Abstract
Bacterial pathogenicity islands (PAI) often encode both effector molecules responsible for disease and secretion systems that deliver these effectors to host cells. Human enterohemorrhagic Escherichia coli (EHEC), enteropathogenic E. coli, and the mouse pathogen Citrobacter rodentium (CR) possess the locus of enterocyte effacement (LEE) PAI. We systematically mutagenized all 41 CR LEE genes and functionally characterized these mutants in vitro and in a murine infection model. We identified 33 virulence factors, including two virulence regulators and a hierarchical switch for type III secretion. In addition, 7 potential type III effectors encoded outside the LEE were identified by using a proteomics approach. These non-LEE effectors are encoded by three uncharacterized PAIs in EHEC O157, suggesting that these PAIs act cooperatively with the LEE in pathogenesis. Our findings provide significant insights into bacterial virulence mechanisms and disease.
Diarrheagenic enterohemorrhagic Escherichia coli (EHEC), enteropathogenic E. coli (EPEC), and Citrobacter rodentium (CR) are attaching/effacing (A/E) bacterial pathogens that attach to host intestinal epithelium and efface brush border microvilli, forming A/E lesions (1, 2). EHEC and EPEC represent a significant threat to human health. Sequencing the genome of EHEC O157:H7, the causative agent of “Hamburger disease” and the most common serotype associated with food and water poisoning, has identified many putative virulence factors (3). These factors are often encoded by pathogenicity islands (PAI) present in the genomes of pathogenic, but not closely related nonpathogenic, strains (4). However, the functions of the PAIs in virulence have not been systematically analyzed.
Many key virulence factors shared by A/E pathogens reside in the locus of enterocyte effacement (LEE), a PAI essential for A/E lesion formation (5-8). The LEE contains 41 genes and encodes a type III secretion system (TTSS), a common virulence mechanism for many human and plant pathogens (4, 9, 10). TTSSs are conserved organelles that deliver bacterial effector proteins capable of modulating host functions into host cells. The LEE encodes proteins for forming such an organelle (2), but the LEE genes involved in assembling and regulating this apparatus have not been defined.
The LEE also encodes a regulator (Ler), an adhesin (intimin) and its receptor (Tir) responsible for intimate attachment, several secreted proteins, and their chaperones (1, 2). The secreted proteins consist of effectors as well as translocators (EspA, EspD, and EspB) required for translocating effectors into host cells. Five LEE-encoded effectors (Tir, EspG, EspF, Map, and EspH) have been identified, which are involved in modulating host cytoskeleton (2, 11). However, nearly half of the LEE genes have no homologs and have not been functionally studied.
Because EHEC and EPEC are human pathogens, efforts aimed at elucidating the function of the LEE have primarily been restricted to in vitro studies. Animal models, including neonatal calves and weaned rabbits, have been used to study A/E pathogens (12, 13). However, CR, a natural mouse pathogen that possesses a LEE highly similar to that of EHEC and EPEC (7, 14), is the only A/E pathogen for which there is a small animal (mouse) model. All of the EHEC and EPEC LEE-encoded virulence factors tested thus far play equivalent roles in CR virulence (12-18), indicating that CR infection of mice is a relevant animal model for studying EPEC and EHEC.
To gain a comprehensive understanding of LEE function, we undertook a systematic approach by generating a full set of deletion mutants for all 41 CR LEE genes and characterizing the mutants for LEE gene expression, type III secretion (TTS), host actin modulation, and virulence in mice. Our studies led to three significant findings: the LEE encodes two additional regulators and a hierarchical switch for TTS; the LEE-encoded TTSS secretes many effectors encoded by other PAIs outside the LEE; and all of the LEE genes are required for full CR virulence in mice.
Materials and Methods
Strains, Plasmids, and Primers. E. coli and CR strains and plasmids are described in Table 3, which is published as supporting information on the PNAS web site. The primers used are available on request. Bacterial growth conditions were as described (17).
LEE Gene Deletion Mutants. Nonpolar deletion mutants of all 41 CR LEE genes were generated by the sacB-based allelic exchange (19) and lambda Red recombinase (20) systems (Table 4, which is published as supporting information on the PNAS web site). Mutants were verified by PCR. Successful complementation was achieved for Δtir, Δeae, Δler, Δorf11, ΔsepL, Δrorf6, ΔespA, ΔespB, and ΔespD by providing the related genes on a pCR2.1-TOPO- or pACYC184-based plasmid, confirming that the mutations did not affect downstream genes and were nonpolar. All CR mutants grew similarly to WT CR in LB and DMEM.
Protein Assays. Total and secreted proteins of CR strains grown in DMEM were analyzed by SDS/PAGE and Western blot as described (17). Rat antibodies against His-tagged CR Tir and mouse monoclonal antibody against EPEC EspB were used.
CAT Assay. PCR products carrying the upstream regulatory regions of CR ler (LEE1), sepZ (LEE2), and tir (LEE5) as defined for EPEC (21) were digested with BamHI and HindIII and cloned into pKK232-8 carrying a promoterless cat gene (Table 3). CAT activity of the transcriptional fusions was measured in CR strains as described (21).
