Abstract
For almost two decades, breast cancer clinical genetics has operated in an environment where a heritable cause of breast cancer susceptibility is identified in the vast minority of women seeking advice about their personal and/or family history of breast and/or ovarian cancer. A new wave of genetic information is upon us that promises to provide an explanation for the greater proportion of current missing heritability of breast cancer. Whilst researchers refine bioinformatic and analytic methodology necessary to interpret the new genetic data, attention needs to be paid to defining appropriate and coordinated pathways for the translation of this information so that it can be applied in clinical genetic services for the benefit of the majority of women who currently have no explanation for their breast cancer susceptibility. The search for additional breast cancer susceptibility genes remains a very active area of research. Exhausting the power of linkage studies that identified BRCA1 and BRCA2, the research community moved to candidate gene studies that led to the identification of ATM, BRIP1, CHEK2, and PALB2 as so-called “moderate-risk” breast cancer susceptibility genes. Mutations in these genes are rare and although early reports suggested that, on average, they are associated with moderate risks of breast cancer; population-based studies have demonstrated that at least some mutations in these genes are associated with breast cancer risks that are comparable to the average risk associated with BRCA2 mutations. The search for additional breast cancer susceptibility genes has now moved onto research platforms applying massively parallel sequencing capable of sequencing whole human exomes and genomes in single instrument runs. These programs are identifying a large number of additional putative breast cancer susceptibility genes, many of which are currently undergoing validation. It is highly anticipated that the remaining missing heritability of breast cancer will be due to mutations in many different genes, each explaining a small proportion of the currently unexplained heritable breast cancer susceptibility. The characterization of PALB2 as a breast cancer susceptibility gene and subsequent research that has refined our understanding of the prevalence and penetrance of heritable mutations in PALB2 offers a precious opportunity to use the data as a model and develop modes of translation that would be appropriate for the anticipated volume of imminent new information.
Keywords: PALB2, breast cancer risk, clinical genetics, translation
Introduction
Homologous recombination (HR) is a major pathway of DNA double-strand break repair that uses the sister chromatid as a template for high fidelity DNA repair. PALB2 (partner and localizer of BRCA2), was identified via a search for novel components of endogenous BRCA2-containing complexes and is critical for its localization to chromatin and recruitment to double-strand breaks. Soon after its identification, heterozygous germline loss-of-function mutations in PALB2 were recognized to be associated with increased risk of breast cancer1 and biallelic mutations in PALB2 were found to explain an unrecognized Fanconi anemia complementation group, designated subtype N (FANCN), associated with considerable increased risk of childhood cancer.2,3 PALB2 is also recruited by BRCA1 in response to DNA damage and serves as a linker between BRCA1 and BRCA2 necessary for BRCA2-mediated HR repair.4,5
Thus, BRCA1, BRCA2, and PALB2 are key breast cancer susceptibility genes that function together in the same DNA-damage response pathway.4,5 Today, women attending clinical genetic services seeking advice about their personal and/or family history of breast and/or ovarian cancer are routinely offered genetic testing for BRCA1 and BRCA2 mutations but not PALB2 mutations; it is unclear why.
PALB2 structure, function, and protein binding partners
The PALB2 protein is about 130 kDa consisting of 1186 amino acids encoded by 13 exons.6 The functional domains of PALB2 include a coiled-coil structure, an ETGE-type KEAP1 binding motif, a chromatin-association motif (ChAM) at the N-terminus, and a WD repeat motif in the C-terminus.4–7
The WD repeat, a domain commonly involved in protein– protein interactions, is a seven-bladed β-propeller domain8 that provides the binding site for the N-terminus of BRCA2.6 The WD repeat domain of PALB2 has a linear topology and is held together by the seventh blade of the repeat domain.8 Disruption of the final blade of the WD repeat domain by the removal of the last four amino acids as a result of PALB2 c.3459C >G, p.Tyr1183* is associated with breast cancer and Fanconi anemia2 providing evidence for the importance of the final blade in the function of the WD repeat domain. PALB2 amino acids 1019 to 1098 interact with BRCA2 amino acids 21 to 39. PALB2 forms a hydrophobic pocket with the tips of the fourth and fifth blades of the WD repeat domain and is lined by amino acids 1019, 1022, 1025, 1037, 1046, 1047, 1070, 1097, and 1098. BRCA2 amino acids 31, 32, and 35, which project from a short helix into the hydrophobic pocket of PALB2, provide the core of interaction between the two proteins.8
The interaction between PALB2 and BRCA2 is required for localizing and stabilizing of BRCA2 to sites of DNA damage by chromatin association.3,9 Loss of PALB2 has been found to abolish BRCA2 focus formation and the ability of the latter to regulate HR repair of double-strand breaks through its direct interaction with RAD51. Some missense mutations in the PALB2 binding region of BRCA2 have also been shown to disrupt PALB2 binding, BRCA2 HR, and double-strand break repair.6
The coiled-coil domain at the N-terminus of PALB2 (amino acids 6–90) provides a binding site for the coiled-coil domain in BRCA1 (amino acids 1393–1475).4,5 PALB2 is recruited by BRCA1 in response to DNA damage.
