Abstract
We have previously shown that double-stranded RNA-triggered, Toll-like receptor 3 (TLR3)-mediated signaling is independent of MyD88, IRAK4, and IRAK. Instead, TRAF6, TAK1, and TAB2 are recruited to TLR3 on poly(I·C) stimulation. TRAF6-TAK1-TAB2 are then translocated to the cytosol where TAK1 is phosphorylated and activated, leading to the activation of IκB kinase and NFκB. The present study addressed two important questions: (i) How are TRAF6, TAK1, and TAB2 recruited to TLR3? (ii) Are TRAF6, TAK1, and TAB2 also required for TLR3-mediated IRF3 activation? Recently, a novel Toll-IL-1 receptor (TIR)-containing adapter, TIR domain-containing adapter inducing IFN-β (TRIF), was shown to play a critical role in TLR3-mediated activation of NF-κB and IRF3. We found that TLR3 recruits TRAF6 via adapter TRIF through a TRAF6-binding sequence in TRIF (PEEMSW, amino acids 250-255). Mutation of this TRAF6-binding sequence abolished the interaction of TRIF with TRAF6, but not with TLR3. Interestingly, mutation of the TRAF6-binding site of TRIF only abolished its ability to activate NF-κB but not IRF3, suggesting that TLR3-mediated activation of NF-κB and IRF3 might bifurcate at TRIF. In support of this finding, we showed that DN-TRAF6 and DN-TAK1 blocked poly(I·C)-induced NF-κB but not IRF3 activation. Furthermore, whereas poly(I·C)-induced NF-κB activation is completely abolished inTRAF6-/- MEFs, the signal-induced activation of IRF3 is TRAF6 independent. In conclusion, TRIF recruits TRAF6-TAK1-TAB2 to TLR3 through its TRAF6-binding site, which is required for NF-κB but not IRF3 activation. Therefore, double-stranded RNA-induced TLR3/TRIF-mediated NF-κB and IRF3 activation diverge at TRIF.
Members of the Toll-IL-1 receptor superfamily, defined by the presence of an intracellular Toll-IL-1 receptor (TIR) domain, are important in mediating inflammation and immune responses. This superfamily can be divided into two main subgroups, based on the extracellular domains; the Ig domain (Ig)-containing and Leucine-rich repeat (LRR) motif-containing receptors. The Ig domain subgroup includes IL-1R1, IL-18R, T1/ST2, and SIGIRR (1-5). IL-1 has been demonstrated to be one of the key orchestrators of the immune response, eliciting a wide range of biological responses, including fever, lymphocyte activation, and leukocyte infusion to the site of injury and infection (6). IL-18 promotes TH1 cell differentiation and NK cell activation, whereas T1/ST2 has important functions in developing TH2 cell responses. The LRR subgroup consists of at least 10 human Toll-like receptors (TLR), which are important in the recognition of pathogens (1, 7-10). An individual TLR recognizes its own specific pathogen-associated molecular patterns (PAMP). Whereas TLR4 has been genetically identified as a signaling molecule essential for the responses to LPS, a component of Gram-negative bacteria (11), TLR2 responds to mycobacteria, yeast, and Gram-positive bacteria (12-15). TLR6 associates with TLR2 and recognizes lipoproteins from microplasma. Whereas TLR5 mediates the induction of the immune response by bacterial flagellins (16), TLR9 has been shown to recognize bacterial DNA (10), and TLR3 recognizes double-stranded RNA (dsRNA) (17). The natural ligands for TLR7, TLR8, and TLR10 are not known, although a synthetic compound (imidazoquinoline compound R848) with antiviral activity has now been described as a ligand for TLR7 and TLR8 (18, 19).
Because Toll-IL-1 receptors share sequence similarities in their intracellular domain, most of them can activate transcription factor NF-κB through a common signaling pathway, which has been studied extensively for the IL-1 receptor. On the binding of IL-1 to the receptor, the cytosolic adapters MyD88 (20-22) and Tollip (23) are rapidly recruited to the receptor complex (IL-1R/IL-1R-Acp), which then recruits serine-threonine kinases IRAK4 (IL-1 receptor-associated kinase 4) (24, 25) and IRAK (26). IRAK is phosphorylated and mediates the recruitment of TRAF6 to the receptor (25, 27). The IRAK-TRAF6 then forms a complex with another adapter protein, Pellino 1, and leaves the receptor to interact with TAK1 (TGFβ-activated kinase 1), a member of the mitogen-activated protein (MAP) kinase kinase kinase (MAPKKK), on the membrane (27). TAK1 is phosphorylated on the membrane, but activated in the cytosol (27). The activation of TAK1 eventually leads to the activation of IκB kinase (IKK), which in turn leads to the phosphorylation and degradation of IκB proteins, and liberation of NF-κB to activate transcription in the nucleus (28-31). Activated TAK1 has also been implicated in the IL-1-induced activation of MKK6 and c-Jun N-terminal kinase (JNK) (32), leading to the activation of other transcriptional factors, including ATF and AP1, thereby also activating gene transcription.