Primer Extension Assay. It was performed as described by using 5 μg of total RNA isolated from bacteria grown in DMEM (21). Primers complementary to CR ler coding region (positions +53 to +73 with respect to the start codon of ler) or to the 20-bp sequence located downstream of the HindIII site in pKK232-8 were used to determine the 5′ end of the ler or ler-cat transcript, respectively. Constitutively expressed ompA was used as a control.
Analysis of Protein TTS by Epitope Tagging. The coding regions of CR LEE genes espF, espG, espH, map, sepZ, rorf1, cesD2, cesD, cesF, sepL, rorf6, ler, orf10, and orf11 were cloned into pTOPO-2HA or pCRespG-2HA/BglII (Table 3) to create a double hemagglutinin (HA) tag at the C termini. The constructs were introduced into CR WT, ΔescN and ΔescD, and TTS of the tagged proteins was analyzed by Western blot by using mouse monoclonal antibody against HA (Covance, Princeton).
Proteomic Analysis of Secreted Proteins. Proteins secreted by CR strains grown in DMEM were precipitated as described (17), separated by 2D gels according to the manufacturer's instructions (Amersham Pharmacia) and analyzed by mass spectrometry and peptide sequencing (22) as detailed in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Bioinformatic Tools. DNA and protein sequences were analyzed by using databases from the National Center for Biotechnology Information, the Sanger Genome Centre and the SwissProt, and the IslandPath program (www.pathogenomics.sfu.ca/islandpath).
Fluorescent Actin Staining on HeLa Cells. The assay was performed by using a protocol optimized for CR (17).
Virulence Assays. NIH Swiss mice from Harlan Sprague-Dawley (Indianapolis) and C57BL/6 or C3H/HeJ mice from The Jackson Laboratory were infected with CR strains. Infection and pathological analyses were performed as before (17, 23) and detailed in Supporting Materials and Methods.
Results
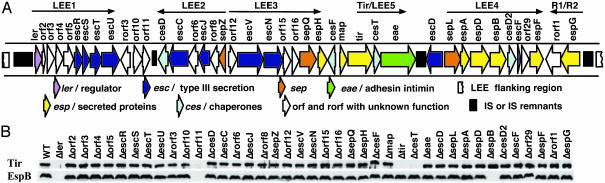
Regulation of LEE Gene Expression. Ler is the only LEE-encoded regulator identified (2). To address whether other LEE genes regulate LEE gene expression, we analyzed all 41 CR LEE mutants for EspB and Tir expression (Fig. 1). Lack of Tir and EspB in Δler confirmed Ler's essential role in LEE gene expression. As expected, Δtir and ΔespB did not produce Tir and EspB, respectively. No Tir was visible in ΔcesT, consistent with CesT's chaperone role for Tir (2). Surprisingly, another LEE-encoded protein, Orf11, was also required for Tir and EspB expression (Fig. 1B). The orf11 gene is highly conserved (5-8), and CR, EHEC, and EPEC orf11 genes all complemented CR Δorf11 (Fig. 2A), indicating that Orf11 is functionally equivalent in all A/E pathogens as a positive regulator.
Fig. 1.
Both Ler and Orf11 are required for expression of LEE genes in CR. (A) Genetic organization of CR LEE (7). (B) Expression of Tir and EspB in WT CR and its 41 LEE mutants. Whole-cell lysates of bacteria grown in DMEM were analyzed by 10% SDS/PAGE and Western blot with anti-Tir and anti-EspB sera.
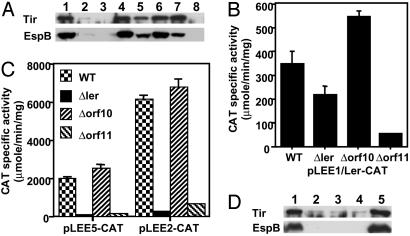
Fig. 2.
Orf11 and Orf10 regulate ler expression in CR. (A) Western blot with anti-Tir and anti-EspB sera of total lysates of bacteria grown in DMEM. Lane 1, WT CR; lane 2, Δorf11; lane 3, ΔlerΔorf11. Also shown are CR Δorf11 complemented by orf11 from CR (pCRorf11, lane 4), EHEC (pEHorf11, lane 5), or EPEC (pEPorf11, lane 6); and CR ΔlerΔorf11 double mutant complemented by CR ler (lane 7) or orf11 (lane 8). (B) Orf11 positively regulates ler expression. The transcriptional activity directed by the ler-cat fusion in pLEE1/Ler-CAT was determined in CR WT, Δler, Δorf10, and Δorf11 grown in DMEM for 6 h. The data are the average of three experiments. (C) Orf11 positively regulates the expression of LEE2 and LEE5 operons by activating ler expression. The activity directed by LEE2 (pLEE2-CAT) and LEE5 (pLEE5-CAT) transcriptional fusions was measured in CR WT, Δler, Δorf10, and Δorf11 as described above. (D) Orf10 acts as a negative regulator of LEE gene expression when expressed from a plasmid. Whole-cell lysates of WT CR carrying pCR2.1-TOPO (the cloning vector, lane 1), pCRorf10-2HA (2HA-tagged orf10, lane 2), pCRorf10 (CR orf10 with its own promoter, lane 3), pCRorf10Plac (Plac-driven CR orf10, lane 4), and pCRorf10orf11 (CR orf10 and orf11 with their own promoter, lane 5) were analyzed as for A.
