | |
| CBS109594 | – | Toronto, High Park, Ontario | infrabuccal pellet of Camponotuspennsylvanicus, in mature Pinus tree | JX975848 | KC018449 | ||
| CBS109595 | – | Zweifaller Wald near Aachen, Germany | infrabuccal pellet of Formica rufa | JX976129 | KC018358 | ||
| CBS109596 | – | St. Andrews, Annesley House, New Brunswick | infrabuccal pellet of Camponotus pennsylvanicus, in Pinus tree | JX975971 | JX976170 | ||
| CBS109597 | – | Scarborough, Ontario | infrabuccal pellet of Camponotus pennsylvanicus, in mature Pinus tree | JX975918 | KC018433 | ||
| CBS109655 | – | Bayerischer Wald, Pfahl bei Viechtach, Germany | infrabuccal pellet of Camponotus herculeanus, in Picea abies | JX975869 | JX976171 | ||
| CBS109658 | – | Zweifaller Wald near Aachen, Germany | infrabuccal pellet of Formica rufa | JX976051 | KC018376 | ||
| CBS109659 | – | Utrecht, Lage Vuursche, Netherlands | infrabuccal pellet of Formica rufa | JX975998 | KC018340 | ||
| CBS123.72, FSU9794 | M. beljakovae | Type of Mortierella beljakovae | Rovensk region, Sarna, Ukraine | soil, coniferous forest | JX976126 | HQ667428.1 | |
| CBS267.71 | – | North Carolina | seedling, Pinus teada | JX976072 | KC018346 | ||
| CBS268.71 | – | North Carolina | seedling, Pinus teada | JX976043 | KC018323 | ||
| CBS274.71 | – | South Carolina | root, Pinus taeda | JX976011 | KC018388 | ||
| CBS275.71 | – | South Carolina | root, Pinus taeda | JX975913 | KC018401 | ||
| CBS276.71 | – | South Carolina | root, Pinus taeda | JX975937 | KC018442 | ||
| CBS806.68 | – | North Carolina | bark of root, Pinus | JX975987 | KC018397 | ||
| Mortierella biramosa | CBS370.95, FSU9795 | M. biramosa | Type of Mortierella wuyishanensis | Wuyi, Fujian, China | forest soil | JX976094 | HQ667389.1 |
| CBS506.81 | – | Odenwald, Oberer Buntsandstein, Germany | decaying fine root, 30 yr old, on acidic loamy soil | JX975963 | KC018407 | ||
| CBS550.80 | – | Odenwald, Germany | rootlet | JX976064 | KC018419 | ||
| Mortierella bisporalis | CBS145.69 | – | Italy | JX975857 | KC018377 | ||
| FSU9675 | M. bisporalis | – | JX975953 | JX976176 | |||
| Mortierella camargensis | CBS110638 | – | Soest, Smickel, Netherlands | thatch of roof | JX976024 | – | |
| CBS221.58, FSU9796 | M. camargensis | Type of Mortierella camargensis | Camargue, Bois des Rièges, France | sandy soil | JX975949 | HQ667408.1 | |
| Mortierella capitata | CBS110640 | – | Berlin, Königin-Luise-Stra ße, near BBA, Germany | soil with Armadillidium | JX975923 | JX976163 | |
| CBS293.96 | – | Naganohara, Gunma, Japan | garden soil | JX976123 | KC018334 | ||
| CBS859.70 | – | North Carolina | pillbug gut | JX976008 | KC018395 | ||
| Mortierella chienii | CBS287.96 | – | Amakubo, Tsukuba, Ibaraki, Japan | soil under Quercus mirsinifolia forest | JX976013 | KC018427 | |
| CBS289.96 | – | Nanamagari, Yokohama, Kanagawa, Japan | soil under Castanopsis sieboldii forest | JX975898 | JX976161 | ||
| CBS290.96 | – | soil under Miscanthus sinensis | JX976075 | KC018373 | |||
| CBS292.96 | M. selenospora | Shitoko, Yakushima Island, Kagoshima, Japan | soil under Ficus microcarpa forest | JX975951 | JX976153 | ||
| CBS554.73 | M. selenospora | Kuang-Miau Co., 16 km E of Tainan, Taiwan | soil from bamboo grove | JX975912 | KC018381 | ||
| Mortierella chlamydospora | CBS120.34, FSU9799 | – | Syntype of Azygozygum chlamydosporum | infected by Rhizoctonia solani | JX975942 | HQ667430.1 | |
| CBS529.75 | – | Netherlands | soil | JX975927 | – | ||
| Mortierella claussenii | CBS790.85 | – | – | JX976012 | JX976159 | ||
| Mortierella clonocystis | CBS357.76, FSU9801 | M. clonocystis | Type of Mortierella clonocystis | Gran Canaria, Spain | soil, under Apollonias canariensis | JX975899 | HQ667395.1 |
| Mortierella cogitans | CBS879.97, FSU9802 | – | Type of Mortierella cogitans | Nagano, Sanada, Sugadaira M.R.C., Japan | decaying tree bark | JX976017 | HQ667360.1 |
| Mortierella cystojenkinii | CBS456.71, FSU9803 | M. cystojenkinii | Type of Mortierella cystojenkinii | Wageningen, Netherlands | agricultural soil | JX976030 | HQ667504.1 |
| CBS660.82 | – | Bakkeveen, Netherlands | Pinus forest | JX975868 | KC018325 | ||
| Mortierella decipiens | CBS873.68 | – | Kiel-Kitzeberg, Germany | wheat field soil | – | JX976173 | |
| Mortierella dichotoma | CBS221.35, FSU9804 | M. dichotoma | Syntype of Mortierella dichotoma | former West-Germany | dung of mouse | JX975842 | HQ667393.1 |
| Mortierella echinosphaera | CBS574.75 | – | near Wageningen, Netherlands | soil | JX976060 | KC018370 | |
| CBS575.75, FSU9805 | M. echinosphaera | Holotype of Mortierella echinosphaera | Aalsmeer, Netherlands | JX976015 | HQ667431.1 | ||
| Mortierella echinula | CBS282.71 | – | Iceland | soil | JX975948 | – | |
| Mortierella elongata | CBS110517 | – | Alti Mountains, South Africa | soil, grassland, summer rainfall region | JX976042 | KC018348 | |
| CBS122.71 | – | Georgia, Monroe, USA | soil, under golf turf-grass | JX976000 | KC018396 | ||
| CBS126.71, FSU823 | M. elongata | Wageningen, Netherlands | agricultural soil | JX976101 | KC018279 | ||
| CBS208.71 | – | Netherlands | greenhouse soil | JX975995 | JX976135 | ||
| CBS276.89 | – | Quebec | (black fly) | JX976111 | KC018452 | ||
| CBS279.62 | – | Kiel-Kitzeberg, Germany | wheat field soil | JX976089 | KC018417 | ||
| CBS344.66 | – | Alaska | tundra soil | JX976081 | KC018322 | ||
| FSU532 | M. elongata | – | JX975976 | KC018281 | |||
| FSU822, CBS125.71 | M. elongata | Wageningen, Netherlands | agricultural soil | JX975978 | KC018282 | ||
| FSU9721 | M. elongata | Münchenroda, Germany | JX975894 | KC018284 | |||
| Mortierella elongatula | CBS488.70, FSU9808 | – | Type of Mortierella elongatula | former West-Germany | municipal waste | JX975967 | HQ667425.1 |
| CBS661.70 | – | Braunschweig, Germany | municipal waste | JX976069 | KC018431 | ||
| Mortierella epicladia | CBS246.75 | – | Suriname | soil, under Elaeis guineensis | JX975890 | KC018361 | |
| CBS355.76, FSU9809 | M. epiclada | Type of Mortierella epicladia | Gran Canaria, Spain | soil, under Apollonias canariensis | JX976130 | HQ667396.1 | |
| CBS356.76 | – | Gran Canaria, Spain | soil, under Apollonias canariensis | JX975972 | – | ||
| CBS555.89 | – | Pará, 200 km SE from Belém, Capitâo Poço, Brasil | rain forest soil | JX975991 | JX976150 | ||
| Mortierella epigama | CBS161.76 | M. epigama | Exeter, Hatherly Laboratories, England | compost heap | JX976109 | JX976158 | |
| CBS489.70, FSU9810 | M. epigama | Type of Mortierella epigama | former West-Germany | municipal waste | JX976057 | HQ667367.1 | |
| CBS881.97 | – | Kagoshima, Kamei, Tokunoshima-Island, Japan | old dung of cow | JX976053 | KC018445 | ||
| Mortierella exigua | CBS358.76 | – | Gran Canaria, Spain | soil, under Apollonias canariensis | JX976113 | KC018439 | |
| CBS510.63 | – | Kiel-Kitzeberg | agricultural soil | JX975863 | JX976134 | ||
| CBS655.68, FSU9811 | M. exigua | Type of Mortierella sterilis | Allahabad, India | farm soil | JX976047 | HQ667406.1 | |
| CBS865.68 | – | Kiel-Kitzeberg, Germany | wheat field soil | JX976070 | – | ||
| Mortierella fatshederae | CBS388.71 | – | Gran Canaria | soil, under Pinus canariensis | JX976003 | JX976136 | |
| Mortierella fimbricystis | CBS943.70 | – | Type of Mortierella fimbricystis | South Patagonia, Puerto Edwards near Beagle Canal, Argentinia | centre of moss cushion, in very wet bog | GU559986.1 | JX976172 |
