Abstract
Much of the CD8+ T cell response in H2b mice with influenza pneumonia is directed at the nucleoprotein366-374 (NP366) and acid polymerase224-233 (PA224) peptides presented by the H2Db MHC class I glycoprotein. These DbNP366- and DbPA224-specific T cell populations are readily analyzed by staining with tetrameric complexes of MHC+ peptide (tetramers) or by cytokine production subsequent to in vitro stimulation with the cognate peptides. The DbPA224-specific CD8+ effector T cells make more tumor necrosis factor (TNF) α than the comparable CD8+DbNP366+ set, a difference reflected in the greater sensitivity of the CD8+DbPA224+ population to TNF receptor (TNFR) 2-mediated apoptosis under conditions of in vitro culture. Freshly isolated CD8+DbNP366+ and CD8+DbPA224+ T cells from influenza-infected TNFR2-/- mice produce higher levels of IFN-γ and TNF-α after in vitro stimulation with peptide, although the avidity of the T cell receptor-epitope interaction does not change. Increased numbers of both CD8+DbPA224+ and CD8+DbNP366+ T cells were recovered from the lungs (but not the spleens) of secondarily challenged TNFR2-/- mice, a pattern that correlates with the profiles of TNFR expression in the TNFR2+/+ controls. Thus, it seems that TNFR2-mediated editing of influenza-specific CD8+ T cells functions to limit the numbers of effectors that have localized to the site of pathology in the lung but does not modify the size of the less activated responder T cell populations in the spleen. Therefore, the massive difference in magnitude for the secondary, although not the primary, response to these DbNP366 and DbPA224 epitopes cannot be considered to reflect differential TNFR2-mediated T cell editing.
Systematic analysis of events occurring in the lung and the lymphoid tissue of C57BL/6J (B6) mice challenged intranasally with the HKx31 (H3N2) influenza A virus has led to the establishment of a highly reproducible model for analyzing the nature of cell-mediated immunity in a localized infectious process (1, 2). Influenza virus replication in the mouse is, because of the requirement for an anatomically restricted substilin-related protease (3) to cleave the surface hemagglutinin (H) molecule, largely restricted to the superficial layer of the respiratory epithelium. Virus-specific CD8+ T cell responses specific for the prominent influenza DbNP366 and DbPA224 epitopes are characterized by massive clonal expansion in the regional lymph nodes and spleen, exit into the circulation, and extravasation into the site of virus-induced pathology (4-6).
Exposure to high levels of antigen in the infected lung leads to further differentiation into potent cytotoxic T lymphocyte (CTL) effectors (1, 7, 8), which in turn function to eliminate the pathogen (9, 10). Subsequent to virus clearance, the resolution of influenza pneumonia is associated with a rapid fall (6) in the numbers of both CD8+DbNP366+ and CD8+DbPA224+ T cells that can be recovered by bronchoalveolar lavage (BAL). This process of contraction proceeds much more slowly for the less activated T cell populations (1, 7, 8) in the spleen and lymph nodes (6).
The CD8+DbNP366+ and CD8+DbPA224+ T cell responses differ in two important ways. The first is that, although the extent of clonal expansion and the magnitude of the CD8+DbNP366+ and CD8+DbPA224+ memory T cell populations generated after primary infection are approximately equivalent in magnitude, the size of the secondary response to DbNP366 is at least 10 times higher than that to DbPA224 (6, 11). The other difference is that the DbPA224-specific set makes more IFN-γ and tumor necrosis factor (TNF) α after short-term in vitro stimulation with the cognate peptide (12), suggesting the possibility that the “functional avidity” (13, 14) of T cell receptor (TCR)-DbPA224 binding is higher than that for the TCR-DbNP366 interaction.
Experiments in cell culture systems have shown that apoptotic elimination of the CD8+ set (15) is mediated by means of a TNF-α/TNF receptor (TNFR) 2 interaction, reflecting that TNFR2 is the major TNFR expressed on activated CD8+ T cells (16). The death pathway engaged after ligation of TNFR2 is regulated differently from the classical pathway signaled by the TNFR1 “death domain” (17). Exposure of antigen-specific CD8+ T cells to IL-2 (18) and/or IFN-γ (19) can sensitize for TNF/TNFR2-mediated apoptosis. Furthermore, high-avidity CD8+ effectors analyzed in vitro have been found to be particularly susceptible to TNFR2-mediated death (20, 21).
The present experiments compare conventional TNFR2+/+ and genetically disrupted TNFR2-/- mice (16) to ask whether this TNF-α-mediated editing process (20) can operate to return the greatly expanded, antigen-driven phase of the influenza virus-specific CD8+ T cell response to the homeostatic equilibrium characteristic of long-term memory (2, 14, 22). We also ask whether such effects are a factor in the dramatic divergence (6, 11) between CD8+DbNP366+ and CD8+DbPA224+ T cell numbers after secondary challenge.
