Abstract
MicroRNAs (miRs) are small noncoding RNAs that have been reported to be promising diagnostic tools. We used quantitative real-time reverse transcription PCR (RT-qPCR) to analyze differentially expressed miRNAs in prostate tumor samples to determine its prognostic value. From 2007 to 2009, tumor tissues were obtained from 73 radical prostatectomy specimens. Differentially expressed miR-96, -145 and -221 were validated by TaqMan RT-qPCR using all 73 tissues. The prognostic value was assessed in terms of biochemical recurrence using Kaplan–Meier and Cox regression analyses. For our patient cohort, the mean age was 64.7 years (50–76 years) and the mean prostate-specific antigen (PSA) was 7.5 ng ml−1. During the follow-up period (mean, 19.4 months), 14 of 73 (19.2%) patients developed biochemical recurrence. Expression of miR-96, -145 and -221 correlated strongly with each other, but there were no correlations between miRNA expression and clinicopathologic parameters. Kaplan–Meier survival curves using the log-rank test showed a decreased biochemical recurrence-free interval with pathologic stage (P<0.001). In addition, patients with Gleason scores over 8, compared with those with a Gleason score of 6, showed a decreased biochemical recurrence-free interval in Kaplan–Meier analysis (P=0.001). However, expression of miR-96, -145 and -221 did not correlate with the biochemical recurrence interval in Kaplan–Meier survival curves or by multivariate analysis using the Cox proportional hazard regression model, either. In conclusion, we did not observe a significant correlation between the expression of miR-96, -145 and -221 and clinicopathologic parameters. To utilize miRNA as a diagnostic tool in clinical practice, more research is needed to understand miRNA mechanisms, identify miRNA targets, and further characterize miRNA function.
Keywords: microRNA, prognosis, prostate cancer, recurrence
Introduction
The natural history of prostate carcinoma (PCa) varies from an indolent tumor to a highly aggressive cancer.1 However, most patients who present with localized prostate cancer may be treated through radical prostatectomy or irradiation, because there is no reliable predictable marker for metastasis or death. PCa has had an increasing socioeconomic impact and a considerable negative impact on health related quality of life.1 There is a critical need to identify a new prognostic marker that can differentiate between tumor and normal tissue, as well as between indolent and aggressive tumors in prostate cancer. Excessive treatment could be avoided if there was an adequate marker or a set of markers that could be used to differentiate an indolent tumor from a highly aggressive cancer.
MicroRNA (miR) is a small noncoding RNA with a length of approximately 22 nucleotides.2 This type of RNA has been recognized as a modulator of gene expression and is especially involved in tumorigenesis.3 miRNA plays an important role in various biological and metabolic processes, including development, differentiation, signal transduction, cell maintenance, diseases and cancers.2 The biogenesis of miRNA has not been fully elucidated, but its function might be mainly determined by the interaction between miRNA and its target mRNA.4 Recent studies on the role of miRNA in breast cancer metastasis gave rise to the notion that miRNA may also be helpful in predicting the progression and metastasis of prostate cancer.5,6,7,8 However, to date, few articles have investigated miRNA regulation in prostate cancer and only five studies have examined miRNA expression in more than 10 samples, with highly inconsistent results.2,9,10,11,12,13
Schaefer et al.4 reported that there is no overlapping subset between the down- and upregulated miRNA patterns of previous studies. In addition, 17 of the 105 types of miRNA even showed an opposite expression pattern.4 Moreover, only a few of these microarray data were validated by quantitative real-time PCR (qRT-PCR) and few studies investigated the association between a characteristic miRNA expression pattern and clinical follow-up data.2 In other words, the problem with miRNA studies in prostate cancer is that the data are inconsistent and there are no overlapping subsets between the down- and up-regulated miRNA patterns. Therefore, we identified the types of miRNA that have been reported to predict clinical recurrence in the literature but never reproduced in other studies with a large number of patients. In this study, we investigated the miR-96, -145 and -221 profiles by qRT-PCR in 73 specimens to determine the association between miRNA expression and clinicopathological data, and to evaluate the diagnostic and prognostic value of these types of miRNA.1,2,4,12
Materials and methods
Patients
This study was approved by the ethical board of the Korea University Anam Hospital and all patients provided written and informed consent. Between 2007 and 2009, paraffin-embedded tumor tissue specimens were collected from 92 radical prostatectomy patients who provided informed consent. The patients had undergone robot-assisted radical prostatectomy at the Department of Urology, Korea University Anam Hospital. Among 92 patients, 19 patients were excluded, because they had undergone adjuvant hormonal therapy after the final pathology was confirmed. Therefore, clinicopathologic parameters of 73 patients were gathered in this study, including age, preoperative prostate-specific antigen (PSA), tumor grading according to the Gleason score based on the whole specimen, tumor stage according to the UICC 2002 TNM system, follow-up time after surgery and PSA concentration during follow-up. Follow-up exams were performed every 3 months for the first 2 years after surgery, every 6 months over the following 3 years, and annually thereafter.