| Mortierella formicicola | CBS109589 | – | Brampton, Ontario | infrabuccal pellet of Camponotus pennsylvanicus, in house (windowsill) | JX975933 | JX976140 | |
| Mortierella gamsii | CBS110630 | – | Boekrijk, Belgium | soil with Porcellio | JX976106 | KC018410 | |
| CBS253.36, FSU9813 | M. gamsii | Syntype of Mortierella spinosa | former West-Germany | forest soil | JX975968 | HQ667415.1 | |
| CBS314.52, FSU9814 | M. cf. gamsii | Syntype of Mortierella spinosa | former West-Germany | forest soil | JX975892 | HQ667384.1 | |
| CBS551.73, FSU824 | M. gamsii | North Carolina | pasture soil | JX976079 | JX976177 | ||
| CBS552.73, FSU825 | M. gamsii | Alleghany County, North Carolina | pasture soil | JX975984 | KC018285 | ||
| CBS749.68, FSU9812 | M. gamsii | Type of Mortierella gamsii | Baarn, Maarschalksbos, Netherlands | soil | HQ667416.1 | ||
| FSU2057 | M. gamsii | – | JX976118 | KC018287 | |||
| Mortierella gemmifera | CBS124.72 | – | Meerdinkbos near Winterswijk, Netherlands | soil, humus layer | JX975909 | KC018390 | |
| CBS134.45, FSU9815 | M. gemmifera | Type of Mortierella gemmifera | near Nottingham, England | soil from pine forest | JX975931 | HQ667371.1 | |
| CBS383.85 | – | Spanderswoud near Bussum, Netherlands | soil, in pine forest | JX976121 | JX976157 | ||
| CBS661.82 | – | Bakkeveen, Netherlands | Endogone lactiflua, Pinus forest | JX975989 | KC018360 | ||
| Mortierella globalpina | CBS226.78 | – | Katwijk, Netherlands | sand dune soil | JX976006 | JX976160 | |
| CBS718.88 | – | Japan | JX975925 | – | |||
| Mortierella globulifera | CBS108.68 | – | Schweden | JX975847 | KC018332 | ||
| CBS746.68 | – | Netherlands | agricultural soil | JX976026 | KC018371 | ||
| CBS857.70, FSU826 | – | England | decaying needle | JX975910 | HQ667369 | ||
| CBS858.70, FSU9817 | M. globulifera | Neotype of Mortierella globulifera | England | decaying root | JX975915 | HQ667368.1 | |
| CBS867.68 | – | Tirol, Obergurgl, Austria | alpine raw humus soil | JX976107 | JX976165 | ||
| Mortierella histoplasmatoides | CBS321.78, FSU9819 | – | Type of Mortierella histoplasmatoides | Louisiana | dung | HQ630309.1 | HQ667386.1 |
| Mortierella horticola | CBS305.52, FSU9820 | M. horticola | Syntype of Mortierella horticola | former West-Germany | JX975874 | HQ667399.1 | |
| CBS869.68 | – | Kiel-Kitzeberg | wheat field soil | JX976058 | JX976138 | ||
| CBS254.76 | – | Wageningen, Netherlands | agricultural soil | JX976021 | JX976166 | ||
| Mortierella humilis | CBS180.72 | – | Piedmont, North Carolina | forest soil | JX976125 | KC018436 | |
| CBS181.72 | – | Piedmont, North Carolina | soil | JX975887 | KC018405 | ||
| CBS222.35, FSU9821 | – | Syntype of Mortierella humilis | Mexico | soil from Pinus forest | HQ630325.1 | HQ667401.1 | |
| CBS363.95 | – | Shennongjia, Hubei, China | forest soil | JX976097 | KC018443 | ||
| CBS443.68, FSU828 | M. humilis | South Carolina | bark of stump | JX976002 | HQ667402 | ||
| CBS745.68, FSU829 | M. humilis | Baarn, Eemnesserweg 90, Netherlands | soil | JX975867 | HQ667403 | ||
| Mortierella hyalina | CBS100563 | – | Schoharie Co., New York | JX976023 | KC018356 | ||
| CBS115655, FSU9822 | M. hyalina | Isotype of Hydrophora hyalina | North of London, Rothamsted, England | roots | HQ630355.1 | HQ667432.1 | |
| CBS117.74 | – | Boekesteyn near ’s-Graveland, Netherlands | JX976083 | KC018392 | |||
| CBS117152 | – | Graz, Austria | soil and chees mixture used as food for mites by E. Ebermann | JX975977 | KC018394 | ||
| CBS166.25 | – | Netherlands | seed | JX975928 | – | ||
| CBS167.25 | – | – | JX975895 | KC018406 | |||
| FSU10532 | M. hyalina | Austria | JX975992 | KC018289 | |||
| FSU509 | M. hyalina | – | JX975981 | KC018291 | |||
| Mortierella hypsicladia | CBS116202, FSU9825 | M. hypsicladia | Type of Mortierella hypsicladia | Kyushu Isl., Kariu Cave, Japan | bat dung in cave | JX975866 | HQ667379.1 |
| CBS116203 | Authentic strain of Mortierella hypsicladia | Kyushu Isl., Kariu Cave, Japan | bat dung in cave | JX975872 | KC018369 | ||
| Mortierella indohii | CBS220.72 | Naaldwijk, Netherlands | greenhouse soil | JX975993 | KC018408 | ||
| CBS331.74, FSU830 | M. indohii | Lienden, Netherlands | root | JX975860 | KC018292 | ||
| CBS460.75, FSU831 | M. indohii | Athens, Georgia | dung of animal | JX975878 | HQ667438 | ||
| CBS478.95 | Chengdu, Sichuan, China | soil | JX975903 | KC018347 | |||
| CBS528.75 | South Africa | bagasse in chicken farm | JX976044 | KC018451 | |||
| CBS665.70 | Wageningen, Netherlands | agricultural soil | JX975956 | KC018357 | |||
| CBS720.71, FSU9826 | M. indohii | Isotype of Mortierella indohii | Athens, Georgia | dung of animal | JX975856 | HQ667377.1 | |
| Mortierella jenkinii | CBS188.73 | Nottingham, England | turf layer of golf green, received fungicidal treatment for long period | JX975999 | KC018389 | ||
| CBS666.75C | Sweden | soil under Picea abies | – | JX975873 | |||
| CBS667.70 | Wageningen, Netherlands | agricultural soil | JX976088 | KC018422 | |||
| CBS850.70 | Wageningen, Netherlands | agricultural soil | JX975849 | KC018352 | |||
| CBS965.73C | Sweden | forest soil | JX976117 | JX976139 | |||
| Mortierella kuhlmanii | CBS157.71, FSU9827 | M. kuhlmanii | Type of Mortierella kuhlmanii | South Carolina, Miley | stump | JX975846 | HQ667372.1 |
| CBS269.71 | stump, Pinus taeda | JX975935 | KC018384 | ||||
| CBS270.71 | Patrick, South Carolina | stump | JX975851 | JX976142 | |||
| CBS271.71 | South Carolina | seedling | JX975883 | KC018338 | |||
| Mortierella lignicola | CBS100594 | – | JX975889 | – | |||
| CBS116.65 | Wageningen, Netherlands | black soil | JX975965 | KC018402 | |||
| CBS207.37, FSU9828 | M. lignicola | Type of Haplosporangium lignicola | Sierra Nevada de Santa Marta, Colombia | rotten wood | JX976095 | HQ667435.1 | |
| CBS313.52, FSU9829 | M. lignicola | Type of Mortierella sepedonioides | former West-Germany | soil under Pinus sylvestris | JX976127 | HQ667434.1 | |
| Mortierella longigemmata | CBS653.93 | Höglwald, Germany | soil | JX976055 | JX976162 | ||
| Mortierella macrocystis | CBS110716 | De Veluwe | oak forest soil | JX976084 | – | ||
| CBS314.85 | former West-Germany | rootlet of gymnosperm | JX975974 | JX976169 | |||
| CBS431.81 | Cundinamarca, páramo Cruz Verde, Colombia | soil | JX975897 | KC018437 | |||
| CBS482.73 | former West-Germany | soil | JX975862 | – | |||
| CBS937.69 | Baarn, Pekingtuin, Netherlands | soil | JX975881 | KC018341 | |||
| Mortierella macrocystopsis | CBS302.87 | South Kingstown, Rhode Island | soil under Pinus resinosa and Pinus strobus | JX975908 | KC018362 | ||
| CBS387.91 | M. cystojenkinii | Norway | soil | JX976105 | JX976144 | ||
| CBS520.88 | Rhode Island | soil | JX976078 | – | |||
| CBS528.87 | South Kingstown, Rhode Island | forest soil, under Pinus resinosa and Pinus strobus | JX975946 | JX976164 | |||
| Mortierella microzygospora | CBS880.97, FSU9831 | M. microzygospora | Type of Mortierella microzygospora | Shiga, Maibara, Japan | soil in hedge | JX976027 | HQ667394.1 |
| Mortierella minutissima | CBS226.35 | former West-Germany | JX976092 | JX976168 | |||
| CBS277.71, FSU832 | M. minutissima | Georgia | forest soil | JX975938 | KC018293 | ||
| FSU2735 | M. zonata | – | JX976103 | KC018318 | |||