Several Toll-IL-1 receptors also use variations of the above common signaling pathway. For example, TLR3 and TLR4 use MyD88-independent pathways to activate transcription factors NF-κB and IRF3 and IFN-β production (13, 33). Recently, the IKK-related kinases IKKε and TANK-binding kinase 1 have been implicated in the phosphorylation and activation of IRF3 (34, 35). TIR domain-containing adapter inducing IFN-β (TRIF) was recently identified as an adapter for TLR3 and TLR4 (36, 37). TRIF-deficient mice were defective in both TLR3- and TLR4-mediated expression of IFN-β and activation of IRF3 (36, 37). Whereas TRIF-deficient mice showed complete loss of TLR3-induced NF-κB activation, TLR4-mediated NF-κB activation was only completely abolished in mice deficient in both MyD88 and TRIF (36, 37).
We have previously shown that dsRNA-triggered, TLR3-mediated signaling is independent of MyD88, IRAK4, and IRAK. Instead, TRAF6, TAK1, and TAB2 are recruited to TLR3 on poly(I·C) stimulation. TRAF6-TAK1-TAB2 are then translocated to the cytosol, where TAK1 is phosphorylated and activated, leading to the activation of IKK and NF-κB (42). In this study, we hypothesized that TRIF plays a critical role in recruiting TRAF6, TAK1, and TAB2 to TLR3 on poly(I·C) stimulation, leading to NF-κB activation. Indeed, we found that TLR3 recruits TRAF6 via adapter TRIF through a TRAF6-binding sequence in TRIF (PEEMSW, amino acids 250-255). Mutation of this TRAF6-binding sequence abolished the interaction of TRIF with TRAF6 but not with TLR3. Interestingly, mutation of the TRAF6-binding site of TRIF only abolished its ability to activate NF-κB but not IRF3, suggesting that TLR3-mediated activation of NF-κB and IRF3 bifurcate at TRIF.
Materials and Methods
Biological Reagents and Cell Culture. Recombinant human IL-1β was provided by the National Cancer Institute. Poly(I·C) was purchased from Amersham Pharmacia Biotech. Anti-flag (M2) was from Sigma. Anti-HA was from Upstate (Charlottesville, VA). HeLa cells, TRAF6+/- and TRAF6-/- MEFs were maintained in DMEM supplemented with 10% FCS, penicillin G (100 μg/ml), and streptomycin (100 μg/ml). 293-TK/Zeo cells, I1A and I3A (38), were maintained in regular DMEM plus 0.4 mg/ml of the neomycin analog G418. 293-TK/Zeo cells stably expressing Flag-TLR3 were maintained in the same media plus hygromycin (0.25 mg/ml).
Recombinant Plasmids. TRIF was amplified by RT-PCR from 293 cells and cloned into pcDNA3.1 (His-TRIF and HA-TRIF). The TRIF mutants including (1) E88A, (2) E252A, (3) E303A, (4) E493A, and (5) E266A and the mutants with combined mutations were generated by the QuikChange site-directed mutagenesis kits with pfu-ultra as the polymerase (Stratagene) and pcDNA3.1-V5-His-TRIF as the template. Mutations were confirmed by sequencing. Expression constructs for Flag-tagged TRAF1, TRAF2, TRAF3, TRAF5, TRAF6, MyD88, and DN-TRAF6 were kindly provided by Holger Wesche (Tularik, South San Francisco, CA). pE-selectin-luc, an NF-κB-dependent E-selectin-luc reporter plasmid, was described by Schindler and Baichwal (39). P561-luc is an IRF3-dependent luciferase construct (40). Dominant-negative TAK1 (TAK1-DN-K66W) was a kind gift from Kunihiro Matsumoto (Nagoya University, Nagoya, Japan).
Coimmunoprecipitation and Immunoblotting. Cells untreated or treated with poly(I·C) (100 μg/ml) were lysed in a Triton-containing lysis buffer (0.5% Triton X-100/20 mM Hepes, pH 7.4/150 mM NaCl/12.5 mM β-glycerophosphate/1.5 mM MgCl2/10 mM NaF/2 mM DTT/1 mM sodium orthovanadate/2 mM EGTA/20 μM aprotinin/1 mM phenylmethylsulfonyl fluoride). Cell extracts were incubated with 1 μg of antibody or preimmune serum (negative control) for 2 h, followed by a 2-h incubation with 20 μl of protein G-Sepharose beads (Amersham Pharmacia Biotech; prewashed and resuspended in PBS at a 1:1 ratio). After incubation, the beads were washed four times with lysis buffer, separated by SDS/PAGE, transferred to Immobilon-P membranes (Millipore), and analyzed by immunoblotting.
Luciferase Reporter Assays. Cells (2 × 105) were transfected by using Lipofectamine-2000 according to the manufacturer's instructions with 200 ng of pE-selectin-luc (for NF-κB activation) or 50 ng of 561-luc (for IRF3 activation), 200 ng of pSV2-β-gal, and indicated amounts of expression constructs. After 24 h, the cells were left untreated or stimulated with poly(I·C) for 6 h before harvest. Luciferase and β-galactosidase activities were determined by using the luciferase assay system and chemiluminescent reagents from Promega.
