Abstract
Single nucleotide polymorphisms (SNPs) in genes that influence development of the male reproductive tract have been associated with male genitourinary abnormalities. However, no studies have tested the relationship between SNPs and intermediate phenotypes such as anogenital distance (AGD), anoscrotal distance (ASD) and penile width (PW). We tested whether 24 common SNPs in eight genes that influence male genital development were associated with intermediate phenotypes in 106 healthy male infants from the Study for Future Families. We used DNA from buccal smears and linear regression models to assess the relationship between anogenital measurements and SNP genotypes with adjustment for covariates. We found that the rs2077647 G allele, located in the coding region of estrogen receptor alpha (ESR1), was associated with a shorter AGD (P=0.02; −7.3 mm, 95% confidence interval (CI): −11.6 to −3.1), and the rs10475 T allele, located in the 3′ untranslated region of activating transcription factor 3 (ATF3), was associated with a shorter ASD (−4.3 mm, 95% CI: −7.2 to −1.4), although this result was not significant (P=0.07) after controlling for multiple comparisons. We observed no association between PW and the SNPs tested. Minor alleles for two SNPs in genes that regulate estrogen signaling during male genital development were associated with AGD and ASD, although the significance of the association was marginal. Our findings suggest that AGD and ASD are influenced by heritable factors in genes known to be associated with frank male genital abnormalities such as hypospadias and cryptorchidism.
Keywords: gene, hypospadias, male, phenotype, polymorphism, reproductive
Introduction
Hypospadias and cryptorchidism are common birth defects affecting approximately 0.5%–4.5% and 2%–8% of infants, respectively, in industrialized countries.1, 2, 3, 4 These conditions are known risk factors for the development of testicular cancer and sperm abnormalities in adult males, suggesting that early programming of genital development can affect male childhood and adult reproductive function.5 Numerous epidemiological and genetic studies of familial and sporadic hypospadias, cryptorchidism and testicular cancer suggest a multifactorial etiology, reflecting the contribution and interaction of both genetic and environmental factors in the development of these conditions.6
Normal male reproductive system development depends on appropriate genetic programming of androgen signaling and functioning during fetal life.7 The androgen receptor (AR) and estrogen receptor genes (alpha (ESR1) and beta (ESR2)) affect development of all male genitourinary structures including the anus, penis, testes and urinary system.8, 9 Insulin-like factor (INSL3) and its receptor LGR8 trigger testicular descent into the scrotum in the third trimester of pregnancy.2, 10 Several other genes have recently been found to be both differentially expressed in penile tissue of boys with hypospadias and to be estrogen responsive: including activating transcription factor 3 (ATF3), connective tissue growth factor (CTGF) and cysteine-rich angiogenic inducer (CYR61).11, 12
Variation in repeat sequence number and single nucleotide polymorphisms in genes controlling development of the male reproductive pathway has been associated with male genital abnormalities. For example, polymorphisms in the ESR1 and ESR2 genes have been associated with both hypospadias and cryptorchidism,9, 13 suggesting that genetic variation can affect normal male genital development. Similarly, polymorphisms in INSL3 and LGR8 have been associated with cryptorchidism.14, 15 In addition, the number of CAG repeats in exon 1 of the AR gene may influence AR transcription with longer CAG repeat length resulting in reduced AR function.16
Whether common variation in genes involved in male reproductive development is associated with intermediate phenotypes such as anogenital distance (AGD), anoscrotal distance (ASD) and penile width (PW) has not been studied. We define intermediate phenotypes to be in the male reproductive developmental pathway and potentially influenced by the same genetic and environmental factors known to affect male genital development. AGD, ASD and PW are markers used in animal studies as reproductive toxicity endpoints reflecting in utero androgen exposure. In animal models, shortened AGD, ASD and smaller PW are associated with decreased androgen exposure and with male reproductive tract birth defects including hypospadias and cryptorchidism.17, 18, 19 In addition, diminished AGD, ASD and PW lengths have been observed in human infants and in the offspring of rats prenatally exposed to some phthalate chemicals, man-made compounds with anti-androgenic properties.20, 21 Similar to animal models, reductions in AGD, ASD and/or PW lengths may be associated with similar birth defects in human males.22 We tested whether polymorphisms in genes known to be involved in the regulation of male genital development are associated with AGD, ASD and PW in male infants and the extent to which such associations observed were modified by prenatal phthalate exposure.
Materials and methods
Participants
Pregnant women were originally recruited in the Study for Future Families I (SFFI), a multicenter pregnancy cohort study from September 1999 to August 2005. Infants born in SFF I were recruited in a follow-up study, Study For Future Families II (SFFII) which examined prenatal environmental exposures and health outcomes. Methods are described in detail elsewhere.23, 24 Eligibility included: pregnancy that was not medically assisted; woman and her partner was >18 years of age; either partner read and/or spoke Spanish or English; and the father was available to participate.