| Mortierella minutissima var. dubia | CBS307.52, FSU9832 | Syntype of Mortierella minutissima var. dubia | former West-Germany | soil | JX976122 | HQ667400.1 | |
| Mortierella nantahalensis | CBS610.70, FSU9834 | M. nantahalensis | Type of Mortierella nantahalensis | Joyce Kilmer Memorial Forest in the Nantahala National Forest, North Carolina | soil | JX976022 | HQ667388.1 |
| Mortierella oligospora | CBS101758 | Pennsylvania | supplement to mushroom culture | JX976032 | KC018327 | ||
| CBS191.79 | Elephant White Nile Island, Sudan | soil | JX975966 | JX976151 | |||
| CBS381.71 | Jaipur, Rambagh Palace Hotel, Rajasthan | soil | JX976033 | KC018368 | |||
| Mortierella paraensis | CBS343.89 | Pará, Capitão Poço, Brazil | forest soil, virgin forest | JX975944 | KC018329 | ||
| CBS547.89, FSU9835 | M. paraensis | Type of Mortierella paraensis | Pará, 200 km SE from Belém, Capitão Poço, Brasil | rain forest soil | HQ630353 | HQ667429.1 | |
| Mortierella parazychae | CBS868.71, FSU9836 | M. parazychae | Type of Mortierella parazychae | Treek near Amersfoort, Netherlands | decaying wood, with Botryobasidium subcoronatum | JX975985 | HQ667362.1 |
| Mortierella parvispora | CBS304.52, FSU9837 | M. parvispora | Syntype of Mortierella gracilis | former West-Germany | soil | JX975859 | – |
| CBS311.52, FSU9838 | Syntype of Mortierella parvispora | former West-Germany | soil | JX976076 | HQ667373.1 | ||
| CBS315.61, FSU834 | M. parvispora | Cheshire, Delamere Forest, England | soil, iron-humus podzol | JX976104 | HQ667374.1 | ||
| CBS316.61, FSU835 | M. parvispora | Cheshire, Delamere Forest, England | soil, iron-humus podzol | JX976029 | HQ667375.1 | ||
| CBS445.68 | Wageningen, Netherlands | beet-field soil | JX976049 | KC018414 | |||
| FSU2736 | M. jenkinii | – | JX976093 | KC018295 | |||
| Mortierella polycephala | CBS227.35 | – | JX976096 | KC018321 | |||
| CBS293.34 | M. hyalina | Netherlands | JX976050 | JX976137 | |||
| CBS327.72, FSU866 | M. polycephala | Lincs., Gibraltar Point, England | salt-marsh soil under Spartina townsendii | JX976085 | JX976175 | ||
| CBS328.72, FSU867 | M. polycephala | UK | soil | JX976102 | KC018296 | ||
| CBS456.66, FSU759 | M. polycephala | near Kiev, Ukraine | dung of wood mouse | JX976034 | KC018297 | ||
| FSU696 | M. polycephala | – | JX976035 | KC018298 | |||
| Mortierella polygonia | CBS248.81 | Sexbierum, Netherlands | clay soil under Solanum tuberosum | JX975891 | JX976145 | ||
| CBS685.71, FSU9839 | Type of Mortierella polygonia | Wageningen, Netherlands | agricultural soil | JX975900 | HQ667378.1 | ||
| Mortierella pseudozygospora | CBS779.86 | Kingston, North Woods, Univ. of Rhode Island Campus, Rhode Island | soil under Quercus-Acer woodland, about sea level, upper 5 cm depth | JX975960 | KC018353 | ||
| CBS780.86 | Peace Dale, Hazard Tract, Rhode Island | soil, under Pinus strobus and Pinus resinosa woodland, from upper 5 cm depth, soil temp. 2.5°C | JX975880 | JX976143 | |||
| Mortierella pulchella | CBS205.86 | Netherlands | root | JX976031 | KC018366 | ||
| CBS312.52, FSU9840 | Authentic strain of Mortierella pulchella | former West-Germany | root | JX976054 | HQ667427.1 | ||
| CBS675.88 | Berlin, Grunewald, Jagen 91, Germany | soil, litter layer | JX976082 | KC018440 | |||
| Mortierella reticulata | CBS110044 | Lanark near Branxholme, Victoria | dung of Perameles gunnii | JX975980 | – | ||
| CBS223.29 | – | JX975973 | – | ||||
| CBS241.33 | – | JX976116 | JX976133 | ||||
| CBS415.81 | Toronto, Ontario | dung of mouse, collected in a house | JX975877 | – | |||
| Mortierella rishikesha | CBS652.68, FSU9842 | Type of Mortierella rishikesha | Rishikesh, India | forest soil | JX976110 | HQ667385.1 | |
| Mortierella rostafinskii | CBS522.70, FSU9844 | Neotype of Mortierella rostafinskii | near Bainbridge, Georgia | soil under Pinus elliottii var. elliottii | JX975885 | HQ667436.1 | |
| Mortierella sarnyensis | CBS122.72, FSU9845 | M. sarnyensis | Type of Mortierella sarnyensis | Rovensk region, near Sarny, Ukraine | coniferous forest | JX975957 | HQ667390.1 |
| Mortierella schmuckeri | CBS156.78 | Madhya Pradesh and Uttar Pradesh regions, India | soil, from ravines | JX975854 | KC018372 | ||
| CBS295.59, FSU9846 | M. schmuckeri | Syntype of Mortierella schmuckeri | Queretaro, Mexico | soil, under Opuntia sp., pH 6.7 | JX976112 | HQ667414.1 | |
| CBS777.86 | Shoshone National Forest, Horse Creek Campground, Wyoming | soil, upper 10 cm, under Pseudotsuga menziesii, alt. 2500 m | JX976099 | KC018413 | |||
| Mortierella sclerotiella | CBS529.68, FSU9847 | M. sclerotiella | Type of Mortierella sclerotiella | Ukraine | dung of mouse | JX975988 | HQ667387.1 |
| Mortierella selenospora | CBS452.88 | Cibodas, Indonesia | soil | JX976037 | KC018429 | ||
| CBS811.68, FSU9848 | M. selenospora | Type of Mortierella selenospora | Horst, Netherlands | mushroom compost, together with Entomophthora coronata and Aphanocladium album | JX975875 | HQ667419.1 | |
| Mortierella simplex | CBS110.68 | Wageningen, Netherlands | oat-field soil | JX975982 | – | ||
| CBS243.82 | Baarn, C. Dopperlaan 18, Netherlands | compost heap | JX975870 | JX976156 | |||
| Mortierella sossauensis | CBS153.76C | Schweden | forest soil under Picea abies | JX976063 | JX976146 | ||
| CBS176.74 | M. clonocystis | Athens, Georgia | Greenhouse soile | JX975926 | KC018428 | ||
| CBS281.71 | South Carolina | root | JX975911 | KC018447 | |||
| CBS890.72 | Ireland | peat soil | JX975865 | KC018385 | |||
| CBS898.68 | Lincs., Gibraltar Point, England | salt-marsh soil | JX975970 | KC018374 | |||
| Mortierella sp. | FSU10519 | M. alpina | Austria | JX975959 | KC018258 | ||
| FSU10520 | M. alpina | Austria | JX975969 | KC018259 | |||
| FSU10522 | M. alpina | Austria | JX975930 | KC018261 | |||
| FSU10523 | M. alpina | Austria | JX976114 | KC018262 | |||
| FSU10551 | M. alpina | Austria | JX975852 | KC018269 | |||
| FSU10555 | M. alpina | Austria | JX975996 | KC018315 | |||
| FSU10558 | M. alpina | Austria | JX975884 | KC018271 | |||
| FSU10683 | M. alpina | Austria | JX976039 | – | |||
| FSU10696 | M. alpina | Austria | JX976108 | – | |||
| FSU10706 | M. alpina | Austria | JX976068 | – | |||
| FSU10715 | M. alpina | Austria | JX976080 | – | |||
| FSU10716 | M. alpina | Austria | JX975879 | – | |||
| FSU8712 | M. alpina | Wehlen, Mosel, Germany | JX975845 | KC018274 | |||
| FSU8722 | M. alpina | Wehlen, Mosel, Germany | JX975961 | KC018275 | |||
| FSU8736 | M. alpina | Wehlen, Mosel, Germany | JX976119 | KC018276 | |||
| FSU8737 | M. alpina | Wehlen, Mosel, Germany | JX975902 | KC018277 | |||
| FSU8738 | M. alpina | Wehlen, Mosel, Germany | JX976010 | KC018278 | |||
| CBS118520 | Græse, Zealand, Denmark | agricultural soil | JX975936 | JX976149 | |||
| FSU10767 | Austria | JX975929 | – | ||||
| FSU10792 | Austria | JX976014 | – | ||||
| FSU10797 | Austria | JX975950 | – | ||||
| FSU10541 | M. elongata | Austria | JX975876 | KC018310 | |||
| FSU10771 | M. elongata | Austria | JX976131 | – | |||
| FSU8711 | M. elongata | Wehlen, Mosel, Germany | JX976071 | KC018283 | |||
| FSU10538 | M. gamsii | Austria | JX975858 | KC018286 | |||
| FSU10535 | M. humilis | Austria | JX976052 | – | |||
| FSU1954 | M. hyalina | – | JX975861 | KC018290 | |||
| FSU10804 | M. minutissima | Austria | JX976020 | – | |||
