Abstract
The existence of a pathway for salvaging the coenzyme B12 precursor dicyanocobinamide (Cbi) from the environment was established by genetic and biochemical means. The pathway requires the function of a previously unidentified amidohydrolase enzyme that converts adenosylcobinamide to adenosylcobyric acid, a bona fide intermediate of the de novo coenzyme B12 biosynthetic route. The cbiZ gene of the methanogenic archaeon Methanosarcina mazei strain Göl was cloned, was overproduced in Escherichia coli, and the recombinant protein was isolated to homogeneity. HPLC, UV-visible spectroscopy, MS, and bioassay data established adenosylcobyric as the corrinoid product of the CbiZ-catalyzed reaction. Inactivation of the cbiZ gene in the extremely halophilic archaeon Halobacterium sp. strain NRC-1 blocked the ability of this archaeon to salvage Cbi. cbiZ function restored Cbi salvaging in a strain of the bacterium Salmonella enterica, whose Cbi-salvaging pathway was blocked. The salvaging of Cbi through the CbiZ enzyme appears to be an archaeal strategy because all of the genomes of B12-producing archaea have a cbiZ ortholog. Reasons for the evolution of two distinct pathways for Cbi salvaging in prokaryotes are discussed.
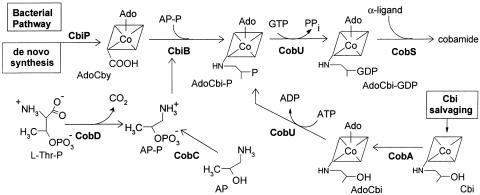
Among vitamins and coenzymes, cobamides (e.g., coenzyme B12) are unique for their structural complexity. Not surprisingly, de novo synthesis of cobamides requires a great deal of genetic information, which is only found in prokaryotes (1-3). The majority of the work on B12 biosynthesis has been performed in bacteria (4-7). In addition to a de novo pathway, bacteria also possess a conserved salvaging pathway for the precursor dicyanocobinamide (Cbi), which is a stable precursor, but is not a true intermediate of the de novo pathway (Fig. 1 and refs. 8 and 9). In bacteria, Cbi salvaging requires attachment of the upper ligand 5′-deoxyadenosine to yield adenosylcobinamide (AdoCbi) (10-13), followed by phosphorylation of AdoCbi to yield AdoCbi-phosphate (AdoCbi-P), which is a true intermediate of the de novo biosynthetic pathway (8). The latter reaction is catalyzed by the kinase activity of a bifunctional ATP:AdoCbi kinase, GTP:AdoCbi-GDP guanylyltransferase enzyme (CobU in Salmonella enterica), which is conserved in cobamide-producing bacteria (14-18).
Fig. 1.
Late steps of cobamide biosynthesis in bacteria. Intermediates are indicated below structures. AP-P, AP phosphate; CobS, cobalamin (5′-P) synthase; CobC, l-threonine kinase; CobA, ATP:co(I)rrinoid adenosyltransferase.
It is clear that some archaea require and synthesize cobamides to live. However, our understanding of how archaea salvage Cbi is limited (8, 19). Analysis of the available archaeal genome sequences revealed the absence of an ortholog of the bacterial bifunctional CobU enzyme, and we recently reported the identification of the gene encoding the nonorthologous replacement of only the GTP:AdoCbi-GDP guanylyltransferase activity in archaea (8, 19). To the best of our knowledge, there have been no reports of ATP:AdoCbi kinase activity in any archaeon. More recent work from our laboratory showed that Halobacterium sp. strain NRC-1 can salvage Cbi, and that Cbi salvaging requires the activity of the cobyric acid synthase (CbiB) enzyme that catalyzes the last step of the de novo corrin ring biosynthetic pathway (20). These findings were consistent with the existence of an alternative Cbi-salvaging pathway in which AdoCbi is converted to adenosylcobyric acid (AdoCby) by a previously unidentified amidohydrolase enzyme. Here, we report the identification of the cbiZ gene as the one encoding the AdoCbi amidohydrolase in the methanogenic archaeon Methanosarcina mazei strain Göl, and in the extremely halophilic archaeon Halobacterium sp. strain NRC-1.
Materials and Methods
Strains and Plasmids. Descriptions of the genotypes of strains and plasmids used in this work, as well as detailed descriptions of plasmid constructions, can be found in Supporting Methods and Table 1, which are published as supporting information on the PNAS web site. A diagram of the Halobacterium sp. NRC-1 DNA included in the most relevant plasmids are included in Fig. 8, which is published as supporting information on the PNAS web site.
Chemicals, Growth Media, Growth Conditions, and Assessment of Viability. Except where noted, all chemicals used in this work were high-purity, commercially available compounds. When added to the medium, corrinoids were present at 100 pM for Halobacterium sp. strain NRC-1 studies and 15 nM for S. enterica studies. All corrinoids were added in their cyano form. AdoCbi was synthesized as described (8); (CN)2Cbi and CNB12 [also known as CNCbl (cobalamin)] were purchased from Sigma; (CN)2Cbi-GDP was synthesized as described (15); (CN)2Cby was a gift from P. Renz (Universität-Hohenheim, Stuttgart, Germany); 5-fluoroorotic acid was purchased from Zymo Research (Orange, CA); and mevinolin was purchased from LKT Laboratories (St. Paul). (R)-1-amino-2-propanol (AP) was purchased from Sigma.