Results
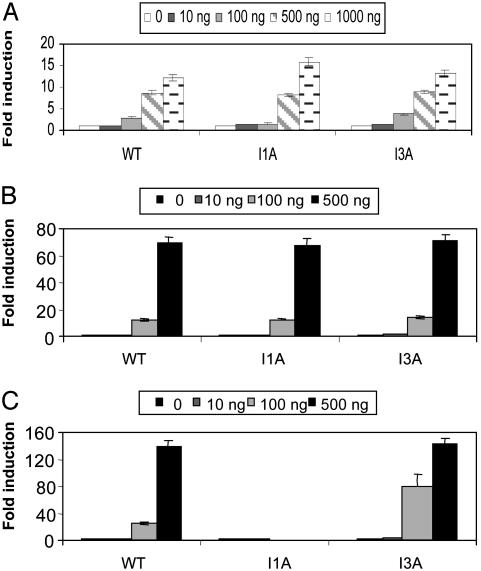
TRIF-Mediated Activation of NF-κB and IRF3 are IRAK- and MyD88-Independent. Through chemical mutagenesis, we previously generated several IL-1-unresponsive cell lines (derived from parental 293 cells) lacking specific components of the pathway. Mutant cell line I1A lacks both IRAK protein and mRNA (38, 40), whereas I3A cells lack MyD88 (Z.J. and X.L., unpublished data). Neither NF-κB nor JNK is activated in IL-1-treated I1A and I3A cells, but these responses are restored in I1A-IRAK and I3A-MyD88 cells, indicating that IRAK and MyD88 are required for both (40). Interestingly, poly(I·C)-induced, TLR3-mediated NF-κB and MAP kinase activation are intact in I1A and I3A mutant cells, indicating that IRAK and MyD88 are not required for TLR3-mediated signaling (42). Adapter molecule TRIF is required for TLR3-mediated signaling (36, 37). We recently examined TRIF-mediated NF-κB and IRF3 activation in I1A and I3A mutant cells, as compared to WT 293 cells. Consistent with its role as an adapter for TLR3, TRIF-induced NF-κB (Fig. 1A) and IRF3 (Fig. 1B) activation are also IRAK- and MyD88-independent. The luciferase reporter constructs driven by NF-κB-dependent E-selectin promoter (27) and IRF3-dependent 561 promoter (43) were used, respectively. As a control, it was shown that MyD88-induced NF-κB activation is IRAK-dependent, whereas MyD88 is incapable of activating IRF3 (Fig. 1C and data not shown).
Fig. 1.
TRIF-mediated activation of NF-κB and IRF3 are IRAK- and MyD88-independent. Increasing amounts of TRIF (0, 10, 100, and 1,000 ng) were cotransfected with E-selectin luciferase reporter construct (200 ng) (A) or 561-luc reporter construct (200 ng) (B) into WT 293, IRAK-deficient (I1A), and MyD88-deficient (I3A) cells. (C) Increasing amounts of MyD88 (0, 10, 100, and 1,000 ng) were cotransfected with E-selectin luciferase reporter construct (200 ng) into WT 293, IRAK-deficient (I1A), and MyD88-deficient (I3A) cells. After 24 h, cells were harvested, followed by luciferase reporter assay. Vector DNA pcDNA3.1 (1 μg) was cotransfected with E-selectin luciferase reporter construct or 561-luc reporter construct (200 ng/each) into these cells as controls. Data are presented as the fold induction of luciferase activity in cells transfected with TRIF, compared with cells transfected with vector controls. Shown are the averages and SD from three independent experiments.
