Abstract
Mammalian airways protect themselves from bacterial infection by using multiple defense mechanisms including antimicrobial peptides, mucociliary clearance, and phagocytic cells. We asked whether airways might also target a key bacterial cell-cell communication system, quorum-sensing. The opportunistic pathogen Pseudomonas aeruginosa uses two quorum-sensing molecules, N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) and N-butanoyl-l-homoserine lactone (C4-HSL), to control production of extracellular virulence factors and biofilm formation. We found that differentiated human airway epithelia inactivated 3OC12-HSL. Inactivation was selective for acyl-HSLs with certain acyl side chains, and C4-HSL was not inactivated. In addition, the capacity for inactivation varied widely in different cell types. 3OC12-HSL was inactivated by a cell-associated activity rather than a secreted factor. These data suggest that the ability of human airway epithelia to inactivate quorum-sensing signal molecules could play a role in the innate defense against bacterial infection.
Quorum sensing is a cell-cell signaling process that allows members of a bacterial population to coordinate their activities (1-5). Many bacteria use this method of communication to regulate genes associated with host colonization and virulence. Pseudomonas aeruginosa is an environmentally ubiquitous opportunistic pathogenic bacterium that causes a variety of infections, including burn infections, and chronic ear and bladder infections (6). Much of the morbidity and mortality in patients with cystic fibrosis is due to P. aeruginosa infections of the pulmonary airways (7). Quorum sensing in P. aeruginosa is mediated by the production, release, and detection of acyl-homoserine lactones (acyl-HSLs) (3, 5, 6, 8). P. aeruginosa produces two acyl-HSL signals, N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) and N-butanoyl-l-homoserine lactone (C4-HSL) (9, 10). These acyl-HSLs are hierarchically regulated in that threshold levels of 3OC12-HSL are required to activate C4-HSL production (11, 12). The 3OC12-HSL and C4-HSL signals regulate hundreds of genes in P. aeruginosa, control the production of a battery of extracellular virulence factors, and play a role in biofilm development (6, 8, 13-16).
Mammalian tissues and cells have developed multiple defense mechanisms as a protection against bacterial infection. These include proteins and peptides, such as lysozyme and defensins, which kill bacteria (17-19). Host defenses also include specific anti-biofilm defense molecules, including lactoferrin (20), which chelates iron and prevents the development of P. aeruginosa biofilms (21). Given the importance of quorum sensing for bacterial virulence, we wondered whether mammalian cells have also developed defense mechanisms that target this communication system. Earlier studies showed that certain bacteria have the ability to disrupt quorum sensing, and this capacity can modulate bacterial infections (22-26). For example, the soil bacterium Bacillus sp. 240B1 produces an enzyme coded by the aiiA gene that hydrolyzes the lactone ring of acyl-HSLs. Recombinant expression of aiiA in plants attenuated quorum-sensing-dependent bacterial infection (22).
Therefore, we asked whether mammalian cells have the ability to disrupt quorum-sensing signals. To answer this question, we tested the persistence of the P. aeruginosa quorum-sensing signals 3OC12-HSL and C4-HSL after exposure to differentiated human airway epithelia.
Materials and Methods
Epithelial Cell Cultures. Primary cultures of human airway epithelia were grown on permeable supports at the air-liquid interface as described (27). Epithelia were used after at least 14 days of culture when they had developed morphologic and functional properties of airway epithelia (27).
Other cell lines were grown as monolayers in 24-well dishes. These cell lines were A549 cells, established from a human bronchoalveolar carcinoma and grown in DMEM/Ham's F-12 medium (1:1) with 10% FCS and 1% l-glutamine; CaCo-2 from a human colon carcinoma grown in Eagle's minimal essential medium (EMEM), 20% FCS, and 1% nonessential amino acids (NEAA); Madin-Darby canine kidney (MDCK) cells from canine kidney grown in DMEM/Ham's F-12 (1:1) and 10% FCS; CHO from Chinese hamster ovary grown in Ham's F-12 and 10% FCS; HeLa from human cervix cultured in EMEM, 10% FCS, and 1% NEAA; 293T from human embryonic kidney and COS-7 from monkey kidney, both grown in DMEM and 10% FCS; and primary cultures of human lung fibroblasts (HLF) grown in DMEM and 20% FCS.