Bacterial Strains Used for Protein Overproduction. Overproduction of native M. mazei CbiZ protein was performed in Escherichia coli strain BL21(λ DE3)-RIL (Stratagene). Overproduction of the CbiZ-chitin-binding protein fusion protein was performed in E. coli strain ER2566 (New England Biolabs).
Growth Studies. Cultures of strains of Halobacterium sp. strain NRC-1 and S. enterica were grown as described (20). The only modification was the addition of 5,6-dimethylbenzimidazole (3 μM) and AP (10 mM) to the S. enterica medium. All plasmids introduced into S. enterica were first passed through a restriction-deficient strain (21).
Generation of Cell-Free Extracts Enriched with CbiZ Protein. The wild-type allele of M. mazei cbiZ was overproduced from the plasmid pMmCBIZ1 in the overproducing strain of E. coli BL21(λ DE3)-RIL (Stratagene). An overnight, 5-ml culture of the overproduction strain carrying pMmCBIZ1 was grown in LB broth containing ampicillin (100 μg/ml) and chloramphenicol (20 μg/ml). The overnight culture was used to inoculate 400 ml of fresh medium. The culture was grown at 30°C with shaking to a cell density of OD650 = 0.55, at which point isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 500 μM. After the addition of isopropyl-β-d-thiogalactopyranoside, the culture was incubated under the same conditions for an additional 3 h. Cells were harvested by centrifugation for 20 min at 10,000 × g at 4°C by using a Beckman-Coulter J21 centrifuge. Cells were resuspended in 4 ml of 50 mM Hepes buffer, pH 7.5, containing NaCl (100 mM) and DTT (5 mM). Cells were broken by two passes through a French press at ≈1 × 105 kPa by using a chilled pressure cell. Cell lysate was clarified by centrifugation for 30 min at 18,000 × g at 4°C using a Beckman-Coulter Avanti J-25I centrifuge. Soluble extract was dialyzed against 1 liter of the resuspension buffer (1:250) in a Slidalyzer (Pierce) cassette (molecular weight cutoff of 10,000) with two buffer changes. As negative control, the same procedure was used to generate cell-free extract from cells harboring the control plasmid pT7-7.
In Vitro CbiZ Amidohydrolase Activity Assay. AdoCbi amidohydrolase activity assays were performed in 100-μl volumes containing 65 μg of cell-free extract protein, 50 mM Na-Hepes buffer, pH 7.5, containing DTT (5 mM) and AdoCbi (30 μM). Reactions were incubated at 37°C for 2 h in dim light and were heat-inactivated at 65°C for 20 min. As a negative control, heat-inactivated cell-free extract was prepared by incubating the extract at 65°C for 20 min. When the assay was performed with highly purified CbiZ, the reactions contained 150 μM AdoCbi, and 4 μg of CbiZ protein was used in lieu of crude cell-free extract.
Detection of Cby. The presence of Cby in reaction mixtures was assessed by means of a bioassay. For this purpose, S. enterica strain JE824 (metE205 cobU330) carrying plasmid pCOBY10 (cobY+) was used as indicator strain. Five microliters of 1:10 dilutions of a reaction mixture was spotted onto an agar overlay containing cells of strain JE824 carrying plasmid pCOBY10. As controls, 5 pmol of authentic (CN)2Cbi and (CN)2Cby were also spotted onto the overlay. Minimal no-carbon E medium was supplemented with glucose, MgSO4, 1,2-propanediol (to induce transcription of the cbi operon) (22), and ampicillin (25 μg/ml). Medium was incubated aerobically at 37°C for 24 h. Under aerobic conditions, de novo corrin ring biosynthesis is blocked in S. enterica, hence making growth dependent on corrinoid intermediates. Cell growth around the application sites indicated the presence of Cby in reaction mixtures.
Overproduction and Purification of Recombinant CbiZ Protein. M. mazei CbiZ protein fused to a C-terminal chitin-binding protein tag was overproduced by using plasmid pMmCBIZ5 in the overproducing strain of E. coli ER2566 (Stratagene). One milliliter of an overnight culture of the overproducing strain carrying plasmid pMmCBIZ5 in LB broth containing ampicillin (100 μg/ml) was used to inoculate two 500-ml batches of fresh medium. Cultures were grown at 30°C with shaking to a cell density of OD600 = 0.55, at which point isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 300 μM. The isopropyl-β-d-thiogalactopyranoside-containing culture was incubated under the same conditions for an additional 3 h. Cells were harvested by centrifugation for 20 min at 8,000 × g at 4°C. Cells were resuspended in 20 ml of 20 mM Hepes buffer, pH 7.5, containing NaCl (500 mM), EDTA (0.1 mM), and Triton X-100 (0.1% vol/vol). Cells were broken by two passes through a French press at ≈1.6 × 105 kPa by using a chilled pressure cell. Cell lysate was clarified by centrifugation for 30 min at 18,000 × g at 4°C. CbiZ protein was purified on chitin beads (New England Biolabs) according to the manufacturer's instructions. The chitin tag was removed from the protein by soaking the chitin beads in the same buffer containing 30 mM DTT for 20 h at 4°C. After purification, 5 ml of the enzyme was dialyzed by using snakeskin-pleated dialysis tubing (molecular weight cutoff of 10,000 (Pierce) at 4°C against 1 liter (1:200) of 50 mM Hepes buffer, pH 7.5, containing NaCl (100 mM) and DTT (5 mM). After two buffer changes, purity of the protein was assessed by SDS/PAGE (23) after staining with Coomasie brilliant Blue R-250 (24). CbiZ protein was stored at -80°C in this buffer after flash-freezing with liquid N2.