Buccal smears from infants were collected in 2007–2008 by inserting the swab into the baby's mouth along the cheek and rubbing vigorously along the cheek and under the tongue to collect cells. Study coordinators rolled the applicator on a collection card for several seconds until saturated with saliva. Cards were allowed to dry overnight at room temperature and then transferred to a standard refrigerator for storage.
Women in SFFI were asked to provide a urine sample at their recruitment visit (average 28 weeks pregnancy). Maternal urine samples were assayed for phthalate metabolites by the Division of Laboratory Sciences, National Center for Environmental Health, Centers for Disease Control and Prevention. The analytical method involved the enzymatic deconjugation of the phthalate metabolites from their glucuronidated form, followed by concentration of the analytes of interest by solid-phase extraction, separation with high-performance liquid chromatography, and detection by isotope-dilution tandem mass spectrometry. Limits of detection are in the low nanogram per milliliter range. Isotopically labeled internal standards and conjugated internal standards were used to increase precision and accuracy of the measurements. Quality control and reagent blank samples were analyzed along with unknown samples to monitor assay performance.25, 26, 27
There were 454 mothers participating in SFFII who gave birth to 223 male infants. A physical exam including measurement of AGD, ASD and PW was conducted on each infant at an average of 12.6 months of age. AGD was measured from the center of the anus to anterior base of the penis. ASD was measured from the center of the anus to posterior portion of rugated scrotum. PW was measured transversely at the head of the phallus. All measurements were made with precision calipers. Anthropometric measurements (weight, length, head circumference) were also obtained. Examiners underwent a standardized training, and inter-rater variability coefficients for all genital measurements were above 80%. All infants were healthy without any genital birth defects and had testes descended into the scrotum. Within the group of mothers with male infants, seven mothers did not consent to buccal smears. Of the remaining 216 of male infants, 106 had full data on maternal prenatal phthalate concentration, full physical exam data with AGD, ASD and PW measurements, and buccal smears that produced usable DNA. We conducted genetic and statistical analyses on all 106 male infants.
Selection of genes and single nucleotide polymorphisms (SNPs)
We chose candidate genes for which either expression or nucleotide polymorphisms had previously been associated with hypospadias and/or cryptorchidism: AR, ESR1, ESR2, INSL3, LGR8, ATF3, CYR61 and CGTF. For each gene, we identified SNPs for genotyping based on either a prior published association with hypospadias or cryptorchidism or a minor allele frequency ≥10% and tagging other SNPs (i.e., tagSNPs) with r2≥0.80 in individuals of European descent from HapMap data. When multiple tagSNPs were available, we preferentially chose SNPs that were more likely to be functional (e.g., demonstrated to have functional effects, location in a protein-coding exon, 3′ untranslated region, 5′ promoter region, exon/intron boundaries) and SNPs that could be assayed on our genotyping platform.
Genotyping methods
Genomic DNA from study subjects was isolated from FTA cards using the QIAamp DNA Investigator Kit (Qiagen, Valencia, CA, USA), followed by genomic amplification using the Qiagen REPLI-g Mini kit (Qiagen) according to the established manufacturer's protocols.
Genotyping was performed by KBiosciences (Hertfordshire, UK) using a fluorescence-based high-throughput allele-specific PCR technique (http://www.kbioscience.co.uk/KASP_manual.pdf). Genotype calling was performed manually by using the KBiosciences SNPviewer software by KBiosciences.
CAG repeat length was assessed at the Functional Genomics Laboratory of the Center for Ecogenetics and Environmental Health at the University of Washington. The number of CAG repeats in the variable tandem number CAG repeat polymorphism in exon 1 of the AR gene was determined by estimating the size of a specific PCR product containing this tri-nucleotide microsatellite. Briefly, fluorescent labeled sense primer FAM-5′-TCCAGAATCTGTTCCAGAGCGTGC-3′ and anti-sense primer 5′-CTCTACGATGGGCTTGGGGAGAAC-3′ were used for PCR with thermocycler conditions of 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 61 °C for 30 s and 72 °C for 30 s. The length of the resultant fluorescent PCR amplicon was determined by using a 377 ABI DNA Sequencer (Applied Biosystems, Foster City, CA, USA) containing fluorescent in-lane size markers according to manufacturer's protocol. The fragment length of the PCR product was called by using the Genotyper software 2.5 (Applied Biosystems). The number of CAG repeats corresponding to the PCR fragment lengths were calculated using several control samples that were previously verified using four-color dye terminator-based DNA sequencing.