Orf11 has 37% identity to a Salmonella protein and 23% to CaiF, a transcriptional activator of the Enterobacteriaceae (24). All three proteins contain a helix-turn-helix motif characteristic of DNA binding proteins (Fig. 5, which is published as supporting information on the PNAS web site). To address the hierarchy of Orf11 and Ler in regulating gene expression, we created a CR double mutant of ler and orf11. Whereas Tir and EspB expression in ΔlerΔorf11 was partially restored by expressing Ler in trans, similarly expressed Orf11 had no such effect (Fig. 2A), suggesting that Orf11 acts upstream of Ler in the regulatory cascade. Orf11's role in regulating ler expression was verified by assaying transcriptional fusions between the cat reporter gene and regulatory regions of the LEE1 (ler) operon and two Ler-dependent operons, LEE2 and LEE5. The activity of the LEE1-cat fusion was decreased in Δorf11 (Fig. 2B), and that of LEE2-cat and LEE5-cat was dramatically reduced in both Δler and Δorf11 (Fig. 2C). Primer extension analysis confirmed that ler expression was reduced in Δorf11 (Fig. 6A, which is published as supporting information on the PNAS web site) and showed that the CR ler promoter is similar to that of EPEC ler as it lacks the proximal promoter of EHEC ler (Fig. 6B). These data suggest that Orf11 is a positive regulator of the expression of Ler, which subsequently facilitates the expression of other LEE operons.
We also observed that plasmids expressing Orf10 dramatically reduced Tir and EspB expression in CR (Fig. 2D) and that ler transcription was increased in Δorf10 as shown by CAT and primer extension assays (Fig. 2 and Fig. 6A), suggesting that orf10 encodes a negative modulator for ler expression. Orf10's inhibitory effect was relieved by coexpressing orf11 (Fig. 2D). Because both Orf10 and Orf11 act upstream of Ler in the regulatory cascade, we propose to name Orf11 GrlA (for global regulator of LEE-activator) and Orf10 GrlR (for global regulator of LEE-repressor).
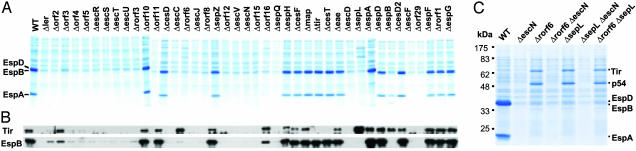
Type III Secretion and Hierarchy. Among the 41 LEE genes (Fig. 1A), 10 (escR, escS, escT, escU, escC, escJ, escV, escN, escD, and escF) encode proteins conserved among TTSSs (2, 4). However, except for escF, escC, escD, escN, and escV (2, 25, 26), the assigned function for these genes was based only on sequence homology. To define the full complement of LEE genes needed for TTS, we analyzed the secretion of translocators EspA, EspB, and EspD and effector Tir in all CR LEE mutants. In addition to the 10 esc genes, 13 other LEE genes were needed for TTS (Fig. 3 and Table 1), with 9 (orf2, orf4, orf5, rorf3, rorf8, orf12, orf15, sepQ, and orf29) required for both translocator and effector secretion and 4 (orf3, rorf6, orf16, and sepL) affecting translocator secretion preferentially. Thus, the LEE encodes 19 proteins essential for TTS. In all LEE mutants defective for TTS, the secretion substrates Tir and EspB were produced in the bacteria (Fig. 1B), indicating a lack of a LEE-encoded feedback inhibitory mechanism seen in the flagellar system (27).
Fig. 3.
Type III secretion by WT CR and its 41 LEE mutants. (A) General protein secretion profile of CR and its mutants. (B) Tir and EspB secretion analyzed by Western blot with anti-Tir and anti-EspB sera. (C) Secretion profile of ΔsepL, Δrorf6, ΔescN (TTS mutant), and their double mutants. Secreted proteins were concentrated from supernatants of bacterial cultures grown in DMEM and analyzed by 12% SDS/PAGE and Coomassie blue G250 staining (A and C) or Western blot (B).
Table 1. Functional characterization of the 41 gene deletion mutants of the locus of enterocyte effacement in C. rodentium.