Acyl-HSL Degradation by Mammalian Cell Cultures. Cells were washed in PBS containing magnesium and calcium (PBS, pH 7.1). Acyl-HSLs in acidified ethyl acetate (EtAc) were dried under nitrogen and dissolved in PBS to give a final concentration of 1 or 10 μM. Acyl-HSLs were added to the apical (150 μl) or basolateral (250 μl) surface of airway epithelia or to the surface of confluent monolayers (250 μl) of cultured cell lines in 24-well culture dishes. Cells were incubated with acyl-HSLs at 37°C in 5.5% CO2 for up to 8 h. The pH of the medium was measured at the start and end of each experiment by using pH indicator strips (EM Reagents, Gibbstown, NJ). At various times, 6-μl samples were collected and added to 100 μl of EtAc. These samples were stored in airtight glass vials at -20°C. Acyl-HSL concentrations of 1-10 μM are within the physiological range of C4-HSL and 3OC12-HSL produced by laboratory cultures of P. aeruginosa (9, 10).
Experiments with Cell Lysates. Cell-free lysates were prepared as follows. After washing a confluent layer of cells with cold PBS, we added 0.5 ml of lysis buffer (50 mM Tris·HCl, pH 6.9/150 mM NaCl/10 μM leupeptin/10 μM aprotinin/1 μM pepstatin A/1 mM phenylmethylsulfonylfluoride/0.1 mg/ml benzamidine). All of the acyl-HSLs used in this study were stable in this buffer at 37°C in 5.5% CO2. After 20 min at 4°C with shaking in lysis buffer, cells were scraped into a microcentrifuge tube and lysed by sonication. Whole cells and other debris were subsequently removed by centrifugation (30 s at 4,500 × g). Centrifugation of the cell lysates (200,000 × g for 20 min at 4°C) was used to separate a soluble and a particulate, membrane-containing fraction. The particulate pellet was suspended in a volume of lysis buffer equal to the volume of the supernatant. Synthetic 3OC12-HSL was dried under nitrogen gas and dissolved in the cell lysate or lysate fractions to a final concentration of 10 μM. The mixtures were incubated at 37°C in 5.5% CO2. Samples were collected and stored as described above.
Quantitative Acyl-HSL Assays. All acyl-HSLs were measured by using quantitative bioassays as described (28). To measure 3OC12-HSL and C12-HSL, we used Escherichia coli MG-4 (pKDT17) (9, 29). For C4-HSL measurements, we used E. coli XL-1 Blue (pECP61) (30). For measuring 3OC6-HSL and C6-HSL, we used E. coli (pHV200I-) (9).
Acyl-HSLs. All acyl-HSLs were chemically synthesized. 3OC6-HSL was purchased from Sigma. All other acyl-HSLs were from Vertex Pharmaceuticals (Coralville, IA).
Results
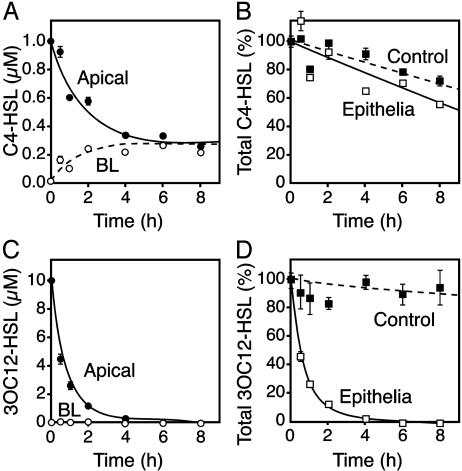
Human Airway Epithelia Rapidly Inactivate 3OC12-HSL but Not C4-HSL. We applied synthetic C4-HSL and 3OC12-HSL to the apical surface of primary cultures of differentiated human airway epithelia grown at the air-liquid interface and measured their concentrations over time. As the concentration of C4-HSL on the apical surface progressively fell, the basolateral concentration increased, reaching equilibrium across the epithelium in 4-6 h (Fig. 1A). A similar equilibration occurred when C4-HSL was added to the basolateral medium (data not shown). In addition, the total amount of C4-HSL showed a relatively small decrease over 8 h (Fig. 1B). These data indicate that airway epithelia were permeable to C4-HSL, but the epithelium showed little or no capacity to degrade the signal.
Fig. 1.