For the selection of tissue specimens, we used the method for embedding tissues into paraffin blocks in duplicate, according to the previously reported method.2,11 A diagnostic hematoxylin and eosin (H&E) section was prepared to verify tumor content and margin status, and to identify areas of normal and tumor tissue. After the pathologist marked the portion that contained more than 90% tumor tissue, another formalin-fixed, paraffin-embedded sample containing this portion was prepared, using a microtome for RNA extraction.
RNA purification from formalin-fixed, paraffin-embedded samples
Five sections of 10-µm thickness and about 100-mm2 surface area formalin-fixed, paraffin-embedded samples were used for total RNA extraction as follows. Samples were deparaffinized by incubation with xylene for 1 h at 60 °C and placed in absolute ethanol for washing three times for 5 min. After air drying for 5 min, tissue sections were completely lysed with 0.8 ml of Trizol reagent (Invitrogen, Carlsbad, CA, USA) and incubated at room temperature for 10 min. With additional 0.2 ml of chloroform, samples were mixed by vortexing and centrifuged for 15 min at 25 199 g. The aqueous phase was transferred to a new 1.5-ml tube and ethanol precipitation was performed. After suspending in 20 µl of RNase-free deionized water, the quality and quantity of isolated total RNA were measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE, USA) spectrophotometer. Purified total RNA samples were stored at −80 °C until use.
Reverse transcription reaction
Total RNA was used for cDNA synthesis in a 20-µl total reaction volume using a Taqman microRNA Reverse Transcription kit (Applied Biosystems, Foster, CA, USA) with specifically designed stem-loop RT primers for each mature miRNA in a multiplexed fashion, according to the instructions provided by the manufacturer.
Quantitative real-time PCR (qRT-PCR)
For detection of mature miRNA, qRT-PCR was performed in duplicate using Taqman universal Mix II and Taqman miRNA assay kit (Applied Biosystems) for quantification of each specific miRNA (miR-96, -145 and -221) and snU6 miRNA. A final reaction volume of 10 µl was used, containing 2 µl of cDNA template, 5 µl of 2× Taqman universal Mix II, 0.5 µl of 20× Taqman miRNA assay and distilled water. The reaction was subjected to denaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 1 min. snU6 was used as the endogenous control for normalization of the expression level. The relative expression level of each miRNA was measured using the ΔCt method in which ΔCt=Ct(miRNA)−Ct(snU6) and Ct represents the threshold cycle number. The fold change in miRNA expression was determined by the 1/2ΔΔCt method as described previously.14
Statistical analysis
Statistical analysis for qRT-PCR was performed with SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). The data collection techniques and the methods of statistical analysis were approved by the Institutional Statistician Review Board of Korea University Anam Hospital. Spearman's rank correlation coefficients were used to evaluate the correlations between parameters. The prognostic value was assessed in terms of biochemical recurrence using Kaplan–Meier survival analysis and Cox's proportional hazard regression analysis using forward stepwise method.
Results
Patients and tumor characteristics
The clinical demographics of the study subjects are summarized in Table 1. In total, 73 patients were included in this study, with a mean age of 64.7 years and a mean preoperative PSA level of 7.5 ng ml−1. Gleason scores were 6 in 27 patients (37.0%), 7 (3+4) in 25 patients (34.2%), 7 (4+3) in 13 patients (17.8%), 8 (4+4) in 6 patients (8.2%) and 9 or higher in 2 patients (2.7%). The pathologic stages were T2a in 14 patients (19.2%), T2b in 4 patients (5.5%), T2c in 30 patients (41.1%), T3a in 22 patients (30.1%) and T3b in 3 patients (4.1%). The mean follow-up period was 19.4 months and a total of 14 patients developed biochemical recurrence. Biochemical progression was defined as PSA≥0.2 ng ml−1 at two consecutive follow-up visits.