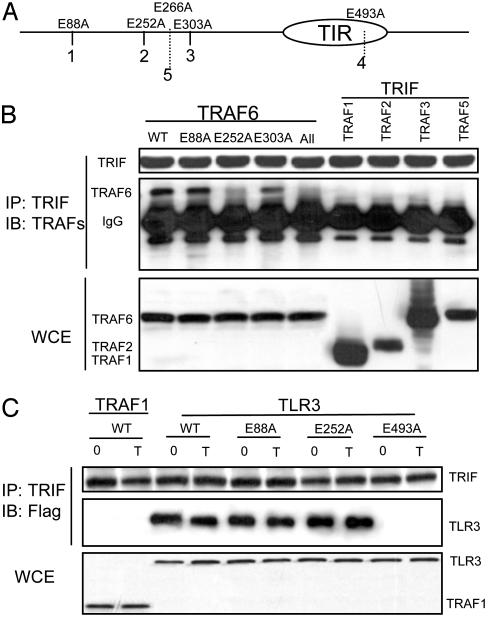
TRIF Specifically Interacts with TRAF6 Through Its TRAF6-Binding Site. We previously showed that TRAF6-TAK1-TAB2 are recruited to TLR3 on poly(I·C) stimulation (42). We hypothesized that TRIF, an adapter for TLR3 plays an important role in recruitment of these signaling components to the receptor. Through coimmunoprecipitation experiments, we found that TRIF specifically interacts with TRAF6 but not with the other TRAF molecules when they are coexpressed in 293 cells (Fig. 2B). Protein sequence analysis of TRIF revealed three typical TRAF6-binding sequences [PxExxD/W/E/F/Y (41), including 1, PEEPPD (amino acids 86-91), 2, PEEMSW (amino acids 250-255), and 3, PVECTE (amino acids 301-306)] (Fig. 2A). TRIF also contains two nontypical TRAF6-binding sequences, numbered as 4, PLESSP (amino acids 491-496), and 5, PPELPS (amino acids 264-269) (Fig. 2A). To determine which TRAF6-binding site is responsible for the interaction of TRIF with TRAF6, we mutated the conserved Glutamic acid (E) to Alanine (A) in the putative TRAF6-binding sites (E88A, E252A, E303A, E493A, and E266A). The TRIF mutants were then transfected into 293 cells and examined for their ability to interact with TRAF6. Mutation on site 2 (E252A), but not other putative TRAF6-binding sequences, greatly reduced the ability of TRIF to interact with TRAF6 (Fig. 2B and data not shown). However, the same mutation did not affect the TLR3-TRIF interaction (Fig. 2C). On the other hand, the mutation on site 4 in the TIR domain of TRIF abolished the interaction of TRIF with TLR3, probably because of the disruption to the TIR domain (Fig. 2C)
Fig. 2.
TRIF specifically interacts with TRAF6 through its TRAF6-binding site. (A) Positions of the three typical putative TRAF6-binding sites, marked as 1, 2, 3, and two untypical sites, marked as 4 and 5 in TRIF are shown (see text for detail). Positions of mutation (Glutamic acid to Alanine) in the putative TRAF6-binding sites are indicated. (B) 293 cells were cotransfected with HA-tagged WT TRIF (WT) and Flag-tagged TRAF 1-6. HA-tagged TRIF mutants described in A were cotransfected with Flag-tagged TRAF6. All, the TRIF mutant that contains mutation in all of the TRAF6-binding sites. Extracts from transfected cells were immunoprecipitated with anti-HA antibody, followed by Western analyses with anti-Flag antibody. Whole cell extract (WCE) was used to check the expression levels of the transfected proteins. (C) 293-TLR3 (Flag-tagged) cells were transfected with HA-tagged TRIF (WT and mutants as described in A) and either untreated (0) or treated with poly(I·C) for 1 h (T), followed by immunoprecipitation with anti-HA antibody and Western analyses with anti-Flag antibody. 293 cells transfected with TRAF1 (instead of TLR3) was used as a control.
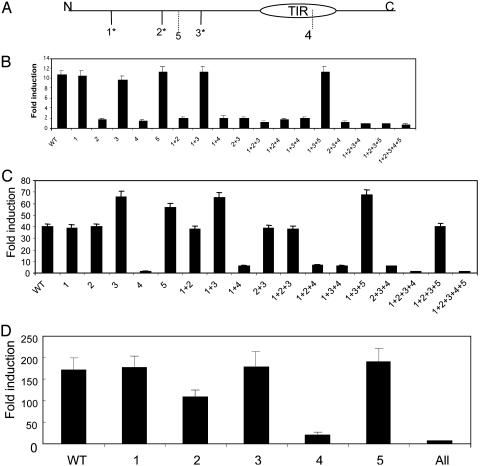
The TRIF-TRAF6 Interaction Is Important for NF-κB but Not IRF3 Activation. We then examined the TRIF mutants for their ability to activate NF-κB and IRF3 by luciferase reporter assay. As shown in Fig. 3B, site 2 (E252A) mutation completely abolished TRIF-induced NF-κB activation. Either individual or combined mutations on this site (E252A) showed complete loss of NF-κB activation. Importantly, the same mutation does not affect IRF3 activation (Fig. 3C). Because the mutation on site 2 abolished the TRIF-TRAF6 interaction (Fig. 2), the above result also implies that the recruitment of TRAF6 to TRIF is probably only important for TRIF to activate NF-κB but not IRF3. On the other hand, the site 4 (E493A) mutation that abolished the TRIF-TLR3 interaction failed to activate NF-κB and IRF3 (Fig. 3 B and C).
Fig. 3.
Mutations of TRIF activate NF-κB and IRF3 differently. (A) Positions of the putative TRAF6-binding sites in TRIF are shown (see text for detail). Two hundred nanograms of WT and TRIF mutants (containing single mutation marked as 1, 2, 3, etc., or combined mutations marked as 1+2, 1+3, 1+4, etc.) were cotransfected with 200 ng of E-selectin luciferase reporter construct (B) or 50 ng of 561-luc reporter construct (C) into WT 293 cells. After 24 h, the transfected cells were harvested, followed by luciferase reporter assay. As controls, vector DNA pcDNA3.1-V5-His (200 ng) was cotransfected with E-selectin luciferase reporter construct (200 ng) or 561-luc reporter construct (50 ng) into 293 cells. Data are presented as the fold induction of luciferase activity of cells transfected with TRIF constructs, compared to cells transfected with vector DNA. Shown are the averages and SD from three independent experiments.