Inactivation of 3OC12-HSL, but not C4-HSL, by human airway epithelia. (A) The concentration of C4-HSL in the apical (filled circles) and basolateral (open circles) solutions of primary cultures of differentiated human airway epithelia over time after addition of 1 μM C4-HSL to the apical surface. (B) The total amount of C4-HSL in the apical and basolateral solutions of the airway epithelium (open squares) and the total amount of C4-HSL in medium without cells (filled squares). Values are percent of the initial C4-HSL. (C) The concentration of 3OC12-HSL in the apical and basolateral solutions of airway epithelia after addition of 10 μM C4-HSL to the apical surface. (D) The total amount of 3OC12-HSL in apical and basolateral solutions and the total amount of 3OC12-HSL in medium without cells as a percent of initial 3OC12-HSL. Data are means ± SEM of three independent experiments.
In contrast to C4-HSL, the concentration of 3OC12-HSL fell rapidly after addition to the apical surface, and there was no accumulation in the basolateral medium (Fig. 1C). Four hours after addition, 3OC12-HSL was no longer detectable in the epithelial medium (Fig. 1D). Likewise, we could not detect 3OC12-HSL in epithelial cell extracts or on the Millipore filter apparatus (data not shown). These results indicate that airway epithelia rapidly inactivated 3OC12-HSL.
Human Airway Epithelia Selectively Inactivate Specific Acyl-HSLs. To determine whether the fatty acid side chain of an acyl-HSL affects degradation by airway epithelia, we applied a variety of acyl-HSLs to the apical surface and measured the total amount remaining after 1 h (Fig. 2). The epithelia selectively inactivated C6-HSL and 3OC12-HSL. The 3OC6-HSL was not inactivated. Acyl-HSLs can be chemically degraded at alkaline pH and acyl-HSLs with short acyl side chains are more sensitive to base hydrolysis than those with longer side chains (31). The pattern of inactivation by airway epithelia did not follow this rule (compare C4-HSL, C6-HSL, and C12-HSL). In addition, the 3O-acyl-HSLs are more sensitive to base hydrolysis than the unsubstituted acyl-HSLs (31). This pattern of degradation was also not observed on airway epithelia (compare C6-HSL and 3OC6-HSL). These results indicate that degradation was not the result of base inactivation of acyl-HSL by the airway epithelia.
Fig. 2.
The specificity of acyl-HSL degradation by human airway epithelia. Shown is the amount of C4-, C6-, 3OC6-, C12-, and 3OC12-HSL remaining in differentiated human airway epithelia after 1 h. Values represent the percent of acyl-HSL remaining compared to the initial level. Data are means ± SEM from three to six independent experiments.
Inactivation of 3OC12-HSL Is Cell-Type-Specific. To learn whether other mammalian cells could inactivate P. aeruginosa acyl-HSLs, we applied C4-HSL and 3OC12-HSL to cultures of several human and nonhuman cells from organs other than lung and measured the amount of acyl-HSL remaining 1 h later (Fig. 3). The pH of the acyl-HSL containing medium on the cells did not change over the 1-h course of these experiments. None of the cells eliminated C4-HSL. However, 3OC12-HSL inactivation varied widely between the different cell types. Cells derived from human epithelia that are exposed to environmental pathogens, such as A549 cells from human lungs and CaCo-2 cells from human colon, showed the greatest amount of 3OC12-HSL inactivation, whereas cells from epithelia not usually exposed to bacteria, such as 293T cells from human kidney, COS-7 cells from monkey kidney, and MDCK cells from canine kidney, showed little or no inactivation of 3OC12-HSL. Primary cultures of human lung fibroblasts had less inactivating activity than epithelia from human lung and CHO cells from hamster ovary failed to inactivate 3OC12-HSL. These data suggest substantial variation in the ability of different mammalian cells to inactivate 3OC12-HSL. Although additional study is required, they also raise the possibility that greater 3OC12-HSL inactivation occurs in cells exposed to the environment where they are likely to come in contact with bacteria.
Fig. 3.
Degradation of 3OC12-HSL by cultured mammalian cells. Shown are the amounts of C4-HSL (gray bars) and 3OC12-HSL (black bars) remaining after a 1-h exposure to various mammalian cells in culture. Cell lines included are A549 cells, established from human bronchoalveolar carcinoma, CaCo-2 from human colon carcinoma, MDCK from canine kidney, CHO from Chinese hamster ovary, 293T from human embryonic kidney, HeLa from human cervix, COS-7 from monkey kidney, and primary cultures of human lung fibroblasts (HLF). Data are means ± SEM (n = 8 replicates).