PALB2 has been shown to bind to DNA via two separate regions in the N-terminus of the protein. One region was localized to amino acids 1 to 200 and the other was localized to amino acids 372 to 561 of PALB2 (referred to as PALB2 truncation 1 [P2T1] and PALB2 truncation 3 [P2T3], respectively).10 P2T3 was later identified to be evolutionarily conserved and contains ChAM within the 395 to 446 amino acids of PALB2.11 PALB2 appears to be able to bind multiple DNA molecules simultaneously. The two regions have varying binding efficiencies to different DNA structures. The binding efficiencies of full length PALB2, and the P2T1 and P2T3 domains were previously compared.10 Full length PALB2 protein was able to bind efficiently to both single-stranded DNA and D loop substrates. Both P2T1 and P2T3 bound efficiently to the D loop substrates but showed reduced single-stranded DNA binding efficiency compared to the full length PALB2 protein. P2T1 and full length PALB2 but not P2T3 were able to bind to Holliday junctions which are structures formed as a result of strand invasion in HR repair. The ability of PALB2 to bind multiple DNA structures might confer flexibility that has been suggested to allow the protein to distinguish between different forms of damaged DNA molecules that could have unusual structures and to target these structures for DNA repair.10
ChAM has not been found to affect the ability of PALB2 to bind other proteins but has been shown to be required for the efficient association of PALB2 to chromatin and for the BRCA1/PALB2/BRCA2 complex to accumulate RAD51 at double-strand break sites.11 ChAM in PALB2 and MORF4L1, an interacting protein of PALB2, have been proposed to be modifiers of the chromatin-association of PALB2, whereas the damage-induced PALB2 focus formation has been found to be primarily modulated by BRCA1.11
MORF4L1 has been found to bind to a region included in amino acids 611–764 of PALB2.12,13 MORF4L1 contains a domain responsible for transcriptional regulation via chromatin remodeling by histone acetylation.14 PALB2 appears to act as a scaffold that links MORF4L1 to the whole BRCA complex that forms in response to DNA damage which includes BRCA1, BRCA2, RAD51, and PALB2.12 Cells depleted of expressed MORF4L1 led to depleted levels of chromatin-associated BRCA2,12 which was similarly observed with PALB2 depletion6 suggesting that MORF4L1 works together with PALB2 to promote BRCA2 functions.
The association of replication protein A (RPA) at single-stranded DNA formed by the resection of double-strand breaks is essential for HR repair by preventing further DNA resection. However, the association between RPA and the single-stranded DNA prevents RAD51 from binding to the single-stranded DNA and hence its recombinase functions. Therefore, the inhibitory effect of RPA has to be overcome with accessory proteins to allow RAD51 binding on the single-stranded DNA. BRCA2 has been found to be one of the accessory proteins.15 Recently, two distinct regions of PALB2, amino acids 101 to 184 and amino acids 853 to 1186, were found to interact directly with the amino acids 184 to 257 of RAD51.10,16 PALB2 was shown to function as an HR mediator by contributing to the alleviation of the inhibition of RPA and promoting RAD51 filament formation.10,16 It has been proposed that PALB2 acts in concert with BRCA2 to firstly overcome the inhibitory effect of RPA and then recruit RAD51 to resected DNA ends to stimulate strand invasion.10
PALB2 also binds to KEAP1 via an ETGE-type KEAP1 binding motif close to the N-terminus. This implicates PALB2 in a cellular redox homeostasis regulatory role as it is able to compete with NRF2 (a transcription factor that activates the expression of antioxidant response element-containing genes) for KEAP1 binding and promote the accumulation of NERF2 in the nucleus. This observation provides an interesting link between oxidative stress and susceptibility to breast cancer and Fanconi anemia.7
The structure of PALB2 and the binding sites of its interacting proteins are depicted in Figure 1.
Figure 1.
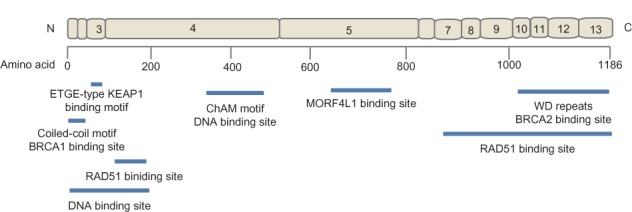
The structure of PALB2 indicating the exonic (exons 1–13) contribution to the protein and the location of the binding sites of its interacting proteins.
Abbreviation: ChAM, chromatin-association motif.
Prevalence of germline PALB2 mutations
Mutations in PALB2 make a small contribution to the heritable breast cancer susceptibility in most populations. PALB2 mutations have been identified in the Australian,17,18 Chinese,19 German,20 Italian,21–23 Dutch,24 North American,25–31 Polish,32 Russian,20 South African,33 and Spanish34 populations. No PALB2 mutations have been observed in the geographically confined population of Iceland.35 The PALB2 germline mutations and their carrier frequencies are listed in Table 1.
Table 1.