Materials and Methods
Mice, Viruses, Infection, and Sampling. Six- to 8-week-old female TNFR2+/+ and TNFR2-/- (16) mice on the C57BL/6J (B6, H2b) background were purchased from The Jackson Laboratory and housed under specific pathogen-free conditions. Some were infected i.p. with 108.5 50% embryonated hen's egg infectious dose (EID50) of the PR8 (H1N1) influenza A virus (1). Naïve and PR8-primed (12-16 weeks previously) mice were anesthetized by i.p. injection of avertin and challenged intranasally with 106.8 EID50 of the HKx31 (H3N2) virus (4, 23). Lymphocytes were isolated from lung by BAL (1), and CD8+ T cells were enriched (24) from single cell preparations of spleen using mAbs (Pharmingen) to CD4 (GK1.5) and MHC class II (TIB120) followed by anti-rat and anti-mouse Ig-coated magnetic beads (Dynal, Oslo).
Intracellular Cytokine Staining. Enriched CD8+ T cells isolated from either the pneumonic lung by BAL or from the spleen of infected mice were stimulated with either 1 μM NP366-374 (ASNENMETM) or PA224-233 (SSLENFRAYV) peptides (11, 25) or graded concentrations of peptides in the presence of brefeldin A and IL-2. For the CD8β blocking experiments, cells were incubated with 10 μg/ml anti-CD8β (clone 53-5.8, Pharmingen) for 30 min before peptide was added. After 5 h the cells were stained with anti-CD8α-peridinin chlorophyll protein-Cy5.5 (Pharmingen), fixed with 1% paraformaldehyde, and permeabilized by washing twice in PBS/0.1% BSA/0.1% saponin. The samples were then stained with anti-IFN-γ-phycoerythrin (PE) (clone XMG1.2, Pharmingen) and anti-TNF-α allophycoerythrin (clone MP6-XT22, Pharmingen), and flow cytometric data were acquired on a FACSCalibur (Becton Dickinson Immunocytometry Systems) by using cellquest software (Becton Dickinson). A minimum of 30,000 lymphoctye/CD8+ events were collected.
Tetramer Staining and Disassociation. Immune CD8+ T cells were stained with the DbNP366-PE (4) or DbPA224-PE tetramers (11). The tetramer dissociation protocol (26) used 5 × 105 BAL lymphocytes stained with either the DbPA224-PE or the DbNP366-PE tetramer at room temperature for 1 h in 0.1% BSA/0.02% sodium azide in PBS (fluorescence-activated cell sorting buffer). The cells were washed three times in fluorescence-activated cell sorting buffer, and then anti-H-2Db (2.5 mg, clone 28-14-8) was added to the cells to neutralize rebinding of the tetramer. Cells were transferred to 37°C, sampled at various times, washed twice, stained with anti-CD8α-FITC, and fixed in paraformaldehyde (1% in PBS). Results are presented as a percentage of maximal mean fluorescence intensity at time 0 before the addition of the anti-H-2Db.
CTL Lines and Cell Death Assays. Splenocytes (3 × 107) isolated from mice previously infected with HKx31 (d8) were cocultured with 3 × 107 irradiated (3,000 rad) syngeneic splenocytes pulsed with 1 μM NP366-374 or PA224-233 peptide in 75-cm2 tissue culture flasks containing RPMI medium 1640 supplemented with 10% FCS, l-glutamine, sodium pyruvate, nonessential amino acids, 5 × 10-5 M 2-mercaptoethanol, Hepes, penicillin, and streptomycin. After 6 days, sensitivity to high-dose peptide stimulation was determined according to Alexander-Miller et al. (20). Briefly, 1 × 105 CTL were restimulated with 5 × 106 syngeneic splenocytes pulsed with either 10 or 0.01 μM specific peptide in 2 ml of complete RPMI as quadruplicate cultures in 24-well plates. After 36-40 h of stimulation, viable cell counts were performed on individual wells by using trypan blue exclusion. Quadruplicate wells were pooled and then stained with tetramer, CD8, and annexin V-FITC. Samples were counterstained with propidium iodide (PI), and the percentage of tetramer+, annexin V+, PI- CTL was determined by using a FACSCalibur (Becton Dickinson) flow cytometer.
Results
The following experiments focus on the comparison of the response characteristics and fate of CD8+DbNP366+ and CD8+DbPA224+ T cells (12) from TNFR2+/+ and TNFR2-/- (16) mice both in vitro and in vivo after primary (naïve) or secondary (PR8, H1N1-primed) challenge with the HKx31 (H3N2) influenza A virus.