These TRIF mutants were examined for their ability to activate the IFNβ promoter, which depends on the activation of both NF-κB and IRF3 (13, 33). Mutation on site 2 (E252A) only partially abolished the ability of TRIF to activate the IFNβ promoter, probably because of the fact that E253A can still activate IRF3 (Fig. 3 C and D). Mutant 4 (E493A) completely failed to activate IFNβ promoter driven luciferase activity, consistent with its inability to activate NF-κB and IRF3 (Fig. 3D).
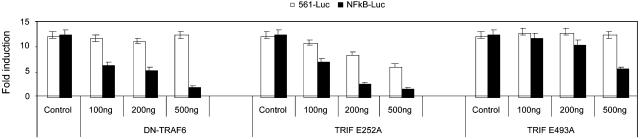
We then examined whether any of these TRIF mutants can function as dominant-negative mutants for TLR3-mediated NF-κB and IRF3 activation (Fig. 4). Interestingly, we found that mutant E252A efficiently inhibited poly(I·C)-induced NF-κB activation in HeLa cells, whereas it had significantly less effect on IRF3 activation in the same cells, confirming that this TRAF6-binding site (site 2) is more critical for TLR3-mediated NF-κB activation than IRF3 activation. The reduction in the fold of induction of poly(I·C)-induced IRF3 activation in HeLa cells transfected with E252A is due to the constitutive IRF3 activation by overexpression of E252A. As a control, mutant E493A did not have any inhibitory role in poly(I·C)-induced TLR3-mediated NF-κB and IRF3 activation.
Fig. 4.
E252A specifically inhibits poly(I·C)-induced NF-κB activation. Increasing amounts of DN-TRAF6, TRIF E252A, and TRIFE493A (0, 100, 200, and 500 ng) were cotransfected with 200 ng of E-selectin-luc or 50 ng of 561-luc construct into HeLa cells. After 24 h, the transfected cells were untreated or stimulated with poly(I·C) for 6 h before harvest. Vector pcDNA3.1-V5-His was used to normalize the total amount of DNA for each transfection. Data are presented as poly(I·C)-induced fold induction of luciferase activity of cells transfected with or without DN-TRAF6, TRIF E252A, and TRIFE493A, compared to cells transfected with vector DNA without stimulation.
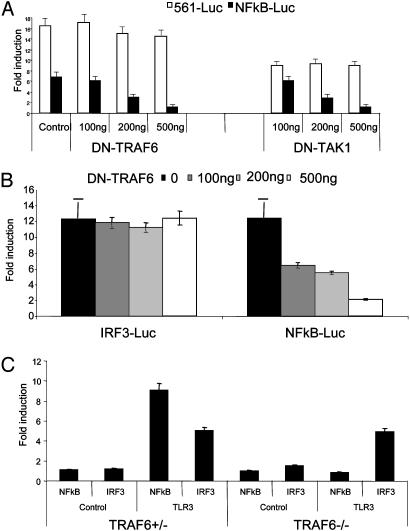
TLR3-Mediated NF-κB and IRF3 Activation Diverges at TRIF. Mutation in the TRAF6-binding site of TRIF (E252A) abolished its ability to activate NF-κB but had no effect on its constitutive activation of IRF3. Furthermore, this E252A mutant efficiently inhibited poly(I·C)-induced NF-κB activation but had much less effect on signal-induced IRF3 activation. Taken together, these results suggest that the TRIF-mediated recruitment of TRAF6 is probably only required for NF-κB but not IRF3 activation, implying that TLR3-mediated NF-κB and IRF3 activation probably bifurcate at TRIF. If that is the case, TRAF6 and TAK1 should only be required for NF-κB but not IRF3 activation. To test this, dominant-negative mutants of TAK1 (DN-TAK1) and TRAF6 (DN-TRAF6) were used to inhibit TRIF-induced signaling. As shown in Fig. 5A, both TAK1 (DN-TAK1, kinase inactive mutant) and TRAF6 (DN-TRAF6) can specifically inhibit TRIF-mediated NF-κB but not IRF3 activation. Furthermore, TAK1 (DN-TAK1, kinase inactive mutant) and TRAF6 (DN-TRAF6) efficiently inhibited poly(I·C)-induced NF-κB but not IRF3 activation in HeLa cells, indicating that these two components are only involved in TLR3-mediated NF-κB but not IRF3 activation (Fig. 5B). This conclusion is supported by the following experiment in TR AF6-/- MEFs. Transfection of TLR3 rendered TRAF6+/- MEFs (TLR3-TRAF6+/- MEFs) responsiveness to poly(I·C) stimulation. Although poly(I·C) induced activation of both NF-κB and IRF3 in TLR3-TRAF6+/- MEFs, it only activated IRF3 in TLR3-TRAF6-/- MEFs, indicating that TRAF6 is required for TLR3-mediated NF-κB but not IRF3 activation (Fig. 5C).