Statistical analysis
Multivariable linear regression was used to examine the association between AGD, ASD or PW (in mm) and genotype. A dominant genetic model was used where subjects who are homozygous or heterozygous for the minor allele are combined for analysis. Infant age, weight and height were assessed at the time of the genital measurements. These measures were associated with infant AGD, ASD and PW, but were highly collinear with one another and thus could not be included simultaneously in the model. Therefore, infant weight percentile (which is a variable that is not collinear with other anthropometric factors) and infant age were used as covariates in analysis; this analytical approach was used in previous analyses of AGD from the SFFII study.21 Study center is a known confounder from previous AGD analyses within the SFF study (likely due to AGD measurement differences between centers),24 and its inclusion in the regression models led to appreciable changes in point estimates for the genital measures in our analysis. The final linear regression model included weight percentile, age and study center as covariates. In addition to estimating the main effects of the genotypes, an exploratory multivariable linear regression analysis was conducted to assess potential gene–environment interaction between prenatal phthalate exposure, genetic variants and AGD, ASD and PW by also including terms representing phthalate concentration and the product of phthalate concentration and minor allele carriership. Log transformation of phthalate concentrations was performed to normalize the distribution. Concentrations below the limit of detection were assigned the specific metabolite limit of detection value divided by the square root of two for statistical analysis, as has been performed previously.28 We examined phthalate metabolites that have individually been associated with AGD 21 and used the sum of log transformed di-ethyl hexyl phthalate (DEHP) phthalate metabolites adjusted for molecular weight, mono(2-ethylhexyl) phthalate (MEHP), mono-(2-ethyl-5-hydroxyhexyl) phthalate (MEHHP) and mono-(2-ethyl-5-oxohexyl) phthalate (MEOHP) to reflect overall DEHP exposure. We examined phthalate interactions for only those SNPs for which there was a statistically significant main effect with either AGD, ASD or PW. In order to account for multiple comparisons, we applied resampling procedures based on non-parametric bootstrap to obtain adjusted P values that control the family error rate at 0.05.29
We examined the CAG repeat length within the AR gene in relation to AGD, ASD and PW using multivariate linear regression models adjusting for infant weight and center. The CAG repeat was examined as a continuous variable and also categorized (<22, 22–25, >25) using cutpoints previously reported in the literature.16
Results
The study population (n=106) included male infants from predominantly Minnesota and Missouri who were 2–34 months of age. Infant weight (5–18.5 kg) and length (57–97.3 cm) varied considerably because of the large infant age range. The majority of mothers were European American and between 25 and 34 years old (Table 1).
Table 1. Demographic characteristics for 106 male infants, Study for Future Families II, 1999–2005.
| n (%) | |
|---|---|
| Center | |
| California | 19 (17.9%) |
| Minnesota | 49 (46.2%) |
| Missouri | 38 (35.8%) |
| Maternal race | |
| Caucasion | 87 (82.1%) |
| Non-White | 18 (17.0%) |
| Unknown | 1 (0.9%) |
| Maternal age category (year) | |
| 18–25 | 21 (19.8%) |
| 25–34 | 61 (57.5%) |
| 35–43.5 | 23 (21.7%) |
| Unknown | 1 (0.9%) |
| Infant age (month) | |
| 2–10 | 22 (20.8%) |
| 10–20 | 44 (41.5%) |
| 20–34 | 40 (37.7%) |
| Infant weight (kg) | |
| 5–8 | 19 (17.9%) |
| 8–12 | 51 (48.1%) |
| 12–18.5 | 36 (34.0%) |
| Infant length (cm) | |
| 57–75 | 26 (24.5%) |
| 75–85 | 37 (34.9%) |
| 85–97.3 | 38 (35.8%) |
| Unknown | 5 (4.7%) |
We submitted a total of 53 SNPs to KBiosciences for genotyping. For approximately 15 SNPs (one in INSL3, two in RXFP2, five in ATF3, three in AR, two in ESR1, one in ESR2 and one in CTGF), no results could be obtained either because an assay could not be designed (and no SNP in tight linkage disequilibrium was available for replacement) or because a designed assay performed poorly (most likely due to poor DNA quality). Given the small sample size, we further excluded results from 14 SNPs (four in INSL3, two in RXFP2, one in ATF3, two in AR, one in ESR1, one in ESR2 and three in CTGF) with a minor allele frequency of less than 10% within our study population. These 14 SNPs included three rare SNPs previously reported only in cases of cryptorchidism.2 Therefore, a total of 24 SNPs were available for testing (Table 2).
Table 2. Single nucleotide polymorphisms analyzed in 106 male infants from the Study for Future Families II, 1999–2005.