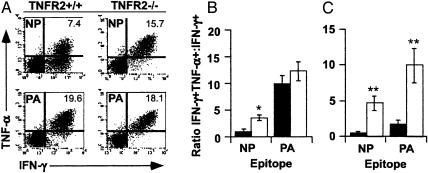
Cytokine Profiles of CD8+ T Cells from TNFR2+/+ and TNFR2-/- Mice. Virus-specific CD8+ T cells were isolated from the lung (by BAL) or spleen on day 8 after intranasal challenge with the HKx31 influenza A virus, then stimulated for 5 h in vitro with 1 μM NP366-374 or PA224-233 peptide. The DbPA224 > DbNP366 hierarchy for prevalence of TNF-α+ cells within the IFN-γ+ set described in ref. 12 was confirmed for the TNFR2+/+ BAL population (Fig. 1A). By contrast, the proportion of IFN-γ+TNF-α+ T cells was approximately equivalent for the CD8+DbNP366+ and CD8+DbPA224+ effectors recovered from the TNFR2-/- mice (Fig. 1A). This pattern of greatly enhanced TNF-α staining (Fig. 1A) for the TNFR2-/- CD8+DbNP366+ set is illustrated by the convergence of IFN-γ+TNF-α+/IFN-γ+ ratios for the TNFR2+/+ and TNFR2-/- BAL (Fig. 1B, P < 0.005) and spleen (Fig. 1C, P < 0.001) populations. Comparable differences in the IFN-γ/TNF-α ratio between the TNFR2+/+ and TNFR2-/- sets were also found for CD8+DbPA224+ T cells recovered from the spleen (Fig. 1C, P < 0.001) but not from the virus-infected respiratory tract (Fig. 1B). However, the latter simply reflects that most of the CD8+DbPA224+ T cells in the TNFR2+/+ BAL population are already IFN-γ+TNF-α+ (Fig. 1A), presumably as a consequence of the higher antigenic load in the lung. The overall pattern is thus that the CD8+DbPA224+ T cells normally make more TNF-α than the CD8+DbNP366+ set after in vitro stimulation with the cognate peptide. However, the level of TNF-α production is increased for both T cell populations in TNFR2-/- mice, perhaps reflecting the absence of TNFR2-mediated feedback control.
Fig. 1.
Prevalence of IFN-γ+TNF-α+ T cells in TNFR2+/+ and TNFR2-/- mice. Lymphocytes were isolated from the spleen and the pneumonic lung (by BAL) on day 8 after primary Hkx31 challenge infection of TNFR2+/+ and TNFR2-/- mice, then stained for IFN-γ and TNF-α expression after 5 h of in vitro stimulation with 1 μM peptide. (A) Typical flow cytometry plots for peptide-stimulated DbNP366- and DbPA224-specific CD8+ T cells in the BAL. (B and C) IFN-γ+TNF-α+/IFN-γ+ ratios for BAL (B) and spleen (C) populations from the TNFR2+/+ (filled bars) and TNFR2-/- (open bars) mice. The data in B and C are expressed as mean ± SD, and statistical significance was determined by using Student's t test (*, P < 0.005; **, P < 0.001; n = 5).
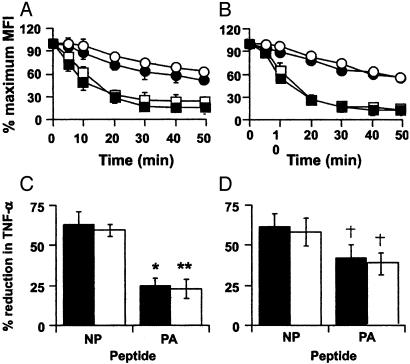
Analysis of TCR Avidity. Do these patterns of differential TNF-α production for CD8+DbNP366+ and CD8+DbPA224+ T cells from TNFR2+/+ and TNFR2-/- mice (Fig. 1) reflect changing profiles of TCR avidity? The characteristics of TCR binding were thus compared for TNFR2+/+ and TNFR2-/- CD8+DbNP366+ and CD8+DbPA224+ T cells isolated from the pneumonic lung (Fig. 2A and C) or spleen (Fig. 2 B and D). As expected from previous experiments with TNFR2+/+ mice, the avidity of TCR binding measured by either tetramer elution (26, 27) or by blocking of peptide-induced cytokine production in the concurrent presence of a mAb to CD8β (28, 29) was higher for DbPA224 than for DbNP366 (Fig. 2). However, there was no TNFR2-related difference in the rate of tetramer disassociation for CD8+DbNP366+ and CD8+DbPA224+ T cells recovered from the BAL and spleen (Fig. 2 A and B). Similarly, the capacity of TNFR2+/+ and TNFR2-/- CD8+DbNP366+ and CD8+DbPA224+ T cells to produce TNF-α (Fig. 2 C and D) was inhibited to an equivalent extent in the presence of anti-CD8β. Therefore, the absence of TNFR2 did not obviously modify the characteristics of peptide stimulation (Fig. 2 C and D) for these two antigen-specific T cell sets or change the DbPA224 > DbNP366 hierarchy in tetramer binding/elution (Fig. 2 A and B). The difference in TNF-α production (Fig. 1) found for the TNFR2-/- mice is not reflected in any changes in the measures of T cell avidity used here (Fig. 2).
Fig. 2.
TCR-epitope binding characteristics for TNFR2+/+ and TNFR2-/- mice. Lymphocyte populations were recovered from the pneumonic lung by BAL (A) or from the spleen (B) of either TNFR2+/+ (filled symbols) or TNFR2-/- (open symbols) mice on day 8 of a primary HKx31 response. The populations were labeled with anti-CD8α-FITC and either the DbNP-PE (▪) or DbPA-PE (•) tetramers at room temperature for 60 min in medium containing azide, then incubated at 37°C in the presence of anti-H2Db to block any rebinding of eluted tetramer. The data are expressed as a percentage of the initial percentage of CD8+ tetramer+ at various times after transfer to 37°C. (C and D) Other cells from the same BAL (C) and spleen (D) population of TNFR2+/+ (filled bars) or TNFR2-/- (open bars) mice were incubated with anti-CD8β, then stimulated with 1 μM NP366 or PA224 peptide. The level of TNF-α production was determined by intracellular cytokine staining, and the data are presented as a percentage of the maximum TNF-α response observed in the absence of anti-CD8β. Statistical differences between the NP366- and PA224-specific responses were determined by using Student's t test (*, P < 0.007; **, P < 0.0001; †, P < 0.005; n = 5).