Table 1. Clinical demographics of patients with prostate carcinoma (n=73).
| Clinical demographics | Value |
|---|---|
| Age, Mean(range), year | 64.7 (50–76) |
| Preoperative PSA, mean±s.d., ng ml−1 | 7.5±3.8 |
| Gleason score, n (%) | |
| 6 | 27 (37.0%) |
| 7 (3+4) | 25(34.2%) |
| 7 (4+3) | 13 (17.8%) |
| 8(4+4) | 6 (8.2%) |
| 9 | 2 (2.7%) |
| T stage, n (%) | |
| T2a | 14 (19.2%) |
| T2b | 4 (5.5%) |
| T2c | 30 (41.1%) |
| T3a | 22 (30.1%) |
| T3b | 3 (4.1%) |
| Biochemical recurrence, n (%) | 14 (19.2%) |
| Follow-up, mean±s.d., months | 19.4±9.8 |
Abbreviation: PSA, prostate-specific antigen.
The correlation between miRNA expression and clinicopathologic parameters
We evaluated whether the expression of miR-96, -145 and -221 was correlated with the Gleason score, pathologic stage and preoperative PSA level. The correlation between the expression of the measured miRNA was determined by Spearman's rank correlation analysis. The expression of miR-96 and -145 showed a fair correlation with each other (rs=0.590, P<0.001), the expression of miR-145 and miR-221 showed a very strong correlation with each other (rs=0.834, P<0.001) and the expression of miR-96 and -221 showed a moderate strong correlation with each other (rs=0.690, P<0.001) (Table 2).
Table 2. Spearman's rank correlations between miRNA and clinicopathologic parameters.
| miR-96 | miR-145 | miR-221 | PSA | GS | Stage | |||
|---|---|---|---|---|---|---|---|---|
| Spearman's rho | miR-96 | Correlation Coefficient | 1 | 0.590 | 0.690 | 0.115 | 0.179 | −0.006 |
| P value | — | 0.000 | 0.000 | 0.367 | 0.157 | 0.962 | ||
| miR-145 | Correlation Coefficient | 1 | 0.834 | 0.041 | −0.116 | −0.142 | ||
| P value | — | 0.000 | 0.751 | 0.363 | 0.263 | |||
| miR-221 | Correlation Coefficient | 1 | −0.010 | −0.071 | −0.090 | |||
| P value | — | 0.939 | 0.579 | 0.479 | ||||
| PSA | Correlation Coefficient | 1 | 0.175 | 0.311 | ||||
| P value | — | 0.139 | 0.007 | |||||
| GS | Correlation Coefficient | 1 | 0.355 | |||||
| P value | — | 0.002 | ||||||
| Stage | Correlation Coefficient | 1 | ||||||
| P value | — |
Abbreviations: GS, gleason score; miRNA, microRNA; PSA, prostate-specific antigen; —, no value.
Preoperative PSA level and Gleason score were not correlated significantly with each other (rs=0.175, P=0.139); pathologic stage and Gleason score showed a fair correlation with each other (rs=0.355, P=0.002); and PSA and pathologic stage showed a fair correlation with each other (rs=0.311, P=0.007). However, we did not find a correlation between miRNA expression and clinicopathologic parameters. miR-96 expression was not correlated with PSA, Gleason score or pathologic stage (P=0.367, 0.157 and 0.962 respectively). miR-145 expression was not correlated with PSA, Gleason score or pathologic stage (P=0.751, 0.363 and 0.263, respectively). miR-221 expression was not correlated with PSA, Gleason score or pathologic stage (P=0.939, 0.579 and 0.479, respectively) (Table 2).
miRNA as a prognostic marker
Sixteen patients experienced a biochemical relapse according to the criteria.
The prognostic value of parameters was tested by Kaplan–Meier survival analysis and Cox proportional hazard regression. The recurrence-free interval was significantly reduced with increasing pathologic stage (P<0.001) and patients with Gleason scores over 8 showed a decreased biochemical recurrence-free interval (P=0.001) compared with a Gleason score of 6 in Kaplan–Meier analysis, showing that our study cohort was representative (Figure 1).
Figure 1.