PALB2 germline mutations and carrier frequencies
| Populations | PALB2 mutations | Case–carrier frequency (%) | Control-carrier frequency (%) | Reference | Study designa |
|---|---|---|---|---|---|
| Australia | PALB2 c.196C >T, p.Gln66* | 1/70 (1.4) | – | 18 | Familial breast cancer |
| 1/747 (0.1%) | – | 36 | Familial breast cancer | ||
| PALB2 c.3113G >A, p.Trp1038* | 1/70 (1.4) | – | 18 | Familial breast cancer | |
| 5/1403 (0.004%) | 0/764 | 17 | Population-based | ||
| 2/66 (3%) | 17 | Early-onset multiple-case breast cancer | |||
| 8/871 (0.9%) | – | 41 | Familial breast cancer | ||
| 7/747 (0.9%) | – | 36 | Familial breast cancer | ||
| PALB2 c.1947_1948 insA, p.Glu650fs*13 | 1/747 (0.1%) | – | 36 | Familial breast cancer | |
| PALB2 c.2982_2983 insT, p.Ala995fs*16 | 1/747 (0.1%) | – | 36 | Familial breast cancer | |
| Canada | PALB2 c.229delT, p.Cys77fs*100 | 1/68 (1.5) | – | 29 | Familial breast cancer |
| PALB2 c.2323C >T, p.Gln775* | 3/406 (0.7) | 0/6440 | 38 | Early-onset and familial breast cancer | |
| 1/71 (1.4) | – | 70 | Familial breast cancer | ||
| 1/491 (0.2) | – | 70 | Ovarian cancer cases | ||
| 1/564 (0.2) | 0/6439 | 27 | Early-onset breast cancer | ||
| PALB2 c.3201_3561del, p.Met1067_Ser1186del | 1/228 (0.4) | – | 28 | Pancreatic cancer cases | |
| People’s Republic of China | PALB2 c.751C >T, p.Gln251* | 2/360 (0.6) | 0/864 | 19 | Familial breast cancer |
| PALB2 c.1050_1051delAAinsTCT, p.Gln350fs*11 | 1/360 (0.3) | 0/864 | 19 | Familial breast cancer | |
| Finland | PALB2 c.1592delT, p.Leu531fs*30a | 17/1918 (0.9) | 6/2501 (0.2) | 39,42 | Population-based |
| 19/947 (2) | 2/1079 (0.2) | 40 | Familial breast cancer | ||
| 8/1274 (0.6) | 40 | Sporadic breast cancer | |||
| Germany/Russia | PALB2 c.5089delAG, p.Arg170fs*8 | 1/81 (1.2) | – | 51 | Pancreatic cancer familiesb |
| PALB2 c.509_510delGA, p.Arg170fs*14 | 1/203 (0.49) | – | 20 | Bilateral breast cancer cases | |
| PALB2 c.758 insT, p.Leu273fs*4 | 1/40 (2.5)c | – | 71 | Triple negative breast cancer | |
| PALB2 c.1240C >T, p.Arg414* | 1/203 (0.49) | – | 20 | Bilateral breast cancer cases | |
| 1/81 (1.2) | – | 51 | Pancreatic cancer familiesb | ||
| PALB2 c.1633G >T, p.Glu545* | 1/203 (0.49) | – | 20 | Bilateral breast cancer cases | |
| PALB2 c.2761C >T, p.Gln921* | 1/203 (0.49) | – | 20 | Bilateral breast cancer cases | |
| PALB2 c.3116delA, p.Asn1039fs*2 | 1/81 (1.2) | – | 51 | Pancreatic cancer familiesb | |
| Italy | PALB2 c.72delG, p.Leu24fs*9 | 1/62 (1.6) | – | 21 | Breast–pancreatic cancer families |
| PALB2 c.1027C >T, p.Gln343* | 1/62 (1.6) | – | 21 | Breast–pancreatic cancer families | |
| PALB2 c.1317delG, p.Gly439fs*13 | 1/95 (1.0) | – | 22 | Familial breast cancer | |
| PALB2 c.2257C >T, p.Arg753* | 1/132 (0.8) | 0/300 | 23 | Familial breast cancer | |
| PALB2 c.3497delG, p.Gly1166fs*21 | 1/62 (1.6) | – | 21 | Breast–pancreatic cancer families | |
| Netherlands | PALB2 c.509_510delGA, p.Arg170fs*14 | 1/110 (1)d | – | 24 | Breast (including male)–pancreatic cancer families |
| Poland | PALB2 c.509_510delGA, p.Arg170fs*14 | 2/339 (0.6) | 1/1310 (0.08) | 31 | Ovarian cancer cases |
| 4/648 (0.6) | 31 | Familial breast cancer | |||
| South Africa | PALB2 c.697delG, p.Val233fs*5 | 1/48 (2.0) | 0/75 | 32 | Unselected early-onset breast cancer |
| Spain | PALB2 c.1056_1057delGA, p.Glu352fs*8 | 1/797 (0.12) | – | 33 | Familial breast cancer |
| United Kingdom | PALB2 c.2386G >T, p.Gly796* | 1/923 (0.1) | 0/1084 | 2 | Familial breast cancer |
| PALB2 c.2982_2983 insT, p.Ala995fs*16 | 1/923 (0.1) | 0/1084 | 2 | Familial breast cancer | |
| PALB2 c.3113G >A, p.Trp1038* | 2/923 (0.2) | 0/1084 | 2 | Familial breast cancer | |
| PALB2 c.3116delA, p.Asn1039fs*2 | 3/923 (0.3) | 0/1084 | 2 | Familial breast cancer | |
| PALB2 c.3549C >G, p.Tyr1183* | 3/923 (0.3) | 0/1084 | 2 | Familial breast cancer | |
| USA | PALB2 c.172_175delTTGT, p.Leu58fs*10 | 1/972 (0.1) | 0/960 | 29 | Familial breast cancer |
| PALB2 c.196C > T, p.Gln66* | 2/972 (0.2) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.509_510delGA, p.Arg170fs*14 | 7/972 (0.7) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.757_758delCT, p.Leu253fs*3 | 4/972 (0.4) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.1240C >T, p.Arg414* | 3/972 (0.3) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.1592delT, p.Leu531fs*30a | 1/559 (0.2) | 0/565e | 36 | Contralateral breast cancer cases | |
| PALB2 c.1653T >A, p.Tyr551* | 1/972 (0.1) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.2386G >T, p.Gly796* | 1/972 (0.1) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.2559C >T, p.Gly853fs*21f | 1/972 (0.1) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.2686_2987 insT, p.Ser896fs*32 | 3/972 (0.3) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.2718G >A, p.Trp906* | 1/972 (0.1) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.28351G >C | 1/559 (0.2) | 0/565e | 36 | Contralateral breast cancer cases | |
| PALB2 c.2920_2921delAA, p.Lys974fs*5 | 1/972 (0.1) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.3026delC, p.Pro1009fs*6 | 2/972 (0.2) | 0/960 | 29 | Familial breast cancer | |
| PALB2 c.3113G >A, p.Trp1038* | 5/972 (0.5) | 0/960 | 29 | Familial breast cancer | |
| 1/559 (0.2) | 0/565e | 36 | Contralateral breast cancer cases | ||
| PALB2 c.3202 + 1G >C | 1/559 (0.2) | 0/565e | 36 | Contralateral breast cancer cases | |