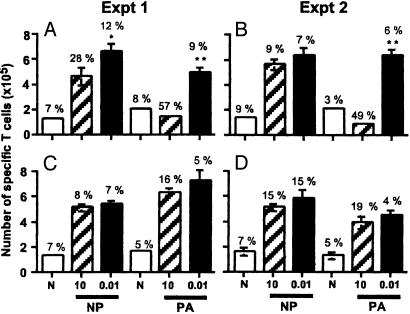
Sensitivity of Cultured T Cells to TNF-Induced Apoptosis. Is the DbPA224 → DbNP366 hierarchy in TNF-α production (Fig. 1) for conventional TNFR2+/+ CD8+ effectors a determinant of altered patterns of T cell survival? Previous studies have demonstrated that high-avidity T cells are sensitive to TNFR2-mediated editing after stimulation with supraoptimal concentrations of peptide (16). Immune spleen populations from TNFR2+/+ and TNFR2-/- mice were incubated for 36-40 h with high (10.0 μM) and low (0.01 μM) concentrations of the NP366-374 or PA224-233 peptide, then analyzed for the numbers of viable cells per well (Fig. 3). After counting, the replicate cultures were pooled and stained with PI (to gate out dead PI+ cells), the relevant tetramer, and mAbs to CD8α and annexin V. The percentage tetramer+/annexin V+ was determined for the PI-CD8+ set, and taken as a measure of the still-viable cells undergoing apoptosis.
Fig. 3.
Differential TNFR2-mediated loss after in vitro stimulation. Enriched CD8+ T cells (1 × 105) from TNFR2+/+ (A and B) or congenic TNFR2-/- (C and D) H2b mouse spleen were cultured without peptide (open bars) or with 10 μM (hatched bars) or 0.01 μM (filled bars) peptide. The results are from two independent experiments (Exp. 1 in A and C and Exp. 2 in B and D). Viable lymphocytes (by trypan blue exclusion) were counted after 36-40 h. The histograms show the mean ± SD cell counts for CD8α-allophycocyanin+ T cells staining with the DbNP366-PE or DbPA224-PE tetramer. These values were compared by Student's t test (*, P < 0.01; **, P < 0.002). The same populations were stained with annexin V-FITC, and the necrotic cells were gated out by PI staining. Above the histograms are the percentages of tetramer+annexin V+/PI-, which can be considered to measure the extent of apoptosis in these still-viable (PI-) lymphocytes.
Exposure to 0.01 μM peptide (Fig. 3, filled bars) was associated with a uniform pattern of expansion for both the DbPA224- and DbNP366-specific T cells in cultures established from both the TNFR2+/+ and TNFR2-/- mice. However, stimulation with 10.0 μM peptide led to a dramatic diminution in the magnitude of the TNFR2+/+ CD8+DbPA224+ set (Fig. 3 A and B, hatched bars). This decrease was associated with a concomitant rise in the percentage of TNFR2+/+ CD8+DbPA224+ T cells staining with annexin V (Fig. 3 A and B, numerical values). Some loss was also seen for the TNFR2+/+ DbNP366-specific population (P < 0.05) in the first experiment (Fig. 3A), but this did not repeat (Fig. 3B). These effects were not apparent for the TNFR2-/- CD8+DbNP366+ and CD8+DbPA224+ populations (Fig. 3 C and D), indicating that cell loss associated with high-dose peptide stimulation (Fig. 3 A and B) is indeed TNF-mediated. Thus, although the TNFR2-/- T cells produce more TNF-α (Fig. 1), this has no effect on T cell viability in the absence of TNFR2-mediated effector mechanisms (Fig. 3 C and D).
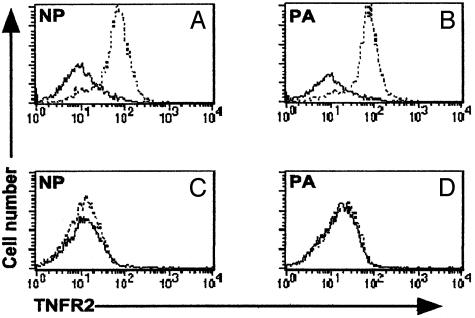
Profiles of in Vivo TNFR2 Expression. Given that TNF-α-mediated cell loss in vitro depends on the capacity to express TNFR2 (Fig. 3), it would seem reasonable to think that any TNF-α-mediated editing process operating in TNFR2+/+ mice would be limited to situations in which there is TNFR2 expression on the responding lymphocyte populations. Both the CD8+DbNP366+ and CD8+DbPA224+ populations obtained by BAL on day 8 after primary HKx31 infection were uniformly positive for TNFR2 (Fig. 4 A and B), although there was little evidence of staining for the same sets of T cells in the spleen (Fig. 4 C and D). This pattern of TNFR2 expression is likely to reflect the level of antigen stimulation (16, 30). The lung is a site of high virus load, whereas there is minimal evidence of productive infection in the lymphoid tissue of HKx31-infected B6 mice (1, 5).