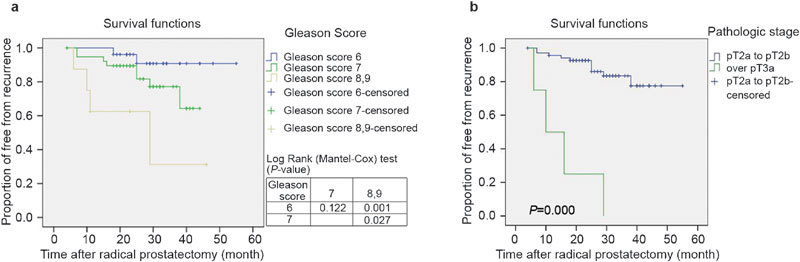
Kaplan–Meier analysis of recurrence free survival according to (a) Gleason score and (b) pathologic stage. Patients with Gleason scores over 8 compared with a Gleason score of 6 showed a decreased biochemical recurrence-free interval (P=0.001). The recurrence-free interval was significantly reduced with increasing pT stage (P<0.001). pT, pathologic T stage.
For analysis, miRNA expression was dichotomized by the median. However, the recurrence-free interval was not significantly different among patients with miR-96, -145 and -221 expression in the tumor samples (P=0.554, 0.503 and 0.348, respectively) (Figure 2).
Figure 2.
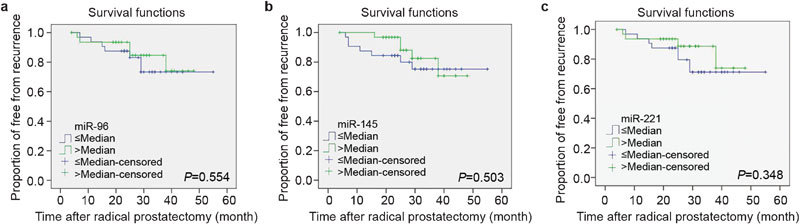
Kaplan–Meier analysis of recurrence-free survival according to (a) miR-96, (b) miR-145 and (c) miR-221. The recurrence-free interval was not significantly reduced with miR-96 (P=0.554), miR-145 (P=0.503) and miR-221 (P=0.348), respectively. miR, microRNA.
In the univariate analysis, pathologic stage and preoperative PSA were significant predictors of clinical recurrence (pathologic stage hazard ratio =13.850 (95% confidence interval: 4.206–45.603); P<0.001 and preoperative PSA hazard ratio=1.245 (95% confidence interval: 1.108–1.399); P<0.001) (Table 3). However, miR-96, -145 and -221 were not significant predictors of clinical recurrence (P=0.560, 0.510 and 0.570, respectively). Three clinicopathologic parameters and miRNA variables were assessed according to their prognostic performance in multivariate Cox proportional hazard regression using the forward stepwise method. Multivariate Cox proportional hazard regression showed that dichotomized miR-96, -145 and -221 were not significant predictors of clinical recurrence; in contrast, the preoperative PSA level and tumor stage were significant predictive factors for clinical recurrence (P=0.038 and 0.018, respectively).
Table 3. Results of univariate and multivariate Cox proportional hazard analyses of clinicopathologic parameters and several miRNAs were differentially expressed with regard to the recurrence-free interval after radical prostatectomy.
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Gleason score 7 | 3.113 (0.660–14.684) | 0.151 | ||
| Gleason score ≥8 | 11.579 (2.099–63.872) | 0.005 | ||
| Tumor stage | 13.850 (4.206–45.603) | <0.001 | 5.609 (1.348–23.330) | 0.018 |
| Preoperative PSA | 1.245 (1.108–1.399) | <0.001 | 1.181 (1.009–1.383) | 0.038 |
| miR-96 | 0.711 (0.225–2.242) | 0.560 | ||
| miR-145 | 0.679 (0.215–2.143) | 0.510 | ||
| miR-221 | 0.360 (0.171–1.896) | 0.570 | ||
Gleason score is ordinal scale variable with the subgroup 6, 7 and 8. Here, reference value of Gleason score is 6. Dichotomized variables are tumor stage (pT2 vs. pT3) and miRNAs (according to the medians). Abbreviations; CI, confidence interval; HR, hazard ratio; miRNA, microRNA; PSA, prostate-specific antigen.
Discussion
Prostate cancer has a diverse natural disease progression and the prognosis of patients is also quite variable. However, the current known prognostic factors for treatment efficacy and tumor recurrence, including the PSA level at diagnosis, staging and the Gleason score have limitations in predicting the prognosis of each patient exactly. Therefore, it is imperative to develop new prognostic tools or markers for differentiating between high and low risk cancer or predicting patient prognosis.
