Abstract
Androgen and androgen receptor (AR) play important roles in male spermatogenesis and fertility, yet detailed androgen/AR signals in Sertoli cells remain unclear. To identify AR target genes in Sertoli cells, we analyzed the gene expression profiles of testis between mice lacking AR in Sertoli cells (S-AR−/y) and their littermate wild-type (WT) mice. Digital gene expression analysis identified 2276 genes downregulated and 2865 genes upregulated in the S-AR−/y mice testis compared to WT ones. To further nail down the difference within Sertoli cells, we first constructed Sertoli cell line TM4 with stably transfected AR (named as TM4/AR) and found androgens failed to transactivate AR in Sertoli TM4 and TM4/AR cells. Interestingly, additional transient transfection of AR-cDNA resulted in significant androgen responsiveness with TM4/AR cells showing 10 times more androgen sensitivity than TM4 cells. In the condition where maximal androgen response was demonstrated, we then analyzed gene expression and found the expression levels of 2313 genes were changed more than twofold by transient transfection of AR-cDNA in the presence of testosterone. Among these genes, 603 androgen-/AR-regulated genes, including 164 upregulated and 439 downregulated, were found in both S-AR−/y mice testis and TM4/AR cells. Using informatics analysis, the gene ontology was applied to analyze these androgen-/AR-regulated genes to predict the potential roles of androgen/AR in the process of spermatogenesis. Together, using gene analysis in both S-AR−/y mice testis and TM4/AR cells may help us to better understand the androgen/AR signals in Sertoli cells and their influences in spermatogenesis.
Keywords: androgen receptor, conditional knockout mice, Sertoli cells, testis, TM4 cells
Introduction
Androgens are critical sex hormones that control the process of spermatogenesis. Their actions are mediated by androgen receptor (AR), a member of the nuclear receptor superfamily. Spermatogenesis, a process of generating mature sperm with half the number of chromosomes (haploid) produced from germ cell precursors (diploid), encompasses a series of precisely timed cellular events that take place in a highly organized tissue, the seminiferous epithelium. Within each cross-section of this tissue, spermatogonia, spermatocytes and spermatids are in intimate physical association with somatic Sertoli cells.1 Sertoli cells provide nutritional as well as morphogenetic support for germ cells during spermatogenesis.
There is some evidence that AR in the Sertoli cells, and not in germ cells, may regulate spermatogenesis as early data demonstrated that deletion of AR gene in mouse germ cells failed to affect spermatogenesis and male fertility,2 but lacking AR in Sertoli cells led to spermatogenesis arrest at the diplotene primary spermatocyte stage.3, 4, 5 Therefore, Sertoli cells are usually considered to be one of the important mediators of androgen/AR action on spermatogenesis. The detailed mechanisms by which AR supports germ cell development, however, remain unclear, although a few AR-regulated genes, such as Rhox5 (also known as Pem) and c-myc, have been identified in Sertoli cells in vivo.6, 7
Using a Cre-Lox conditional knockout strategy, we generated a tissue-specific knockout mouse with the AR gene deleted only in Sertoli cells (S-AR−/y) by crossing mice with a floxed exon 2 of the AR with mice expressing Cre under the control of the anti-Mullerian hormone promoter (AMH-Cre).4, 8, 9 Phenotype analyses show that the S-AR−/y mice were indistinguishable from wild-type (WT) AR mice (B6 AR+/y) with the exception of testes, which were significantly atrophied. S-AR−/y mice were infertile, with spermatogenic arrest predominately at the diplotene premeiotic stage and almost no sperm detected in the epididymides.4 For identifying the AR targets responsible for these phenotypes, we compared the gene expression profiles of adult whole testis mRNA from S-AR−/y mice and their WT littermates.
Although the cell-specific gene knockout mouse is a good model to study gene function, it still remains not easy to further analyze gene expression in a given cell type (such as Sertoli cell) due to the multiple cell composition existing within the testis. Other studies using AR hypomorphs mouse10 and Sertoli cell-selective AR knockout-juvenile spermatogonial depletion (SCARKO-jsd) mouse11 also faced similar difficulties in identifying AR target genes in a given cell within testis. For distinguishing AR target genes in Sertoli cells, we used an in vitro cultured Sertoli cell line TM4, which was isolated from the testis of Balb/c mice and expresses the AR,12 and could therefore potentially be used to determine androgen-regulated genes.
Using these two different systems, 603 genes were found to be potential androgen-/AR-regulated genes in Sertoli cells. Among these androgen-/AR-regulated genes, genes involved in ras protein signal transduction and ubiquitin-protein ligase were clearly overrepresented.
Materials and methods
Generation of Sertoli cell-specific AR−/y mice
To obtain S-AR−/y animals, transgenic AMH-Cre (C57-B6/SJL) male mice expressing Cre recombinase, under the control of the AMH gene promoter, were mated with floxed AR (C57-B6/129 SEVE) female mice as previously described.4 These AMH-Cre transgene animals were also used as a control for S-AR−/y animals. Whole testes were obtained from animals at 10-week-old. Immediately after removal, testis samples were stored in RNAlater RNA Stabilization Reagent (Qiagen, Chatsworth, CA, USA) before being used for total RNA isolation.
Cell culture, transient transfection and stable transfection
The TM4 cell line was obtained from the American Type Culture Collection. The cells were maintained in DMEM/F12 medium (Life Technologies, Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS), 100 U ml−1 penicillin and 100 µg ml−1 streptomycin in 25-cm2 culture flasks at 37 °C in humidified atmosphere with 5% CO2. Before treating with androgens testosterone (T) or dihydrotestosterone (DHT), cells were maintained in DMEM/F12 supplemented with charcoal-treated FBS (androgen-free; Thermo Fisher Scientific, Waltham, MA, USA) for 48 h. Transient transfection was carried out by using Lipofectamine 2000 (Life Technologies) performed according to the manufacturer's instructions. The efficiency of transient transfection on TM4 cells is about 5%–10% as judged by transfection with a pEGFP-C1 vector (Clontech, Palo Alto, CA, USA). A recombinant vector expressing mouse AR, pcDNA3.1-AR, was constructed by cloning mouse AR open reading fragment into pcDNA3.1 vector (Life Technologies), with T7 promoter just upstream of the cloned gene. After 2 days of pcDNA3.1-AR transfection to prepare stable transfected clones, cells were split into 100-mm dishes with selective medium containing antibiotic Geneticin (G418) at 100 µg ml−1. Most of the non-transfected TM4 cells in these groups died, whereas G418-resistant colonies appeared after 14 days of cultivation. The required monoclonal cells were cultured for 1 week to establish a stable transfected pcDNA3.1-AR cell line (TM4/AR) under the selective pressure of 100 µg ml−1 G418. All cells to be tested in the following assays had a passage number of 3–6.