| Strain or mutant | LEE gene expression* | Type III secretion† | Pedestal formation‡ | Virulence in mice§ | Predicted function for the encoded protein |
|---|---|---|---|---|---|
| WT | + | + | + | +++ | Not applicable |
| Δler | − | −/No expression | − | − | Positive regulator |
| Δorf2 | + | − | − | − | TTSS |
| Δorf3 | + | ±EspA, EspB | − | − | Secretion of EspA and EspB |
| Δorf4 | + | − | − | − | TTSS |
| Δorf5 | + | − | − | − | TTSS |
| ΔescR | + | − | − | − | TTSS |
| ΔescS | + | − | − | − | TTSS |
| ΔescT | + | − | − | − | TTSS |
| ΔescU | + | − | − | − | TTSS |
| Δrorf3 | + | ± | ± | ± | Assembly of TTSS apparatus? |
| Δorf10/grIR | ++ | + | + | ++ | Negative regulator |
| Δorf11/grIA | − | −/No expression | − | − | Positive regulator |
| ΔcesD | + | −/EspD | ± | − | Secretion of EspD |
| ΔescC | + | − | − | − | TTSS |
| Δrorf6 | + | −/Translocators | − | − | Switch/secretion of translocators |
| ΔescJ | + | − | − | − | TTSS |
| Δrorf8 | + | − | − | − | TTSS |
| ΔsepZ | + | + | + | ± | Unknown |
| Δorf12 | + | − | − | − | TTSS |
| ΔescV | + | − | − | − | TTSS |
| ΔescN | + | − | − | − | TTSS |
| Δorf15 | + | − | − | − | TTSS |
| Δorf16 | + | ±/Translocators | ± | ± | Secretion of translocators |
| ΔsepQ | + | − | − | − | TTSS |
| Δorf18/espH | + | + | + | ++ | Secreted protein/effector |
| Δrorf10/cesF | + | ±/EspF? | + | + | Chaperone for EspF |
| Δorf19/map | + | + | + | ++ | Secreted protein/effector |
| Δtir | + | + | − | − | Secreted protein/effector |
| ΔcesT | + | ±/Tir | − | − | Chaperone for Tir |
| Δeae | + | + | − | − | Adhesin (intimin) |
| ΔescD | + | − | − | − | TTSS |
| ΔsepL | + | −/Translocators | − | − | Switch/secretion of translocators |
| ΔespA | + | + | − | − | Secreted protein/translocator |
| ΔespD | + | + | − | − | Secreted protein/translocator |
| ΔespB | + | + | − | − | Secreted protein/translocator |
| Δorf27/cesD2 | + | ±/EspD? | + | ± | Chaperone for EspD? |
| ΔescF | + | − | − | − | TTSS |
| Δorf29 | + | − | − | − | TTSS |
| ΔespF | + | + | + | + | Secreted protein/effector |
| Δrorf1 | + | + | + | + | Unknown |
| Δrorf2/espG | + | + | + | + | Secreted protein/effector |
Normal (+), no (−), or increased (++) LEE gene expression.
Normal (+), no (−), or attenuated (±) type III secretion. The protein substrate(s) were indicated after ”/” if the mutation affected the secretion of only specific protein(s).
Normal (+), no (−), or weak (±) fluorescent actin staining beneath the attaching bacteria.
Virulence for the WT strain (+++), slightly attenuated or more variable in colonization and disease (++), attenuated in early bacterial colonization and colonic hyperplasia (+), severely attenuated colonization and no apparent colonic hyperplasia (±), and avirulent with no disease (−). This qualitative designation was based on virulence tests of C. rodentium strains in three strains of mice. See Tables 5 and 6 and Fig. 9 for the quantitative presentation of the virulence data.
Type III chaperones are critical for secretion of their substrates (9, 27). In CR, CesT was needed for Tir stability and secretion, and CesD was essential for EspD secretion (Fig. 3 and Fig. 7, which is published as supporting information on the PNAS web site), like EPEC CesT and CesD (2, 28). However, unlike the EPEC cesD2 mutant that has reduced EspD secretion (29), CR ΔcesD2 secreted EspD normally (Fig. 7). The role of CesF in EspF secretion was reported for EPEC (2) and was not tested here.
The mutants of orf3, orf16, rorf6, and sepL affected secretion of translocators and effectors differentially (Figs. 3 and 7). Δorf3 and Δorf16 secreted Tir normally. However, Δorf3 secreted normal EspD but much less EspA and EspB whereas Δorf16 secreted greatly reduced EspA, EspB, and EspD, indicating that they modulate translocator secretion preferentially. ΔsepL and Δrorf6 did not secrete detectable EspA, EspD, and EspB although the translocators were produced (Fig. 1B). Interestingly, both ΔsepL and Δrorf6, as well as the double mutant Δrorf6ΔsepL, had greatly enhanced secretion of Tir and a 54-kDa protein (p54) (Fig. 3C). The secretion of Tir and p54 was by means of the LEE-encoded TTSS because double mutants ΔsepLΔescN and Δrorf6ΔescN did not secrete both proteins. This result suggests that SepL and Rorf6 may act as a molecular switch controlling secretion hierarchy of translocators and effectors.
Identification of Effectors Secreted by LEE-Encoded TTSS. The main function of TTSS is to deliver effectors into host cells, and effector genes can be located both within and outside PAIs encoding TTSS (10, 30, 31). Five LEE-encoded effectors have been identified in EPEC (2, 11). To define the effectors encoded by CR LEE, we tagged all 14 LEE-encoded proteins (EspF, EspG, EspH, Map, SepZ, Rorf1, CesD, CesD2, CesF, SepL, Rorf6, Ler, GrlA, and GrlR) that are not involved in TTS or host cell adhesion with a 2HA epitope at the C termini and analyzed their secretion in WT CR and TTS mutant ΔescN. Although all of the tagged proteins were expressed and stable, only Tir, EspG, EspF, EspH, and Map were type III secreted by CR (data not shown), suggesting that CR LEE encodes only five effectors, similar to EPEC LEE (2, 11).