| Gene | Minor allele | SNP rs number* | Minor allele frequency (%) | Context |
|---|---|---|---|---|
| Relaxin/insulin like family receptor (RXFP2) | A | rs7325513 | 49 | Coding-synonymous |
| G | rs17076657 | 13 | Coding-nonsynonymous | |
| A | rs7331833 | 25 | ||
| Activating transcription factor (ATF3) | T | rs3125289 | 47 | Intron |
| T | rs1126526 | 18 | mrRNA-untranslated | |
| T | rs10475 | 26 | mrRNA-untranslated | |
| Androgen receptor (AR) | T | rs1204038 | 17 | Intron |
| T | rs1204039 | 17 | Intron | |
| C | rs2767564 | 16 | Intron | |
| G | rs4827546 | 17 | Intron | |
| C | rs4827547 | 15 | Intron | |
| C | rs5918758 | 14 | Intron | |
| G | rs5919395 | 14 | Intron | |
| C | rs5919402 | 19 | Intron | |
| G | rs7061037 | 15 | Intron | |
| Estrogen receptor 1 (ESR1) | G | rs2077647 | 49 | Coding-synonymous |
| A | rs1062577 | 10 | mRNA-untranslated | |
| C | rs3798577 | 33 | mRNA-untranslated | |
| Estrogen receptor 2 (ESR2) | G | rs1255998 | 15 | mRNA-untranslated |
| G | rs928554 | 43 | mRNA-untranslated | |
| C | rs1256120 | 16 | mRNA-untranslated | |
| Cysteine-rich angiogenic inducer (CYR61) | A | rs2297141 | 43 | Intron |
| C | rs9658584 | 21 | Intron | |
| Connective tissue growth factor (CTGF) | T | rs9399005 | 27 | Intron |
Abbreviation: SNP, single nucleotide polymorphism.
*dbSNP reference SNP.
The G allele of rs2077647 (located in a coding region of ESR1) was associated with a shorter AGD (−7.3 mm, 95% CI: −11.6 to −3.1) (Figure 1). In addition, the T allele of rs10475 (located in the 3′ untranslated region of ATF3) was associated with a shorter ASD (−4.3 mm, 95% CI: −7.2 to −1.4) (Figure 2). We observed no significant association between PW and any SNP tested (Figure 3). After resampling to control for multiple comparisons, we found that the adjusted P value for the association between the minor allele of rs2077647 and AGD was 0.02 and the adjusted P value for the association between the minor allele of rs10475 and ASD was 0.07.
Figure 1.
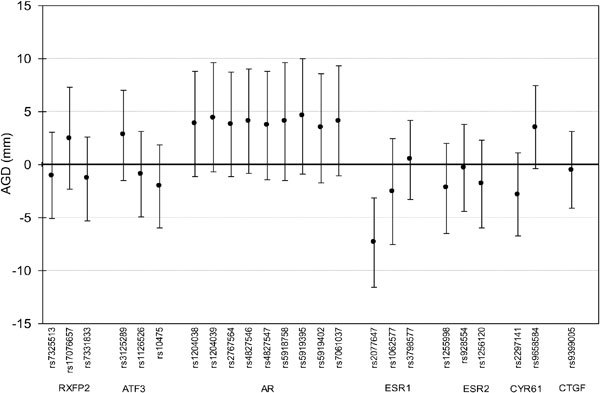
Difference in anogenital distance (AGD) (mm), 95% confidence interval, associated with minor allele carriership (adjusted for study center, age and infant weight percentile), Study for Future Families II, 1999–2005.
Figure 2.
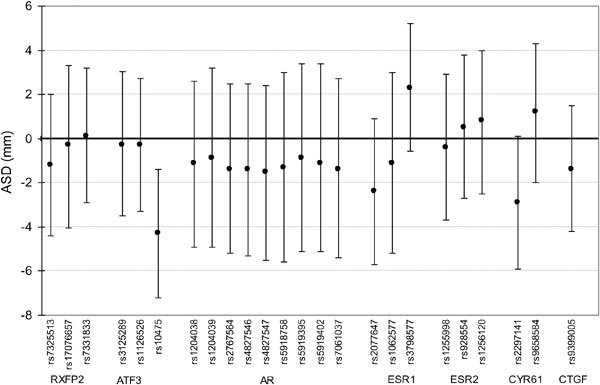
Difference in anoscrotal distance (ASD) (mm), 95% confidence interval, associated with minor allele carriership (adjusted for study center, age and infant weight percentile), Study for Future Families II, 1999–2005.
Figure 3.
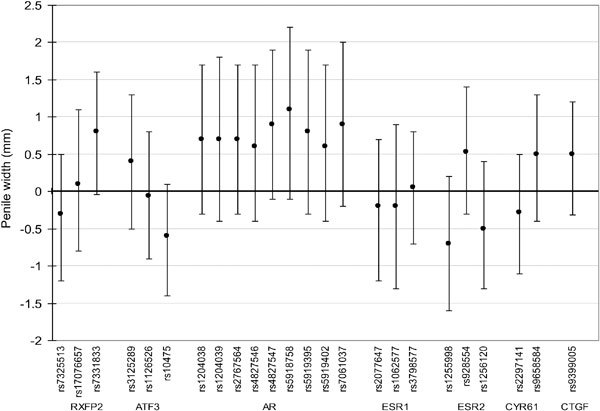
Difference in penile width (PW) (mm), 95% confidence interval, associated with minor allele carriership (adjusted for study center, age and infant weight percentile), Study for Future Families II, 1999–2005.
We examined whether maternal phthalate exposure modified the relationship between the SNPs rs2077647 and rs10475 and AGD and ASD, respectively. The coefficients for the interactions terms were small in absolute value and not statistically significant in multivariate linear regression analyses (Table 3).