| PALB2 c.3549C >G, p.Tyr1183* | 1/94 (1) | – | 30 | Breast–pancreatic cancer families | |
| 1/559 (0.2) | 0/565e | 36 | Contralateral breast cancer cases | ||
| 1/97 (1) | – | 24 | Male breast cancer cases | ||
| PALB2 c.2962C >T, p.Gln988* | 1/94 (1) | – | 30 | Breast–pancreatic cancer families |
Notes:
Guide only, see reference for full details
study includes EUROPAC
carrier also BRCA1 mutation carrier
carrier is a male breast cancer case
controls are unilateral breast cancer case
mutation identified via reverse transcription PCR.
Abbreviations: EUROPAC, The European Registry of Hereditary Pancreatitis and Familial Pancreatic Cancer; PCR, polymerase chain reaction.
Similar to the mutation spectrum observed in BRCA1 and BRCA2, protein truncating mutations in PALB2 are distributed throughout the coding region but in contrast to its binding partners, there is no evidence that missense mutations in PALB2 play a significant role of breast cancer predisposition.36,37 Four PALB2 mutations are of note in terms of multiple observations, PALB2 c.509_510delGA (p.Arg170fs*14) has been observed in seven unrelated women in Poland,32 PALB2 c.2323C >T (p.Gln775*) in French Canadians,27,29,38 PALB2 c.1592delT (p.531fs*30) found in approximately 2% of women affected with breast cancer associated with a strong family history in Finland,39,40 and PALB2 c3113G >A (p.Trp1038*) identified in affected women in the United Kingdom,1 the United States,30 and Australia, where it has been observed in approximately 1% of affected women with a family history of breast cancer.17,18,36,41
Breast cancer risk
The rarity of mutations in PALB2 has made it challenging to estimate the associated breast (and other) cancer risk. The first study that reported an association between PALB2 mutations and breast cancer risk came from a large case-control mutation screening initiative from the UK involving familial breast cancer cases and unaffected controls from the UK. Using only some information obtained from just ten mutation carrying families who carried five different protein truncating mutations (see Table 1), and under strong modeling assumptions, the average relative risk associated with these mutations was estimated indirectly to be 2.3-fold (95% confidence interval [CI], 1.4–3.9).1
Subsequent population-based studies have enabled mutation-specific estimates to be made and these risk estimates have been higher than the initial report. For example, PALB2 c.1592delT was identified in 18/1918 (0.9%) breast cancer cases from Northern Finland, unselected for family history compared with 6/2,501 (0.2%) unaffected controls (odds ratio [OR] 3.94; 95% CI,1.5–12.1)42 and another study in Helsinki identified 19 PALB2 c.1592delT carriers in 947 (2%) familial breast cancer cases (OR 11.03; 95% CI, 2.65–97.78; P < 0.0001).40
Other studies have provided further insights into the breast cancer risk associated with carrying protein truncating mutations in PALB2. Of note is the work done within the Women’s Environment, Cancer, and Radiation Epidemiology (WECARE) Study, a nested case-control study of the Contralateral Breast Cancer study,43 involving women from the United States and Denmark. This work screened 559 women with contralateral breast cancer and 565 matched women with unilateral breast cancer. Five PALB2 mutations were identified, all in women with contralateral breast cancer (0.9%). The first-degree female relatives of the five carriers demonstrated significantly higher incidence of breast cancer than relatives of non-carriers cases and were estimated to confer a 5.3-fold increase in risk of breast cancer (95% CI, 1.8–13.2).37
Due to the limited number of studies conducted within unselected breast cancer cases, estimation of the age-specific cumulative risk (penetrance) of breast cancer associated with PALB2 mutations has been limited. Using the family histories of the case carriers unselected for age or family history reported in Northern Finland,39 PALB2 c.1592delT was estimated to be associated with a 40% (95% CI, 17%–77%) risk of breast cancer to the age of 70 years.42 Similarly, in a population-based case-control-family study of Australian women, PALB2 c.3113G >A was identified in 5/1403 (0.4%) breast cancer cases and 0/764 (0%) unaffected controls. Using the family histories of the five carrier cases, the estimated cumulative risk for PALB2 c.3113G >A was 91% (95% CI, 44%–100%) to the age of 70 years.17 Therefore, population-based studies of breast cancer that have directly used the family history data have indicated that at least some PALB2 mutations are associated with a breast cancer risk (penetrance) comparable to that of the average pathogenic mutation in BRCA2: 45% (95% CI, 31%–56%)44. There is no evidence to suggest that different PALB2 mutations differ in terms of breast cancer risk although there is evidence that some PALB2 mutation-positive patient-derived cell lines have a measurable three-dimensional nuclear organization differences (telomere counts, chromosomal rearrangements, and centromere distribution).45
Risk of other cancers
Mutations in BRCA1 and BRCA2 have considerable risk for several other cancers and, as these gene products function together in the same DNA-damage response pathway as PALB2, specific attention has been paid to the possibility that PALB2 mutations could predispose to a similar group of cancer types.