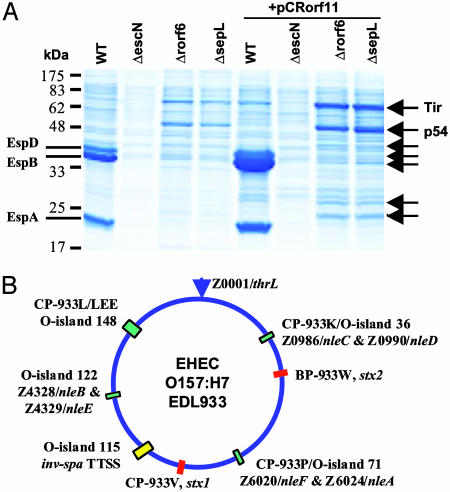
As shown in Fig. 3C, ΔsepL and Δrorf6 did not secrete translocators but had enhanced secretion of effector Tir and p54 by means of the LEE-encoded TTSS. p54 likely represents a secreted protein encoded outside the LEE. To identify p54 and other non-LEE-encoded effectors in CR, we used GrlA overexpressed from a plasmid to increase LEE gene expression and TTS. CR overexpressing GrlA secreted more (>300%) EspA, EspB, and EspD than WT, and the same plasmid greatly enhanced (by >400%) Tir and p54 secretion in ΔsepL and Δrorf6, with no translocators secreted (Fig. 4A). At least six additional proteins were secreted by ΔsepL and Δrorf6, but not by TTS mutant ΔescN (Fig. 4A), and they were characterized by proteomic analysis (Table 2 and Fig. 8, which is published as supporting information on the PNAS web site). This analysis confirmed that the 5 LEE-encoded effectors were type III secreted by ΔsepL. In addition, we identified 7 non-LEE-encoded secreted proteins (Table 2). Because ΔsepL and Δrorf6 did not secrete translocators but secreted effectors preferentially, these 7 secreted proteins likely represent potential effectors and were designated NleA (p54), NleB, NleC, NleD, NleE, NleF, and NleG (for non-LEE-encoded effectors) (Table 2). We have since shown that NleA is a translocated effector targeted to the host cell Golgi (32).
Fig. 4.
Identification of both LEE- and non-LEE-encoded proteins secreted by the LEE-encoded TTSS. (A) Effect of overexpressing CR orf11 on TTS in WT CR and its ΔsepL or Δrorf6 mutants. Secreted proteins were analyzed by 15% SDS/PAGE and Coomassie blue staining. The additional type III secreted proteins by ΔsepL and Δrorf6 carrying pCRorf11 are indicated by arrows and were characterized by proteomic analyses (Table 2 and Fig. 8). (B) A diagram showing locations of the O-islands encoding the six identified non-LEE effectors in the EHEC O157:H7 genome (3). Also shown are the locations of the Shiga toxin genes (stx), the LEE, the inv-spa-like TTSS, and the associated prophages (CP- and BP-933).
Table 2. Effectors and putative effectors secreted by the LEE-encoded TTSS in C. rodentium.
| Serial number | Proposed name | Estimated kDa | Estimated pl | Gene location | Homologues in EHEC and other pathogens by BLASTP searches |
|---|---|---|---|---|---|
| 5 | Tir | 68 | 5.0 | LEE | Tir, conserved in all A/E pathogens. |
| 10 | EspG | 44 | 7.3 | LEE | EspG, conserved in all A/E pathogens. |
| C1&C2 | Map | 23 | 9.0 | LEE | Map, conserved in all A/E pathogens. |
| C3 | EspF | 31 | 11.0 | LEE | EspF, conserved in all A/E pathogens. |
| C5&C6 | EspH | 21 | 8.7 | LEE | EspH, conserved in all A/E pathogens. |
| 7 | NleA | 54 | 5.8 | Non-LEE | EHEC Z6024 in O-island 71 near prophage CP-933P. |
| 12 | NleB | 39 | 5.9 | Non-LEE | EHEC Z4328 in O-island 122, REPEC LEE-associated RorfE, and S. typhimurium STMF1. Also homologous to Z0985 of O-island 36. |
| 13 | NleC | 40 | 4.6 | Non-LEE | EHEC Z0986 in O-island 36 near prophage CP-933K. |
| 14 | NleD | 28 | 7.1 | Non-LEE | EHEC Z0990 in O-island 36, in the same O-island as Z0985 and Z0986. Also similar to P. syringae effector HopPtoH. |
| 17 | NleE | 27 | 6.3 | Non-LEE | EHEC Z4329, in the same O-island 122 as Z4328. Also similar to REPEC LEE-associated RorfD, and S. flexneri ORF122. |
| 19 | NleF | 24 | 4.7 | Non-LEE | EHEC Z6020, in the same O-island 71 as Z6024. Similar to hypothetical proteins in Yersinia pestis and Helicobacter pylori. |
| 20 | NleG | 26 | 5.8 | Non-LEE | No homologue found. Peptide sequence identified: QQENAPSS(I/L)QTR. |
Because the CR genome is not yet sequenced, it is unclear how the new effector genes are organized. Of the seven Nle proteins, only NleG may be unique to CR based on peptide sequences, and NleA-F are highly conserved in EHEC O157 (Table 2). The EHEC NleA-F homologs are encoded by genes clustered in three discrete regions (O-islands 36, 71, and 122) of the genome (3), with each region encoding at least two Nle proteins (Fig. 4B). Homologs of all six EHEC effector genes are also present and similarly organized in the partially sequenced EPEC genome, showing 89-95% nucleotide identity. Some of them also have homologs in other pathogens, such as rabbit EPEC, Pseudomonas syringae, Shigella flexneri, and Salmonella typhimurium (Table 2) (8, 30-33), suggesting the importance of these newly identified non-LEE-encoded effectors in virulence.