Fig. 4.
Cell surface expression of TNFR2 on virus-specific CD8+ T cells recovered from the lung but not the spleen. The CD8+ set was enriched from cell populations obtained by BAL (A and B) or disruption of the spleen (C and D) on day 7 after secondary HKx31 → PR8 challenge and stained with the DbNP366 (A and C) or DbPA224 (B and D) tetramers, anti-CD8α-FITC, and biotinylated anti-mouse TNFR2 (broken line). The-TNFR2 staining was developed with streptavidin-allophycocyanin. The negative control was an irrelevant primary isotype-specific Ab (bold line). The fluorescence-activated cell sorting profiles are representative of three separate experiments.
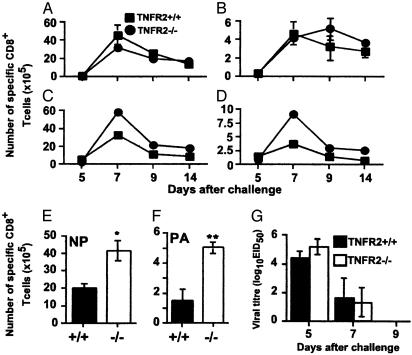
Effect of TNF Editing on the in Vivo Response. The CD8+DbNP366+ and CD8+DbPA224+ populations recovered from the spleen and BAL after primary HKx31 infection did not obviously differ in magnitude for the TNFR2-/- and TNFR2+/+ mice (data not shown). The same was true for the spleen after secondary challenge (Fig. 5 A and B), although counting pooled BAL samples suggested that there were more CD8+DbNP366+ (Fig. 5C) and CD8+DbPA224+ T cells (Fig. 5D) in the lungs of the TNFR2-/- group. Repeating this analysis for individual BAL samples taken on day 7 of the secondary response showed that the CD8+DbNP366+ and CD8+DbPA224+ sets were larger by a factor of 2 (P < 0.005) and 4 (P < 0.0001), respectively, in the TNFR2-/- mice (Fig. 5F). Furthermore, calculating the results (Fig. 5 E and F) as the ratio of CD8+DbNP366+/CD8+DbPA224+ T cells within individual BAL populations gave a 2-fold higher value for the TNFR2-/- mice (P < 0.05, data not shown), indicating that the extent of TNF editing was greater for DbPA224-specific set. However, these differences were not associated with any obvious enhancement of virus clearance (Fig. 5G). Thus, it seems that TNF-α editing mediated by means of cell-surface TNFR2 plays a part in reducing the numbers of highly activated, virus-specific CD8+ T cells in the respiratory tract of mice with influenza pneumonia. Although the magnitude of this editing process shows some correlation with TCR avidity profiles measured in vitro (Fig. 2), the effect is seen for both CD8+DbNP366+ and CD8+DbPA224+ effector T cells.
Fig. 5.
Virus-specific CD8+ T cell numbers in BAL populations from TNFR2-/- mice. Secondarily challenged (HKx31 → PR8) TNFR2+/+ (▪) and TNFR2-/- (•) mice were sampled at various time points, and CD8+ T cell populations from five mice were enriched from individual spleens (A and B) or pooled BALs (C and D), then stained with anti-CD8α and the DbNP366 (A and C) or DbPA224 (B and D) tetramers. The numbers of virus-specific CD8+ T cells (E and F) were determined from the percentage of cells staining and the total cell counts (data not shown). The analysis of the BAL was repeated on day 7 for the TNFR2+/+ (filled bars) and TNFR2-/- (open bars) to give the numbers of CD8+DbNP366+ (E) and CD8+DbPA224+ (F) T cells. Statistical significance was determined by Student's t test (*, P < 0.007; **, P < 0.001; n = 5). (G) Lung homogenates from the TNFR2+/+ and TNFR2-/- mice were titrated in embryonated hen's eggs, and the 50% egg infectious dose (EID50) was determined by the capacity of infected allantoic fluid to agglutinate chicken erythrocytes.
Discussion
These experiments establish that TNFR2-mediated editing operates to modulate influenza virus-specific CD8+ effector T cell numbers by means of antigen dose and TCR avidity-dependent mechanisms. This correlates with the expression of cell-surface TNFR2 and operates in the “high antigen” milieu of the infected lung but not in the “low antigen” environment of the secondary lymphoid tissue. Increased cell counts in BAL populations recovered from TNFR2-/- mice are found for both the CD8+DbNP366+ and CD8+DbPA224+ sets, with the effect being marginally more apparent for the high-avidity CD8+DbPA224+ T cells.