Fig. 5.
Activation of NF-κB, but not IRF3, was blocked by DN-TRAF6 and DN-TAK1. (A) Increasing amounts of DN-TRAF6 and DN-TAK1 (0, 100, 200, and 500 ng) were cotransfected with 100 ng of TRIF and 200 ng of E-selectin luciferase reporter construct or 50 ng of 561-luc construct into 293 cells. After 48 h, transfected cells were harvested, followed by luciferase reporter assay. Vector pcDNA3.1-V5-His was used to normalize the total amount of DNA for each transfection. Vector DNA (600 ng) and E-selectin-luc (200 ng) or 561-luc (50 ng) were used as control. Data are presented as the fold induction of luciferase activity of cells transfected with TRIF with or without dominant-negative mutants of TRAF6 and TAK1, compared to cells transfected with vector DNA. (B) Increasing amounts of DN-TRAF6 (0, 100, 200, and 500 ng) were cotransfected with 200 ng of E-selectin-luc or 50 ng of 561-luc construct into HeLa cells. After 24 h, the transfected cells were untreated or stimulated with poly(I·C) for 6 h before harvest. Vector pcDNA3.1-V5-His was used to normalize the total amount of DNA for each transfection. Data are presented as poly(I·C)-induced fold induction of luciferase activity of cells transfected with or without DN-TRAF6, compared to cells transfected with vector DNA without stimulation. (C) Two hundred nanograms of TLR3 or control vector DNA was cotransfected with 200 ng of E-selectin-luc or 200 ng of 561-luc into TRAF6+/- and TRAF6-/- MEFs. After 24 h, the transfected cells were untreated or treated with poly(I·C) for 6 h before harvest, followed by luciferase reporter assay. Data are presented as the PolyI·C-induced fold induction of luciferase activity of cells transfected with TLR3, compared to cells transfected with vector DNA without stimulation. Shown are the averages and SD from three independent experiments.
Discussion
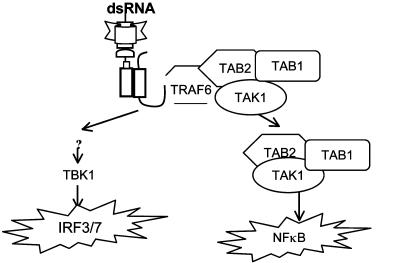
Based on the data presented in this article and in published studies (34-37, 42), we propose a model for TLR3-mediated signaling (Fig. 6). On poly(I·C) stimulation, adapter molecule TRIF is recruited to TLR3 through their TIR-TIR domain interaction. TRIF then recruits TRAF6 to TLR3 through its TRAF6-binding site, followed by recruitment of TAK1 and TAB2 via their interaction with TRAF6. TRAF6-TAK1-TAB2 are then dissociated from the receptor and translocated to the cytosol, where TAK1 is activated. The activated TAK1 leads to the activation of IKK and NF-κB. Independent of TRAF6-TAK1-TAB2, through interaction with unknown intermediate signaling components, TRIF leads to the activation of TBK1/IKKε, resulting in IRF3 activation.
Fig. 6.
Model of TLR3-mediated signaling (see text).
Sarkar et al. (43) recently reported that TRIF has distinct binding sites for TRAF6 and TBK1, consistent with our hypothesis that TRIF is the divergent point in TLR3-mediated NF-κB and IRF3 activation. However, in the same manuscript, they also showed that a C-terminal deletion of TRIF led to further reduction in NF-κB activity, and concluded that TRIF can activate NF-κB through both TRAF6-dependent and -independent mechanisms. In our study, we found that the E252A mutation alone can completely abolish NF-κB activity, indicating that TRIF-mediated NF-κB activation is solely TRAF6-dependent. Our conclusion is strongly supported by the fact that TLR3-mediated NF-κB activation is completely abolished in TRAF6-/- MEFs.
TRIF-deficient mice were defective in both TLR3- and TLR4-mediated expression of IFN-β and activation of IRF3 (36, 37). Whereas TRIF-deficient mice showed complete loss of TLR3-induced NF-κB activation, TLR4-mediated NF-κB activation was only completely abolished in mice deficient in both MyD88 and TRIF (36, 37). The fact that both poly(I·C)-induced NF-κB and IRF3 activation are completely abolished in TRIF-deficient mice suggests that TLR3 probably only mediates TRIF-dependent pathways (36, 37). In support of this conclusion, we have previously shown that IL-1 receptor proximal signaling components (including MyD88, IRAK4, and IRAK) are not involved in TLR3-mediated signaling pathway (ref. 42; Z.J. and X.L., unpublished data). Therefore, unlike the IL-1R and other TLRs (including TLR4) that they all use the common MyD88-dependent pathway, TLR3 seems to only employ MyD88-independent TRIF-dependent pathways. One possible explanation is that the conserved proline residue within the cytoplasmic domain of the murine and human TLR proteins that is mutated in LPS-hyporesponsive C3H/HeJ mice (P712H) is represented by an alanine residue in TLR3.