Table 3. Linear regression analysis of joint relationship of genotypes and maternal urinary phthalate with genital distance measures (adjusted for center, age and infant weight percentile), Study for Future Families II, 1999–2005.
| Genotype | Measure | Phthalate | Phthalate* genotype interaction term coefficient (95% CI) | P value | |
|---|---|---|---|---|---|
| rs2077647 | AA or AG | AGD | Sum DEHP Metabolites (MEHP/MEOHP/MEHHP) | −1.3 (−6.0, 3.4) | 0.58 |
| MEP | −2.0 (−4.8, 0.8) | 0.16 | |||
| MBP | 0.1 (−4.4, 4.5) | 0.97 | |||
| rs10475 | TT or TC | ASD | Sum DEHP metabolites (MEHP/MEOHP/MEHHP) | −0.8 (−4.2, 2.7) | 0.67 |
| MEP | 0.01 (−2.1, 2.1) | 0.99 | |||
| MBP | −0.7 (−4.2, 2.9) | 0.71 |
Abbreviations: AGD, anogenital distance; ASD, anoscrotal distance; MBP, mono-butyl phthalate; MEHHP, mono-(2-ethyl-5-hydroxyhexyl) phthalate; MEHP, mono(2-ethylhexyl) phthalate; MEOHP, mono-(2-ethyl-5-oxohexyl) phthalate; MEP, mono-ethyl phthalate.
*Refers to a statistical interaction term to examine how phthalate modify the relationship between genotype and outcome.
The mean CAG repeat length was 23 with a range of 10–29. We observed a 2.95-mm increase in ASD in infants with >25 CAG repeats as compared to infants with <22 CAG repeats (P=0.80). We did not observe an association between CAG repeat length and AGD or PW (Table 4).
Table 4. Associations (regression coefficients, 95% confidence intervals (CI)) between AR CAG repeat length and anogenital measures, Study for Future Families II, 1999–2005*.
| AGD (mm) | ASD (mm) | PW (mm) | |||||
|---|---|---|---|---|---|---|---|
| n | β | 95% CI | β | 95% CI | β | 95% CI | |
| AR CAG repeat length | |||||||
| <22 | 53 | Referent | Referent | Referent | |||
| 22–25 | 33 | −2.39 | −6.56, 1.78 | −0.34 | −3.57, 2.88 | −0.36 | −1.23, 0.50 |
| >25 | 16 | 0.48 | −4.83, 5.80 | 2.95 | −1.16, 7.06 | 0.21 | −0.89, 1.32 |
| Per one repeat increase | −0.14 | −0.80, 0.52 | 0.06 | −0.45, 0.57 | 0.003 | −0.14, 0.14 | |
Abbreviations: AGD, anogenital distance; ASD, anoscrotal distance; PW, penile width.
*All analyses adjusted for center, age and weight percentile.
Discussion
We conducted a pilot study to test the hypothesis that common variation in genes previously reported to influence hormonally-mediated male genital development is associated with the intermediate phenotypes AGD, ASD and PW. We found two SNPs that were independently associated with AGD and ASD. ESR1 is well known to play an important role in early genital development, and recent studies suggest that ATF3 may affect differentiation of penile tissues but the extent to which ATF is involved in male reproductive development remains to be determined.
Appropriate concentration and timing of hormonal signaling during fetal life are critical for normal male reproductive development.10, 30 Specifically, increased estrogen exposure during fetal life can lead to an increased incidence of hypospadias and cryptorchidism in mice.31 Estrogen signaling is predominantly mediated by ESR1 and ESR2, both of which are expressed in fetal male genital tissues. Polymorphisms within ESR1 have been associated with hypospadias, male infertility and sperm parameters including decreased sperm density and abnormal sperm motility and morphology.13, 32 We found that the rs2077647 G allele was associated with a shorter AGD. Several previous studies have found this allele to be associated with a variety of disease phenotypes including: increased risk of Alzheimer's disease,33 increased risk of colon, bladder and prostate cancers,34, 35 and increase in child onset mood disorders.36 All of these disorders are thought to be in part hormonally mediated, potentially through the actions of ESR1.
ATF3 is an estrogen responsive gene that is expressed within the genital tubercle during mouse sexual differentiation and has been found to be upregulated in foreskin tissue in human patients with hypospadias.37, 38 ATF3 is involved in suppression of cell cycling, and hypothesized to inhibit cell growth in urethral formation.11 In a cohort of 41 boys with hypospadias, 10% carried rare ATF3 alleles as compared to none of thirty controls.39 Beleza-Meireles et al.40 found that the odds of having the individual minor alleles of three common, unlinked ATF SNPs (rs3125289, rs1877474, rs11119982) spanning a 16-kb region in intron 1 were increased in 330 cases of hypospadias boys as compared to 380 healthy controls in Sweden. No published studies have found an association between the rs10475 polymorphism and male reproductive outcomes. We found that ASD was decreased in infants with the rs10475 T allele, suggesting that ATF3 may be involved in the formation of male reproductive tissues more generally and not limited to the urethral plate. This SNP is located in the 3′ untranslated region of ATF3, and could be functionally important if it alters the binding of regulatory miRNAs. We used the TargetScan database (http://www.targetscan.org) to assess whether this SNP, or a strongly correlated SNP, is located within a predicted miRNA binding site, but found no such evidence.
