Abstract
In cells infected with herpes simplex virus 1, the RNA encoded by the stress-inducible immediate early response gene IEX-1 was up-regulated immediately after infection. However, the accumulated RNA was degraded 3′-5′, and the protein was detectable only at very early times after infection. The degradation was dependent on the UL41 gene encoding the virion host shutoff (vhs) protein and resulted in the accumulation of truncated RNA containing the 5′-end portion of the transcript. IEX-1 contains an AU-rich element (ARE) in its 3′-untranslated domains known to regulate negatively the RNA lifespan. To examine the role of ARE in signaling the degradation, we compared the stability of several RNAs up-regulated during infection to WT virus. These were ARE-containing RNAs encoding IEX-1, c-fos, and IκBα and the non-ARE-containing RNAs GADD45β and tristetraprolin. We report that the ARE-containing RNAs exemplified by IEX-1 RNA are deadenylated and cleaved in the ARE within the 3′ UTR in a UL41-dependent manner. In contrast, Northern blot hybridizations and analyses of poly(A) tails revealed no evidence of degradation of GADD45β RNA. GADD45β protein was detected in WT virus-infected cells. These results indicate that the degradation of RNAs and the mechanism by which cellular RNAs are degraded are selective and may be sequence specific. The persistence of partially degraded ARE-containing RNAs may reflect specific targeting of the vhs proteins to the ARE and the modification of the RNA degradation machinery of the cell induced by the presence of the vhs protein.
One of the key early events in the replication of herpes simplex virus 1 (HSV-1) is the diminished incorporation of amino acids into cellular proteins. This process was mapped to a gene designated UL41, and the protein product was designated virion host shutoff or vhs (1-3). vhs is an Mr 58,000 polypeptide, which is packaged in the tegument of HSV virions, and its activity has been extensively investigated. The current model of vhs function is that, on infection, vhs is released into the cytoplasm and acts as an RNase in conjunction with the translational factor eIF4H (4). It has been reported that vhs degrades sequences near the 5′ end of mRNA more rapidly than those at the 3′ end (5). The nucleolytic activity of vhs is specific for mRNA. Host rRNA and tRNA are unaffected, whereas viral and host mRNAs are rapidly degraded. Because viral mRNA is transcribed at a higher rate than cellular mRNA, viral proteins do accumulate. At middle or late times after infection, vhs interacts with the α-trans-inducing factor also known as VP16 encoded by the UL48 ORF and it becomes inactive (6). The function of vhs appears to be twofold, to enable efficient transition from α to β and γ protein synthesis and to block host responses to infection. Indeed, ΔUL41 mutants are more sluggish in their replication and replicate poorly in immunocompetent experimental animal systems (7).
The problem addressed in this report stems from two observations. The first involved comparisons of host cell RNAs induced in WT virus-infected cells and in cells infected with a ΔUL41 mutant (8). The expectation was that the number of cellular genes whose transcripts would be up-regulated and the level of accumulated mRNAs in ΔUL41-mutant-infected cells would be vastly higher than those in WT virus-infected cells. This was not the case. The second observation involved a thorough analysis of one cellular RNA, encoded by the stress-inducible immediate-early response gene IEX-1, and revealed that although the RNA was indeed up-regulated after HSV-1 infection, it was rapidly degraded as a result of a 3′-5′ RNA decay in a UL41-dependent manner (9). These results appeared to challenge both the specificity and the mechanism of vhs-dependent degradation of cellular RNAs.
In an attempt to resolve the apparent discrepancy in the results published to date, we examined several more cellular RNAs with respect to their stability and expression in WT virus-infected cells. In this instance, the selection of the RNAs was not random. It is now well established that specific regulatory sequences, often located within the 3′ UTR of the mRNA, function to promote stability or instability of the transcript (10). The AU-rich element (ARE), an adenylate- and uridylate-rich element of the sequence AUUUA found in many unstable mRNAs, is the most widespread determinant of RNA instability, although the mere presence of the pentanucleotide does not predict instability. The AREs differ in size, AU content, and number of AUUUA motifs; a recently updated ARE database (http://rc.kfshrc.edu.sa/ARED) lists ≈900 ARE-containing mRNAs (11). IEX-1 mRNA is listed in this database. The published data on the properties of ARE-containing RNAs indicate that the products of degradation in infected cells do not linger for extended periods of time (12, 13). This is in stark contrast to the observed persistence of truncated IEX-1 RNAs in HSV-1-infected cells. To investigate this phenomenon further, we selected two ARE-containing RNAs, those encoding IκBα and c-fos proteins, and two RNAs not containing ARE (11), i.e., those encoding GADD45β and tristetraprolin (TTP).