In this respect, new molecular markers such as miRNA seem to be attractive markers for prostate cancer. miRNA may be helpful for discriminating not only between normal and cancer tissue, but also differentiating between poorly differentiated tumors from insignificant tumors according to the expression profiles.4,15 Recent studies in breast cancer have investigated the role of miRNA in mediating metastasis, and a genetic expression assay using miRNA has been recommended to improve risk classification and recurrence prediction.16 Similarly, it is expected that miRNA might also play an important role in determining the best treatment strategy or predicting the prostate cancer patient prognosis.
However, to date, the data from previous studies have been inconsistent, which has made it difficult to make conclusions regarding the use of miRNA as a diagnostic biomarker, although several studies in other types of cancers have demonstrated the usefulness of miRNA measurements.4 Schaefer et al.4 indicated that there was no overlapping of miRNA expression in prostate cancer, possibly because of differences in the samples used for the miRNA expression analysis. In other words, a tumor tissue sample may include normal or benign prostate hyperplasia tissue even though a microdissection was performed to include over 90% of the tumor based on H&E stain preparation.10,11 In our study, we also tried to include over 90% of the tumor through H&E stain preparation.
There are few reports demonstrating a correlation between miRNA expression and clinicopathologic parameters with a sufficient sample size. According to a previous study, at least 70 samples are necessary to obtain a power of at least 80% for predicting tumor recurrence.2 Schaefer et al.2 used radical prostatectomy specimens from 79 patients to evaluate the miRNA as a prognostic marker. Spahn et al.1 used specimens from 92 patients to analyze miR-221 expression. We also used radical prostatectomy specimens from 73 patients to analyze the relationship between miRNA expression and clinicopathologic parameters. Finally, previous miRNA studies in prostate cancer have produced inconsistent results and few studies have done with a sufficient sample size. Therefore, we performed a literature review to identify suitable miRNA and chose miR-96, -145 and -221, which are proven diagnostic markers based on association studies with clinicopathologic data.1,2,12 RT-PCR was used to validate our findings using a sufficient sample size.
Schaefer et al.2 performed miRNA profiling in 79 PCa tissues by RT-qPCR to evaluate the potential of using miRNA as a prognostic marker. They reported that expression of miR-96 was associated with Gleason score and biochemical tumor recurrence after radical prostatectomy. They showed that increased miR-96 expression was significantly associated with Gleason score in the Cox model (P=0.052). It was the first report to indicate its association with prostate cancer. In other words, their result was not consistent with previous miRNA results. They mentioned that the recurrence-free interval was significantly reduced with higher Gleason score and pT stage, indicating the representativeness of their cohort. In addition, the failure rate was comparable with generally observed results. In our study, the recurrence-free interval was significantly decreased with higher Gleason score and pathologic T stage and the recurrence rate corresponded with that observed in previous studies. The Gleason score, preoperative PSA level and pathologic stage were strongly correlated with each other. Our results indicated that our study group was also representative. However, in our study, we found that miR-96 was not a prognostic indicator in Kaplan–Meier and Cox proportional hazard regression models.
Until the study of Schaefer et al.2, only one prior study identified a characteristic expression pattern after examining the relationship between miRNA and clinical data. Tong et al.12 investigated the association between the miRNA expression profile and clinical data using 40 prostatectomy specimens, although the sample size was smaller than that of Schaefer et al.2 and our study.2,12 They found five downregulated types of miRNA, including miR-145, and emphasized that the finding of decreased miR-145 expression was consistent with the finding that miR-145 was one of the most frequently downregulated types of miRNA in 16 prostatectomy specimens in the study by Ozen et al.10 Decreased miR-145 expression has been described in lung caner, breast cancer and hepatocellular carcinoma.17,18 In addition, the expression of miR-145 has consistently been found to be decreased in the adenomatous and cancer stages of colorectal cancer.19,20 Porkka et al.11 reported that miR-145 was downregulated in PCa compared with BPH. Ozen et al.10 also reported that miR-145 was significantly downregulated in prostate cancer as determined by miRNA microarrays. However, we did not find an association between the expression of miRNA-145 and clinicopathologic parameters in prostate cancer.