Corrinoid Analysis. Corrinoids present in the reaction mixture were derivatized to their cyano form by adding 10 μl of 100 mM KCN and incubating in the light at room temperature for 30 min. Samples were filtered by using Corning Spin-X centrifuge filters. Corrinoids were separated by using a Waters HPLC system equipped with a Luna (Phenomenex) 5-μ C18 column (150 × 4.6 mm) developed with a modification of the system reported elsewhere (25) at a flow rate of 1 ml/min-1. The column was equilibrated with a buffer system containing 98% A/2% B. One minute after injection of the sample, the column was developed for 5 min with a linear gradient until the final composition of the buffer system was 75% A/25% B. A second linear gradient of 15 min developed the column to a final buffer composition of 65% A/35% B; solvent A = 100 mM phosphate buffer, pH 6.5, 10 mM KCN; solvent B = 100 mM phosphate buffer, pH 8.0, 10 mM KCN:acetonitrile (1:1). Corrinoids were identified by their spectra by using a Waters photodiode array detector. Authentic (CN)2Cbi and (CN)2Cby were used as standards.
MS. The HPLC-purified product of the CbiZ reaction was dried under vacuum by using a Savant concentrator, was resuspended in 5 ml of ddH2O, and was loaded onto a 1-ml LiChroprep RP-8 (EM Separations) column equilibrated with ddH2O. The column was washed with 30-ml of ddH2O, and corrinoid was eluted with 5 ml of methanol. The eluted sample was dried under vacuum, resuspended in 1 ml of ddH2O, filtered in Spin-X filters, and again dried under vacuum. The sample was submitted for analysis to the MS facility at the University of Wisconsin-Madison Biotechnology Center. The mass spectrum was obtained by using a Bruker Daltronics (Billerica, MA) BIFLEX III matrix-assisted laser desorption/ionization-time-of-flight mass spectrometer.
Results
Identification of the Gene Encoding the Archaeal AdoCbi Amidohydrolase Enzyme. A comparative genomics approach was used to identify genes that could encode AdoCbi amidohydrolase activity (i.e., putative hydrolases or conserved hypothetical protein ORFs near known Cob genes). This approach allowed for the identification of an uncharacterized conserved protein sometimes flanking identified ORFs involved in the late steps of cobamide biosynthesis. ORF Vng1583C (gi 15790554) (hereafter referred to as cbiZ) of Halobacterium sp. strain NRC-1 is the last gene of a putative four-gene operon (cobSYDcbiZ) encoding orthologs of bacterial functions known to catalyze late steps of coenzyme B12 synthesis (Fig. 8).
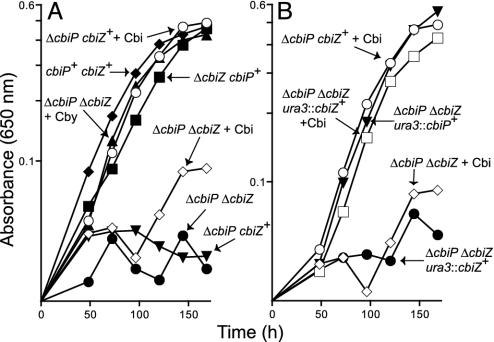
cbiZ (ORF Vng1583C) Function Is Required for Cbi Salvaging in Halobacterium. To determine whether CbiZ was required for Cbi salvaging, a derivative of strain JE6738 (ΔcbiP) was constructed. Strain JE6812 (ΔcbiP Δ cbiZ) carried an in-frame deletion of the gene encoding the Cbi acid synthase (CbiP), the enzyme that catalyzes the second-to-last step of the corrin ring biosynthesis. A mutation in cbiP blocks de novo corrin ring synthesis in Halobacterium (20), thus demanding salvaging of precursors present in the medium. Strain JE6812 (ΔcbiP ΔcbiZ) was tested for its ability to salvage different corrinoids. Like strain JE6738 (ΔcbiP cbiZ+), strain JE6812 (ΔcbiP ΔcbiZ) failed to grow in chemically defined medium lacking corrinoids (Fig. 2A, •), and the addition of (CN)2Cby [a derivatized de novo pathway intermediate (Fig. 1)] restored wild-type growth (Fig. 2A, ▴; doubling time = 27 h). Addition of either (CN)2Cbi-GDP [a derivatized de novo pathway intermediate (Fig. 1)] or CNB12 also restored wild-type growth of both strains (data not shown). However, whereas the addition of (CN)2Cbi to the medium allowed wild-type growth of JE6738 (ΔcbiP) (doubling time = 24 h), it did not support growth of strain JE6812 (ΔcbiP ΔcbiZ) (Fig. 2 A, ○ vs. ⋄). These data established a strong correlation between the loss of cbiZ function and a block in Cbi salvaging under conditions that demanded salvaging of this precursor.