| FSU10552 | M. parvispora | Austria | JX976009 | KC018294 | |||
| FSU10712 | Austria | JX975941 | – | ||||
| FSU10730 | Austria | JX975916 | – | ||||
| FSU10753 | Austria | JX976016 | – | ||||
| FSU10758 | Austria | JX976005 | – | ||||
| FSU10759 | M. parvispora | Austria | JX975934 | – | |||
| FSU10789 | Austria | JX976065 | – | ||||
| FSU10530 | Austria | JX975893 | KC018306 | ||||
| FSU10540 | Austria | JX975986 | KC018309 | ||||
| FSU10557 | Austria | JX975932 | – | ||||
| FSU2188 | – | JX975945 | KC018316 | ||||
| FSU10534 | M. verticillata | Austria | JX975914 | KC018317 | |||
| Mortierella strangulata | CBS455.67, FSU9849 | M. strangulata | Neotype of Mortierella strangulata | Baarn, Groeneveld, Nmetherlands | dung, of fox ? | JX975997 | HQ667437.1 |
| Mortierella stylospora | CBS211.32, FSU9850 | M. stylospora | Type of Mortierella stylospora | Victoria | sandy loam | JX976086 | HQ667359.1 |
| Mortierella turficola | CBS430.76 | Heseper Veen near Coevorden, Netherlands | decaying Sphagnum recurvum | JX975919 | KC018444 | ||
| CBS431.76 | Heseper Veen near Coevorden, Netherlands | decaying Sphagnum recurvum | JX976025 | KC018333 | |||
| CBS432.76, FSU9851 | M. turficola | Neotype of Mortierella turficola | Heseper Veen near Coevorden, Netherlands | decaying Sphagnum recurvum | JX975952 | HQ667426.1 | |
| CBS433.76 | Heseper Veen near Coevorden, Netherlands | decaying Sphagnum recurvum | JX975939 | KC018424 | |||
| CBS547.76 | Cauca en Huila, Cordillera Central, Parque Nacional del Puracé, 3100 m alt., Colombia | soil from mountain forest under Weinmannia etc. | JX975896 | KC018339 | |||
| CBS581.80 | Netherlands | Trio compost | JX976040 | KC018409 | |||
| Mortierella verticillata | CBS130.66 | Lancashire, Freshfield, England | sandy forest soil | JX976007 | KC018326 | ||
| CBS131.66 | Lancashire, Freshfield, England | sandy forest soil | JX975886 | KC018446 | |||
| CBS220.58, FSU9853 | M. verticillata | Type of Haplosporangium fasciculatum | Fontainebleau, France | soil under Betula sp. | JX975905 | JN940873.1 | |
| CBS225.35, FSU9854 | M. verticillata | Syntype of Mortierella marburgensis | former West-Germany | JX975940 | JQ040251.1 | ||
| CBS279.71 | South Carolina | root | JX975917 | KC018426 | |||
| CBS280.71 | South Carolina | root | JX976066 | KC018404 | |||
| CBS315.52, FSU9856 | M. verticillata | Syntype of Mortierella marburgensis | former West-Germany | forest soil | JX975943 | – | |
| CBS346.66, FSU9852 | Alaska | tundra soil | JX975855 | HQ667397.1 | |||
| CBS374.95, FSU9855 | M. verticillata | Type of Haplosporangium attenuatis- simum | Wuyi, Fujian, China | forest soil | JX976077 | HQ667398.1 | |
| Mortierella wolfii | CBS614.70, FSU9860 | M. cf. wolfi | Matamata, New Zealand | decayed hay | JX975975 | HQ667420.1 | |
| CBS209.69, FSU9858 | M. wolfii | Keele, England | coal spoil tip soil | HQ630303.1 | HQ667380.1 | ||
| CBS611.70, FSU9857 | Morrinsville, New Zealand | lung, dying from mycotic pneumonia | HQ630306.1 | HQ667383.1 | |||
| CBS612.70, FSU9859 | New Zealand | decayed hay | HQ630304.1 | HQ667381.1 | |||
| CBS651.93, FSU9862 | M. wolfii | Limburg, Horst, Netherlands | compost for mushrooms | JX975904 | HQ667382.1 | ||
| Mortierella zonata | CBS228.35, FSU9863 | M. zonata | Type of Mortierella zonata | former West-Germany | JX975983 | HQ667433.1 | |
| CBS615.70 | Braunschweig-Völkenrode, Germany | soil | JX975958 | KC018434 | |||
| CBS617.76 | Cordillera, Central Parque Nacional del Puracé, 3900 m alt. | páramo soil, open vegetation with extensive pasture | JX976028 | JX976141 | |||
| CBS863.68 | Ringwood, New Forest, UK | forest soil | JX975888 | KC018335 | |||
| Mortierella zychae | CBS102879 | Toronto High Park, Ontario | pellet of Camponotus pennsylvanicus (carpenter ant) | JX976074 | |||
| CBS109599 | El Yunque, Rio Blanco Trail, Puerto Rico | infrabuccal pellet of ant | JX975882 | ||||
| CBS143.91 | former West-Germany | JX976091 | |||||
| CBS316.52, FSU9864 | M. zychae | Type of Mortierella zychae | Allgäu, Germany | decaying wood | JX975979 | HQ667407.1 | |
| CBS531.81 | former West-Germany | mushroom casing soil | JX975962 | KC018421 | |||
| FSU719 | M. zychae | – | JX976128 | KC018319 | |||
| Umbelopsis isabellina | NRRL1757, CBS100559 | Wisconsin | soil | JN943789.1 | JN940879.1 |
Preparation of genomic DNA, PCR amplification and DNA sequencing
Genomic DNA was prepared from mycelia grounded to a fine powder in liquid nitrogen followed by purification (Cenis 1992) or living cultures alternatively, using the Jetquick general DNA clean up kit (Genomed) or a high-throughput 96-well plate extraction (Ivanova et al. 2006) following the given protocols. The PCR for the amplification of the ITS1-5.8S-ITS2 nuclear ribosomal DNA region uses ITS5/ITS1 and ITS4 under standard or semi-nested conditions (White et al. 1990, Stielow et al. 2009). PCR for amplifying the partial 28S rDNA (LSU) was done using the standard primers LR0R and LR5 or the NL-primer (http://www.biology.duke.edu/fungi/mycolab/primers.htm). The primers differ only in their annealing temperature (55 °C or 60 °C). Increasing cycle extension time (90 s/cycle) was done in some cases to improve amplification. PCR products were directly purified using FastAP thermosensitive alkaline phosphatase and shrimp alkaline phosphatase (Fermentas, Thermo Scientific) or using the GeneClean protocol (Vogelstein & Gillespie 1979). The cycle-sequencing reaction was set up using ABI big dye terminator v. 3.1, following the manufactures instructions or by using a quarter of the suggested volumes (modified manufactures protocol), followed by bidirectional sequencing with a laboratory capillary electrophoresis system (Life Technologies 3730XL DNA analyser). Sequences were evaluated with Chromas Lite (Technelysium Pty. Ltd.). Sequencing primers were the same as used for PCR. Manually correction and assembling of forward and reverse sequences was done using the Biolomics database (www.bio-aware.com) (Vu et al. 2012) or Seqman (v. 7.2.1). Sequences were deposited at NCBI GenBank (Table 2).
Alignments and phylogenetic analyses
A total of 364 sequences of ITS and 213 sequences of LSU were generated in this study. For the extension of the dataset additional sequences were retrieved from GenBank (Table 2). A total of 15 sequences were excluded and 562 were subjected to further analyses (298 ITS and 263 LSU sequences). Alignments were performed with MAFFT v. 6.833 (Katoh 2008) as implemented in EPoS (Griebel et al. 2008). Maximum Likelihood analyses were carried out using RAxML (Stamatakis 2006) provided by the CIPRES Science Gateway v. 3.2 (http://www.phylo.org). RAxML was run under the default settings with the following adjustments: GTRGAMMA for bootstrapping and final tree inference with 1 000 bootstrap iterations. The resulting phylogenetic trees which based on the LSU sequences were used to identify clusters of strains. For these clusters MAFFT alignments of the ITS region were computed and RAxML analyses performed. Subsequent alignments are crucial since ITS is in general highly diverse on higher level classification. If a group of sequences contains a high number of a repetitive species not all sequences were included in the ITS tree. Alignments and trees are deposited in TreeBASE2 under http://purl.org/phylo/treebase/phylows/study/TB2:S13827.