Both DbNP366 and DbPA224 are recognized by a diverse spectrum of TCRs (31, 32), which might be expected to vary from the aspect of the avidity of individual TCR-epitope interactions. However, TNFR2-mediated editing of highly activated CD8+DbNP366+ and CD8+DbPA224+ T cells did not obviously lead to the elimination of the “best binders.” There were no detectable differences in TCR avidity profiles for the TNFR2+/+ and TNFR2-/- BAL populations, although it is possible that the tetramer elution and peptide stimulation (in the presence of a mAb to CD8β) protocols that were used would not have been sufficiently sensitive to measure such a selective process.
Also, like the change in functional avidity (13) described for the progressive “maturation” of virus-specific CD8+ memory T cell populations, altered profiles of peptide-induced cytokine production in vitro do not necessarily reflect the preferential survival of a higher-avidity set. Longitudinal TCR CDR3β sequence analysis of CD8+DbNP366+ (K. Kedzierska, P.C.D., and S.J.T., unpublished observations) and CD8+DbPA224+ (32) T cells in the BAL and spleen and of individual TNFR2+/+ mice has not shown any obvious divergence in clonotype distribution profiles, either during the acute response or through to long-term memory. Therefore, the TNFR2-mediated editing that we have shown here to operate in the BAL does not seem to be characterized by the enrichment (or exclusion) of particular TCRβ clonotypes (refs. 31 and 32 and K. Kedzierska, P.C.D., and S.J.T., unpublished observations).
The extent of TNFR2-mediated cell death induced by in vitro stimulation with 10 μM (but not 0.01 μM) cognate peptide was much greater for the high-avidity CD8+DbPA224+ T cells than for the lower-avidity CD8+DbNP366+ set. The fact that this TCR avidity-related effect was less apparent for the comparable inflammatory T cell populations recovered from the infected lung may reflect that the in vivo antigen dose is likely to be higher for DbNP366 than for DbPA224. Probing freshly isolated antigen-presenting cells from influenza virus-infected B6 mice with DbNP366-specific and DbPA224-specific hybridoma lines indicates that DbPA224 is expressed only on dendritic cells, the “professional” antigen-presenting cells, whereas DbNP366 can also be detected on macrophages and epithelial cells (33). This correlates with the fact that, although both DbNP366-specific and DbPA224-specific CTL activity measured by 51Cr release can be demonstrated for peptide-pulsed EL4 cells, only the CD8+DbNP366+ effectors are lytic for virus-infected EL4 targets (11). Thus, it is likely that the DbPA224 epitope is not expressed on productively infected lung epithelium, although there are many dendritic cells in the pneumonic lung (33-35). Any protection associated with the CD8+DbPA224+ T cell response (10) may thus be mediated by means of mechanisms other than the direct recognition of virus-infected respiratory epithelium (9). However, the present experiments with TNFR2-/- mice do not necessarily rule out a role for TNF-α (36) in such a process, because the CD4+ T helper-dependent Ab response can also operate to clear influenza A viruses in the complete absence of CD8+ T cell effector function (37-39).
The observation that TNFR2-mediated editing is apparent for the TNFR2+ cells isolated by BAL but not for the TNFR2- spleen populations is supported by evidence that TNFR2 is the major TNFR expressed on activated CD8 T cells (15, 16, 40). The increased TNF-α production observed in TNFR2-/- T cells may reflect increased functional differentiation of antigen-specific cells that are normally sensitive to TNFR2-mediated apoptosis. The localized effect of the TNFR2-mediated editing also means that selective TNF-α-mediated deletion of the higher-avidity CD8+DbPA224+ set cannot possibly account for the 10-fold difference in magnitude between the secondary responses to DbNP366 and DbPA224 in TNFR2+/+ mice. Only a minority of antigen-specific T cells ever enter the site of virus-induced pathology: the number of virus-specific CD8+ effectors that localize to the infected lung are very small when compared with the size of the CD8+DbNP366+ and CD8+DbPA224+ populations in the responding lymphoid tissue (4, 6). The reason that the primary responses to DbNP366 and DbPA224 are comparable in magnitude may reflect that naïve T cells are driven to an inexorable differentiation and proliferation program by relatively short-term exposure to antigen presented on dendritic cells (41, 42), whereas the more readily activated memory set may be induced to replicate longer as a consequence of (perhaps suboptimal) stimulation by a variety of cell types. The latter situation applies only for DbNP366 and not for DbPA224, which is restricted to the dendritic cells (33). It is also the case that the structural NP is made in the order of 30-fold greater abundance than the PA enzyme (43), although there is no necessary correlation between protein abundance and the magnitude of the consequent epitope-specific CD8+ T cell response (44, 45).
Given that the limited TNF-α-mediated editing process described here is peptide dose-dependent, it cannot be a major factor in the progressive return of responding T cell numbers to homeostatic equilibrium after viral antigen is eliminated, a process that is completed within 14 days of the initial influenza challenge (33, 46). There are alternatives. Several distinct cell death pathways have been found to operate in the resolution phase of antigen-specific CD8+ T cell responses (47), although, perhaps because the damaged cells are rapidly phagocytosed and destroyed, it is difficult to find apoptotic lymphocytes in lymphocyte populations recovered directly from the in vivo situation (48). The proapoptotic bcl2 family member bim (49, 50) seems to be particularly important in this regard. However, although mechanisms involving TNF-mediated elimination may not be important in the maturation of influenza-specific CD8+ T cell memory, the situation could be quite different for pathogens (such as lymphocytic choriomeningitis virus) that replicate extensively in lymphoid tissue (51).