Fig. 2.
Nutritional analyses of Halobacterium strains. Shown is B12-dependent growth of Halobacterium strains in defined liquid medium at 37°C. Strains are indicated by their genotype. Corrinoids added to the medium are indicated next to each genotype. Strains used were JE6738, ΔcbiP cbiZ+; JE6811, cbiP+ ΔcbiZ; JE6812, ΔcbiP ΔcbiZ; JE7002, ΔcbiP ΔcbiZ ura3::cbiP+; and JE7210, ΔcbiP ΔcbiZ ura3::cbiZ+. All corrinoids were present in the medium at 100 pM.
cbiZ Function Is Necessary and Sufficient for Cbi Salvaging in Halobacterium. The observed block in Cbi salvaging in strain JE6812 (ΔcbiP ΔcbiZ) was corrected when a wild-type allele of cbiZ was reintroduced into the chromosome. Strain JE7210 (ΔcbiP ΔcbiZ ura3::cbiZ+) grew in chemically defined medium supplemented with Cbi with a doubling time of 22 h (Fig. 2B, □), but did not grow without corrinoids due to the lack of cbiP function (Fig. 2B, •). Hence, cbiZ+ was necessary and sufficient to restore Cbi salvaging in the mutant strain. To demonstrate that cbiZ function was not required for de novo corrin ring synthesis, the de novo pathway of strain JE6812 (ΔcbiP ΔcbiZ) was restored by reintroducing a wild-type cbiP allele into the chromosome. Strain JE7002 (ΔcbiP ΔcbiZ ura3::cbiP+) grew in medium without any corrinoids (Fig. 2B, ▾; doubling time = 27 h). It was concluded that cbiP function was necessary and sufficient to restore de novo cobamide synthesis in strain JE7002 even in the absence of cbiZ.
cbiZ Is Not Involved in de Novo Biosynthesis in Halobacterium. To determine whether strain JE6811 (ΔcbiZ) was deficient in de novo cobamide biosynthesis, growth was assessed in chemically defined liquid medium under conditions where cobamides were essential for growth. The kinetics of growth of strain JE6811 (ΔcbiZ) in chemically defined medium lacking corrinoids was very similar to that of the wild-type strain (Fig. 2A, ▪ vs. ♦ with doubling times of 30 and 34 h, respectively.
An Archaeal cbiZ Gene Restores Cbi Salvaging in an S. enterica Mutant Strain. To lend support to the conclusion that CbiZ was involved in Cbi salvaging in archaea, the ability of an archaeal cbiZ+ gene to complement an S. enterica strain defective in Cbi salvaging was tested. In previous experiments in our laboratory, Halobacterium sp. strain NRC-1 genes have failed to complement S. enterica mutants, presumably because of the severe difference in internal salt concentrations. Instead, we focused on the archaeal ortholog of cbiZ [ORF Mm0173 (gi 21226275)] in M. mazei Göl, a mesophilic, methanogenic archaeon whose genes have been successfully expressed in S. enterica (20). The CbiZ-dependent Cbi salvaging pathway is also expected to exist in M. mazei Göl, because it is predicted to have orthologs to proteins required for this pathway (20, 26).
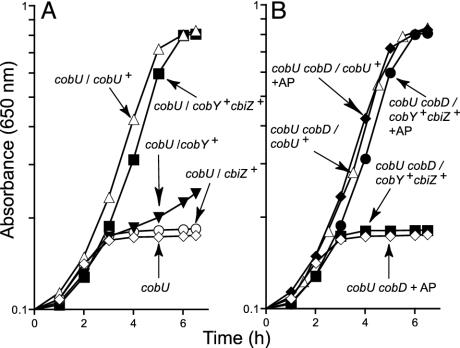
S. enterica strain JE824 (metE205 cobU330) was used to test for MmcbiZ+ function. Growth of this strain depends on cobamide-dependent methylation of homocysteine by the cobamide-dependent methionine synthase enzyme (27). Because the de novo pathway in S. enterica is inactive under aerobic conditions, growth in defined medium lacking methionine requires corrinoid salvaging. Mutation cobU330 eliminates both the ATP:AdoCbi kinase and the GTP:AdoCbi-P guanylyltransferase activities and blocks de novo corrin ring biosynthesis and Cbi salvaging in this bacterium (28). To restore CbiZ-, Cbi-dependent growth of strain JE824, an NTP:AdoCbi-P nucleotidyltransferase was provided. For this purpose, the M. mazei cobY+ gene was introduced into strain JE824. Plasmid pJO52 (cobU+) was used as positive control, whereas plasmid pT7-7 was used as vector-only, negative control. Plasmids pMmCBIZ1 (cbiZ+), pMmCBIZ2, (cbiZ+ cobY+), or pCOBY10 (cobY+) were introduced into strain JE824. Resulting strains were grown aerobically in medium supplemented with Cbi. Under the conditions used, growth depended on Cbi salvaging. Cbi-dependent growth was only observed when either S. enterica cobU+ (Fig. 3A, ▵) or M. mazei cbiZ+ and cobY+ were provided in trans (Fig. 3A, ▪). These data supported the conclusion that cbiZ function was required for Cbi salvaging.