Western blot analysis
Cell lysates of TM4 cells were separated by SDS–PAGE (10% gels) and electroblotted onto polyvinylidine difluoride membrane (Immobilon P, 0.45-µm-pore-size; Millipore, Bedford, MA, USA). Blots were blocked with Tris-buffered saline with 0.5% Tween 20 plus 5% dry nonfat milk and antibodies were diluted in this buffer as suggested by the manufacturers. The primary antibody was the rabbit polyclonal anti-AR antibody (sc-816; Santa Cruz Biotechnology, Inc., Santa Cruz, USA), and the secondary antibody was a goat anti-rabbit IgG conjugated to horseradish peroxidase (1∶3000 dilution; Pierce, Rockford, IL, USA). Blots were incubated in primary antibodies overnight at 4 °C, washed three times in Tris-buffered saline with 0.5% Tween 20, and then incubated with secondary antibodies for 1 h at room temperature. Glyceraldehyde-3-phosphate dehydrogenase was detected by a monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase antibody (sc-25778; Santa Cruz Biotechnology, Inc.) as an internal control for equivalent protein loading. Immunoblots were visualized on photographic films using the SuperSignal West Pico chemiluminescent substrate (Pierce) following the manufacturer's instructions.
Constructs and luciferase assay
The mouse mammary tumor virus-luciferase (MMTV-Luc) construct contains the promoter of the mouse mammary tumor virus long terminal repeat.13 The prostate-specific antigen (PSA)-Luciferase construct contains a 600-bp fragment of the PSA promoter with an additional 2.4-kb enhancer sequence cloned upstream of luciferase.14 Androgen response element-luciferase was constructed by inserting two synthetic tandem repeats of the androgen-responsive element (ARE) in multiple cloning region of pGL3-Promoter vector (Promega, Madison, WI, USA). Primers (5'-CAG AAC AGC ATG TTC TGA ATT CCA AGA ACA GCA TGT TCT C-3' and 5'-TCG AGA GAA CAT GCT GTT CTT GGA ATT CAG AAC ATG CTG TTC TGG TAC-3') were used. These two strands were allowed to form a duplex prior to ligation within the multiple cloning site upstream of the luc reporter gene in the linearized vector.
A day before transfection, ∼6000 TM4 or TM4/AR cells were seeded in 96-well plate. The constructs (0.1 µg) were then cotransfected with 0.1 µg of pSV-β-Galactosidase control vector and pcDNA3.1-AR at indicated quantity to 60%–70% confluent TM4 or TM4/AR cells. After 6 h, medium was changed. The cells were treated for 24 h with indicated concentrations of androgen (T or DHT) or antiandrogen (flutamide) in fresh DMEM/F12 medium containing charcoal-treated FBS. Transfection experiments were done independently for both cell lines two times in triplicate wells.
After 24 h incubation, cells were harvested for luciferase assay and β-galactosidase (β-gal) assay as described in the protocol provided by Promega. Briefly, we added 50 µl lysis buffer per well and perform a single freeze–thaw cycle to ensure complete lysis, mixed 20 µl cell lysate with 100 µl luciferase assay reagent and read light intensity in Turner BioSystems Modulus Single Tube Multimode Reader (Thermo Fisher Scientific). Luciferase activity was normalized for transfection efficiency using corresponding β-gal activity. The ratios of luciferase/β-gal activity were used as indicators for transcriptional activity of different promoters.
Total RNA extraction and digital gene expression (DGE) sequencing and statistical analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer's instructions. RNA quality and concentration were determined using a 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). Total RNA (4 µg) of each sample was used in DGE sequencing. Briefly, after double-stranded cDNA synthesis using oligo(dT)18, the cDNA was digested with NlaIII and ligated to the first adapter, GEX adapter 1, containing a recognition site for the MmeI. The second adapter, GEX index adapter, was ligated after MmeI digestion. Tags flanked by both adapters were enriched by PCR using Gex PCR primers 1 and 2 following the manufacturer's protocol. Finally, DGE libraries were performed to cluster generation and sequencing on the Illumina Cluster Station and Genome Analyzer II (Illumina Inc., San Diego, CA, USA) following the manufacturer's protocol.
For statistical analysis, we filtered the potentially erroneous tags (only one copy number, only adaptor sequences, or containing unknown sequences ‘N') to get the clean tags. All the possible 17 bases length sequences next to the NlaIII restriction site of the reference sequences mouse genome (mm9) and four bases plus sequences were extracted and constructed as a reference database. Two kinds of tags (unambiguous and ambiguous tags) were obtained when all clean tags were mapped to the reference database using SOAP V2.0 (Beijing Genomics Institute at Shenzhen, Shenzhen, China)15 allowing no more than one base mismatch. Only unambiguous tags, which mapped to only one gene, were used for gene expression analysis. Transcripts per million clean tags were applied to compare gene expression between different samples.
RT-PCR and quantitative real-time PCR analysis (qRT-PCR)
Total RNA was reverse-transcribed into cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). PCR was performed in triplicate using the SYBR Green PCR Master Mix (Invitrogen) with cDNA (3 µl) and gene-specific primers (300 nmol l−1 each) in a 50 µl (final volume) reaction mixture on an ABI 7000 Real-Time PCR System (Life Technologies). The primer sequences were shown in Table 1. An RT-minus control was included to detect any contaminating genomic DNA. To exclude the occurrence of primer dimers and non-specific PCR products, melting curve analysis was performed using the ABI PRISM 7000 Sequence Detection System Software version 1.0.1 (Applied Biosystems, Foster City, CA, USA). The relative expression level of each gene was normalized to a housekeeping gene, peptidylprolyl isomerase A (Ppia).