While our study supports a relationship between common genetic variants and male reproductive phenotypes, it has several limitations, the most notable of which is a relatively small sample size given the effect size for which we were looking. Additionally, a substantial fraction of genotypes failed to be called suggesting that the original samples, and thus, the DNA quality, had degraded. We conducted 27 independent tests for each of the intermediate phenotypes. We used a resampling procedure based on bootstrap to account for multiple comparisons and found that adjusted P values were very similar to those within the original analysis.29
We did not observe a statistically significant interaction between prenatal phthalate exposure, genetic variants and the three measures we examined. This result was to be expected given the small sample size. Nevertheless, this is an important environmental exposure to consider in future studies. Phthalates are synthetic chemicals produced in large volumes and used in a variety of industrial and common products.41 It is well demonstrated that prenatal phthalate exposure decreases early life testosterone concentrations leading to a variety of adverse reproductive defects in animal offspring,18 but the mechanisms by which this occurs are still unclear. DEHP and dibutyl phthalate have been found to decrease INSL3 mRNA expression and decrease testosterone concentrations in pregnant dams,42 but it is likely that other mechanisms are at play. These may involve a gene–environment interaction in which risk alleles increase an individual's susceptibility to phthalate exposure and subsequent health impacts. Future studies with larger sample sizes will be needed to adequately test if and how phthalates modify male reproductive development.
Our study is novel in that it is the first study to use AGD and ASD, intermediate phenotypes for male reproductive abnormalities, in an attempt to identify genetic risk variants that might also be important in birth defects such as hypospadias and cryptorchidism. Our results suggest that variation in ESR1 and ATF3 are associated with the hormone-sensitive male phenotypes, AGD and ASD. These results are consistent with the roles of ESR1 and ATF3 in estrogen mediated signaling of genital development, and suggest that development of estrogen responsive male reproductive tissues may be influenced by heritable factors. Using novel intermediate phenotypes to understand the genetic contribution to hormone male reproductive development will help elucidate the causes of abnormalities in male reproductive programming.
Author contributions
SS (first author) wrote the grant for the initial study, conducted the study with team members, conducted analysis and wrote the manuscript. SMS (last author) mentored the entire project and was involved in conception, study design, analysis and write-up and review. SHS was the PI on the parent grant that collected all of the specimens used for the study. She reviewed and edited the final manuscript. FMF and HWW were responsible for DNA extraction and genetic analyses. MB contributed to final review and edits of the manuscript. CZ contributed to statistical analysis of the data. RG contributed to project conception and reviewed/edited the final manuscript.
Acknowledgments
This work was supported by the UW NIEHS sponsored Center for Ecogenetics and Environmental Health (Grant No.: NIEHS P30ES07033) and a NIH NICHD K-12 Award HD053984-02. The Study for Future Families was supported by NIH grants R01-ES09916 to the University of Missouri, MO1-RR00400 to the University of Minnesota, MO1-RR0425 to Harbor-UCLA Medical Center and Grant 18018278 from the State of Iowa to the University of Iowa and USEPA R-82943601.
The authors have no competing financial interests to declare.