Spahn et al.1 investigated whether miR-221 has potential as a prognostic marker in prostate cancer using the largest clinical sample size to date. They found that miR-221 was strongly downregulated in prostate cancer compared with the benign control in a microarray study. In addition, the overall expression of miR-221 was downregulated in primary prostate cancer and metastasis samples in real-time RT-PCR analysis. However, comparison of BPH samples and non-cancerous peripheral zone prostatic tissue did not show any difference. They used RT-PCR to analyze miR-221 expression in a cohort consisting of 92 patients. In their study, Kaplan–Meier analysis and Cox proportional hazard analysis showed that lower miR-221 expression was associated with tumor progression and recurrence in a high-risk prostate cancer cohort. In addition, downregulation of miR-221 has been confirmed by other miRNA studies in prostate cancer and has been shown to be a relatively consistent event in prostate cancer.9,10,11 However, we did not find an association between clinicopathologic parameters and miR-221 expression in our study.
In our study, we did not observe an association between miR-96, -145 or -221 expression and clinicopathologic parameters of prostate cancer, indicating that these types of miRNA do not have prognostic value for prostate cancer patients. These results are paradoxically consistent with the inconsistency of results obtained from previous studies regarding miRNA expression in prostate cancer. As previously mentioned, results may vary depending on the technical methods used to obtain the samples, such as microdissection or H&E preparation to verify tumor content and margin status.10,11 Further, there is variation across studies in the techniques used for miRNA preparation (total RNA or purified miRNA with partial detection of premature miRNA), as well as with miRNA measurement platforms.10,13,21,22 And more importantly, we do not know the exact mechanisms by which miRNA is regulated or processed in tumors.4 In addition, it is widely accepted that one miRNA can target many mRNA sequences responsible for several different proteins.23 In other words, it is not a ‘one-to-one' relationship between miRNA expression and target mRNA expression and the relationship depends on orchestral and dynamic regulation.24 In the future, more objective methods should be developed to study miRNA, and more research is needed to recognize the targets of miRNA and to explore their complex molecular effects. At the same time, consistent results about previously reported miRNA are needed to develop diagnostic markers for clinical use.
We did note a significant correlation among the expression of the three types of miRNA in our study. Thus, it seems that there might be characteristic miRNA expression in each patient, but it is unclear if the expression profiles are due to tumor differentiation or behavior. In addition, a previous comparative study with mRNA showed that miRNA expression patterns provide more precise diagnostic results.21 Based on the successful use of mRNA expression profiles in breast cancer treatment, the results of the comparative study seem to be encouraging for prostate cancer. While we did not observe a significant relationship between the expression of miR-96, -145 and -221 and clinicopathologic parameters for prostate cancer, future studies should continue to work toward identifying and validating types of miRNA that have prognostic value for prostate cancer.
Conclusions
We analyzed the prognostic values of miR-96, -145 and -221 in prostate cancer. We did not find an association between the expression of these types of miRNA and clinicopathologic parameters of a cohort of prostate cancer patients. Although miRNA has many advantages as a diagnostic tool, more research is needed to identify the mechanisms of miRNA and its targets for further characterizing the function of the miRNA. In addition, the identification of new types of miRNA through microarray analysis needs to be validated by data from other studies, as well as studies that demonstrate a correlation between miRNA expression and clinicopathologic parameters.
Author contributions
SGK participated in the design of the study, the analysis and interpretation of the data, and the drafting of the manuscript. YRH and SJK helped in the experiment. SHK and HSP helped the data analysis. JGL, JC participated in the design of the study. CHK participated in the design of the study and helped the experiment and the manuscript drafting.
Acknowledgments
This study was supported by a research grant (No. K1220301) from Korea University College of Medicine (Seoul, Korea).
The authors declare no competing financial interests.
References
- Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. International journal of cancer. J Int Cancer. 2010;127:394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. International journal of cancer. J Int Cancer. 2010;126:1166–76. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Kristiansen G, Lein M, Schrader M, et al. MicroRNAs and cancer: current state and future perspectives in urologic oncology. Urol Oncol. 2010;28:4–13. doi: 10.1016/j.urolonc.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi XB, Tepper CG, White RW. MicroRNAs and prostate cancer. J Cell Mol Med. 2008;12:1456–65. doi: 10.1111/j.1582-4934.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Tong AW, Fulgham P, Jay C, Chen P, Khalil I, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–16. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Cowland JB, Hother C, Gronbaek K. MicroRNAs and cancer. APMIS. 2007;115:1090–106. doi: 10.1111/j.1600-0463.2007.apm_775.xml.x. [DOI] [PubMed] [Google Scholar]
- Benowitz S. Revised guidelines signal that gene expression profiles are coming of age. J Natl Cancer Inst. 2008;100:916–7. doi: 10.1093/jnci/djn228. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–7. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]