Pedestal Formation. The LEE is sufficient for inducing A/E lesions and actin-rich pedestals (2). We analyzed the ability of all 41 CR LEE mutants to induce pedestal formation on HeLa cells. As shown in Table 1, genes required for LEE expression (ler and orf11) and for TTS/translocation were all essential for pedestal formation, as were tir, cesT, and eae. The orf16 and cesD mutants induced sporadic pedestals and were much less efficient than WT, consistent with their role in TTS. Genes grlR, sepZ, espH, cesF, map, cesD2, espF, rorf1, and espG were not needed for pedestal formation, suggesting that Tir is the only LEE-encoded effector essential for this function.
Virulence in Mice. Because EPEC and EHEC are human pathogens, identification of LEE-encoded virulence factors has progressed slowly. To date, the role of only eight LEE genes (eae, espA, espB, tir, espG, escD, map, and cesD2) has been tested in humans or animal models (2, 12-18, 29, 34). We capitalized on the CR-mouse infection model and tested the virulence of all 41 CR LEE mutants in mice. Our results not only confirmed the role of the 8 known virulence factors, but also determined the role in virulence of the other 33 proteins encoded by the LEE (Table 1).
The degree of importance of a given LEE gene in disease varies with its function (Table 1 and Tables 5 and 6 and Fig. 9, which are published as supporting information on the PNAS web site). The genes for activating LEE gene expression (ler and grlA) were absolutely required for CR virulence, highlighting the central role of Ler and GrlA-regulated genes in pathogenesis. The negative regulator GrlR also played a role, with ΔgrlR showing a minor but significant defect in colonization and colonic hyperplasia. This finding indicates that coordinated expression of LEE genes in vivo is critical for full CR virulence. Genes encoding the TTS/translocation function were all essential. The effect on virulence was more diverse for effectors and chaperones. Tir was the only essential LEE-encoded effector. The phenotype of Δeae was similar to that of Δtir, consistent with the essential role of Tir and intimin in bacterial colonization and disease (13-15,17). Although ΔespF and ΔespG showed moderate attenuation, Δmap and ΔespH were only slightly attenuated. The phenotype of mutants for type III chaperones correlated with that of their cognate substrates. ΔcesT was severely attenuated in virulence, similar to Δtir. ΔcesF showed attenuation similar to ΔespF. Like ΔespD, ΔcesD displayed little virulence. However, ΔcesD2 was only moderately attenuated because it still colonized mice and induced mild disease, suggesting that the two EspD chaperones play different roles.
Some CR LEE mutants (Δrorf3, Δorf16, ΔcesD2, and ΔsepZ), although still able to colonize NIH Swiss mice, did not induce severe colonic hyperplasia. Several other mutants (ΔgrlR, Δmap, ΔcesF, Δrorf1, ΔespG, ΔespF, and ΔespH) displayed only slight attenuation in virulence in NIH Swiss or C57BL/6 mice, with Δrorf1 and ΔespG showing attenuated colonization and disease at early time points (Tables 5 and 6). We further characterized these mutants in the more susceptible C3H/HeJ mice (23). Although infection by WT resulted in 100% mortality between day 6 and 10 postinfection, C3H/HeJ mice infected by Δrorf3, Δorf16, ΔcesD2, and ΔsepZ survived, indicating that these mutants are attenuated in virulence (Fig. 9). Mice infected by Δrorf1, ΔespF, and ΔcesF survived 2-3 days longer than mice infected by WT. Mutations in grlR, map, espG, and espH did not alter CR's lethality in C3H/HeJ mice, but these mutants showed more mouse to mouse variation than WT in colonization and colonic hyperplasia. Collectively, our results indicate that remarkably all of the LEE genes contribute to full CR virulence in mice.
Discussion
CR infection of mice offers many advantages as an animal model for studying the LEE function of A/E pathogens. To gain a global view of LEE's function as a PAI, we used a systematic approach to analyze all 41 CR LEE genes and functionally categorized their roles in virulence. Our results demonstrate that the entire LEE is needed for complete CR virulence in mice, in contrast to the redundancy of PAI genes in Salmonella and other pathogens (10).
In addition, our functional studies of CR LEE have yielded several significant findings. Besides Ler, the LEE encodes another positive regulator, GrlA, as well as a negative regulator, GrlR, indicating that regulation of LEE gene expression is much more complex than previously anticipated (2). Our results suggest that both GrlA and GrlR act upstream of Ler in the regulatory cascade (Fig. 2). GrlA shares homology with CaiF, a known DNA-binding protein involved in transcriptional activation (24). GrlR represents a negative regulator that is not homologous to any known transcriptional factors. GrlR likely regulates LEE gene expression by modulating GrlA activity (Fig. 2D). In support of this view, GrlR has been shown to interact with GrlA (35).