RESULTS AND DISCUSSION
Phylogenetic analyses and relationships within the Mortierellales based on single-locus analyses
According to previous studies (White et al. 2006, Petkovits et al. 2011), the major genus of the Mortierellales, Mortierella, appears as paraphyletic genus since the genera, Dissophora, Gamsiella and Lobosporangium are nested within. Since there is no sequence data or living material available for Aquamortierella and Modicella (White et al. 2006) these genera were not included. Due to lacking species material the newly proposed and described genus Echinochlamydosporium (Jiang et al. 2011) was also excluded from the current analysis. Although the pre-molecular classification schemes defined morphologically well-supported clades (Linnemann 1941, Zycha et al. 1969, Gams 1977) these clades could not be retained in any molecular based analyses (White et al. 2006, Petkovits et al. 2011, this study). The present study extended a previous study by addition of sequence information for 407 specimens. One isolate, Mortierella mutabilis, was excluded due to miss-fitting morphological characteristics. The morphology of M. mutabilis is in contradiction with its original description (Linnemann 1941) and resembles Gamsiella multidivaricata in all morphological features as well its molecular data. Since only one isolate is available, we postpone its phylogenetically analysis till additional material is available. Nineteen species were additionally included with a total of 115 sequences. Out of these sequences 57 sequences were generated for ITS, 58 for LSU and 1 ITS sequence was retrieved from GenBank.
Out of 421 specimens in total, 213 sequences for LSU and 364 sequences for ITS were generated. The dataset was supplemented with additional sequences form GenBank (69 LSU and 11 ITS sequences) (Table 2).
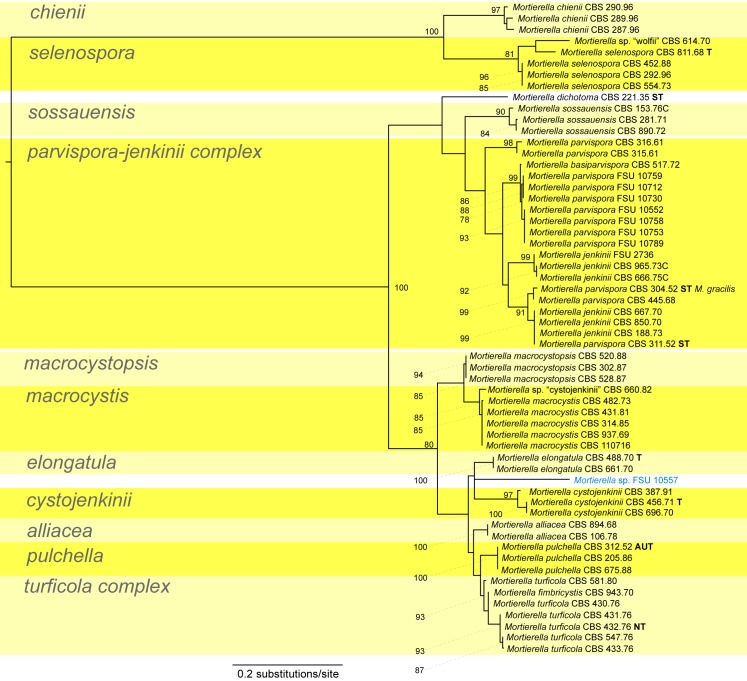
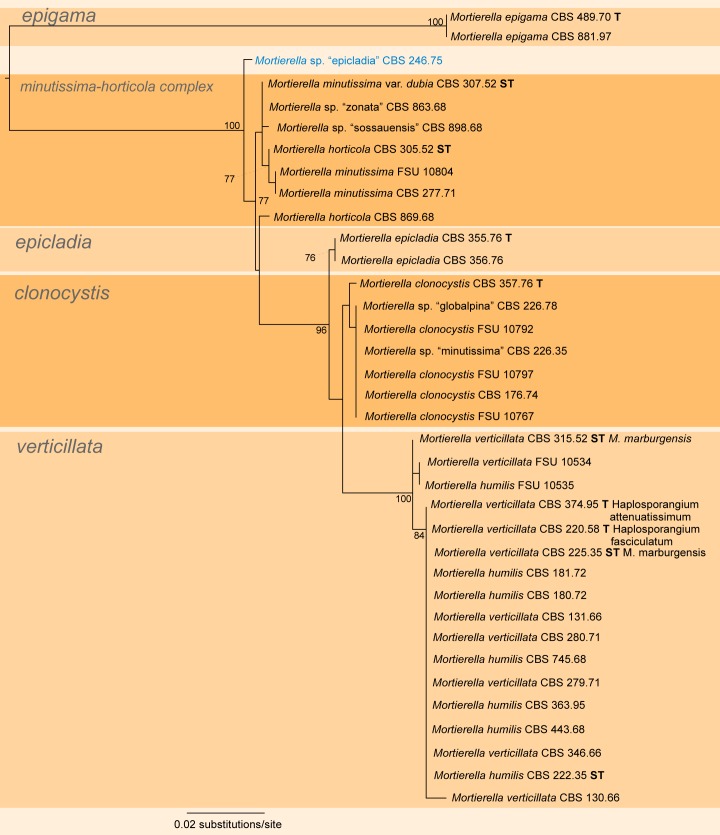
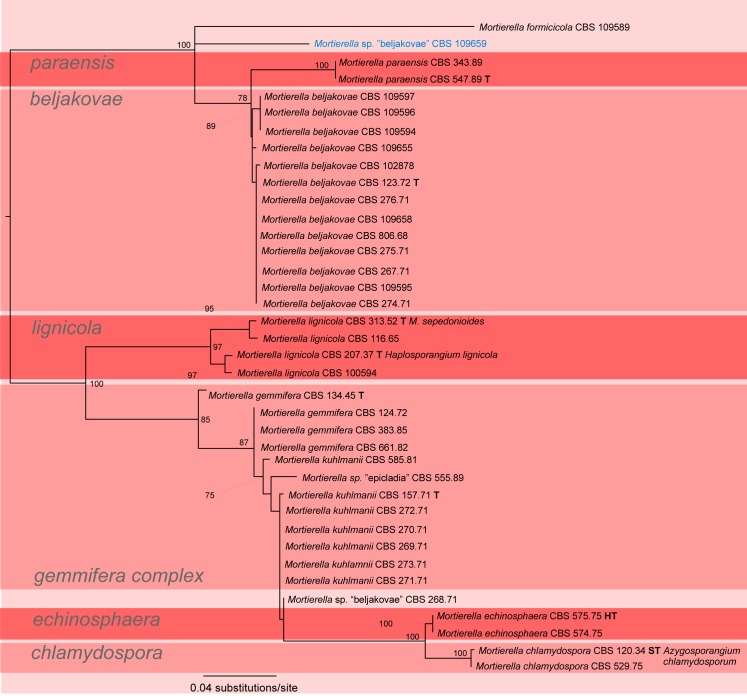
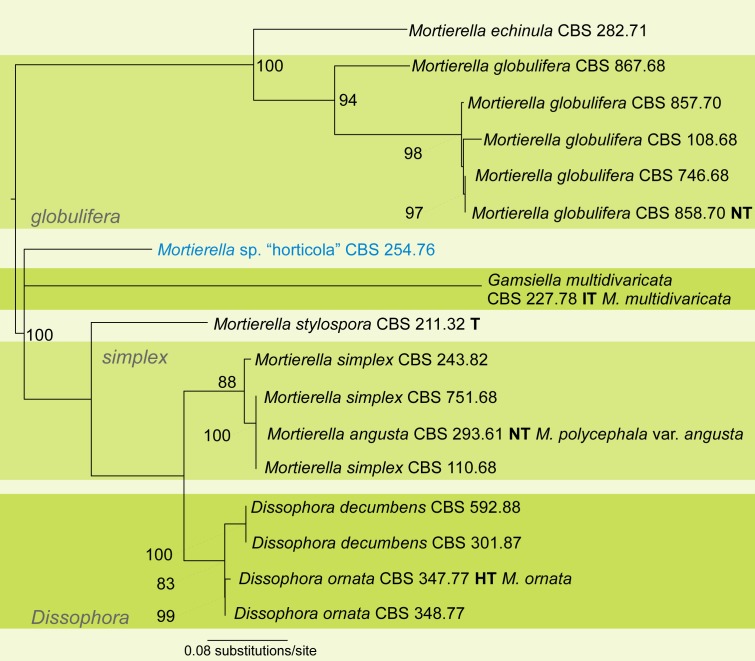
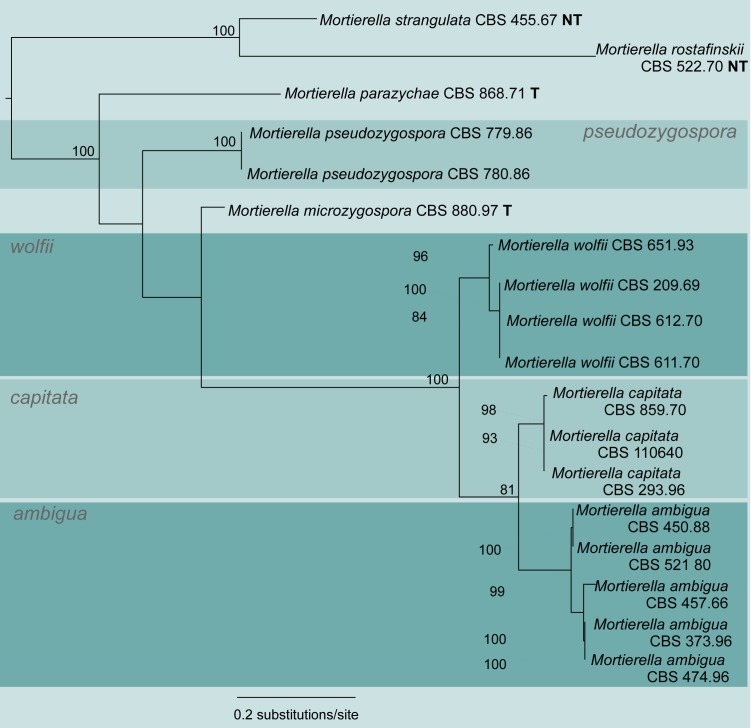
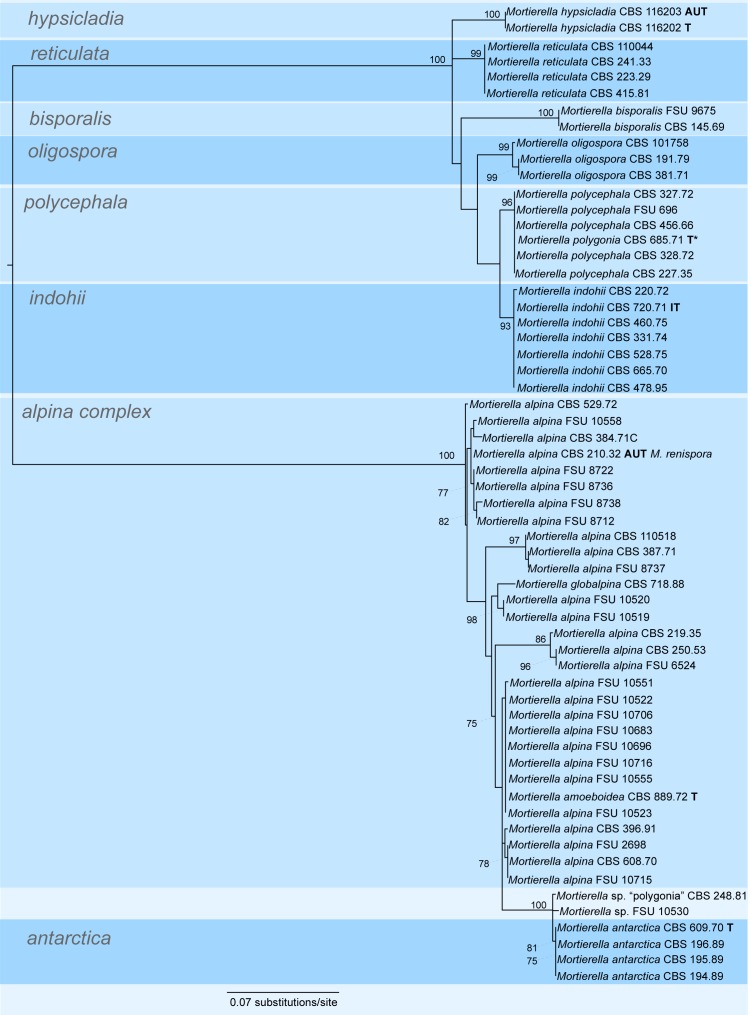
A first phylogenetic tree based on LSU sequences from 266 taxa was generated to define placement and relationships of all sequences generated in this study (data not shown). A subset of all relevant groups and isolates was taken for the final tree of the LSU dataset (Fig. 3, just for better overview). The final alignment contains 781 characters and 101 taxa. For subsequent deep-level analyses seven artificial subsets out of eight clades of this tree were defined referring to the previously published group delimitations (Petkovits et al. 2011). For each group the ITS1-5.8S rDNA-ITS2 sequences were aligned and analysed with Maximum Likelihood although the backbone of the underlying LSU tree is not resolved (Fig. 3). Groups are mainly located on one branch (‘monophyletic’) except for the under-represented chienii/selenospora-group which was combined and aligned together with the most basal group. Taking these groups as single taxa sets allows alignments providing phylogenetic signals with higher resolution on deep level classification. The alignments of the subsets consists of the following numbers of taxa and characters: subset 1: 58/816 (means 58 taxa and 816 characters, Fig. 4); subset 2: 36/636 (Fig. 5); subset 3: 38/701 (Fig. 6); subset 4: 17/710 (Fig. 7); subset 5: 18/761 (Fig. 8); subset 6: 60/703 (Fig. 9); subset 7: 73/688 (Fig. 10).
Fig. 3.
Maximum Likelihood analysis based on 781 aligned nucleotides of the D1/D2 domain of the large subunit (LSU, 28S) rDNA from 101 taxa (100 ingroup taxa of the Mortierellales and 1 outgroup taxon Umbelopsis as member of the Mucorales, Meyer & Gams 2003). The phylogram based on a MAFFT-Alignment (L-ins-I). Node supports above 75 % is given. The tree defines 7 groups: groups 1–7, which are more profoundly analysed in individual analyses based on the ITS1-5.8S-ITS2 shown in Fig. 4–10. The strains named Mortierella sp. ‘epithet’ are strains with an originally different assignment based on morphology. Blue marked strains are potential new species.