Acknowledgments
We thank Dr. Katherine Kedzierska for critical review of the manuscript, Dr. W. Xie and Dr. J. Lin for the DbNP and DbPA tetramers, Dr. Richard Cross and Jennifer Hoffrage for help with cell sorting, Elvia Olivas and Twala Hogg for excellent technical assistance, and Vicki Henderson and Phyllis Halliday for help with preparation of the manuscript. These experiments were supported by U.S. Public Health Service Grants AI29579 and CA21765, the American Lebanese Syrian Associated Charities, and Australian National Health and Medical Research Council Burnet Fellowship (to P.C.D.).
Abbreviations: TNF, tumor necrosis factor; TNFR, TNF receptor; CTL, cytotoxic T lymphocyte; TCR, T cell receptor; PI, propidium iodide; BAL, bronchoalveolar lavage; PE, phycoerythrin.
References
- 1.Allan, W., Tabi, Z., Cleary, A. & Doherty, P. C. (1990) J. Immunol. 144, 3980-3986. [PubMed] [Google Scholar]
- 2.Doherty, P. C. & Christensen, J. P. (2000) Annu. Rev. Immunol. 18, 561-592. [DOI] [PubMed] [Google Scholar]
- 3.Walker, J. A., Molloy, S. S., Thomas, G., Sakaguchi, T., Yoshida, T., Chambers, T. M. & Kawaoka, Y. (1994) J. Virol. 68, 1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn, K. J., Belz, G. T., Altman, J. D., Ahmed, R., Woodland, D. L. & Doherty, P. C. (1998) Immunity 8, 683-691. [DOI] [PubMed] [Google Scholar]
- 5.Flynn, K. J., Riberdy, J. M., Christensen, J. P., Altman, J. D. & Doherty, P. C. (1999) Proc. Natl. Acad. Sci. USA 96, 8597-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall, D. R., Turner, S. J., Belz, G. T., Wingo, S., Andreansky, S., Sangster, M. Y., Riberdy, J. M., Liu, T., Tan, M. & Doherty, P. C. (2001) Proc. Natl. Acad. Sci. USA 98, 6313-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelso, A., Groves, P., Ramm, L. & Doyle, A. G. (1999) Int. Immunol. 11, 617-621. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, B. J., Costelloe, E. O., Fitzpatrick, D. R., Haanen, J. B., Schumacher, T. N., Brown, L. E. & Kelso, A. (2003) Proc. Natl. Acad. Sci. USA 100, 2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topham, D. J., Tripp, R. A. & Doherty, P. C. (1997) J. Immunol. 159, 5197-5200. [PubMed] [Google Scholar]
- 10.Webby, R. J., Andreansky, S., Stambas, J., Rehg, J. E., Webster, R. G., Doherty, P. C. & Turner, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 7235-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belz, G. T., Xie, W., Altman, J. D. & Doherty, P. C. (2000) J. Virol. 74, 3486-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belz, G. T., Xie, W. & Doherty, P. C. (2001) J. Immunol. 166, 4627-4633. [DOI] [PubMed] [Google Scholar]
- 13.Slifka, M. K. & Whitton, J. L. (2001) Nat. Immunol. 2, 711-717. [DOI] [PubMed] [Google Scholar]
- 14.Sprent, J. & Surh, C. D. (2002) Annu. Rev. Immunol. 20, 551-579. [DOI] [PubMed] [Google Scholar]
- 15.Sarin, A., Conan-Cibotti, M. & Henkart, P. A. (1995) J. Immunol. 155, 3716-3718. [PubMed] [Google Scholar]
- 16.Zheng, L., Fisher, G., Miller, R. E., Peschon, J., Lynch, D. H. & Lenardo, M. J. (1995) Nature 377, 348-351. [DOI] [PubMed] [Google Scholar]
- 17.Pimentel-Muinos, F. X. & Seed, B. (1999) Immunity 11, 783-793. [DOI] [PubMed] [Google Scholar]
- 18.Lenardo, M. J. (1991) Nature 353, 858-861. [DOI] [PubMed] [Google Scholar]
- 19.Refaeli, Y., Van Parijs, L., Alexander, S. I. & Abbas, A. K. (2002) J. Exp. Med. 196, 999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander-Miller, M. A., Leggatt, G. R., Sarin, A. & Berzofsky, J. A. (1996) J. Exp. Med. 184, 485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derby, M. A., Snyder, J. T., Tse, R., Alexander-Miller, M. A. & Berzofsky, J. A. (2001) Eur. J. Immunol. 31, 2951-2959. [DOI] [PubMed] [Google Scholar]
- 22.Tanchot, C., Fernandes, H. V. & Rocha, B. (2000) Philos. Trans. R. Soc. London B Biol. Sci. 355, 323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilbourne, E. D. (1969) Bull. W. H. O. 41, 643-645. [PMC free article] [PubMed] [Google Scholar]