Another interesting finding is a LEE-encoded secretion hierarchy between translocators and effectors. Because translocators are needed to translocate effectors into host cells, translocators ought to be secreted ahead of effectors. Yet, this hierarchy of secretion remains an open question (9). We have shown that the LEE encodes four proteins (Orf3, Orf16, SepL, and Rorf6) that modulate the secretion of translocators and effectors differentially. Orf3 and Orf16 affect translocator secretion only because their mutants secrete effectors normally (Fig. 3). How Orf16 functions is not clear, but there is evidence that Orf3 is a chaperone for EspA and EspB because Orf3 interacts with both EspA and EspB (35). The function of SepL and Rorf6 is different from that of Orf3 and Orf16 because ΔsepL and Δrorf6 mutants secrete no translocators but increased effectors (Fig. 4A), suggesting that SepL and Rorf6 control the switch for translocator and effector secretion. Consistent with our data, SepL has been shown to interact with Rorf6 (35). The secretion profile of CR ΔsepL resembles that of an EHEC or EPEC sepL mutant, which secretes no translocators but increased amounts of Tir and p54 (ref. 36 and unpublished data), indicating that the same mechanism operates in other A/E pathogens. In addition, Salmonella pathogenicity island 2 (SPI2) encodes a SepL homolog (SsaL), and there is evidence that such a switch exists in TTSS encoded by Salmonella SPI1 and S. flexneri (10, 30, 37).
Type III effectors secreted by both plant and animal pathogens mediate many aspects of disease (4, 9, 10). The LEE encodes five effectors (Table 2) (2, 11), a small number compared with other pathogens (10, 30, 31). The LEE is sufficient for pedestal formation, and many LEE-encoded effectors are involved in modulating host cytoskeleton (2). However, the repertoire of LEE-encoded effectors does not explain the full spectrum of host disease symptoms incurred by A/E pathogens, such as intestinal inflammation and diarrhea, suggesting that the LEE-encoded TTSS also secretes non-LEE-encoded effectors. There is evidence that A/E pathogens can counteract host defense by delivering effectors to inhibit host phagocytosis and to suppress NF-κB activation and proinflammatory cytokine expression (38, 39).
Our discovery of GrlA and the SepL/Rorf6 secretion hierarchy switch led us to design a proteomics-based screen for effectors secreted by means of the LEE-encoded TTSS, identifying seven potential non-LEE-encoded effectors in CR (Table 2). Six of them are highly conserved in EHEC and EPEC, and several also show homology to proteins encoded by other human and plant pathogens. In EHEC, these effectors are encoded outside the LEE by three PAIs that are present in many A/E pathogens (Fig. 4B) (3, 8, 33). Their genes have dinucleotide bias and low G+C% contents, hallmarks of PAIs (4). They are either associated with prophages or flanked by mobile insertion sequences and are absent from the genome of nonpathogenic E. coli (3), suggesting acquisition via horizontal transfer. Our data offer compelling evidence that the repertoire of virulence factors used by A/E pathogens is significantly larger than originally thought and that at least three PAIs act cooperatively with the LEE in pathogenesis.
In conclusion, our analysis of the LEE has led us to discover previously uncharacterized mechanisms governing TTS and gene regulation in A/E pathogens. Our finding of a large repertoire of non-LEE-encoded effectors indicates that diseases mediated by A/E pathogens may require coordinated action of effectors encoded by the LEE and at least three other PAIs. The challenges now are to elucidate how each effector modulates host cellular processes and to establish the link between effectors and disease. In this regard, we have shown that the non-LEE-encoded NleA is a type III translocated effector in CR, EHEC, and EPEC. Although NleA does not affect pedestal formation, it still plays a critical role in CR virulence in mice (32). It therefore seems that these non-LEE-encoded effectors hold additional keys to our understanding of EHEC- and EPEC-mediated diseases.
Supplementary Material
Acknowledgments
We thank R. Fernandez, M. Wickham, B. Coombes, P. Hardwidge, N. Strynakda, and E. Frey for reviewing the manuscript and R. A. Edwards and B. L. Wanner for strains and plasmids. B.B.F. is supported by the Canadian Institutes of Health Research, the Howard Hughes Medical Institute, and the Natural Sciences and Engineering Research Council (Canada). J.L.P. is funded by Dirección General de Asuntos del Personal Académico, Consejo Nacional de Ciencia y Tecnología (Mexico), and the Howard Hughes Medical Institute.
Abbreviations: PAI, pathogenicity island; EPEC, enteropathogenic Escherichia coli; EHEC, enterohemorrhagic E. coli; CR, Citrobacter rodentium; A/E, attaching/effacing; LEE, locus of enterocyte effacement; TTSS, type III secretion system; TTS, type III secretion; HA, hemagglutinin.