Fig. 4.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 1. The phylogram was constructed from a MAFFT-Alignment of 816 aligned nucleotides of 58 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Fig. 5.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 2. The phylogram was constructed from a MAFFT-Alignment of 636 aligned nucleotides of 36 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Fig. 6.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 3. The phylogram was constructed from a MAFFT-Alignment of 701 aligned nucleotides of 38 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Fig. 7.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 4. The phylogram was constructed from a MAFFT-Alignment of 710 aligned nucleotides of 17 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Fig. 8.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 5. The phylogram was constructed from a MAFFT-Alignment of 761 aligned nucleotides of 18 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Fig. 9.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 6. The phylogram was constructed from a MAFFT-Alignment of 703 aligned nucleotides of 60 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Fig. 10.
Maximum Likelihood analysis based on the ITS1-5.8S-ITS2 dataset for clade 7. The phylogram was constructed from a MAFFT-Alignment of 688 aligned nucleotides of 73 taxa. Node support above 75 % is given. The phylogram is midpoint rooted.
Our results do not allow for the revelation of the natural relationships between different species or between groups of species since the clades are poorly supported in the LSU tree. But definition of boundaries between the species/species groups is possible and the presented species groups are in full accordance with the twelve large clades distinguished in a previous study (Petkovits et al. 2011). Because the current dataset is more comprehensive, we will keep, but also extend some of the groups.
Group 1 – selenospora and parvispora (Fig. 4, some morphological features are displayed in Fig.1j, 2i) contains the two most basal groups of the LSU tree (Fig 3). Mortierella selenospora clusters well with M. chienii (Bootstrap support BS = 100 %). Mortierella chienii was not included in the previous study (Petkovits et al. 2011). In cases where the morphological identification does not match the position of the strain in the ITS tree the strains were designated as Mortierella sp. with the epithet in quotation marks. Strains which are very distinct, not part of a clade and consequently might represent undescribed species are highlighted in blue. The selenospora clade also contains the questionable M. wolfii CBS 614.70 which shows different characteristics (e.g. no thermotolerance) to the original M. wolfii strains although the sporangiospores are ellipsoidal to kidney-shaped like those of M. wolfii. A detailed analysis of the morphology and several molecular markers is needed to clarify the status of this particular strain. The other group termed ‘parvispora’ contains also the species M. alliacea, M. basiparvispora, M. fimbricystis, M. jenkinii, M. macrocystis, M. macrocystopsis, M. sossauensis in addition to the previously included species (M. cystojenkinii, M. dichotoma, M. elongatula, M. parvispora, M. pulchella, M. turficola; Petkovits et al. 2011). Mortierella alliacea, M. chienii, M. cystojenkinii, M. elongatula, M. macrocystis, M. macrocystopsis, M. pulchella and M. sossauensis form well-supported clades and the morphologically defined species boundaries are well reflected in the ITS tree (Fig. 4). The parvispora-jenkinii-complex consists predominantly of strains morphologically identified as M. jenkinii or M. parvispora. These two species differ mainly by the shape of their sporangiospores: ellipsoidal for M. jenkinii and globose for M. parvispora. This distinction is not supported by the ITS tree, mixing both types of spores. The strain M. basiparvispora CBS 517.72 is also clustering in this complex, but is differing morphologically from the ex-type strain of this species, which was not included in this study (Gams 1976). A detailed revision of this species in relation to Mortierella will be needed.