Fig. 3.
Nutritional analyses of S. enterica strains. Cbi-salvaging-dependent growth of S. enterica strains in chemically defined liquid medium at 37°C. (A) All strains were derivatives of strain JE824 (metE205 cobU330). (B) All strains were derivatives of strain JE6894 (metE205 cobU330 cobD1272). Strains are indicated by their genotype. Plasmids used were pT7-7, control vector; pJO52, cobU+; pCOBY10, cobY+; pMmCBIZ1, cbiZ+; and pMmCBIZ2, cbiZ+ cobY+. In all cases, (CN)2Cbi was added to 15 nM.
cbiZ Restores Cbi Salvaging Via a Pathway Different from the One Found in Bacteria. Although CbiZ restored Cbi salvaging in strain JE824, these results did not shed any insights into how Cbi was salvaged. To identify the entry point for Cbi, we used an S. enterica strain carrying a mutation in the l-threonine decarboxylase (CobD) enzyme. A block at this step in the pathway would not affect Cbi salvaging if the entry point was AdoCbi-P, as expected if CobU were functional (Fig. 1). If, however, Cbi was converted to an earlier intermediate, CobD function would be required for Cbi salvaging.
Plasmid pMmCBIZ2 (cobY+ cbiZ+) failed to restore Cbi salvaging in strain JE6984 (metE205 cobU330 cobD1272) (Fig. 3B, ▪); the control strain carrying plasmid pJO52 (cobU+) salvaged Cbi (Fig. 3B, ▵). Cbi salvaging by the strain carrying plasmid pMmCBIZ2 was restored when AP was added to the medium (Fig. 3B, •). This result was expected because the addition of AP compensates for the lack of CobD function (9, 29). These results were consistent with the idea that CbiZ-dependent Cbi salvaging occurs via an alternative pathway that converts Cbi to an intermediate before AdoCbi-P.
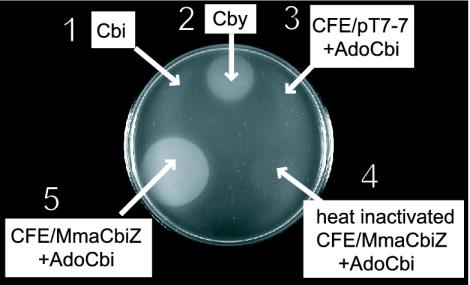
Cell-Free Extracts Enriched for CbiZ Contain an AdoCbi Amidohydrolase Activity. We considered the possibility that CbiZ was an amidohydrolase enzyme that converted Cbi to Cby by removing the AP propanol moiety of Cbi. To test this idea, M. mazei CbiZ was overproduced from plasmid pMmCBIZ1 (cbiZ+) in E. coli overproducing strain BL21(λDE3)-RIL. AdoCbi was incubated with cell-free extract enriched for CbiZ, and amidohydrolase activity was measured by using a bioassay that detected the presence of Cby in the reaction mixture. If Cbi were converted to Cby by CbiZ, the cobamide auxotrophy of strain JE824 carrying plasmid pMmCOBY10 (cobY+) would be corrected, resulting in growth around the application point of the sample. Cby synthesis was only detected in reaction mixtures that contained cell-free extract enriched with CbiZ protein (Fig. 4, spot 5), suggesting the CbiZ protein had AdoCbi amidohydrolase enzyme activity.
Fig. 4.
Bioassay for the detection of Cby synthesized in vitro by cell-free extract of E. coli cells overproducing CbiZ protein. Shown is the response of indicator strain JE824 (metE205 cobU330) with plasmid pCOBY10 (cobY+) to 5 μl of 1:10 dilutions of the deproteinated reaction mixtures and 5 pmol of standards. Growth around the area of application indicates the presence of Cby in the reaction mixture. Shown are the (CN)2Cbi standard (spot 1), the (CN)2Cby standard (spot 2), vector-only control (spot 3), heat-inactivated control (spot 4), and the complete reaction mixture (spot 5).