Our previous studies implicated the role of TRAF6, TAK1, and TAB2 in TLR3-mediated signaling, showing that these signaling components are recruited to TLR3 on poly(I·C) stimulation. In this paper, we have provided a mechanism for how TRIF may help to link TRAF6, TAK1, and TAB2 to TLR3. Furthermore, our results suggest that TRIF-mediated recruitment of TRAF6 to TLR3 and the subsequent activation of TAK1 are probably only required for TLR3-induced NF-κB but not IRF3 activation. Therefore, despite the differences in the receptor proximal signaling events, TLR3 shares similar downstream signaling components with the IL-1R-mediated pathway to activate NF-κB, including TRAF6, TAK1, TAB2, and IKK (Fig. 6). Whereas we showed that TRIF recruits TRAF6 to TLR3 through a TRAF6-binding site, it is likely that TAK1 and TAB2 are recruited to TLR3 through TRAF6, probably similar to how TAK1 and TAB2 interact with TRAF6 in the IL-1 pathway (27). Future studies are required to understand how TRAF6, TAK1, and TAB2 are dissociated from TLR3 and how TAK1 is activated in the cytosol. Although it is possible that TRIF directly interacts with TBK1/IKKε to activate IRF3, TRIF may recruit additional intermediate components to TLR3. Future research should focus on the identification of novel signaling molecules that participate in TLR3-TRIF-mediated IRF3 activation.
Acknowledgments
We thank Dr. Holger Wesche for the cDNA constructs of TRAF 1-6 and Dr. Kunihiro Matsumoto for the TAK1 constructs. This work was supported by National Institutes of Health Grant GM 600020 (to X.L.).
Abbreviations: IKK, IκB kinase; TIR, Toll-IL-1 receptor; TLR, Toll-like receptor; TRIF, TIR domain-containing adapter inducing IFN-β.
References
- 1.Rock, F. L., Hardiman, G., Timans, J. C., Kastelein, R. A. & Bazan, J. F. (1998) Proc. Natl. Acad. Sci. USA 95, 588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitcham, J. L., Parnet, P., Bonnert, T. P., Garka, K. E., Gerhart, M. J., Slack, J. L., Gayle, M. A., Dower, S. K. & Sims, J. E. (1996) J. Biol. Chem. 271, 5777-5783. [DOI] [PubMed] [Google Scholar]
- 3.Parnet, P., Garka, K. E., Bonnert, T. P., Dower, S. K. & Sims, J. E. (1996) J. Biol. Chem. 271, 3967-3970. [DOI] [PubMed] [Google Scholar]
- 4.Lovenberg, T. W., Crowe, P. D., Liu, C., Chalmers, D. T., Liu, X. J., Liaw, C., Clevenger, W., Oltersdorf, T., De Souza, E. B. & Maki, R. A. (1996) J. Neuroimmunol. 70, 113-122. [DOI] [PubMed] [Google Scholar]
- 5.Thomassen, E., Renshaw, B. R. & Sims, J. E. (1999) Cytokine 11, 389-399. [DOI] [PubMed] [Google Scholar]
- 6.Dinarello, C. A. (1996) Blood 87, 2095-2147. [PubMed] [Google Scholar]
- 7.Medzhitov, R., Preston-Hurlburt, P. & Janeway, C. A., Jr. (1997) Nature 388, 394-397. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi, O., Kawai, T., Sanjo, H., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Takeda, K. & Akira, S. (1999) Gene 231, 59-65. [DOI] [PubMed] [Google Scholar]
- 9.Chuang, T. H. & Ulevitch, R. J. (2000) Eur. Cytokine Network 11, 372-378. [PubMed] [Google Scholar]
- 10.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K. & Akira, S. (2000) Nature 408, 740-745. [DOI] [PubMed] [Google Scholar]
- 11.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Huffel, C. V., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. (1998) Science 282, 2085-2088. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi, O., Hoshino, K., Kawai, T., Sanjo, H., Takada, H., Ogawa, T., Takeda, K. & Akira, S. (1999) Immunity. 11, 443-451. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi, O., Kaufmann, A., Grote, K., Kawai, T., Hoshino, K., Morr, M., Muhlradt, P. F. & Akira, S. (2000) J. Immunol. 164, 554-557. [DOI] [PubMed] [Google Scholar]
- 14.Underhill, D. M., Ozinsky, A., Hajjar, A. M., Stevens, A., Wilson, C. B., Bassetti, M. & Aderem, A. (1999) Nature 401, 811-815. [DOI] [PubMed] [Google Scholar]
- 15.Underhill, D. M., Ozinsky, A., Smith, K. D. & Aderem, A. (1999) Proc. Natl. Acad. Sci. USA 96, 14459-14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., Eng, J. K., Akira, S., Underhill, D. M. & Aderem, A. (2001) Nature 410, 1099-1103. [DOI] [PubMed] [Google Scholar]