Table 1. Primers for quantitative RT-PCR.
| Symbol | Transcript ID | Primers |
|---|---|---|
| Akap1 | NM_009648 | F: 5′-CACCACTGCCTTCGCTGGCA-3′R: 5′-TGGGGTGTGTGTGCTGCTGG-3′ |
| Cst3 | NM_009976 | F: 5′-GCAGCTCGTGGCTGGAGTGAA-3′R: 5′-AGTGTGTGCCTTTCCAGGGCAC-3′ |
| Iapp | NM_010491 | F: 5′-TGCCTCGGACCACTGAAAGGGA-3′R: 5′-TGGCCGTGTTGCACTTCCGT-3′ |
| Mybbp1a | NM_016776 | F: 5′-CCGCCTGGTCAGCCTTGTGG-3′R: 5′-CCACGGTTGCGGTCGTCCAA-3′ |
| Sord | NM_146126 | F: 5′-GCTGGGAAGCAAGCCGGAGG-3′R: 5′-AATGGCTGCGTGCACCAGGG-3′ |
| St3gal5 | NM_011375 | F: 5′-GGCGATCCGCCTTTGCGATAC-3′R: 5′-CGGGTGTACCATTGCAGGGAGG-3′ |
| Ndufa1 | NM_019443 | F: 5′-CCGGGGTGTCCACTGCGTACA-3′R: 5′-CGCGTTCCATCAGATACCACTGGT-3′ |
| Prelid1 | NM_025596 | F: 5′-AGTGTTCGCCGCCTTCTGGC-3′R: 5′-CGCTCTGCCCAACGTGGCAT-3′ |
| Tmem86a | NM_026436 | F: 5′-TCCTGGTGGTGCAACCCGGA-3′R: 5′-CGCTGACCCACGATGGGCTG-3′ |
| Phkg2 | NM_026888 | F: 5′-ACGTGGGGCCGGAGGATGAA-3′R: 5′-CACAGCGGCGGACCACAGAG-3′ |
| Nup37 | NM_028334 | F: 5′-AAGGCCAGGAGCTCGCAAGC-3′R: 5′-AGTTTCCTCGGGATGCCAGCA-3′ |
| Cox6b1 | NM_025628 | F: 5′-GCAGCTTCCGGCCAGTCGGAA-3′R: 5′-GGTTGGGGAAGCGGCTGTCAAA-3′ |
| Mrps6 | NM_080456 | F: 5′-TCACAGCCAGCAGCACAGCC-3′R: 5′-ACCACGTCAATGTCTCGCGCC-3′ |
| Ccdc136 | NM_145574 | F: 5′-CCGTGCAGGAGCAGCTGG-3′R: 5′-GCCAAGAAAACCATAGGCTTCCACA-3′ |
| Ppia | NM_008907 | F: 5′-ACACGCCATAATGGCACTGG-3′R: 5′-ATTTGCCATGGACAAGATGCC-3′ |
Abbreviations: F, forward primer; R, reverse primer.
Bioinformatics analysis
Gene ontology was predicted by DAVID Bioinformatics Resources (http://www.david.abcc.ncifcrf.gov).
Results
Construction of TM4/AR cell line with higher androgen responsiveness than parental TM4 cells
We have stably transfected pcDNA3.1-AR into TM4 cells by Lipofectamine and obtained four individual cell clones with resistance to G418. We tested the expression of AR mRNA and protein in the G418-resistant populations by RT-PCR and Western blot, respectively. The results showed that the transcriptional mRNA and translational protein level of AR were significantly higher in the stably transfected TM4/AR cells than those found in the parental TM4 cells (P<0.01, Figure 1).
Figure 1.
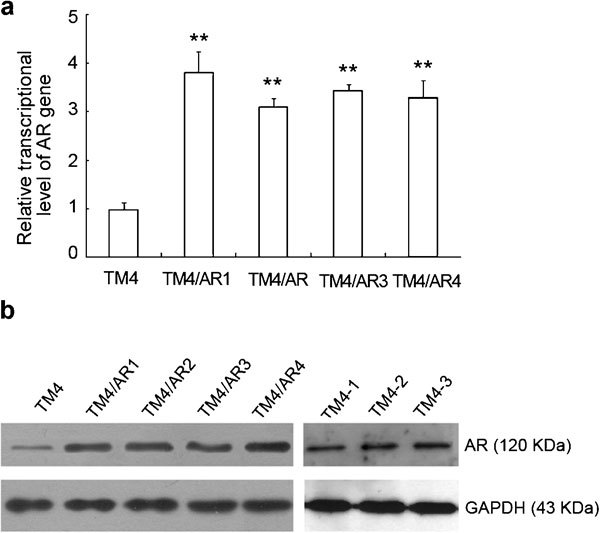
The gene expression of AR in TM4 cells and four stably transfected TM4/AR cell lines. (a) The relative transcriptional activity of AR detected by qRT-PCR. **P<0.01 compared with TM4. (b) The relative expression of AR protein in TM4 and four TM4/AR cell lines detected by Western blot. GAPDH was used as a loading control. AR, androgen receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qRT-PCR, quantitative reverse-transcription PCR analysis; TM4-1, TM4/AR1 cells; TM4-2, TM4/AR1 cells with AR-cDNA transient transfected; TM4-3, TM4/AR1 cells with AR-cDNA transient transfected and T treatment.
To assay the potential androgen responsiveness in these Sertoli cells, we transfected TM4 and TM4/AR cells with three ARE reporter constructs, MMTV-Luc, prostate-specific antigen-Luciferase and Androgen response element-luciferase, containing luciferase gene that could be activated by AR in the presence of T. In TM4 and four TM4/AR cell lines, both androgens, T and DHT, fail to induce the expression of luciferase reporter gene, which is under the control of ARE, at 10 nmol l−1, a concentration of T in mouse serum, or at 10 µmol l−1, a concentration of T in mouse testis (Figure 2).
Figure 2.