Group 2 – verticillata-humilis (Fig. 5, some morphological features are displayed in Fig. 1c, 2a, g, r) is a group that also contains the genera M. clonocystis, M. epicladia, M. epigama, M. horticola and M. minutissima. The topology is similar to the one previously published (Petkovits et al. 2011) but includes some morphologically misidentified specimens. Mortierella zonata CBS 863.68 and M. sossauensis CBS 898.68 are well separated from any other members of their species. The main cluster of M. sossauensis is closely related to the parvispora-jenkinii complex (Fig. 4) while the type strain of M. zonata is related to M. hyalina and M. bainieri (Fig. 10). After a profound morphological revision M. zonata CBS 863.68 and M. sossauensis CBS 898.68 should be renamed and included in the M. minutissima-M. horticola complex, which makes this phylogenetic group of M. minutissima-M. horticola indistinguishable by ITS sequences although both species could be distinguished by the number of their spores in the sporangiola. While M. minutissima develops few-spored sporangiola, M. horticola produces single-spored sporangiola. This suggests that the number of spores per sporangium is not strictly fixed in this group and is therefore not of taxonomic relevance. The single specimen CBS 246.75 resembles M. epicladia but it clusters distantly from the ex-type material CBS 355.76 which is close to M. clonocystis (Fig. 5). Since no other known species group together with CBS 246.75, this might be a so far undescribed species. CBS 226.78 was originally deposited as M. globalpina and CBS 226.35 as M. minutissima but molecular data of both species currently resembles M. clonocystis, indicating an original misapplication or a contamination. Morphology of both species was checked twice and both species were finally assigned to M. clonocystis. The morphospecies M. clonocystis, M. epicladia and M. epigama are well recognized by the ITS tree while M. verticillata and M. humilis form another species complex. Another apparent cluster, the M. verticillata-M. humilis cluster, contains strains including type strains of both species. Based on ITS sequences, a differentiation is not possible. Sequences are similar between 98–100 %. Both species are morphologically similar without any significant differences. Consequently both species should be synonymized.
Group 3 – lignicola (Fig. 6, some morphological features are displayed in Fig. 1n, y, 2j, l, s, w). This group contains the species Mortierella beljakovae, M. chlamydospora, M. echinosphaera, M. formicicola, M. gemmifera, M. kuhlmanii, M. lignicola and M. paraensis. Several of the morphologically defined species, namely M. beljakovae, M. chlamydospora, M. echinosphaera, M. formicicola, M. lignicola and M. paraensis, are nicely detected by the molecular data. Mortierella chlamydospora and M. echinosphaera appear to be closely related as they are sister groups (BS = 100 %). The species M. gemmifera and M. kuhlmanii are morphologically very similar (complex is supported by BS = 85 %) and differ just gradually by spore shape and chlamydospores. The ex-type strains of both species differ just by 12 different base pairs in the ITS sequences (= 98 %). The original morphological identification of strain CBS 268.71 could not be verified because it did not sporulate under different conditions, but its molecular data places it between the gemmifera-complex, M. chlamydospora and M. echinosphaera. The strains CBS 109659 and CBS 555.89 were not examined morphologically and assigned as Mortierella sp. since their original descriptions does not correspond with the molecular data.
Group 4 – mutabilis, globulifera and angusta (Fig. 7, some morphological features are displayed in Fig. 1e, s, v, x, 2v). This group contains two of the three included non-Mortierella genera: Gamsiella and Dissophora. The genus Gamsiella does not cluster with any other mortierellean species, although it was reported to be sister with M. mutabilis (Petkovits et al. 2011). A revision of the morphology revealed different features for M. mutabilis as originally described. Mortierella mutabilis should develop explicitly branched sporangiophores with globose sporangia containing globose to subglobose sporangiospores, for example. But the observed morphology resembles that of Gamsiella. Furthermore, LSU and ITS sequences are similar with 100 and 99.8 %, respectively. Based on these data, we are rejecting the previous group named mutabilis (Petkovits et al. 2011). For the final placement of M. mutabilis, additional strain material is necessary.
The angusta group is extended by M. simplex and consists of the subclades M. angusta-M. simplex (BS = 88 %) and the subclade Dissophora with D. decumbens and D. ornata (BS = 100 %). Mortierella simplex could not by differentiated from M. angusta by significant features, suggesting an upcoming synonymization of both species. The globulifera group contains exclusively M. globulifera (BS = 94 %). The strain CBS 254.76 formerly identified as M. horticola might represent a new species because of its distinct ITS sequence. The ITS sequences of true M. horticola strains belong to group 2 (Fig. 5) where the ex-syntype of this species is located.
Group 5 – strangulata and wolfii (Fig. 8, some morphological features are displayed in Fig. 1q, r, 2c, t) contains only few species, which could all be identified by molecular data. The wolfii group (BS = 100 %) is extended in this study by M. ambigua (clade support BS = 99 %). Mortierella ambigua is sister clade (BS = 81 %) to M. capitata (BS = 98 %) and both clades are sister group to M. wolfii (BS = 96 %). The strangulata group is retained, containing M. strangulata and M. rostafinskii (BS = 100 %). Mortierella microzygospora, M. parazychae and M. pseudozygospora were not assigned to any defined group.
Group 6 – alpina and polycephala (Fig. 9, some morphological features are displayed in Fig. 1b, g, h, k, o, w, 2d, m, n, p). The polycephala group harbours the type species of the whole genus Mortierella: M. polycephala. Therefore, this clade resembles the core group of the genus Mortierella. Related to M. polycephala and well supported in LSU (BS = 99 %) and ITS (BS = 100 %) are the species M. bisporalis, M. hypsicladia, M. indohii, M. oligospora, M. polygonia and M. reticulata. Except for the ex-type strain of M. polygonia CBS 685.71 which clusters within the M. polycephala, all species form well supported clades (Fig. 9). But judging from the different observed morphology of M. polygonia, which is that of M. polycephala instead of that originally described (Gams 1976), this strain should be treated as such. Although the strain is sterile, it shows the typical stylospores of M. polycephala. A second isolate of M. polygonia (CBS 248.81) could not be confirmed as ‘true’ M. polygonia since it does not sporulate, displaying only untypical stylospores and clusters within the alpina-complex (Fig. 9). Therefore the status of this species seems doubtful. Mortierella alpina is one of the major species isolated and identified from our environmental samples collected in Austria. Mortierella alpina forms a heterogeneous cluster with the two species M. antarctica and M. amoeboidea. For M. amoeboidea again is the observed morphology not identical with the described one and resembles the species indicated by molecular data. This justifies M. amoeboidea W. Gams 1976 to be treated as synonym of M. alpina Peyronel 1913. One isolate of M. globalpina (CBS 718.88) is placed within the alpina complex and one isolate (CBS 226.78) is located in the M. clonocystis clade (Fig. 5). Verification by inclusion of the type strain is not possible since this particular strain seems to be dead now.
Group 7 – gamsii (Fig. 10, some morphological features are displayed in Fig. 1a, d, f, p, u, 2b, e, f, h, k, o, q, u) is the largest group in this and our previous study containing 73 taxa. The previous dataset (Petkovits et al. 2011) with the species Mortierella acrotona, M. armillariicola, M. biramosa, M. camargensis, M. cogitans, M. elongata, M. exigua, M. gamsii, M. histoplasmatoides, M. hyalina, M. nantahalensis, M. rishikesha, M. sarnyensis, M. schmuckeri, M. sclerotiella, M. zonata and M. zychae was extended by M. bainieri, M. claussenii, M. fatshederae and M. longigemmata. Mortierella armillariicola, M. bainieri, M. fatshederae, M. hyalina and M. zychae form monophyletic clades supported by the coherence of several strains (Fig. 10). Mortierella exigua, M. gamsii and M. zonata are polyphyletic. Strains identified as these species appear in different places of the tree. None of the strains of M. exigua clusters together with the ex-type strain. For M. gamsii at least three divided clusters are present. One sequence of an ex-type strain is placed in the elongata-complex. Mortierella schmuckeri forms one monophyletic clade together with M. claussenii and M. camargensis (BS = 97 %). Due to a lack of sufficient amounts of strains neither the phylogenetic position nor the species coherence of M. acrotona, M. cogitans, M. histoplasmatoides, M. longigemmata, M. nantahalensis, M. sclerotiella and M. zonata could be confirmed.