- 24.Hou, S., Hyland, L., Ryan, K. W., Portner, A. & Doherty, P. C. (1994) Nature 369, 652-654. [DOI] [PubMed] [Google Scholar]
- 25.Townsend, A. R., Rothbard, J., Gotch, F. M., Bahadur, G., Wraith, D. & McMichael, A. J. (1986) Cell 44, 959-968. [DOI] [PubMed] [Google Scholar]
- 26.Kalergis, A. M., Boucheron, N., Doucey, M. A., Palmieri, E., Goyarts, E. C., Vegh, Z., Luescher, I. F. & Nathenson, S. G. (2001) Nat. Immunol. 2, 229-234. [DOI] [PubMed] [Google Scholar]
- 27.Whelan, J. A., Dunbar, P. R., Price, D. A., Purbhoo, M. A., Lechner, F., Ogg, G. S., Griffiths, G., Phillips, R. E., Cerundolo, V. & Sewell, A. K. (1999) J. Immunol. 163, 4342-4348. [PubMed] [Google Scholar]
- 28.Cawthon, A. G. & Alexander-Miller, M. A. (2002) J. Immunol. 169, 3492-3498. [DOI] [PubMed] [Google Scholar]
- 29.Fahmy, T. M., Bieler, J. G., Edidin, M. & Schneck, J. P. (2001) Immunity 14, 135-143. [PubMed] [Google Scholar]
- 30.Alexander-Miller, M. A., Derby, M. A., Sarin, A., Henkart, P. A. & Berzofsky, J. A. (1998) J. Exp. Med. 188, 1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deckhut, A. M., Allan, W., McMickle, A., Eichelberger, M., Blackman, M. A., Doherty, P. C. & Woodland, D. L. (1993) J. Immunol. 151, 2658-2666. [PubMed] [Google Scholar]
- 32.Turner, S. J., Diaz, G., Cross, R. & Doherty, P. C. (2003) Immunity 18, 549-559. [DOI] [PubMed] [Google Scholar]
- 33.Crowe, S. R., Turner, S. J., Miller, S. C., Roberts, A. D., Rappolo, R. A., Doherty, P. C., Ely, K. H. & Woodland, D. L. (2003) J. Exp. Med. 198, 399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt, P. G., Stumbles, P. A. & McWilliam, A. S. (1999) J. Leukocyte Biol. 66, 272-275. [DOI] [PubMed] [Google Scholar]
- 35.Stumbles, P. A., Strickland, D. H., Pimm, C. L., Proksch, S. F., Marsh, A. M., McWilliam, A. S., Bosco, A., Tobagus, I., Thomas, J. A., Napoli, S., et al. (2001) J. Immunol. 167, 228-234. [DOI] [PubMed] [Google Scholar]
- 36.Guidotti, L. G., Borrow, P., Brown, A., McClary, H., Koch, R. & Chisari, F. V. (1999) J. Exp. Med. 189, 1555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eichelberger, M., Allan, W., Zijlstra, M., Jaenisch, R. & Doherty, P. C. (1991) J. Exp. Med. 174, 875-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topham, D. J. & Doherty, P. C. (1998) J. Virol. 72, 882-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerhard, W. (2001) Curr. Top. Microbiol. Immunol. 260, 171-190. [DOI] [PubMed] [Google Scholar]
- 40.Ware, C. F., Crowe, P. D., Vanarsdale, T. L., Andrews, J. L., Grayson, M. H., Jerzy, R., Smith, C. A. & Goodwin, R. G. (1991) J. Immunol. 147, 4229-4238. [PubMed] [Google Scholar]
- 41.van Stipdonk, M. J., Lemmens, E. E. & Schoenberger, S. P. (2001) Nat. Immunol. 2, 423-429. [DOI] [PubMed] [Google Scholar]
- 42.Kaech, S. M. & Ahmed, R. (2001) Nat. Immunol. 2, 415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamb, R. A. & Krug, R. M. (1996) in Fields Virology, eds. Fields, B. N., Howley, P. M. & Knipe, D. M. (Lippincott, Philadelphia), Vol. 1, pp. 1353-1395. [Google Scholar]
- 44.Sant, A. & Yewdell, J. (2003) Curr. Opin. Immunol. 15, 66-68. [DOI] [PubMed] [Google Scholar]
- 45.Yewdell, J. W. & Bennink, J. R. (1999) Annu. Rev. Immunol. 17, 51-88. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton-Easton, A. & Eichelberger, M. (1995) J. Virol. 69, 6359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Kappler, J. & Marrack, P. (2002) Curr. Opin. Immunol. 14, 354-359. [DOI] [PubMed] [Google Scholar]
- 48.Belz, G. T., Altman, J. D. & Doherty, P. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13812-13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Bouillet, P., Strasser, A., Kappler, J. & Marrack, P. (2002) Immunity 16, 759-767. [DOI] [PubMed] [Google Scholar]
- 50.Pellegrini, M., Belz, G., Bouillet, P. & Strasser, A. (2003) Proc. Natl. Acad. Sci. USA 100, 14175-14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Boer, R. J., Oprea, M., Antia, R., Murali-Krishna, K., Ahmed, R. & Perelson, A. S. (2001) J. Virol. 75, 10663-10669. [DOI] [PMC free article] [PubMed] [Google Scholar]