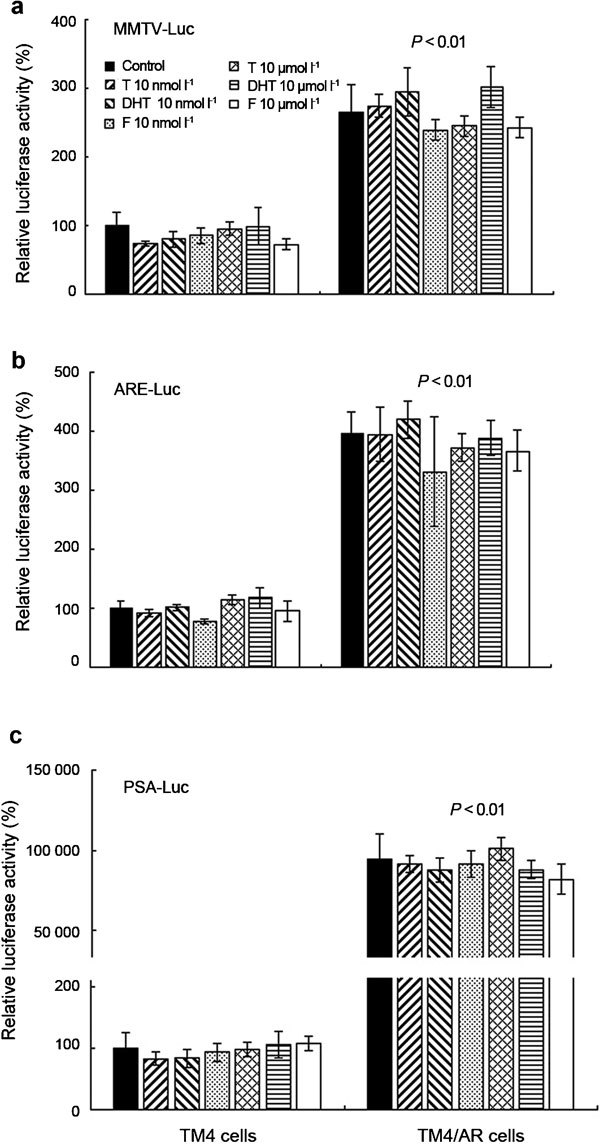
The effect of androgens (T and DHT) and antiandrogen flutamide on the expression level of reporter genes under the control of androgen response element in TM4 or TM4/AR cells. Luciferase activity was normalized to β-galactosidase activity and then shown relative to that of TM4 cells without androgen or antiandrogen treatment, which was set to 100%. Each value represents the mean±s.d. of experiments performed in triplicate. The data of TM4/AR cells are representative of four TM4/AR cell lines. ARE-Luc, androgen response element-luciferase; DHT, dihydrotestosterone; F, flutamide; MMTV-Luc, mouse mammary tumor virus-luciferase; PSA-Luc, prostate specific antigen-Luciferase; T, testosterone.
To examine if the saturated androgens in culture medium is the reason for no androgen responsiveness in the system, we added two concentrations of antiandrogen flutamide into the culture medium and found neither 10 nmol l−1 nor 10 µmol l−1 (Figure 2) of flutamide could significantly change the activity of luciferase. These results suggested that the transcriptional activity of AR in TM4 or TM4/AR cells was not induced by androgens. Although androgen responsiveness could not be detected in TM4/AR cells, we found the expression of reporter gene in TM4/AR cells was much higher than that in TM4 cells (P<0.01, Figure 2), suggesting that transfection of additional AR in TM4/AR cells did result in some changes, although it still failed to respond significantly to androgen treatment.
In contrast to above results, androgen responsiveness was detected when the ARE reporter constructs were cotransfected with AR-cDNA expression vector into TM4 or TM4/AR cells. In the TM4 cells, a significant response to androgens became measurable after adding 5 ng AR-cDNA construct and a maximal response was observed after adding 50 ng AR-cDNA construct (Figure 3a). In TM4/AR cells, the androgen response was detected after adding 1 ng AR-cDNA construct and a maximal response was observed after adding 5 ng AR-cDNA construct (Figure 3a). When the same concentration of AR-cDNA vector was cotransfected, luciferase activity reached its maximal value at 10 nmol l−1 of T in TM4 cells and 100 nmol l−1 in TM4/AR cells (Figure 3b). The fact that TM4/AR cells need less cotransfected AR-cDNA for inducing luciferase activity than TM4 cells suggests that AR, via stably transfected AR-cDNA in TM4/AR cells, did play some roles in the androgen responsiveness upon cotransfection of AR-cDNA.
Figure 3.
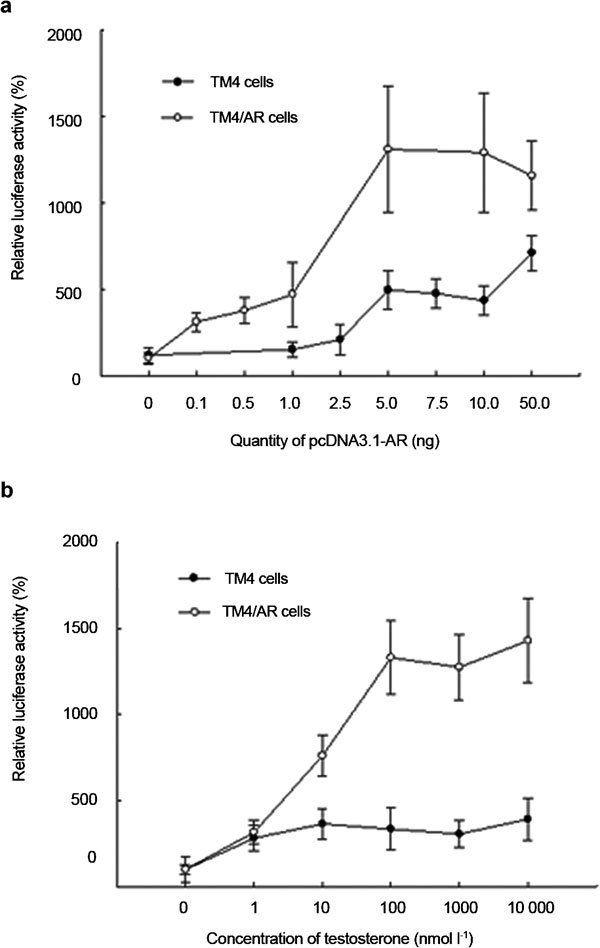
The effect of transient expression of AR and treatment of T on reporter gene expression controlled by androgen-responsive element in TM4 or TM4/AR cells. The MMTV-Luc luciferase activity in TM4 or TM4/AR cells was induced by cotransfecting AR-cDNA dose dependently in the presence of 100 nmol l−1 T (a) and induced by T in dose-dependent manner when cotransfected with 0.1 µg AR-cDNA (b). Luciferase activity was normalized to β-galactosidase activity and then shown relative to that of no treatment, which was set to 100%. Each value represents the mean±s.d. of experiments performed in triplicate. The data of TM4/AR cells are representative of four TM4/AR cell lines. AR, androgen receptor; MMTV-Luc, mouse mammary tumor virus-luciferase; T, testosterone.