Male breast cancer is a striking feature of the BRCA2-mutation associated syndrome.46–48 A small number of the pedigrees reported in the literature to carry protein truncating PALB2 mutations contained cases of male breast cancer,1,34 although the actual carrier status of these men was not often known. Through extensive screening in a variety of settings, several studies that have investigated the prevalence of PALB2 mutations in male breast cancer have found little evidence that PALB2 mutations are associated with increased risk of male breast cancer,39,49 although some individually striking pedigrees have been reported.24,25
A study of familial pancreatic cancer that applied exome-capture followed by massively parallel sequencing identified a PALB2 germline mutation and found an additional three mutation carriers when they sequenced PALB2 in an additional 96 highly selected pancreatic cancer families.50 Subsequent follow-up screening in less highly selected pancreatic cancer families has found PALB2 germline mutations to be very rare in pancreatic cancer cases28 although there is some evidence that the mutation rate may be slightly higher in families with both pancreatic and breast cancer. In a collection of 94 women with breast cancer and a personal or family history of pancreatic cancer, Hofstatter et al31 identified two (2.1%) protein truncating PALB2 mutations. A study of 81 European familial pancreatic cancer families identified three (3.7%) protein truncating PALB2 mutations, all occurring in families that also had histories of breast cancer and a study of 62 Italian breast–pancreatic cancer families identified three (4.8%) protein truncating PALB2 mutations.21,31,51 A study of 56 pancreatic cancer families including 28 that also had cases of breast cancer from The Netherlands, a study of 26 Italian pancreatic cancer cases (selected for their personal and family history of pancreatic, breast, and ovarian cancer) and a study of 77 breast-pancreatic cancer families attending Memorial Sloan-Kettering Cancer Center (USA), failed to find any PALB2 germline mutation carriers.52–54
There is little data and no evidence supporting an association between PALB2 mutations and prostate cancer risk39,55 or ovarian cancer risk,32,39 although a few mutation carrying pedigrees have been presented in the literature that might stimulate further research in this area.32,39
Making more precise estimates of the risk of these cancers associated with PALB2 germline mutations is challenged by several factors including the rarity of PALB2 mutations overall and the rarity of the cancers under consideration and the setting in which the majority of these studies have been conducted (ie, in highly selected families).
PALB2 and fanconi anemia
Shortly after the first description of PALB2, biallelic mutations in PALB2 were found to explain an unrecognized FANCN and associated with considerable increased risk of childhood cancer.2,3 Interestingly, the cancer spectrum and ages of onset of biallelic PALB2 mutations are similar to that of biallelic BRCA2 (FANCD1)56,57 mutation carriers again indicating the close functional connection of PALB2 and BRCA2 in tumorigenic pathways.58
Using pathology to identify carriers
Breast cancer tumor morphology can be suggestive of underlying familial, if not heritable, risk. In a population-based sample of 375 women with early-onset breast cancer cases with no known high-risk mutation in a breast cancer susceptibility gene, minimal sclerosis, presence of circumscribed growth, extensive intraductal carcinoma, and lobular growth patterns were independent predictors of increased breast cancer risk for their first-degree female relatives (2.0-fold to 3.3-fold increased risk for relatives, P < 0.02 for all listed features). Relatives of the 128 (34%) index cases with none of these four features were at population risk (standardized incidence ratio 1.03; 95% CI, 0.57–1.85), while relatives of the 37 (10%) index cases with two or more features were at high risk (standardized incidence ratio, 5.18; 95% CI, 3.22–8.33).59
Breast cancer morphological features can be used to identify women most likely to carry germline mutations in known breast cancer susceptibility genes. It has been known for some time that some morphological features are more common in cancers arising in BRCA1 mutation carriers.60 These features have been identified by studying carriers across a wide range of ages at diagnosis and ascertained either because of their strong family cancer history or through population-based sampling. Lack of estrogen receptor (ER) and progesterone receptor (PR) expression has also been reported to improve prediction of BRCA1 mutation status based on family history.61–63 A population-based sample of 452 young women with breast cancer, found that just two breast tumor morphological features (trabecular growth pattern and high mitotic index) were sufficient to 28 of 29 (97%) BRCA1 mutation carriers in the study.64 Moreover, prediction of mutation status using these two features was more sensitive and specific than using family history alone, and when combined, the area under the receiver operator curve was in excess of 0.9.