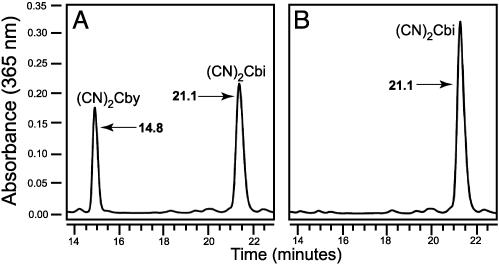
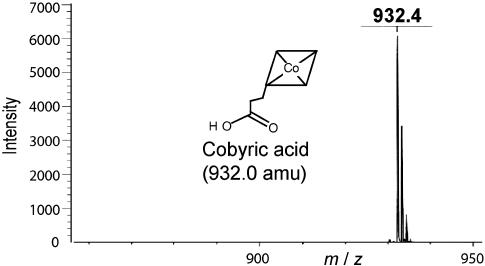
CbiZ-Dependent Conversion of Cbi to Cby. The CbiZ protein was purified to homogeneity by using a C-terminal chitin-binding protein tag, which was subsequently cleaved (data not shown). Purified CbiZ enzyme (>95 homogeneity by SDS/PAGE) was tested for AdoCbi amidohydrolase activity by incubating the protein with AdoCbi and monitoring the formation of the product Cby by using HPLC protocols described above. A signal for Cby was clearly detectable in the complete reaction mixture (Fig. 5A), but was absent when CbiZ protein was inactivated before incubation with the substrate (Fig. 5B). Under these conditions, a specific activity of 6.4 μ mol per min per mg of protein was calculated. When (CN)2Cbi was used as substrate, the specific activity was reduced 3-fold (2.1 μmol per min per mg of protein). To demonstrate the reduced activity was not due to inhibitory effects of the CN anion, substrate levels of KCN were added to the reaction mixture. KCN did not significantly affect the specific activity of the protein (data not shown). HPLC-purified reaction product was analyzed by MS. The signal with an m/z = 932.4 signal that was consistent with the expected molecular mass of Cby without any ligands (932.0 atomic mass units; Fig. 6). A bioassay confirmed the presence of Cby in the HPLC-purified peak (data not shown). These results confirmed that Cby was a product of the CbiZ reaction; i.e., that CbiZ had AdoCbi amidohydrolase enzyme activity.
Fig. 5.
HPLC analysis of the CbiZ reaction. Chromatograms of components of reaction mixture monitored at 365 nm (A) and the heat-inactivated control (B). Numbers represent times (in minutes) of elution after injection.
Fig. 6.
MS analysis of the product of the CbiZ reaction. Shown is the matrix-assisted laser desorption/ionization-time-of-flight MS anaysis of the HPLC-purified product of the CbiZ reaction. The signal with the m/z value of 932.4 was consistent with the molecular mass of Cby (without ligands) where z = +1. No significant signals were detected above an m/z value of 950.
Discussion
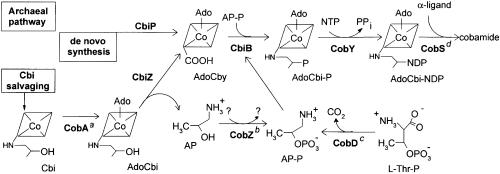
Archaea and Bacteria Salvage Cbi Via Two Distinct Pathways. The genetic and biochemical evidence reported here and elsewhere (20) supports the conclusion that prokaryotes have evolved at least two distinct pathways for salvaging the precursor Cbi from the environment. Information currently available from genome databases suggests that these Cbi-salvaging pathways evolved and remained segregated in separate domains of life; i.e., in the Archaea or the Bacteria. Both pathways accomplish the same goal, which is to convert AdoCbi to AdoCbi-P, a true intermediate of the de novo biosynthetic pathway (8, 9). The differences between the bacterial and archaeal Cbi salvaging pathways are illustrated in Figs. 1 and 7. The chief difference is the point of entry for AdoCbi. In this model, we assumed that archaea convert Cbi to AdoCbi, however, the identity of the ATP:co(I)rrinoid adenosyltransferase in archaea has yet to be established experimentally. At this point, it is unclear whether the substrate for CbiZ needs to be adenosylated in the cell. Under the in vitro conditions described in this paper, it is clear that (CN)2Cbi can be used as a substrate by CbiZ and the activity is only reduced 3-fold. Further characterization of the CbiZ protein may provide insight to when the corrin ring is adenosylated during salvaging in archaea.
Fig. 7.
Late steps of cobamide biosynthesis in archaea. Intermediates are indicated below structures. CobY, NTP:AdoCbi-P nucleotidyltransferase. a, The putative archaeal ortholog to the bacterial CobA protein. b, CobZ is the archaeal nonorthologous replacement of the bacterial CobC protein (C. L. Zayas, J.D.W., and J.C.E.-S., unpublished results). c, The archaeal CobD is the ortholog to the bacterial CobD protein (C. L. Zayas, J.D.W., and J.C.E.-S., unpublished results).d, The archaeal CobS is the ortholog to the bacterial CobS protein (32).