TM4/AR cells were used in the following DGE analysis in the condition (transient transfection with 5 ng AR-cDNA construct and treatment with 100 nmol l−1 T) where maximal androgen response was demonstrated.
Identification of T- and AR-regulated genes in S-AR−/y mouse testes and TM4/AR cells
To identify AR-regulated genes in Sertoli cells, gene expression patterns in testes of S-AR−/y mice and control WT littermates were compared. DGE analysis showed that 5141 differentially expressed genes. Of these genes, 2865 were expressed at a higher level in S-AR−/y compared with WT control mice (referred to as downregulated by AR) and 2276 were expressed at a lower level in S-AR−/y compared with WT control mice (referred to as upregulated by AR) (Supplementary Table 1).
To avoid confounding effects of differences in testicular cell composition occurring in S-AR−/y mice, gene expression patterns in cultured TM4/AR cells were also compared with or without T/AR treatment. We conducted DGE sequencing analysis of total RNA from TM4/AR cells (TM4-1), TM4/AR cells with AR-cDNA transiently transfected (TM4-2), and TM4/AR cells with AR-cDNA transiently transfected and T treatment (TM4-3). In TM4-2 cells, 125 transcripts were upregulated and 28 downregulated compared with TM4-1 cells (Supplementary Table 2), suggesting that adding AR alone, in the absence of androgen, can also modulate some selective mRNA expressions. The altered mRNA expression in TM4-3 cells that demonstrated greater than twofold change compared with mRNA expression in TM4-1 cells are much higher (Supplementary Table 3): 1354 genes increased and 960 decreased relative to TM4-1 cells, suggesting most of AR-modulated genes are androgen-dependent. We also compared the difference between TM4-3 and TM4-2 cells and found the expression of 481 mRNAs were increased and 781 decreased, further suggesting that the majority of AR modulated gene expressions are androgen dependent (Supplementary Table 4).
There were 603 regulated genes, including 164 upregulated and 439 downregulated by androgen and AR, in the overlap part of two sets of differential expression genes in S-AR−/y mice testis and TM4/AR cells (Supplementary Table 5). The 14 genes strongly regulated and highly expressed both in S-AR−/y and TM4/AR cells are listed in Table 2.
Table 2. Some of the genes most strongly regulated and highly expressed both in S-AR−/y and TM4/AR cells.
| Symbol | Description | Log2 ratio by DGE | Log2 ratio by qPCR* | ||
|---|---|---|---|---|---|
| TM4-3/TM4-1 | S-AR−/y/WT | TM4-3/TM4-1 | S-AR−/y/WT | ||
| Upregulated by testosterone and androgen receptor | |||||
| Akap1 | A kinase (PRKA) anchor protein 1 | 1.40 | −3.33 | 1.13±0.15 | −3.12±0.11 |
| Phkg2 | Phosphorylase kinase, γ2 (testis) | 2.55 | −3.33 | 0.87±0.21 | −3.71±0.36 |
| Sord | Sorbitol dehydrogenase | 1.52 | −3.92 | 1.22±0.13 | −4.70±0.25 |
| Ccdc136 | Coiled-coil domain containing 136 | 1.68 | −3.46 | 0.83±0.21 | −3.59±0.47 |
| Downregulated by testosterone and androgen receptor | |||||
| Nup37 | Nucleoporin 37 | −5.92 | 4.01 | −6.59±0.21 | 4.94±0.23 |
| Cst3 | Cystatin C | −4.47 | 4.82 | −3.41±0.19 | 3.54±0.32 |
| Iapp | Islet amyloid polypeptide | −6.04 | 4.05 | −1.45±0.44 | 3.58±0.26 |
| Mybbp1a | MYB binding protein (P160) 1a | −5.50 | 3.36 | −4.31±0.47 | 2.54±0.19 |
| St3gal5 | ST3 β-galactoside α-2,3 -sialyltransferase 5 | −3.78 | 3.58 | −2.58±0.12 | 2.19±0.24 |
| Ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | −3.95 | 3.57 | −3.64±0.25 | 4.43±0.17 |
| Prelid1 | PRELI domain containing 1 | −5.31 | 4.30 | −3.32±0.21 | 2.01±0.20 |
| Tmem86a | Transmembrane protein 86A | −4.73 | 4.67 | −2.17±0.19 | 3.09±0.18 |
| Cox6b1 | Cytochrome c oxidase, subunit vib polypeptide 1 | −4.70 | 4.49 | −1.17±0.19 | 3.81±0.21 |
| Mrps6 | Mitochondrial ribosomal protein S6 | −5.99 | 4.60 | −3.26±0.18 | 4.12±0.24 |
Abbrevations: DGE, digital gene expression; qPCR, quantitative PCR; S-AR−/y, S-AR−/y mouse testis; TM4-1, TM4/AR cells; TM4-2, TM4/AR cells with AR-cDNA transient transfected; TM4-3, TM4/AR cells with AR-cDNA transient transfected and T treatment; WT, wild-type mouse testis.
*mean±s.d.
Confirmation of DGE data
To validate results obtained from DGE analysis, the qRT-PCR was performed in the 14 genes that appeared to be the most strongly regulated and highly expressed (Table 2). The results showed that 12 of 14 mRNAs demonstrated more than twofold changes by qRT-PCR. The two exceptions are Phkg2 and Ccdc136 in TM4/AR cells treated with AR and T, with the same variation trends, but less than twofold change.