CONCLUSIONS
In order to study and evaluate the monophyly of Mortierella, and to address the phylogenetic relationships of other genera in the Mortierellales, we analysed one of the largest datasets of LSU and ITS sequences for this order. The genera Dissophora, Gamsiella and Lobosporangium are placed within the genus Mortierella. This suggests either a polyphyly of Mortierella with the necessity to establish additional genera or the necessity to reduce the existing genera to one. Although our study contains a comprehensive dataset it is still not possible to elucidate all species and species groups of the Mortierellales. It was already proposed that additional molecular markers are necessary for a profound phylogenetic study (Petkovits et al. 2011). But our study supports existing and reveals new contradictions to the traditional morphology based classifications (Linnemann 1941, Zycha et al. 1969, Gams 1977). Several species, originally iden-tified as one, appear on different places in the phylogenetic analyses. This might originate either from simple misapplications or from the observed phenomenon of dependency of the phenotype on culture conditions (Petkovits et al. 2011). Furthermore, names of new genera and species published just recently may be superfluous at a nomenclatural level because their respective phylogenetic markers were not compared with the full molecular dataset of the Mortierellales, e.g. Echinochlamydosporium variabile (Jiang et al. 2011), which may turn out to be a micromorphologically degenerate Mortierella stylospora. Here we present the most comprehensive molecular dataset of the Mortierellales which is available up to date and facilitates revision of existing and validation of upcoming names. Finally, all these actions will lead to several species name changes and synonymizations. Nevertheless, several species or even groups of species seem to be distinguishable by morphology and phylogeny. The monophyletic clade of Mortierella s.str. contains the type species of the genus, M. polycephala Coem. 1863. Whether additional species are related to this group and therefore belonging to the genus Mortierella needs to be evaluated in further studies. Current data (Petkovits et al. 2011) are contradictory with regard to relationships of species and species groups. Due to the lack of suitable morphological criteria the following species and species groups were misapplied and require taxonomic revision, where indicated nomenclatural synonymization. These are: M. angusta, M. basiparvispora, M. carmagensis, M. fimbricystis, M. gamsii, M. gemmifera, M. globalpina, M. horticola, M. humilis, M. jenkinii, M. kuhlmanii, M. minutissima, M. parvispora, M. rishikesha, M. schmuckeri, M. simplex, M. sossauensis, M. turficola, M. verticillata and M. zonata.
Underrepresented in this study, but due to the lack of comprehensive additional material, are the species: M. acrotona, M. angusta, M. dichotoma, M. epicladia, M. exigua, M. fimbricystis, M. formicicola, M. longigemmata, M. microzygospora, M. nantahalensis, M. parazychae, M. rishikesha, M. rostafinskii, M. sclerotiella and M. strangulata.
Table 3.
Summary of isolates which were revised and assigned to different species within this study.
| Strain number | Original name | Revised name |
|---|---|---|
| CBS585.81 | M. alpina | M. kuhlmanii |
| CBS696.70 | M. alpina | M. cystojenkinii |
| CBS272.71 | M. bainieri | M. kuhlmanii |
| CBS273.71 | M. bainieri | M. kuhlmanii |
| CBS292.96 | M. chienii | M. selenospora |
| CBS554.73 | M. chienii | M. selenospora |
| CBS387.91 | M. macrocystopsis | M. cystojenkinii |
| FSU2736 | M. parvispora | M. jenkinii |
| CBS293.34 | M. polycephala | M. hyalina |
| CBS176.74 | M. sossauensis | M. clonocystis |
Acknowledgments
This research was supported by an international cooperation grant of the German and Hungarian Research Foundations (DFG Vo 772/9-1 and OTKA NN106394) and the Hungarian grant TÉT_10-1-2011-0747. Tamas Petkovits was supported by the European Union and by the European Social Fund (project number: TÁMOP-4.2.2/B-10/1-2010-0012). We like to express our gratitude to Martin Kirchmair (University of Innsbruck, Austria) for collecting and providing environmental strains of Mortierella from the alpine region. Also we would like to thank Domenica Schnabelrauch (Max Planck Institute for Chemical Ecology Jena, Germany) and the members of the molecular barcoding team at the CBS Utrecht for technical support in DNA sequencing.
REFERENCES
- Cavalier-Smith T.1998. A revised six-kingdom system of life. Biological Reviews of the Cambridge Philosophical Society 73, 3: 203–266 [DOI] [PubMed] [Google Scholar]
- Cenis JL.1992. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Research 20, 9: 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalabuda TV.1968. Systemica familiae Mortierella. Novosti Sistematiki Nizshikh Rastenii 5: 120–131 [Google Scholar]
- Chesters CGC.1933. Azygozygum chlamydosporum nov. gen. et sp. A phycomycete associated with a diseased condition of Antirrhinum majus. Transactions of the British Mycological Society 18, 3: 199–214 [Google Scholar]
- Coemans E.1863. Quelques hyphomycetes nouveaux. 1. Mortierella polycephala et Martensella pectinata. Bulletin de l’Académie Royale des Sciences de Belgique Classe des Sciences 2, ser. 15: 536–544 [Google Scholar]
- Dewèvre A.1893. Contribution a l’étude des Mucorinées, avec essai d’une monographie de ces champignons. Grevillea 22, 101: 1–8 [Google Scholar]
- Fischer A.1892. Mortierellaceae. In: Rabenhorst L, Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz. Band 1, Abth. IV. Kummer E, Leipzig, Germany: 268–283 [Google Scholar]
- Gams W.1969. Gliederungsprinzipien in der Gattung Mortierella. Nova Hedwigia 18, 1: 30–44 [Google Scholar]
- Gams W.1976. Some new or noteworthy species of Mortierella. Persoonia 9, 1: 111–140 [Google Scholar]
- Gams W.1977. A key to the species of Mortierella. Persoonia 9, 3: 381–391 [Google Scholar]
- Griebel T, Brinkmeyer M, Böcker S.2008. EPoS: a modular software framework for phylogenetic analysis. Bioinformatics 24: 2399–2400 [DOI] [PubMed] [Google Scholar]
- Hawksworth DL.2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycological Research 105, 12: 1422–1432 [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509–547 [DOI] [PubMed] [Google Scholar]
- Higashiyama K, Fujikawa S, Park EY, Shimizu S.2002. Production of arachidonic acid by Mortierella fungi. Biotechnology and Bioprocess Engineering 7: 252–262 [Google Scholar]
- Hoffmann K, Voigt K, Kirk PM.2011. Mortierellomycotina subphyl. nov., based on multi-gene genealogies. Mycotaxon 115, 1: 353–363 [Google Scholar]
- Holland HL.2001. Biotransformation of organic sulfides. Natural Product Reports 18: 171–181 [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MJ.2009. Atlas of clinical fungi, 3rd ed Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands: / Universitat Rovira i Virgili, Reus, Spain [Google Scholar]
- Ivanova NV, Dewaard JR, Hebert PDN.2006. An inexpensive, automation-friendly protocol for recovering high quality DNA. Molecular Ecology Notes 6: 998–1002 [Google Scholar]
- Jiang XZ, Yu HY, Xiang MC, Liu XY, Liu XZ.2011. Echinochlamydosporium variabile, a new genus and species of Zygomycota from soil nematodes. Fungal Diversity 46, 1: 43–51 [Google Scholar]
- Katoh T.2008. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 2008, 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA.2008. Ainsworth & Bisby’s dictionary of the fungi. 10th ed CAB International, Wallingford, UK [Google Scholar]
- Linnemann G.1941. Die Mucorineen-Gattung Mortierella Coemans. Pflanzenforschung 23: 1–64 [Google Scholar]
- Malloch D.1967. A new genus of Mucorales. Mycologia 59: 326–329 [Google Scholar]
- Meyer W, Gams W.2003. Delimitation of Umbelopsis (Mucorales, Umbelopsidaceae fam. nov.) based on ITS sequence and RFLP data. Mycological Research 107: 339–350 [DOI] [PubMed] [Google Scholar]
- Nagy LG, Petkovits T, Kovács GM, Voigt K, Vágvölgyi C, Papp T.2011. Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytologist 191, 3: 789–794 [DOI] [PubMed] [Google Scholar]
- Nirenberg HI.1981. A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany 59: 1599–1609 [Google Scholar]
- Novotelnova1950. Notulae systematicae e sectione cryptogamica. Instituti Botanici Nominee V.L. Komarovii l’Académie des Sciences de l’URSS 6: 160 [Google Scholar]
- Petkovits T, Nagy LG, Hoffmann K, Wagner L, Nyilasi I, et al. 2011. Data partitions, Bayesian analysis and phylogeny of the zygomycetous fungal family Mortierellaceae, inferred from nuclear ribosomal DNA sequences. PLoS ONE 6, 11: e27507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyronel B.1913. I germi astmosferici dei fungi con micelio: 17 Dissertation; Padua, Italy [Google Scholar]
- Stamatakis A.2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stielow B, Bubner B, Hensel G, Munzenberger B, Hoffmann P, et al. 2009. The neglected hypogeous fungus Hydnotrya bailii Soehner (1959) is a widespread sister taxon of Hydnotrya tulasnei (Berk.) Berk. & Broome (1846). Mycological Progress 9: 195–203 [Google Scholar]
- Thaxter R.1914. New or peculiar Zygomycetes. 3: Blakeslea, Dissophora and Haplosporangium, nova genera. Botanical Gazette Crawfordsville 58: 353–366 [Google Scholar]
- Vogelstein B, Gillespie D.1979. Preparative and analytical purification of DNA from agarose. Proceedings of the National Academy of Sciences of the United States 76, 2: 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TD, Eberhardt U, Szöke S, Groenewald M, Robert V.2012. A laboratory information management system for DNA barcoding workflows. Integrative Biology 4, 7: 744–755 [DOI] [PubMed] [Google Scholar]
- White MM, James TY, O’Donnell K, Cafaro MJ, Tanabe Y, Sugiyama J.2006. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia 98: 872–884 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J.1990. Amplication and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, California [Google Scholar]
- Zycha H, Siepmann R, Linnemann G.1969. Mucorales. Eine Beschreibung aller Gattungen und Arten dieser Pilzgruppe. Cramer, Lehre [Google Scholar]