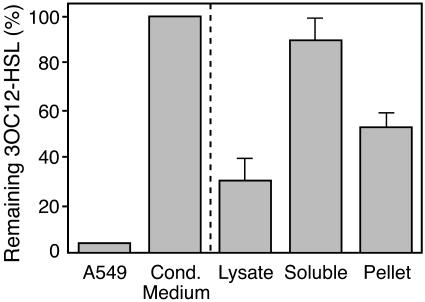
The 3OC12-HSL Degrading Activity Is Associated with Cell Membranes. To determine whether a cell-secreted factor inactivates 3OC12-HSL, we removed medium from monolayers of A549 cells and incubated it with 3OC12-HSL. One hour later, we recovered nearly all of the 3OC12-HSL from the conditioned medium, whereas none was recovered after addition to cells (Fig. 4). This finding suggests that 3OC12-HSL inactivation was cellular and not secreted into the medium in significant quantities.
Fig. 4.
Inactivation of 3OC12-HSL by A549 cell lysates. Shown is the amount of 3OC12-HSL remaining after a 1-h exposure to A549 cells; conditioned medium collected after a 1-h incubation on A549 cells (Cond. Medium); cell lysate clarified by low-speed centrifugation; and the supernatant or the particulate (Pellet) fraction of the cell lysate after ultracentrifugation. Values are the percentage of 3OC12-HSL remaining after 1 h compared to the initial concentration. Data are means ± SEM from three independent experiments.
To further localize 3OC12-HSL degradation, we disrupted A549 cells and incubated 3OC12-HSL with the cell lysate. Fig. 4 shows that the lysate contained 3OC12-HSL-degrading activity. In contrast, lysates prepared from CHO cells did not degrade 3OC12-HSL. Separation of the A549 lysate into soluble and particulate fractions showed that the majority of the 3OC12-HSL inactivating activity was in the cell-membrane-containing particulate fraction. Together, these studies suggest the degrading activity was present in the cells rather than secreted, and this activity was membrane-associated.
Lysates of A549 cells degraded 3OC12-HSL in a time-dependent manner (Fig. 5A), and the amount of 3OC12-HSL remaining after 1 h depended on the amount of A549 cell lysate added (Fig. 5B). Boiling the extract abolished 3OC12-HSL inactivation, suggesting a heat-sensitive factor (Fig. 5A). These results raise the possibility that 3OC12-HSL was degraded by an enzyme present in A549 cells.
Fig. 5.
The dependence of 3OC12-HSL degradation by A549 cell lysates on time and lysate dilution. (A) Time course of 3OC12-HSL degradation. 3OC12-HSL (10 μM) was added to A549 cell extracts for the indicated times. Data are percent of 3OC12-HSL remaining. The black circle shows the percent of 3OC12-HSL remaining in boiled A549 cell extract after 1 h. (B) A lysate dilution series (the incubation time was 1 h). Data are means ± SEM of three independent experiments; in some cases, error bars are hidden by symbols.
Discussion
In P. aeruginosa, 3OC12-HSL is a signal that regulates virulence (1, 4, 6, 8, 13). Interestingly, 3OC12-HSL also signals mammalian host cells. This quorum-sensing molecule increased production of cyclooxygenase-2 in human lung fibroblasts (32) and several proinflammatory chemokines, including IL-8, in human bronchiolar epithelial cells (33). 3OC12-HSL also induced T cell production of IFN-γ, which can activate macrophages and initiate a proinflammatory response (34). In addition, macrophages and neutrophils treated with 3OC12-HSL displayed morphologies and gene expression profiles characteristic of apoptosis (35). Our data indicate that mammalian cells have also developed the ability to inactivate this bacterial signal.
Because P. aeruginosa quorum-sensing signals regulate virulence, 3OC12-HSL inactivation could serve an important role in the innate defense against certain bacterial infections. Consistent with this hypothesis, transgenic expression of the bacterial acyl-HSL lactonase AiiA in tobacco plants attenuated leaf infections by Erwinia carotovora, a plant pathogen that uses quorum sensing for regulation of virulence. In addition, expressing recombinant AiiA in potatoes reduced Erwinia soft rot (22). The implication of these studies in plants is that degradation of quorum-sensing signals by eukaryotic cells protects the host from infection by attenuating virulence of the pathogen.