For the spermatogenic arrest predominately at the diplotene premeiotic stage and almost no mature sperm produced in S-AR−/y mice, the genes specifically expressed in mature sperm were differently expressed in S-AR−/y and WT mice testis (Table 3). From another point of view, this also confirmed the DGE data, although it would make identification of AR target genes harder.
Table 3. Some of the genes most strongly regulated and highly expressed in S-AR−/y mice, but not in TM4/AR cells.
| Symbol | Description | TPM | ||||
|---|---|---|---|---|---|---|
| WT | S-AR−/y | TM4-1 | TM4-2 | TM4-3 | ||
| Spata3 | Spermatogenesis associated 3 | 745.54 | 1.36 | — | — | — |
| Hils1 | Histone H1-like protein in spermatids 1 | 2527.18 | 4.89 | 0 | 0 | 0.56 |
| Spem1 | Sperm maturation 1 | 1309.14 | 2.72 | — | — | — |
| Spatc1 | Spermatogenesis and centriole associated 1 | 397.57 | 1.09 | — | — | — |
| Spata19 | Spermatogenesis associated 19 | 869.26 | 2.99 | — | — | — |
| Spaca3 | Sperm acrosome associated 3 | 67.38 | 0.27 | — | — | — |
| Txndc2 | Thioredoxin domain containing 2 (spermatozoa) | 191.91 | 0.81 | 0 | 0 | 0.28 |
| Gapdhs | Glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 1480.03 | 6.25 | 0.26 | 0 | 0 |
| Odf3l1 | Outer dense fiber of sperm tails 3-like 1 | 217.25 | 1.09 | — | — | — |
| Spata9 | Spermatogenesis associated 9 | 46.36 | 0.27 | 0.26 | 0.28 | 0 |
| Spata18 | Spermatogenesis associated 18 | 197.3 | 1.36 | — | — | — |
| Odf3b | Outer dense fiber of sperm tails 3B | 38.54 | 0.27 | — | — | — |
| Spam1 | Sperm adhesion molecule 1 | 35.58 | 0.27 | — | — | — |
| Txndc3 | Thioredoxin domain containing 3 (spermatozoa) | 32.07 | 0.27 | — | — | — |
| Spert | Spermatid associated | 117.52 | 1.09 | — | — | — |
| Spata20 | Spermatogenesis associated 20 | 462.26 | 6.79 | — | — | — |
| Spaca4 | Sperm acrosome associated 4 | 220.75 | 3.26 | — | — | — |
| Catsper3 | Cation channel, sperm associated 3 | 48.79 | 0.81 | 0 | 0 | 0.28 |
| Izumo1 | Izumo sperm-egg fusion 1 | 73.31 | 1.36 | — | — | — |
| Spaca1 | Sperm acrosome associated 1 | 146.36 | 3.26 | — | — | — |
Abbrevations: TPM, normalized expression of gene; SAR–/y, S-AR–/y mice testis; TM4-1, TM4/AR cells; TM4-2, TM4/AR cells with AR-cDNA transient transfected; TM4-3, TM4/AR cells with AR-cDNA transient transfected and T treatment; WT, wild-type mice testis; –, no signal.
Bioinformatics analysis
The genes regulated by T and AR encoded proteins encompassing a broad range of molecules. Gene ontology was predicted by using DAVID 2.1 Beta (Laboratory of Immunopathogenesis and Bioinformatics, Frederick, MD, USA). Among the regulated genes resulting from the absence of AR in Sertoli cells and T treatment for TM4 cells, there was an overrepresentation of genes involved in Ras protein signal transduction (Baiap2, Kifap3, Sdcbp, Ctnnal1, Cdc42ep4, Nras, Farp2, LOC100048021, Lat, Grb2; P=8.6×10−5) classified by their biological process; mitochondrion (81 genes, P=8.1×10−10, Supplemental Table 6) by their cellular component or nucleotide binding (100 genes, P=1.1×10−5, Supplementary Table 7), and ubiquitin-protein ligase activity (Fbxo6, G2e3, Skp2, Chfr, Herc5, Ube2e3, Mdm2, Ube3a, Ube2b, Ube4b; P=4.2×10−3) by their molecular function.
Discussion
In this study, we used adult mice with AR-specific knockout from Sertoli cells (S-AR−/y) as an in vivo model system for identifying the T-/AR-regulated genes in adult Sertoli cells that may be involved in the control of spermatogenesis. As many as 5141 genes were differentially expressed in S-AR−/y testes vs. those from WT testes. There were different cell components in testes from S-AR−/y and WT because S-AR−/y was atrophied and spermatogenesis was arrested predominately at the diplotene premeiotic stage. For the purpose of distinguishing the genes regulated by AR in Sertoli cells, we constructed an in vitro system, Sertoli TM4/AR cell line. The genes regulated in S-AR−/y mice in vivo were compared to the T-/AR-regulated genes in TM4/AR cells in vitro, and 603 T-/AR-regulated genes were found both in S-AR−/y and TM4/AR cells. First, we removed the genes differentially expressed in S-AR−/y mice testis because of different cell compositions between S-AR−/y and WT mice testis. Second, 24-h treatment with T on TM4/AR cells will help us in identifying directly regulated genes by AR, but not indirectly regulated genes.
For construction of androgen response Sertoli cells line, we first detected the expression level of AR and androgen responsiveness of TM4 cells. Despite the presence of detectable endogenous AR in the original TM4 cells or stably transfected AR-cDNA in TM4/AR cells, we were surprised to find that androgens still failed to significantly induce AR transactivation in these cells. However, androgen responsiveness was restored by adding AR via transient transfection of AR-cDNA. These data suggested that the amount of AR expressed in these Sertoli cells played essential roles. Our data also showed the luciferase activity in TM4/AR cells is much higher than that found in TM4 cells in the absence or presence of the same amount of androgens. This induction in the absence of exogenous T suggests that the additional AR (via stably transfected AR-cDNA) could function in an T-independent manner, which is in agreement with an early report showing transfected AR-cDNA alone might lead to increased Rhox5 Pp transcripts expression in MSC1 cells without treatment with androgens.6 These data suggest that some ligands other than T might exist to stimulate AR transcriptional activity in TM4/AR Sertoli cells. This hypothesis can also be used to explain why the same amount of T in TM4/AR cells can induce more AR transcriptional activity than in TM4 cells (Figure 3).