References
- 1.Frankel, G., Phillips, A. D., Rosenshine, I., Dougan, G., Kaper, J. B. & Knutton, S. (1998) Mol. Microbiol. 30, 911-921. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, S. C., Haigh, R. D., Freestone, P. P. E. & Williams, P. H. (2003) Clin. Microbiol. Rev. 16, 365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perna, N. T., Plunkett, G., 3rd, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 4.Hacker, J. & Kaper, J. B. (2000) Annu. Rev. Microbiol. 54, 641-679. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, S. J., Wainwright, L. A., McDaniel, T. K., Jarvis, K. G., Deng, Y. K., Lai, L. C., McNamara, B. P., Donnenberg, M. S. & Kaper, J. B. (1998) Mol. Microbiol. 28, 1-4. [DOI] [PubMed] [Google Scholar]
- 6.Perna, N. T., Mayhew, G. F., Posfai, G., Elliott, S., Donnenberg, M. S., Kaper, J. B. & Blattner, F. R. (1998) Infect. Immun. 66, 3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, W., Li, Y., Vallance, B. A. & Finlay, B. B. (2001) Infect. Immun. 69, 6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tauschek, M., Strugnell, R. A. & Robins-Browne, R. M. (2002) Mol. Microbiol. 44, 1533-1550. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G. R. (2002) J. Cell Biol. 158, 401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galan, J. E. (2001) Annu. Rev. Cell Dev. Biol. 17, 53-86. [DOI] [PubMed] [Google Scholar]
- 11.Tu, X., Nisan, I., Yona, C., Hanski, E. & Rosenshine, I. (2003) Mol. Microbiol. 47, 595-606. [DOI] [PubMed] [Google Scholar]
- 12.Dean-Nystrom, E. A., Bosworth, B. T., Moon, H. W. & O'Brien, A. D. (1998) Infect. Immun. 66, 4560-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marches, O., Nougayrede, J. P., Boullier, S., Mainil, J., Charlier, G., Raymond, I., Pohl, P., Boury, M., De Rycke, J., Milon, A., et al. (2000) Infect. Immun. 68, 2171-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schauer, D. B. & Falkow, S. (1993) Infect. Immun. 61, 4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnenberg, M. S., Tzipori, S., McKee, M. L., O'Brien, A. D., Alroy, J. & Kaper, J. B. (1993) J. Clin. Invest. 92, 1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe, A., Heczko, U., Hegele, R. G. & Finlay, B. B. (1998) J. Exp. Med. 188, 1907-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng, W., Vallance, B. A., Li, Y., Puente, J. L. & Finlay, B. B. (2003) Mol. Microbiol. 48, 95-115. [DOI] [PubMed] [Google Scholar]
- 18.Mundy, R., Pickard, D., Wilson, R. K., Simmons, C. P., Dougan, G. & Frankel, G. (2003) Mol. Microbiol. 48, 795-809. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, R. A., Keller, L. H. & Schifferli, D. M. (1998) Gene 207, 149-157. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustamante, V. H., Santana, F. J., Calva, E. & Puente, J. L. (2001) Mol. Microbiol. 39, 664-678. [DOI] [PubMed] [Google Scholar]
- 22.Houthaeve, T., Gausepohl, H., Mann, M. & Ashman, K. (1995) FEBS Lett. 376, 91-94. [DOI] [PubMed] [Google Scholar]
- 23.Vallance, B. A., Deng, W., Jacobson, K. & Finlay, B. B. (2003) Infect. Immun. 71, 3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchet, A., Nasser, W., Eichler, K. & Mandrand-Berthelot, M.-A. (1999) Mol. Microbiol. 34, 562-575. [DOI] [PubMed] [Google Scholar]
- 25.Kresse, A. U., Schulze, K., Deibel, C., Ebel, F., Rohde, M., Chakraborty, T. & Guzman, C. A. (1998) J. Bacteriol. 180, 4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gauthier, A., Puente, J. L. & Finlay, B. B. (2003) Infect. Immun. 71, 3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aldridge, P. & Hughes, K. T. (2001) Trends Microbiol. 9, 209-214. [DOI] [PubMed] [Google Scholar]
- 28.Wainwright, L. A. & Kaper, J. B. (1998) Mol. Microbiol. 27, 1247-1260. [DOI] [PubMed] [Google Scholar]
- 29.Neves, B. C., Mundy, R., Petrovska, L., Dougan, G., Knutton, S. & Frankel, G. (2003) Infect. Immun. 71, 2130-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchrieser, C., Glaser, P., Rusniok, C., Nedjari, H., D'Hauteville, H., Kunst, F., Sansonetti, P. & Parsot, C. (2000) Mol. Microbiol. 38, 760-771. [DOI] [PubMed] [Google Scholar]
- 31.Collmer, A., Lindeberg, M., Petnicki-Ocwieja, T., Schneider, D. J. & Alfano, J. R. (2002) Trends Microbiol. 10, 462-469. [DOI] [PubMed] [Google Scholar]
- 32.Gruenheid, S., Sekirov, I., Thomas, N. A., Deng, W., O'Donnell, P., Goode, D., Li, Y., Frey, E. A., Brown, N. F., Metalnikov, P., et al. (2004) Mol. Microbiol. 51, 1233-1249. [DOI] [PubMed] [Google Scholar]
- 33.Morabito, S., Tozzoli, R., Oswald, E. & Caprioli, A. (2003) Infect. Immun. 71, 3343-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallance, B. A., Deng, W., De Grado, M., Chan, C., Jacobson, K. & Finlay, B. B. (2002) Infect. Immun. 70, 6424-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creasey, E. A., Delahay, R. M., Daniell, S. J. & Frankel, G. (2003) Microbiology 149, 2093-2106. [DOI] [PubMed] [Google Scholar]
- 36.Kresse, A. U., Beltrametti, F., Muller, A., Ebel, F. & Guzman, C. A. (2000) J. Bacteriol. 182, 6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubori, T. & Galan, J. E. (2002) J. Bacteriol. 184, 4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celli, J., Olivier, M. & Finlay, B. B. (2001) EMBO J. 20, 1245-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauf, N. & Chakraborty, T. (2003) J. Immunol. 170, 2074-2082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.