In an earlier publication (9), we reported that IEX-1 RNA is degraded in a 3′-5′ direction and that the truncated 5′ domains persist and are readily detected for many hours in the infected cells. We have extended these findings to show that in addition the RNA is deadenylated, that an endonucleolytic cleavage occurs within the ARE, and that all of these events are UL41 gene dependent. Neither IEX-1 nor IκBα proteins accumulate in appreciable quantities. IκBα and c-fos RNAs undergo similar patterns of degradation. In stark contrast, analyses of GADD45β and TTP RNAs yielded no evidence of persistence of deadenylated RNAs or preferential 3′-5′ RNA decay, and moreover, these proteins accumulate in infected cells. These results indicate that the degradation of RNAs and the mechanism by which cellular RNAs are degraded are selective and may be sequence-specific.
Materials and Methods
Cells and Viruses. SK-N-SH and HeLa cells obtained from the American Type Culture Collection were propagated in DMEM supplemented with 5% newborn calf serum. Telomerase-transformed primary human foreskin fibroblasts (14), a kind gift of T. Shenk (Princeton University, Princeton) were cultured in DMEM supplemented with 10% FCS. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (15). The ΔUL41-mutant virus, R2621, was reported elsewhere (16).
Isolation of RNA. Total RNA was extracted with the aid of TRIzol reagent (Life Technologies, Rockville, MD) according to the manufacturer's instructions. DNase treatment (Life Technologies), phenol/chloroform extraction, and ethanol precipitation (Fisher Scientific) were carried out to remove possible DNA contamination. Cytoplasmic RNA was isolated with the aid of RNeasy mini kit according to the protocol suggested by the manufacturer (Qiagen).
Deadenylation of RNA. Total cytoplasmic RNA was isolated at various times from mock- and HSV-1(F)-infected cells. Poly(A)- RNA was prepared in vitro by treating RNA samples with oligo(dT) and RNase H (17). RNA (12 μg) was mixed with 0.5 μg of oligo(dT)15 (Promega) in annealing buffer (20 mM Tris, pH 8.0/5 mM MgCl2/2 mM DTT/0.006% BSA) and then digested with Escherichia coli RNase H (Ambion) at 37°C for 30 min. RNA was phenol extracted, ethanol precipitated, electrophoresed in a 1.5% agarose gel, and analyzed by Northern blot hybridizations.
Northern Blots. Cytoplasmic (8 μg) or total (15 μg) RNA was loaded onto denaturing formaldehyde gel and transferred onto a nylon membrane. Prehybridization and hybridization were done with the ULTRAhyb buffer (Ambion) supplemented with 200 μg of denatured salmon sperm DNA per ml (Stratagene). The prehybridization and hybridization steps were carried out at 42°C for DNA probe and at 68°C for RNA probes. The membranes were rinsed as suggested by the manufacturer of ULTRAhyb and exposed to film for signal detection. For c-fos detection, an oligoprobe antisense (5′-TGCGGGTGATGGTAGTAAGAGAGGCTATCCCC-3′) was 32P-labeled by using [γ-32P]ATP and T4 polynucleotide kinase. For IκBα, GADD45β, and TTP mRNA detection, the specific coding sequences were amplified by RT-PCR of total RNA purified from HSV-1-infected human-foreskin fibroblasts as described (9) and by using the following pairs of primers: IκBα forward-EcoRI (5′-CGAATTCGCGCCATGTTCCAGGCG-3′) and IκBα reverse-NotI (5′-ATGTTCTTGCGGCCGCTT TGCACTCA-3′); GADD45β forward-EcoRI (5′-CCGAATTCATGAC GCTGGAAGAGCTC-3′) and GADD45β reverse-NotI (5′-ATAAGAATGCGGCCGCTCGAGGTCAGC GTTCCTGAAGA-3′); and TTP forward-EcoRI (5′-AACGAATTCACCATGGATCTGACTGCCATC-3′) and TTP reverse-NotI (5′-CTGACCGGGCGGCCGCTTTGTCACTCAGAAAC-3′). The generated fragments were ligated into pcDNA3.1(+) (Invitrogen) after cleavage by EcoRI and NotI restriction enzymes and subsequently sequenced. To generate the probes for Northern blot hybridizations, the fragments containing the sequence of interest were cut from their vectors, using the appropriate restriction enzymes, and subsequently 32P-labeled by using Prime-a-Gene labeling system (Promega), according to the manufacturer's instructions. For the IEX-1 RNA 3′-UTR analyses, two RNA probes antisense to the 3′ UTR were generated by transcription in vitro. The Ribo1 template was amplified by PCR from HSV-1-infected human-foreskin fibroblasts cDNA by using a forward primer encompassing the stop codon of IEX-1 coding sequence (nucleotides 491-510 of the published sequence, GenBank accession no. NM003897) (Ribo1 forward: 5′-CGCCTTCTAACTGTGACTCC-3′) and a reverse primer located 300 bp downstream from the stop codon (nucleotides 781-800) and containing the T7 promoter sequence at its own 5′-end (Ribo1 reverse: 5′-TAATACGACTCACTATAGGTCACCTAGGAGGACGTACAT-3′). For Ribo2 template amplification, a forward primer located in the same position of Ribo1 reverse primer (Ribo2 forward: 5′-ATGTACGTCCTCCTAGGTGA-3′) and a reverse primer complementary to the region located between nucleotides 1,056 and 1,075 of IEX-1 RNA and containing the T7 promoter sequence at its own 5′ end (Ribo2 reverse: 5′-TAATACGACTCACTATAGGCGCCGAAGTCTCACACAGTA-3′) were used. PCR products were used directly as templates in the in vitro transcription reaction in presence of [α-32P]UTP and T7 polymerase with the aid of the MAXIscript in vitro transcription kit (Ambion).