A detailed analysis of the morphological features of PALB2 mutation-associated breast cancers has not been previously conducted. Characterization of the morphology of breast cancers arising in PALB2 mutation carriers offers the possibility of identifying tumor morphological features predictive of an underlying germline PALB2 mutation. Such predictive features would be useful at the time of breast cancer diagnosis, enabling focused and rapid genetic testing that could also facilitate personalized treatment strategies, as well as enabling identification of those relatives who have also inherited a similar high breast cancer risk.
Some information about the general morphology of breast tumors arising in PALB2 mutation carriers is available from work studying breast tumors carrying the Finnish founder mutation PALB2 c.1592delT. Heikkinen et al,40 identified 27 PALB2 mutation carriers and found that carriers with a family history of breast cancer were more likely to have “triple negative” tumors (P < 0.0001) more often of higher grade (P = 0.0027) and to have greater expression of Ki67 (P = 0.0004), when compared to familial non-PALB2 mutation-associated breast cancers.40 Most other reports have been relatively small but have demonstrated that the PALB2 mutation-associated tumor phenotype is variable ranging from basal-like triple negative high grade breast cancers through to grade 1 invasive ductal carcinomas that express both ER and PR.1,19,23,29,32–34,36,38–41
The phenotypes of the small number of reported PALB2 mutation-associated breast cancers suggest that some may resemble the specific phenotype well described for BRCA1 mutation-associated breast tumors60,64 particularly apparent in PALB2 c.1592delT mutation carrying breast cancers. The phenotype is also clearly variable, as observed for BRCA2 mutation-associated breast cancers.60,65 Given the diversity of PALB2 binding partners, the functional connection of PALB2, BRCA2, and BRCA1, the plausibility of PALB2 mutation-specific phenotypes and the possibility of mutation-targeted personalized therapies, much more work is needed to examine the important question of PALB2 mutation-associated tumor phenotype.
International efforts
Mutations in PALB2 are rare (varying from 0.1% to 2.7% depending on the population, see above) but for women carrying them, and their relatives who might also be mutation carriers, knowing their mutation status has the potential to be clinically important as carriers are at high risk of breast cancer. Identified mutation carriers could be informed of optimal, risk appropriate clinical screening, and treatment.
Increasing the precision of risk estimates, further characterizing the tumor phenotype, the underlying biological and the tumorigenic pathways has been predominantly limited to small studies of highly selected women and sometimes their families. The PALB2 Interest Group has been established to facilitate the international coordination and collaboration required to extend our understanding of PALB2 in breast cancer susceptibility and tumor progression and to try to overcome some of the challenges presented by the rarity of mutations in most populations. Work conducted within the Breast Cancer Association Consortium (BCAC) (http://ccge.medschl.cam.ac.uk/consortia/bcac/links/links.html) and the Collaborative Oncological Gene-environment Study (COGS; www.cogseu.org/) could also assist in this regard in the near future.
Time for translation?
Today, women attending clinical genetic services seeking advice about their personal and/or family history of breast and/or ovarian cancer are routinely offered genetic testing for BRCA1 and BRCA2 mutations but not PALB2 mutations; why?
The reasons are many and varied and some are locally determined. Genetic counseling and genetic testing for BRCA1 and BRCA2 mutations were enthusiastically embraced and rapidly incorporated into clinical genetic services worldwide very soon after the genes were identified. Through experience, the discipline is now much more aware of the complexities of genetic testing in this area including genetic data interpretation, health economic impact, limited risk reduction strategies, psychosocial impacts, and many others; consequently, clinicians are now generally more cautious about the consideration of utilizing new genetic information. In many systems there are no clear pathways of translation and no clear indications of a minimal dataset, or what threshold defines a new breast cancer susceptibility gene as “clinically actionable”. It is time that this issue is addressed as PALB2 leads the initiative of new genetic information relevant to women with breast cancer, which is likely to be very large.
The characterization of PALB2 as a breast cancer susceptibility gene and subsequent research that has refined our understanding of the prevalence and penetrance of heritable mutations in PALB2 (as described above) offers a very important opportunity to use the data as a model. We can develop modes of translation that is appropriate for PALB2 and in so doing create a scaffold to support the translation of the anticipated volume of new information that will be available in the near future via the application of new genetic technology.66–68
Personalized medicine for women carrying PALB2 germline mutations is also likely to be feasible in the short term. PALB2-deficient cells have been shown to be sensitive to PARP inhibitors10 and a dramatic response to mitomycin C in a patient with advanced pancreatic cancer69 demonstrates the utility of these drugs in identified PALB2 mutation carriers. Thus, identifying women with breast cancers that are deficient in PALB2 is likely to be of significant clinical relevance especially for providing tailored therapy and improving outcomes.