The experiments with recombinant plants have also suggested that administering agents that interfere with quorum sensing could be a valuable therapeutic strategy. Our data suggest that at least some mammalian tissues already possess such mechanisms. Despite this activity in human airway epithelia, significant levels of 3OC12-HSL have been measured in the sputum of P. aeruginosa-infected cystic fibrosis patients (36). Moreover, the concentrations of acyl-HSLs within the lungs of mice infected with P. aeruginosa were sufficient to activate promoters controlled by quorum-sensing signals in recombinant E. coli (37). In light of these studies, we speculate that inadequacy or a failure of acyl-HSL inactivation might contribute to the persistence of acyl-HSLs and biofilm production in chronic lung infections. Hence, augmenting the innate mechanisms that inactivate quorum-sensing signals or supplying new inactivating mechanisms could be of value in treating chronic infections.
Our study leaves unanswered the question of what process inactivates 3OC12-HSL. However, we speculate that an enzymatic activity is responsible. This hypothesis is consistent with several of our findings: inactivation was selective for signals with certain acyl side chains; the capacity for inactivation varied widely in different cell types; inactivation occurred in cell-free lysates and the ability of a lysate to inactivate 3OC12-HSL depended on the amount of lysate used; it was time dependent; and heating abolished the inactivation. Because bacterial enzymes that degrade acyl-HSL quorum-sensing signals are lactonases that open the HSL ring (22, 23, 25) or acylases that cleave the acyl side chain from the HSL ring (24, 26), perhaps a mammalian cell factor will operate by one or both of these processes.
Acknowledgments
We thank Philip Karp, Pary Weber, and Tamara Nesselhauf for excellent assistance. This work was supported by National Institute of General Medicine Grant GM59026 (to E.P.G.), National Heart, Lung, and Blood Institute Grant HL61234 (to J.Z. and M.J.W), and the W. M. Keck Foundation (E.P.G.). The cell cultures were provided by the University of Iowa In Vitro Models and Cell Culture Core, which was supported in part by National Heart, Lung, and Blood Institute Grants HL61234 and HL51670, National Institutes of Diabetes and Digestive and Kidney Diseases Grant DK54759, and the Cystic Fibrosis Research Development Program. M.J.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: CHO, Chinese hamster ovary; EMEM, Eagle's minimal essential medium; HSL, homoserine lactone.
References
- 1.Fuqua, C. & Greenberg, E. P. (2002) Nat. Rev. Mol. Cell Biol. 3, 685-695. [DOI] [PubMed] [Google Scholar]
- 2.Fuqua, C., Parsek, M. R. & Greenberg, E. P. (2001) Annu. Rev. Genet. 35, 439-468. [DOI] [PubMed] [Google Scholar]
- 3.Parsek, M. R. & Greenberg, E. P. (2000) Proc. Natl. Acad. Sci. USA 97, 8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J. & Salmond, G. P. (2001) FEMS Microbiol. Rev. 25, 365-404. [DOI] [PubMed] [Google Scholar]
- 5.Whitehead, N. A., Byers, J. T., Commander, P., Corbett, M. J., Coulthurst, S. J., Everson, L., Harris, A. K., Pemberton, C. L., Simpson, N. J., Slater, H., et al. (2002) Antonie Leeuwenhoek 81, 223-231. [DOI] [PubMed] [Google Scholar]
- 6.Smith, R. S. & Iglewski, B. H. (2003) Curr. Opin. Microbiol. 6, 56-60. [DOI] [PubMed] [Google Scholar]
- 7.Welsh, M. J., Ramsey, B. W., Accurso, F. & Cutting, G. R. (2001) in Metabolic and Molecular Basis of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B. & Vogelstein, B. (McGraw-Hill, New York), 8th Ed., Vol. 3, Part 21, pp. 5121-5189. [Google Scholar]