Real-Time PCR. Real-time PCRs were performed on a Prism 7000 sequence detection system (Applied Biosystems) with SYBR-green chemistries as described (9). The IEX-1 primer sets were reported (9). The following primers were also used: 5′-FOS 5′-GCAGCGAGCAACTGAGAA-3′ (forward) and 5′-AACATCATCGTGGCGGTTA-3′(reverse); 3′-FOS 5′-AGGACCTTATCTGTGCGTGAAAC-3′ (forward) and 5′-CCGGAAGAGGTAAGGACTTGAG-3′ (reverse); 5′-GAD 5′-CTCGCCAAGGACTTTGCAATAT-3′ (forward) and 5′-GCAAAATCCGAGCCAGAGA-3′(reverse); rt-GAD 5′-TCGGCCAAGTTGATGAATGTG-3′ (forward) and 5′-GGATGAGCGTGAAGTGGATTTG-3′ (reverse); and 3′-GAD 5′-CCCTCGACAAGACCACACTTT-3′ (forward) and 5′-TCGCTCTCAGTGGTTCGAATA-3′ (reverse).
Immunoblot of GADD45β Cells were collected, rinsed once with cold PBS, and lysed in lysis buffer [100 mM NaCl/50 mM Tris·HCl, pH 7.5/0.5% Nonidet P-40/1 mM sodium orthovanadate and protease inhibitor mixture (Complete protease mixture, Roche Diagnostics)]. Proteins (100 μg) were separated on a denaturing 15% polyacrylamide gel and electrically transferred to a nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with 10% nonfat dry milk in TBS (10 mM Tris·HCl, pH 8/150 mM NaCl) and reacted with the GADD45β-specific mAb (kindly provided by G. Franzoso, University of Chicago) for 1 h at room temperature and then overnight at 4°C. The primary Ab was diluted in TBS-T (TBS plus 0.1% Tween 20) containing 1% nonfat dry milk. The membrane was rinsed three times with TBS-T and then exposed to secondary Ab at room temperature for 1.5 h. Secondary Ab was peroxidase-conjugated goat anti-mouse (Sigma), diluted 1:1,000 in TBS-T containing 5% nonfat dry milk. To develop peroxidase-conjugated secondary Ab, the immunoblot was reacted with enhanced chemiluminescence Western blotting detection reagents, according to the manufacturer's instructions (Amersham Pharmacia).
Results
Several HSV-1-Induced mRNAs but Not All Are Degraded After HSV-1 Infection. In an earlier report, we showed that IEX-1 mRNA was up-regulated after HSV-1 infection, but the synthesis of the protein was precluded by several mechanisms, including the vhs-dependent 3′-end degradation of its own RNA (9). A striking feature of the results was the presence of truncated forms of IEX-1 RNA in the cytoplasm of infected cells. To determine whether this phenomenon was specific for IEX-1 or a more general mechanism, the RNA accumulations of several up-regulated genes during HSV-1 replication were analyzed by Northern blot hybridizations. We observed two entirely distinct patterns of accumulation of cellular RNAs.
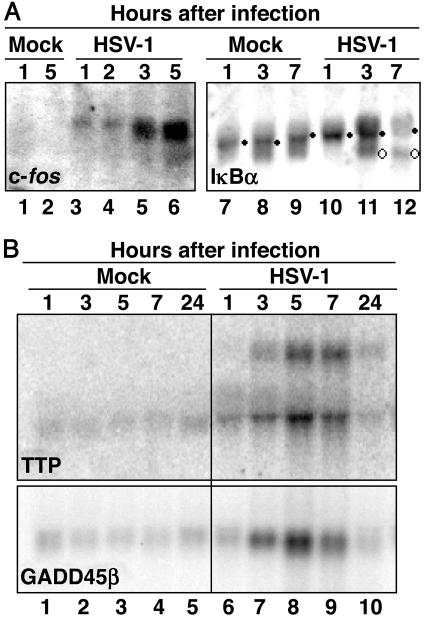
The first pattern is exemplified by IEX-1 reported in the earlier publication (9). A pattern similar to that of IEX-1 RNA was observed for c-fos and IκBα RNA (Fig. 1A). Thus, in both instances, bands of progressively smaller RNAs were detected in the cytoplasm of cells harvested at 3 h after HSV-1 infection (Fig. 1A, lanes 5 and 11). Furthermore, as predicted from the studies on the expression of IEX-1, the IκBα protein disappeared very quickly during the infection (18).