Conclusion
PALB2, along with BRCA1 and BRCA2, are key breast cancer susceptibility genes that function together in the same DNA-damage response pathway. Biallelic mutations in PALB2 (FANCN) explain a Fanconi Anemia complementation group that has similar clinical features as the group explained by BRCA2 (FANCD1). Morphological and immunohistochemical features of PALB2 mutation-associated breast tumors are shared with tumors arising in women with BRCA1 and BRCA2 germline mutations, although both the PALB2 and BRCA2 mutation-associated morphological features appear quite varied.
Mutations in PALB2 are rare (varying from 0.1% to 2.7% of affected women from multiple-case breast cancer families) but the risk of breast cancer, at least for some PALB2 mutations, is high and comparable to that of the average pathogenic mutation in BRCA2. International efforts are moving the field forward via extensive collaboration to address some of the challenges presented by the rarity of the mutations.
Data supports the progress of information about PALB2 into a clinical translation phase and offers the discipline an opportunity to prepare a path suitable for the translation of future genetic information. This potential for treatment-focused testing is relevant to families with breast cancer in their risk management and targeted cancer treatment, and offers promise in genetically-targeted cancer prevention in the future.
Footnotes
Disclosure
The authors report no conflict of interest in this work.
References
- 1.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 3.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F, Ma J, Wu J, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106(17):7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Cai H, Wu T, et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol. 2012;32(8):1506–1517. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10(9):990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sy SM, Huen MS, Zhu Y, Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol Chem. 2009;284(27):18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buisson R, Dion-Côté AM, Coulombe Y, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17(10):1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleuyard JY, Buisson R, Masson JY, Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13(2):135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayakawa T, Zhang F, Hayakawa N, et al. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123(Pt 7):1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284(32):21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266(1–2):111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dray E, Etchin J, Wiese C, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17(10):1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southey MC, Teo ZL, Dowty JG, et al. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12(6):R109. doi: 10.1186/bcr2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong MW, Nordfors C, Mossman D, et al. BRIP1, PALB2, and RAD51C mutation analysis reveals their relative importance as genetic susceptibility factors for breast cancer. Breast Cancer Res Treat. 2011;127(3):853–859. doi: 10.1007/s10549-011-1443-0. [DOI] [PubMed] [Google Scholar]
- 19.Cao AY, Huang J, Hu Z, et al. The prevalence of PALB2 germline mutations in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res Treat. 2009;114(3):457–462. doi: 10.1007/s10549-008-0036-z. [DOI] [PubMed] [Google Scholar]
- 20.Bogdanova N, Sokolenko AP, Iyevleva AG, et al. PALB2 mutations in German and Russian patients with bilateral breast cancer. Breast Cancer Res Treat. 2011;126(2):545–550. doi: 10.1007/s10549-010-1290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterlongo P, Catucci I, Pasquini G, et al. PALB2 germline mutations in familial breast cancer cases with personal and family history of pancreatic cancer. Breast Cancer Res Treat. 2011;126(3):825–828. doi: 10.1007/s10549-010-1305-1. [DOI] [PubMed] [Google Scholar]
- 22.Balia C, Sensi E, Lombardi G, Roncella M, Bevilacqua G, Caligo MA. PALB2: a novel inactivating mutation in a Italian breast cancer family. Fam Cancer. 2010;9(4):531–536. doi: 10.1007/s10689-010-9382-1. [DOI] [PubMed] [Google Scholar]
- 23.Papi L, Putignano AL, Congregati C, et al. A PALB2 germline mutation associated with hereditary breast cancer in Italy. Fam Cancer. 2010;9(2):181–185. doi: 10.1007/s10689-009-9295-z. [DOI] [PubMed] [Google Scholar]
- 24.Adank MA, van Mil SE, Gille JJ, Waisfisz Q, Meijers-Heijboer H. PALB2 analysis in BRCA2-like families. Breast Cancer Res Treat. 2011;127(2):357–362. doi: 10.1007/s10549-010-1001-1. [DOI] [PubMed] [Google Scholar]
- 25.Ding YC, Steele L, Kuan CJ, Greilac S, Neuhausen SL. Mutations in BRCA2 and PALB2 in male breast cancer cases from the United States. Breast Cancer Res Treat. 2011;126(3):771–778. doi: 10.1007/s10549-010-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding YC, Steele L, Chu LH, et al. Germline mutations in PALB2 in African-American breast cancer cases. Breast Cancer Res Treat. 2011;126(1):227–230. doi: 10.1007/s10549-010-1271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghadirian P, Robidoux A, Zhang P, et al. The contribution of founder mutations to early-onset breast cancer in French-Canadian women. Clin Genet. 2009;76(5):421–426. doi: 10.1111/j.1399-0004.2009.01277.x. [DOI] [PubMed] [Google Scholar]
- 28.Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137(3):1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA. 2007;104(16):6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casadei S, Norquist BM, Walsh T, et al. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71(6):2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofstatter EW, Domchek SM, Miron A, et al. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer. 2011;10(2):225–231. doi: 10.1007/s10689-011-9426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dansonka-Mieszkowska A, Kluska A, Moes J, et al. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20. doi: 10.1186/1471-2350-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sluiter M, Mew S, van Rensburg EJ. PALB2 sequence variants in young South African breast cancer patients. Fam Cancer. 2009;8(4):347–353. doi: 10.1007/s10689-009-9241-0. [DOI] [PubMed] [Google Scholar]