References
- Main KM, Skakkebaek NE, Virtanen HE, Toppari J. Genital anomalies in boys and the environment. Best Pract Res Clin Endocrinol Metab. 2010;24:279–89. doi: 10.1016/j.beem.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Virtanen HE, Toppari J. Epidemiology and pathogenesis of cryptorchidism. Hum Reprod Update. 2008;14:49–58. doi: 10.1093/humupd/dmm027. [DOI] [PubMed] [Google Scholar]
- Paulozzi LJ. International trends in rates of hypospadias and cryptorchidism. Environ Health Perspect. 1999;107:297–302. doi: 10.1289/ehp.99107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulozzi LJ, Erickson JD, Jackson RJ. Hypospadias trends in two US surveillance systems. Pediatrics. 1997;100:831–4. doi: 10.1542/peds.100.5.831. [DOI] [PubMed] [Google Scholar]
- Skakkebaek NE, Rajpert-De Meyts E, Main KM. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum Reprod. 2001;16:972–8. doi: 10.1093/humrep/16.5.972. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E. Developmental model for the pathogenesis of testicular carcinoma in situ: genetic and environmental aspects. Hum Reprod Update. 2006;12:303–23. doi: 10.1093/humupd/dmk006. [DOI] [PubMed] [Google Scholar]
- Moore KL. The Developing Human: Clinical Oriented Embryology. 4th ed. Philadelphia, PA; WB Saunders; 1988. pp. 246–85. [Google Scholar]
- Watanabe M, Yoshida R, Ueoka K, Aoki K, Sasagawa I, et al. Haplotype analysis of the estrogen receptor 1 gene in male genital and reproductive abnormalities. Hum Reprod. 2007;22:1279–84. doi: 10.1093/humrep/del513. [DOI] [PubMed] [Google Scholar]
- Beleza-Meireles A, Kockum I, Lundberg F, Söderhäll C, Nordenskjöld A. Risk factors for hypospadias in the estrogen receptor 2 gene. J Clin Endocrinol Metab. 2007;92:3712–8. doi: 10.1210/jc.2007-0543. [DOI] [PubMed] [Google Scholar]
- Nation TR, Balic A, Southwell BR, Newgreen DF, Hutson JM. The hormonal control of testicular descent. Pediatr Endocrinol Rev. 2009;7:22–31. [PubMed] [Google Scholar]
- Willingham E, Baskin LS. Candidate genes and their response to environmental agents in the etiology of hypospadias. Nat Clin Pract Urol. 2007;4:270–9. doi: 10.1038/ncpuro0783. [DOI] [PubMed] [Google Scholar]
- Wang Z, Liu BC, Lin GT, Lin CS, Lue TF, et al. Up-regulation of estrogen responsive genes in hypospadias: microarray analysis. J Urol. 2007;177:1939–46. doi: 10.1016/j.juro.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Ban S, Sata F, Kurahashi N, Kasai S, Moriya K, et al. Genetic polymorphisms of ESR1 and ESR2 that may influence estrogen activity and the risk of hypospadias. Hum Reprod. 2008;23:1466–71. doi: 10.1093/humrep/den098. [DOI] [PubMed] [Google Scholar]
- El Houate B, Rouba H, Sibai H, Barakat A, Chafik A, et al. Novel mutations involving the INSL3 gene associated with cryptorchidism. J Urol. 2007;177:1947–51. doi: 10.1016/j.juro.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Feng S, Cortessis VK, Hwang A, Hardy B, Koh CJ, et al. Mutation analysis of INSL3 and GREAT/LGR8 genes in familial cryptorchidism. Urology. 2004;64:1032–6. doi: 10.1016/j.urology.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Lim HN, Nixon RM, Chen H, Hughes IA, Hawkins JR. Evidence that longer androgen receptor polyglutamine repeats are a causal factor for genital abnormalities. J Clin Endocrinol Metab. 2001;86:3207–10. doi: 10.1210/jcem.86.7.7674. [DOI] [PubMed] [Google Scholar]
- Gray LE, Jr, Ostby JS, Kelce WR. Developmental effects of an environmental antiandrogen: the fungicide vinclozolin alters sex differentiation of the male rat. Toxicol Appl Pharmacol. 1994;129:46–52. doi: 10.1006/taap.1994.1227. [DOI] [PubMed] [Google Scholar]
- Gray LE, Jr, Ostby J, Furr J, Price M, Veeramachaneni DN, et al. Perinatal exposure to the phthalates DEHP, BBP, and DINP, but not DEP, DMP, or DOTP, alters sexual differentiation of the male rat. Toxicol Sci. 2000;58:350–65. doi: 10.1093/toxsci/58.2.350. [DOI] [PubMed] [Google Scholar]
- Foster PM, Mylchreest E, Gaido KW, Sar M. Effects of phthalate esters on the developing reproductive tract of male rats. Hum Reprod Update. 2001;7:231–5. doi: 10.1093/humupd/7.3.231. [DOI] [PubMed] [Google Scholar]
- Gray LE, Jr, Wilson VS, Stoker T, Lambright C, Furr J, et al. Adverse effects of environmental antiandrogens and androgens on reproductive development in mammals. Int J Androl. 2006;29:96–104. doi: 10.1111/j.1365-2605.2005.00636.x. [DOI] [PubMed] [Google Scholar]
- Swan SH. Environmental phthalate exposure in relation to reproductive outcomes and other health endpoints in humans. Environ Res. 2008;108:177–84. doi: 10.1016/j.envres.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Breyer BN, Eisenberg ML, Baskin LS. Associations among hypospadias, cryptorchidism, anogenital distance, and endocrine disruption. Curr Urol Rep. 2008;9:137–42. doi: 10.1007/s11934-008-0025-0. [DOI] [PubMed] [Google Scholar]