Cobamide-producing bacteria evolved a conserved multifunctional enzyme that can use AdoCbi as substrate and convert it to AdoCbi-P (CobU in S. enterica) in a single catalytic step (Fig. 1). Archaea, on the other hand, convert AdoCbi to AdoCbi-P in two steps. First, the amidohydrolase activity of CbiZ cleaves off the aminopropanol moiety of AdoCbi yielding AdoCby; second, AdoCby is converted to AdoCbi-P by the action of the AdoCbi-P synthase (CbiB) enzyme (Fig. 7). Results from nutritional analysis of cbiP and cbiB mutants of Halobacterium sp. strain NRC-1 and complementation studies of S. enterica mutants unable to salvage Cbi using archaeal genes (20), strongly support the Cbi salvaging pathway delineated in Fig. 7 for archaea. Here and elsewhere (8, 19, 20), we have shown the existence of this pathway in euryarchaeotes. Whether this pathway is present in other archaea needs to de investigated. All available archaeal genome sequences contain orthologs of CbiZ and other proteins known or predicted to be required for the salvaging of Cbi (CobADSTY and CbiB), making it likely that the CbiZ-dependent Cbi-salvaging pathway is conserved among all archaea.
Although the bacterial CobU and the archaeal CbiZ enzymes are both used by cells to salvage Cbi, the enzymes share no sequence similarity, and in fact CbiZ does not share homology to any previously characterized proteins and contains no obvious motifs. The two enzymes, however, use AdoCbi as substrate. It will be interesting to see what, if any, structural similarities exist between how the two enzymes bind AdoCbi. No evidence for an ATP requirement was obtained. The structural analysis of the CbiZ protein warrants future study.
Although archaea and bacteria seem to have separate pathways for the salvaging of Cbi, putative orthologs of the CbiZ protein exist in Bacillus halodurans (30) and Bacillus subtilis (31). In both organisms, the CbiZ protein appears to be fused to the C terminus of the BtuD protein, the ATP-binding component of the B12 ABC-transport system. Because the CbiZ enzyme appears to be primarily a Cbi-salvaging function in archaea, a close association with the transport system should not be surprising. What is intriguing, however, is the role that CbiZ may be playing in B. halodurans, especially because this bacterium already has a putative ortholog to the bacterial ATP:AdoCbi kinase enzyme used by B12-producing bacteria to salvage Cbi. In the case of B. subtilis, the role of CbiZ is even more obscure because this bacterium lacks any of the B12 biosynthetic enzymes. In the absence of other Cob enzymes, Cbi salvaging would be unlikely. CbiZ may be playing a different role in these bacteria.
Why Are Two Cbi-Salvaging Pathways Involved? It is unclear what selective pressures directed the evolution of two different Cbi-salvaging pathways in prokaryotes, and why these pathways were segregated to either bacteria or archaea. One could speculate that prokaryotes that constitutively express genes encoding de novo biosynthetic enzymes might have evolved the amidohydrolase route, in response to the stability of the intermediate to which AdoCbi would be converted. It is possible that AdoCbi-P (the result of a hypothetical kinase) would not be stable enough for the next enzyme of the pathway to use it as substrate, whereas AdoCby, the product of CbiZ, could be.
Prokaryotes that conditionally express the corrin ring biosynthetic functions, but constitutively express functions required for the assembly of the nucleotide loop (e.g., S. enterica) face a different problem. Under some conditions, these organisms could find Cbi in the environment, but whether such growth conditions were not conducive for the expression of the AdoCbi-P synthase (CbiB) enzyme (e.g., presence of oxygen), the organism would be unable to make coenzyme B12 from AdoCbi, whether salvaging of the latter depended on the activity of an amidohydrolase enzyme. This problem would be circumvented through the evolution of a kinase enzyme to convert AdoCbi to AdoCbi-P by direct phosphorylation. The fact that such AdoCbi kinase activity has only been found in a protein that also has the next catalytic activity (GTP:AdoCbi-P guanylyltransferase) of the pathway suggests that AdoCbi-P may be too unstable to be released from the enzyme.
Supplementary Material
Acknowledgments
We thank P. Renz for his gift of Cby and G. Gottschalk for his gift of M. mazei chromosomal DNA. This work was supported by National Institutes of Health Grant GM40313 (to J.C.E.-S.). J.D.W. was supported in part by the Ira L. Baldwin and Jerome Stefaniak Predoctoral Fellowships.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cbi, dicyanocobinamide; CbiB, cobyric acid synthase; AdoCbi, adenosylcobinamide; Cby, cobyric acid; AdoCby, adenosylCby; AdoCbi, adenosylCbi; AdoCbi-P, adenosylCbi-phosphate; AP, (R)-1-amino-2-propanol; CobU, GTP:AdoCbi-GDP guanylyltransferase enzyme; CobD, l-threonine decarboxylase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. GI15790554 (Halobacterium sp. strain NRC-1 ORF Vng1583C) and GI21226275 (Methanosarcina mazei ORF Mm0173)].