- 8.De Kievit, T. R. & Iglewski, B. H. (2000) Infect. Immun. 68, 4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson, J. P., Gray, K. M., Passador, L., Tucker, K. D., Eberhard, A., Iglewski, B. H. & Greenberg, E. P. (1994) Proc. Natl. Acad. Sci. USA 91, 197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson, J. P., Passador, L., Iglewski, B. H. & Greenberg, E. P. (1995) Proc. Natl. Acad. Sci. USA 92, 1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latifi, A., Foglino, M., Tanaka, K., Williams, P. & Lazdunski, A. (1996) Mol. Microbiol. 21, 1137-1146. [DOI] [PubMed] [Google Scholar]
- 12.Pesci, E. C. & Iglewski, B. H. (1997) Trends Microbiol. 5, 132-134. [DOI] [PubMed] [Google Scholar]
- 13.Donabedian, H. (2003) J. Infect. 46, 207-214. [DOI] [PubMed] [Google Scholar]
- 14.Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W. & Greenberg, E. P. (1998) Science 280, 295-298. [DOI] [PubMed] [Google Scholar]
- 15.Schuster, M., Lostroh, C. P., Ogi, T. & Greenberg, E. P. (2003) J. Bacteriol. 185, 2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I. & Iglewski, B. H. (2003) J. Bacteriol. 185, 2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boman, H. G. (2003) J. Int. Med. 254, 197-215. [DOI] [PubMed] [Google Scholar]
- 18.Ganz, T. (2003) Nat. Rev. Immunol. 3, 710-720. [DOI] [PubMed] [Google Scholar]
- 19.Hancock, R. E. W. & Scott, M. G. (2000) Proc. Natl. Acad. Sci. USA 97, 8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnaud, S. & Evans, R. W. (2003) Mol. Immunol. 40, 395-405. [DOI] [PubMed] [Google Scholar]
- 21.Singh, P. K., Parsek, M. R., Greenberg, E. P. & Welsh, M. J. (2002) Nature 417, 552-555. [DOI] [PubMed] [Google Scholar]
- 22.Dong, Y. H., Wang, L. H., Xu, J. L., Zhang, H. B., Zhang, X. F. & Zhang, L. H. (2001) Nature 411, 813-817. [DOI] [PubMed] [Google Scholar]
- 23.Dong, Y. H., Xu, J. L., Li, X. Z. & Zhang, L. H. (2000) Proc. Natl. Acad. Sci. USA 97, 3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Y. H., Xu, J. L., Hu, J., Wang, L. H., Ong, S. L., Leadbetter, J. R. & Zhang, L. H. (2003) Mol. Microbiol. 47, 849-860. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, H. B., Wang, L. H. & Zhang, L. H. (2002) Proc. Natl. Acad. Sci. USA 99, 4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leadbetter, J. R. & Greenberg, E. P. (2000) J. Bacteriol. 182, 6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karp, P. H., Moninger, T. O., Weber, S. P., Nesselhauf, T. S., Launspach, J. L., Zabner, J. & Welsh, M. J. (2002) Methods Mol. Biol. 188, 115-137. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer, A. L., Hanzelka, B. L., Eberhard, A. & Greenberg, E. P. (1996) J. Bacteriol. 178, 2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passador, L., Tucker, K. D., Guertin, K. R., Journet, M. P., Kende, A. S. & Iglewski, B. H. (1996) J. Bacteriol. 178, 5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsek, M. R., Schaefer, A. L. & Greenberg, E. P. (1997) Mol. Microbiol. 26, 301-310. [DOI] [PubMed] [Google Scholar]
- 31.Yates, E. A., Philipp, B., Buckley, C., Atkinson, S., Chhabra, S. R., Sockett, R. E., Goldner, M., Dessaux, Y., Camara, M., Smith, H. & Williams, P. (2002) Infect. Immun. 70, 5635-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, R. S., Kelly, R., Iglewski, B. H. & Phipps, R. P. (2002) J. Immunol. 169, 2636-2642. [DOI] [PubMed] [Google Scholar]
- 33.DiMango, E., Zar, H. J., Bryan, R. & Prince, A. (1995) J. Clin. Invest. 96, 2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, R. S., Harris, S. G., Phipps, R. P. & Iglewski, B. H. (2002) J. Bacteriol. 184, 1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tateda, K., Ishii, Y., Horikawa, M., Matsumoto, T., Miyairi, S., Pechere, J. C., Standiford, T. J., Ishiguro, M. & Yamaguchi, K. (2003) Infect. Immun. 71, 5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erickson, D. L., Endersby, R., Kirkham, A., Stuber, K., Vollman, D. D., Rabin, H. R., Mitchell, I. & Storey, D. G. (2002) Infect. Immun. 70, 1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, H., Song, Z., Hentzer, M., Andersen, J. B., Heydorn, A., Mathee, K., Moser, C., Eberl, L., Molin, S., Høiby, N. & Givskov, M. (2000) Microbiology 146, 2481-2493. [DOI] [PubMed] [Google Scholar]