In the past reports, for identifying AR target genes in Sertoli cells, several AR knockout mice models were used. Only three of the 40 potential AR target genes in prepubertal SCARKO mice testes,16 Rhox5, Stard10 and Ccdc70 were also in the list of AR-regulated genes in our S-AR−/y mouse testis, but none of these three potential AR target genes were found to be regulated by AR in TM4/AR cells. Although the same mice model was used, prepubertal and adult testes have different gene expression profiles, which could be the main reason for the lack of consistent data.
Although adult animals involved in Arinvflox(ex1−neo)/Y; Tg(Amh-Cre),10 testicular feminized mice17 and SCARKO-jsd (Utp14bjsd/jsd, juvenile spermatogonial depletion)11 systems were used, there were still much inconsistent data. Of the 62 AR-regulated transcripts found in Arinvflox(ex1−neo)/Y; Tg(Amh-Cre) system,10 there are four overlapping genes, including s100a10, pepd, acly and sc5d, with 603 T-/AR-regulated genes in this study. Of the 20 upregulated genes and six downregulated genes reported in testicular feminized mice,17 only Nudt19 was found regulated in S-AR−/y mice testis and TM4/AR cells. Recently, 157 genes were reported as downregulated and 197 genes as upregulated in the adult SCARKO-jsd mice compared to jsd mice.11 Ripk3 is the only overlapping gene between the list in SCARKO-jsd and our list in S-AR−/y and TM4/AR cells. A conspicuous reason is that the gene expression profiles of testes from Arinvflox(ex1−neo)/Y and jsd have been intensively changed. The juvenile spermatogonial depletion mutation in the Utp14b gene affected the gene expression in mice testis and complicated the analysis of AR target genes although the trouble because of different germ cell compositions in the testis was solved.
Inconsistent data were also reported on how androgens could modulate their target genes in the Sertoli cell line. Kasumi et al.18 reported that androgen treatment of TM4 cells induced the expression of Mif and S100a11; however, the similar induced patterns were not detected in our TM4/AR system and the transwell-cultured TM4 cells by Prante et al.19 Nine genes, including Rhox5 and Dab2, were regulated by T in the transwell-cultured TM4 cells.19 A similar change of Dab2 expression was demonstrated in our DGE data. However, it was reported that AR and the androgen, R1881, only weakly induced the activity of Rhox5 promoter in 15P1 and TM4 Sertoli cell lines.20 While we were able to demonstrate the increased luciferase activity induced by T in our TM4 and TM4/AR cells (Figure 3), we were unable to detect the expression of Rhox5 after T treatment (data not shown), because of low levels of GATA factors.20 Rhox5 was the only gene identified in common with the studies on AR knockout mice, but its expression was lost in our Sertoli cell line TM4 and TM4/AR. Knockout of Rhox5 gene has only limited effects on fertility of male mice compared with knockout of AR gene.21 Therefore, some other genes must be involved in the AR signals controlling spermatogenesis. These complicating and inconsistent data could be largely due to differences in culture conditions and experimental set-up of the Sertoli cell lines. In addition, they could be related to the complicated mechanism of AR signal in Sertoli cells.
A conspicuous characteristic of our 603 regulated genes is that there are many more T-/AR-downregulated genes than upregulated genes. There were a few reports about the regulation of androgen on AR expression, which is different in individual cells, such as smooth-muscle cells,22 prostate cancer cells,23, 24 osteoblast-like cells,25 bone cells26 and several rat organs.27 In our TM4/AR cells, AR protein level decreased when treated with T (data not shown), although the mRNA levels of AR were constant. The low AR protein level would result in decreased expression of AR target genes. Indeed, these AR target genes are not inhibited, but activated by AR. For this reason, the T/AR-downregulated genes should be interpreted with caution.
The dramatic overrepresentation of genes involved in ubiquitin-protein ligase activity among these 603 genes is possibly related with the fact that large amounts of protein must be degraded in Sertoli cells. Ubiquitination of histone is one of the epigenetic factors that regulate genes expression. Therefore, genes expression in Sertoli cells could be regulated by AR via changing the ubiquitination of histone.
In our previous study, 2058 genes in a gradually increased manner and 903 genes in a gradually decreased manner in the developmental stages of mice testis were identified, which is probably involved in spermatogenesis.28 By comparisons of these genes, 22 of 2058 genes and 28 of 903 genes were in the list of 603 T-/AR-regulated genes in this study (Supplementary Table 8). These genes, representing the most potentially important roles in spermatogenesis, are worth further studying.
In conclusion, the present study has identified a subset of genes that are differentially expressed in adult testes with or without an active AR in Sertoli cells and also in TM4/AR cells with or without testosterone treatment. Of particular interest is the finding that some genes that may be related to ubiquitin-protein ligase activity. If ubiquitination of histone was affected, expression of many genes could be changed.
Author contributions
QXZ carried out total RNA extraction and DGE analysis, participated in all experiments, and drafted the manuscript. XYZ carried out Western blot analysis and participated in cell culture. ZMZ participated in total RNA extraction. WL participated in RT-PCR and quantitative real-time PCR analysis. LL participated in luciferase assay. GL participated in design of the study and coordination. ZMC participated in design of the study and coordination. YTG gave final approval of the version to be published. CSC conceived of the study and gave final approval of the version to be published.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 30971636), and the George H. Whipple Professorship Endowment, and National Science Council, Taiwan, China (96-2314-B-182A-023-MY2 and 97-2314-B-182A-077-MY3).
The authors declare no competing financial interest.
Footnotes
Note: Supplementary Information accompanies the paper on Asian Journal of Andrology website (http://www.nature.com/aja).