- 17.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732-738. [DOI] [PubMed] [Google Scholar]
- 18.Hemmi, H., Kaisho, T., Takeuchi, O., Sato, S., Sanjo, H., Hoshino, K., Horiuchi, T., Tomizawa, H., Takeda, K. & Akira, S. (2002) Nat. Immunol. 3, 196-200. [DOI] [PubMed] [Google Scholar]
- 19.Jurk, M., Heil, F., Vollmer, J., Schetter, C., Krieg, A. M., Wagner, H., Lipford, G. & Bauer, S. (2002) Nat. Immunol. 3, 499 (lett.). [DOI] [PubMed] [Google Scholar]
- 20.Lord, K. A., Hoffman-Liebermann, B. & Liebermann, D. A. (1990) Oncogene 5, 1095-1097. [PubMed] [Google Scholar]
- 21.Wesche, H., Henzel, W. J., Shillinglaw, W., Li, S. & Cao, Z. (1997) Immunity 7, 837-847. [DOI] [PubMed] [Google Scholar]
- 22.Adachi, O., Kawai, T., Takeda, K., Matsumoto, M., Tsutsui, H., Sakagami, M., Nakanishi, K. & Akira, S. (1998) Immunity 9, 143-150. [DOI] [PubMed] [Google Scholar]
- 23.Burns, K., Clatworthy, J., Martin, L., Martinon, F., Plumpton, C., Maschera, B., Lewis, A., Ray, K., Tschopp, J. & Volpe, F. (2000) Nat. Cell Biol. 2, 346-351. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, N., Suzuki, S., Duncan, G. S., Millar, D. G., Wada, T., Mirtsos, C., Takada, H., Wakeham, A., Itie, A., Li, S., et al. (2002) Nature 416, 750-756. [DOI] [PubMed] [Google Scholar]
- 25.Li, S., Strelow, A., Fontana, E. J. & Wesche, H. (2002) Proc. Natl. Acad. Sci. USA 99, 5567-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao, Z., Xiong, J., Takeuchi, M., Kurama, T. & Goeddel, D. V. (1996) Nature 383, 443-446. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, Z., Ninomiya-Tsuji, J., Qian, Y., Matsumoto, K. & Li, X. (2002) Mol. Cell. Biol. 22, 7158-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercurio, F., Zhu, H., Murray, B. W., Shevchenko, A., Bennett, B. L., Li, J., Young, D. B., Barbosa, M., Mann, M., Manning, A. & Rao, A. (1997) Science 278, 860-866. [DOI] [PubMed] [Google Scholar]
- 29.Regnier, C. H., Song, H. Y., Gao, X., Goeddel, D. V., Cao, Z. & Rothe, M. (1997) Cell 90, 373-383. [DOI] [PubMed] [Google Scholar]
- 30.Woronicz, J. D., Gao, X., Cao, Z., Rothe, M. & Goeddel, D. V. (1997) Science 278, 866-869. [DOI] [PubMed] [Google Scholar]
- 31.Zandi, E., Rothwarf, D. M., Delhase, M., Hayakawa, M. & Karin, M. (1997) Cell 91, 243-252. [DOI] [PubMed] [Google Scholar]
- 32.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z. & Matsumoto, K. (1999) Nature 398, 252-256. [DOI] [PubMed] [Google Scholar]
- 33.Kawai, T., Adachi, O., Ogawa, T., Takeda, K. & Akira, S. (1999) Immunity 11, 115-122. [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S.-M. & Maniatis, T. (2003) Nat. Immunol. 4, 491-496. [DOI] [PubMed] [Google Scholar]
- 35.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G.-P., Lin, R. & Hiscott, J. (2003) Science 300, 1148-1151. [DOI] [PubMed] [Google Scholar]
- 36.Yammamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K. & Akira, S. (2003) Science, in press.
- 37.K. Hoebe, X. Du, P. Georgel, E. Janssen, K. Tabeta, S. O. Kim, J. Goode, P. Lin, N. Mann, S. Mudd, K. Crozat, S. Sovath, J. Han & and B. Beutler. (2003) Nature, in press. [DOI] [PubMed]
- 38.Li, X., Commane, M., Jiang, Z. & Stark, G. R. (2001) Proc. Natl. Acad. Sci. USA 98, 4461-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindler, U. & Baichwal, V. R. (1994) Mol. Cell. Biol. 14, 5820-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, X., Commane, M., Burns, C., Vithalani, K., Cao, Z. & Stark, G. R. (1999) Mol. Cell. Biol. 19, 4643-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye, H., Arron, J. R., Lamothe, B., Cirilli, M., Kobayashi, T., Shevde, N. K., Segal, D., Dzivenu, O. K., Vologodskaia, M., Yim, M., et al. (2002) Nature 418, 443-447. [DOI] [PubMed] [Google Scholar]
- 42.Jiang, Z., Zamanian-Daryoush, M., Nie, H., Silva, A. M., Williams, B. R. & Li, X. (2003) J. Biol. Chem. 278, 16713-16719. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar, S. N., Smith, H. L., Rowe, T. M. & Sen, G. C. (2003) J. Biol. Chem. 278, 4393-4396. [DOI] [PubMed] [Google Scholar]