- 34.García MJ, Fernández V, Osorio A, et al. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res Treat. 2009;113(3):545–551. doi: 10.1007/s10549-008-9945-0. [DOI] [PubMed] [Google Scholar]
- 35.Gunnarsson H, Arason A, Gillanders EM, et al. Evidence against PALB2 involvement in Icelandic breast cancer susceptibility. J Negat Results Biomed. 2008;7:5. doi: 10.1186/1477-5751-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teo ZL, Park DJ, Provenzano E, et al. Prevalence of PALB2 mutations in Australasian multiple-case breast cancer families. Breast Cancer Res. 2013;15(1):R17. doi: 10.1186/bcr3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tischkowitz M, Capanu M, Sabbaghian N, et al. Rare germline mutations in PALB2 and breast cancer risk: a population-based study. Hum Mutat. 2012;33(4):674–680. doi: 10.1002/humu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foulkes WD, Ghadirian P, Akbari MR, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9(6):R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erkko H, Xia B, Nikkilä J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446(7133):316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 40.Heikkinen T, Kärkkäinen H, Aaltonen K, et al. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15(9):3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- 41.Teo ZL, Sawyer SD, James PA, et al. The incidence of PALB2 c.3113G >A in women with a strong family history of breast and ovarian cancer attending familial cancer centres in Australia. Fam Cancer. 2013 doi: 10.1007/s10689-013-9620-4. [DOI] [PubMed] [Google Scholar]
- 42.Erkko H, Dowty JG, Nikkilä J, et al. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clin Cancer Res. 2008;14(14):4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein JL, Langholz B, Haile RW, et al. Study design: evaluating gene-environment interactions in the etiology of breast cancer – the WECARE study. Breast Cancer Res. 2004;6(3):R199–R214. doi: 10.1186/bcr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wark L, Novak D, Sabbaghian N, et al. Heterozygous mutations in the PALB2 hereditary breast cancer predisposition gene impact on the three-dimensional nuclear organization of patient-derived cell lines. Genes Chromosomes Cancer. 2013;52(5):480–494. doi: 10.1002/gcc.22045. [DOI] [PubMed] [Google Scholar]
- 46.Couch FJ, Farid LM, DeShano ML, et al. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet. 1996;13(1):123–125. doi: 10.1038/ng0596-123. [DOI] [PubMed] [Google Scholar]
- 47.Thorlacius S, Olafsdottir G, Tryggvadottir L, et al. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet. 1996;13(1):117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 48.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 49.Sauty de Chalon A, Teo Z, Park DJ, et al. Are PALB2 mutations associated with increased risk of male breast cancer? Breast Cancer Res Treat. 2010;121(1):253–255. doi: 10.1007/s10549-009-0673-x. [DOI] [PubMed] [Google Scholar]
- 50.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324(5924):217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78(5):490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 52.Ghiorzo P, Pensotti V, Fornarini G, et al. Contribution of germline mutations in the BRCA and PALB2 genes to pancreatic cancer in Italy. Fam Cancer. 2012;11(1):41–47. doi: 10.1007/s10689-011-9483-5. [DOI] [PubMed] [Google Scholar]
- 53.Harinck F, Kluijt I, van Mil SE, et al. Routine testing for PALB2 mutations in familial pancreatic cancer families and breast cancer families with pancreatic cancer is not indicated. Eur J Hum Genet. 2012;20(5):577–579. doi: 10.1038/ejhg.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stadler ZK, Salo-Mullen E, Sabbaghian N, et al. Germline PALB2 mutation analysis in breast-pancreas cancer families. J Med Genet. 2011;48(8):523–525. doi: 10.1136/jmg.2010.087379. [DOI] [PubMed] [Google Scholar]
- 55.Tischkowitz M, Sabbaghian N, Ray AM, Lange EM, Foulkes WD, Cooney KA. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in hereditary prostate cancer. Prostate. 2008;68(6):675–678. doi: 10.1002/pros.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirsch B, Shimamura A, Moreau L, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103(7):2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- 57.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44(1):1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70(19):7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dite GS, Makalic E, Schmidt DF, Giles GG, Hopper JL, Southey MC. Tumour morphology of early-onset breast cancers predicts breast cancer risk for first-degree relatives: the Australian Breast Cancer Family Registry. Breast Cancer Res. 2012;14(4):R122. doi: 10.1186/bcr3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lakhani SR, Jacquemier J, Sloane JP, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90(15):1138–1145. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- 61.Mavaddat N, Barrowdale D, Andrulis IL, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21(1):134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.James PA, Doherty R, Harris M, et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006;24(4):707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- 63.Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20(9):2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 64.Southey MC, Ramus SJ, Dowty JG, et al. Morphological predictors of BRCA1 germline mutations in young women with breast cancer. Br J Cancer. 2011;104(6):903–909. doi: 10.1038/bjc.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Armes JE, Trute L, White D, et al. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59(8):2011–2017. [PubMed] [Google Scholar]
- 66.Park DJ, Lesueur F, Nguyen-Dumont T, et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90(4):734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruark E, Snape K, Humburg P, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493(7432):406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson JF, Reifenberger JG, Giladi E, et al. Single-step capture and sequencing of natural DNA for detection of BRCA1 mutations. Genome Res. 2012;22(2):340–345. doi: 10.1101/gr.122192.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther. 2011;10(1):3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]