References
- 1.Roth, J. R., Lawrence, J. G. & Bobik, T. A. (1996) Annu. Rev. Microbiol. 50, 137-181. [DOI] [PubMed] [Google Scholar]
- 2.Scott, A. I. & Roessner, C. A. (2002) Biochem. Soc. Trans. 30, 613-620. [DOI] [PubMed] [Google Scholar]
- 3.Warren, M. J., Raux, E., Schubert, H. L. & Escalante-Semerena, J. C. (2002) Nat. Prod. Rep. 19, 390-412. [DOI] [PubMed] [Google Scholar]
- 4.Rondon, M. R., Trzebiatowski, J. R. & Escalante-Semerena, J. C. (1997) Prog. Nucleic Acid Res. Mol. Biol. 56, 347-384. [DOI] [PubMed] [Google Scholar]
- 5.Raux, E., Lanois, A., Warren, M. J., Rambach, A. & Thermes, C. (1998) Biochem. J. 335, 159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santander, P. J., Roessner, C. A., Stolowich, N., Holderman, M. T. & Scott, A. I. (1997) Chem. Biol. 4, 659-666. [DOI] [PubMed] [Google Scholar]
- 7.Blanche, F., Cameron, B., Crouzet, J., Debussche, L., Thibaut, D., Vuilhorgne, M., Leeper, F. J. & Battersby, A. R. (1995) Angew. Chem. Int. Ed. Engl. 34, 383-411. [Google Scholar]
- 8.Thomas, M. G. & Escalante-Semerena, J. C. (2000) J. Bacteriol. 182, 4227-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brushaber, K. R., O'Toole, G. A. & Escalante-Semerena, J. C. (1998) J. Biol. Chem. 273, 2684-2691. [DOI] [PubMed] [Google Scholar]
- 10.Fonseca, M. V. & Escalante-Semerena, J. C. (2000) J. Bacteriol. 182, 4304-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonseca, M. V. & Escalante-Semerena, J. C. (2001) J. Biol. Chem. 276, 32101-32108. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca, M. V., Buan, N. R., Horswill, A. R., Rayment, I. & Escalante-Semerena, J. C. (2002) J. Biol. Chem. 277, 33127-33131. [DOI] [PubMed] [Google Scholar]
- 13.Bauer, C. B., Fonseca, M. V., Holden, H. M., Thoden, J. B., Thompson, T. B., Escalante-Semerena, J. C. & Rayment, I. (2001) Biochemistry 40, 361-374. [DOI] [PubMed] [Google Scholar]
- 14.O'Toole, G. A. & Escalante-Semerena, J. C. (1995) J. Biol. Chem. 270, 23560-23569. [DOI] [PubMed] [Google Scholar]
- 15.Thomas, M. G., Thompson, T. B., Rayment, I. & Escalante-Semerena, J. C. (2000) J. Biol. Chem. 275, 27376-27386. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, T., Thomas, M. G., Escalante-Semerena, J. C. & Rayment, I. (1999) Biochemistry 38, 12995-13004. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, T. B., Thomas, M. G., Escalante-Semerena, J. C. & Rayment, I. (1998) Biochemistry 37, 7686-7695. [DOI] [PubMed] [Google Scholar]
- 18.Blanche, F., Debussche, L., Famechon, A., Thibaut, D., Cameron, B. & Crouzet, J. (1991) J. Bacteriol. 173, 6052-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodson, J. D., Peck, R. F., Krebs, M. P. & Escalante-Semerena, J. C. (2003) J. Bacteriol. 185, 311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodson, J. D., Zayas, C. L. & Escalante-Semerena, J. C. (2003) J. Bacteriol. 185, 7193-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai, S. P., Hartin, R. J. & Ryu, J. (1989) J. Gen. Microbiol. 135, 2561-2567. [DOI] [PubMed] [Google Scholar]
- 22.Rondon, M. R. & Escalante-Semerena, J. C. (1992) J. Bacteriol. 174, 2267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 24.Sasse, J. (1991) in Current Protocols in Molecular Biology, eds. Ausubel, F. A., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), Vol. 1, pp. 10.6.1-10.6.8. [Google Scholar]
- 25.Blanche, F., Thibaut, D., Couder, M. & Muller, J.-C. (1990) Anal. Biochem. 189, 24-29. [DOI] [PubMed] [Google Scholar]
- 26.Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R. A., Martinez-Arias, R., Henne, A., Wiezer, A., Baumer, S., Jacobi, C., et al. (2002) J. Mol. Microbiol. Biotechnol. 4, 453-461. [PubMed] [Google Scholar]
- 27.Drennan, C. L., Huang, S., Drummond, J. T., Matthews, R. G. & Ludwig, M. L. (1994) Science 266, 1660-1674. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole, G. A. (1994) Ph.D. thesis (Univ. of Wisconsin, Madison, WI).
- 29.Grabau, C. & Roth, J. R. (1992) J. Bacteriol. 174, 2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takami, H., Nakasone, K., Takaki, Y., Maeno, G., Sasaki, R., Masui, N., Fuji, F., Hirama, C., Nakamura, Y., Ogasawara, N., et al. (2000) Nucleic Acids Res. 28, 4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessieres, P., Bolotin, A., Borchert, S., et al. (1997) Nature 390, 249-256. [DOI] [PubMed] [Google Scholar]
- 32.Maggio-Hall, L. A. (2001) in Department of Bacteriology (Univ. of Wisconsin, Madison, WI), p. 155.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.