Supplementary Information
References
- Mruk DD, Cheng CY. Sertoli–Sertoli and Sertoli–germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev. 2004;25:747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Yeh SD, Wang RS, Yeh S, Zhang C, et al. Differential effects of spermatogenesis and fertility in mice lacking androgen receptor in individual testis cells. Proc Natl Acad Sci USA. 2006;103:18975–80. doi: 10.1073/pnas.0608565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci USA. 2004;101:1327–32. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, et al. Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc Natl Acad Sci USA. 2004;101:6876–81. doi: 10.1073/pnas.0307306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RS, Yeh S, Tzeng CR, Chang C. Androgen receptor roles in spermatogenesis and fertility: lessons from testicular cell-specific androgen receptor knockout mice. Endocr Rev. 2009;30:119–32. doi: 10.1210/er.2008-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Meistrich ML, Wilkinson MF. Pem homeobox gene promoter sequences that direct transcription in a Sertoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. . Mol Endocrinol. 2003;17:223–33. doi: 10.1210/me.2002-0232. [DOI] [PubMed] [Google Scholar]
- Lim K, Yoo JH, Kim KY, Kweon GR, Kwak ST, et al. Testosterone regulation of proto-oncogene c-myc expression in primary Sertoli cell cultures from prepubertal rats. J Androl. 1994;15:543–50. [PubMed] [Google Scholar]
- Lecureuil C, Fontaine I, Crepieux P, Guillou F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis. 2002;33:114–8. doi: 10.1002/gene.10100. [DOI] [PubMed] [Google Scholar]
- Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, et al. Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci USA. 2002;99:13498–503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker SM, Shima JE, Connolly CM, Sharma M, Holdcraft RW, et al. Transcriptional profiling of androgen receptor (AR) mutants suggests instructive and permissive roles of AR signaling in germ cell development. Mol Endocrinol. 2007;21:895–907. doi: 10.1210/me.2006-0113. [DOI] [PubMed] [Google Scholar]
- Zhou W, Wang G, Small CL, Liu Z, Weng CC, et al. Gene expression alterations by conditional knockout of androgen receptor in adult Sertoli cells of Utp14bjsd/jsd (jsd) mice. Biol Reprod. 2011;84:400–8. doi: 10.1095/biolreprod.110.090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather JP. Establishment and characterization of two distinct mouse testicular epithelial cell lines. Biol Reprod. 1980;23:243–52. doi: 10.1095/biolreprod23.1.243. [DOI] [PubMed] [Google Scholar]
- Ham J, Thomson A, Needham M, Webb P, Parker M. Characterization of response elements for androgens, glucocorticoids and progestins in mouse mammary tumour virus. Nucleic Acids Res. 1988;16:5263–76. doi: 10.1093/nar/16.12.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizokami A, Gotoh A, Yamada H, Keller ET, Matsumoto T. Tumor necrosis factor-alpha represses androgen sensitivity in the LNCaP prostate cancer cell line. J Urol. 2000;164:800–5. doi: 10.1097/00005392-200009010-00053. [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Denolet E, de Gendt K, Allemeersch J, Engelen K, Marchal K, et al. The effect of a sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Mol Endocrinol. 2006;20:321–34. doi: 10.1210/me.2005-0113. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Abel M, Charlton HM, Hu B, Johnston H, et al. Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive tfm mouse testis. Endocrinology. 2007;148:2914–24. doi: 10.1210/en.2006-1412. [DOI] [PubMed] [Google Scholar]
- Kasumi H, Komori S, Sakata K, Yamamoto N, Yamasaki T, et al. Upregulation of macrophage migration inhibitory factor and calgizzarin by androgen in TM4 mouse Sertoli cells. Asian J Androl. 2006;8:549–54. doi: 10.1111/j.1745-7262.2006.00196.x. [DOI] [PubMed] [Google Scholar]
- Prante BC, Garman KL, Sims BN, Lindsey JS. Matrix-coated transwell-cultured TM4 sertoli cell testosterone-regulated gene expression mimics in vivo expression. In Vitro Cell Dev Biol Anim. 2008;44:434–43. doi: 10.1007/s11626-008-9135-8. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Rao MK, Kaur R, Buttigieg MR, Wilkinson MF. GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Mol Cell Biol. 2008;28:2138–53. doi: 10.1128/MCB.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean JA 2 nd, Chen MA, Wayne CM, Bruce SR, Rao M, et al. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–82. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid N, Vernet D, Fuentes Navarro A, Rodriguez JA, Swerdloff RS, et al. Up-regulation of the levels of androgen receptor and its mRNA by androgens in smooth-muscle cells from rat penis. Mol Cell Endocrinol. 1993;90:219–29. doi: 10.1016/0303-7207(93)90155-d. [DOI] [PubMed] [Google Scholar]
- Grad JM, Dai JL, Wu S, Burnstein KL. Multiple androgen response elements and a Myc consensus site in the androgen receptor (AR) coding region are involved in androgen-mediated up-regulation of AR messenger RNA. Mol Endocrinol. 1999;13:1896–911. doi: 10.1210/mend.13.11.0369. [DOI] [PubMed] [Google Scholar]
- Blanchere M, Berthaut I, Portois MC, Mestayer C, Mowszowicz I. Hormonal regulation of the androgen receptor expression in human prostatic cells in culture. J Steroid Biochem Mol Biol. 1998;66:319–26. doi: 10.1016/s0960-0760(98)00056-9. [DOI] [PubMed] [Google Scholar]
- Grad JM, Lyons LS, Robins DM, Burnstein KL. The androgen receptor (AR) amino-terminus imposes androgen-specific regulation of AR gene expression via an exonic enhancer. Endocrinology. 2001;142:1107–16. doi: 10.1210/endo.142.3.8049. [DOI] [PubMed] [Google Scholar]
- Wiren KM, Zhang X, Chang C, Keenan E, Orwoll ES. Transcriptional up-regulation of the human androgen receptor by androgen in bone cells. Endocrinology. 1997;138:2291–300. doi: 10.1210/endo.138.6.5163. [DOI] [PubMed] [Google Scholar]
- Quarmby VE, Yarbrough WG, Lubahn DB, French FS, Wilson EM. Autologous down-regulation of androgen receptor messenger ribonucleic acid. Mol Endocrinol. 1990;4:22–8. doi: 10.1210/mend-4-1-22. [DOI] [PubMed] [Google Scholar]
- Xiao P, Tang A, Yu Z, Gui Y, Cai Z. Gene expression profile of 2058 spermatogenesis-related genes in mice. Biol Pharm Bull. 2008;31:201–6. doi: 10.1248/bpb.31.201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