- Swan SH, Kruse RL, Liu F, Barr DB, Drobnis EZ, et al. Semen quality in relation to biomarkers of pesticide exposure Environ Health Perspect20031111478–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan SH, Main KM, Liu F, Stewart SL, Kruse RL, et al. Decrease in anogenital distance among male infants with prenatal phthalate exposure. Environ Health Perspect. 2005;113:1056–61. doi: 10.1289/ehp.8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Silva MJ, Needham LL, Calafat AM. Determination of 16 phthalate metabolites in urine using automated sample preparation and on-line preconcentration/high-performance liquid chromatography/tandem mass spectrometry. Anal Chem. 2005;77:2985–91. doi: 10.1021/ac0481248. [DOI] [PubMed] [Google Scholar]
- Silva MJ, Slakman AR, Reidy JA, Preau JL, Jr, Herbert AR, et al. Analysis of human urine for fifteen phthalate metabolites using automated solid-phase extraction. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;805:161–7. doi: 10.1016/j.jchromb.2004.02.038. [DOI] [PubMed] [Google Scholar]
- Swan SH. Prenatal phthalate exposure and anogenital distance in male infants. Environ Health Perspect. 2006;114:A88–9. doi: 10.1289/ehp.114-a88b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung R, Reed LD. Estimation of average concentration in the presence of nondetectable values. Appl Occup Environ Hyg. 1990;5:46–51. [Google Scholar]
- Pollard KS, Dudoit S, van der Laan MJ.Multiple Testing Procedures: R multtest Package and Application to Genomics U.C. Berkeley Division of Biostatistics Working Paper Series 2004. Working Paper 164. http://biostats.bepress.com/ucbbiostat/paper164
- Manson JM, Carr MC. Molecular epidemiology of hypospadias: review of genetic and environmental risk factors. Birth Defects Res A Clin Mol Teratol. 2003;67:825–36. doi: 10.1002/bdra.10084. [DOI] [PubMed] [Google Scholar]
- Kim KS, Torres CR, Jr, Yucel S, Raimondo K, Cunha GR, et al. Induction of hypospadias in a murine model by maternal exposure to synthetic estrogens. Environ Res. 2004;94:267–75. doi: 10.1016/S0013-9351(03)00085-9. [DOI] [PubMed] [Google Scholar]
- Safarinejad MR, Shafiei N, Safarinejad S. Association of polymorphisms in the estrogen receptors alpha, and beta (ESR1, ESR2) with the occurrence of male infertility and semen parameters. J Steroid Biochem Mol Biol. 2010;122:193–203. doi: 10.1016/j.jsbmb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Schupf N, Lee JH, Wei M, Pang D, Chace C, et al. Estrogen receptor-alpha variants increase risk of Alzheimer's disease in women with Down syndrome. Dement Geriatr Cogn Disord. 2008;25:476–82. doi: 10.1159/000126495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghel A, Narita D, Seclaman E, Popovici E, Anghel M, et al. Estrogen receptor alpha polymorphisms and the risk of malignancies. Pathol Oncol Res. 2010;16:485–96. doi: 10.1007/s12253-010-9263-9. [DOI] [PubMed] [Google Scholar]
- Sonoda T, Suzuki H, Mori M, Tsukamoto T, Yokomizo A, et al. Polymorphisms in estrogen related genes may modify the protective effect of isoflavones against prostate cancer risk in Japanese men. Eur J Cancer Prev. 2010;19:131–7. doi: 10.1097/CEJ.0b013e328333fbe2. [DOI] [PubMed] [Google Scholar]
- Mill J, Kiss E, Baji I, Kapornai K, Daróczy G, et al. Association study of the estrogen receptor alpha gene (ESR1) and childhood-onset mood disorders. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1323–6. doi: 10.1002/ajmg.b.30751. [DOI] [PubMed] [Google Scholar]
- Liu B, Agras K, Willingham E, Vilela ML, Baskin LS. Activating transcription factor 3 is estrogen-responsive in utero and upregulated during sexual differentiation. Horm Res. 2006;65:217–22. doi: 10.1159/000092402. [DOI] [PubMed] [Google Scholar]
- Liu B, Wang Z, Lin G, Agras K, Ebbers M, et al. Activating transcription factor 3 is up-regulated in patients with hypospadias. Pediatr Res. 2005;58:1280–3. doi: 10.1203/01.pdr.0000187796.28007.2d. [DOI] [PubMed] [Google Scholar]
- Kalfa N, Liu B, Klein O, Wang MH, Cao M, et al. Genomic variants of ATF3 in patients with hypospadias. J Urol. 2008;180:2183–8. doi: 10.1016/j.juro.2008.07.066. [DOI] [PubMed] [Google Scholar]
- Beleza-Meireles A, Töhönen V, Söderhäll C, Schwentner C, Radmayr C, et al. Activating transcription factor 3: a hormone responsive gene in the etiology of hypospadias. Eur J Endocrinol. 2008;158:729–39. doi: 10.1530/EJE-07-0793. [DOI] [PubMed] [Google Scholar]
- Schettler T. Human exposure to phthalates via consumer products. Int J Androl. 2006;29:134–9. doi: 10.1111/j.1365-2605.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- Song XF, Wei GH, Liu X, Zhang DY, Chen X, et al. Effects of diethylhexyl phthalate (DEHP) on INSL3 mRNA expression by Leydig cells derived from mouse embryos and in newborn mice. J Int Med Res. 2008;36:512–21. doi: 10.1177/147323000803600316. [DOI] [PubMed] [Google Scholar]