Fig. 1.
RNA accumulations of c-fos, IκBα, TTP, and GADD45β in cells infected with HSV-1. Cells were mock-infected or infected with 10 plaque-forming units (pfu) of HSV-1(F) per cell. Cytoplasmic RNA was purified and analyzed as described in Materials and Methods. (A Left) Accumulation of c-fos mRNA in cytoplasmic RNA extracted from mock-infected (lanes 1 and 2) or HSV-1-infected (lanes 3-6) HeLa cells. (A Right) Accumulation of IκBα mRNA in cytoplasmic RNA extracted from mock-infected (lanes 7-9) or HSV-1-infected (lanes 10-12) SK-N-SH-infected cells. •, intact RNA; ○, degraded RNA. (B) Accumulation of TTP and GADD45β mRNAs in cytoplasmic RNA extracted from mock-infected (lanes 1-5) or HSV-1-infected (lanes 6-10) HeLa cells.
The second pattern was exemplified by two RNAs, that of TTP and that of the growth arrest and DNA damage-inducible gene 45β (GADD45β) (Fig. 1B). Microarray studies reported elsewhere showed that both genes were up-regulated after HSV-1 infection (8). Northern blot analyses (Fig. 1B) verified the up-regulation of both genes, but in this instance there was no evidence that the RNAs were subject to degradation. As already reported for IEX-1 mRNA (9) and for α-globin RNA (19), the slower-migrating additional cytoplasmic RNA band from infected cells detected with the probe for TTP mRNA (Fig. 1B Upper, lanes 7-10) could represent unspliced TTP RNA whose presence is dependent on the presence of functional ICP27 (20). This band was absent in cells infected with a Δα27-mutant virus (data not shown).
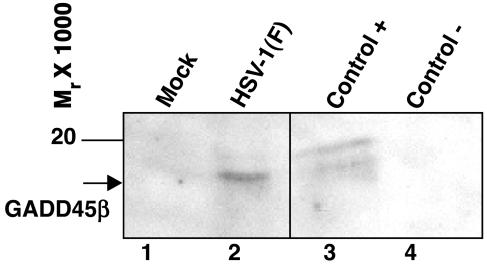
Consistent with the accumulation of RNAs, we observed the accumulation of TTP (data not shown) and GADD45β proteins (Fig. 2) in infected cells. GADD45β protein was clearly overexpressed in HeLa cells harvested 7 h after infection with HSV-1(F) (Fig. 2, lane 2). The 293 cells stably transduced with a GADD45β construct were used as a positive control (Fig. 2, lane 3).
Fig. 2.
Accumulation of GADD45β protein in HeLa cells after HSV-1 infection. HeLa cells were mock-infected (lane 1) or infected with 10 pfu of HSV-1(F) per cell (lane 2) and were collected at 7 h after infection. Cell lysate from 293 cells stably transfected with a GADD45β-flag tagged construct or the empty vector was used as positive (lane 3) and negative (lane 4) controls, respectively.
We conclude from these studies that cellular mRNAs induced during WT virus infection fall into at least two groups. The first is degraded in a UL41 gene-dependent manner, and the translation products are either not made or do not accumulate in amounts detectable in immunoblots. The second group appears to be stable throughout infection and yields demonstrable quantities of translation products.
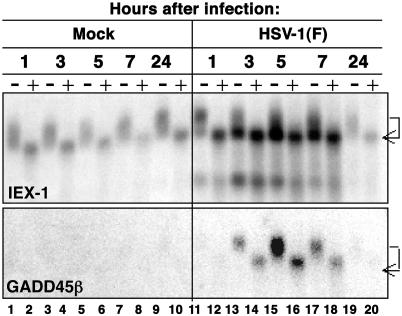
IEX-1 mRNA but Not GADD45β mRNA Is Deadenylated During HSV-1 Infection. Poly(A) tail removal is a critical first step in the decay pathway for many yeast and mammalian RNAs. The objective of the experiments described in this section was to determine whether the cellular RNAs accumulating after infection are deadenylated. In this series of experiments, we selected IEX-1 and GADD45β as representative of the two classes of RNAs described above. Cytoplasmic RNAs were extracted from HeLa cells at different times after mock infection or infection with 10 pfu of HSV-1(F) per cell. Poly(A)- RNAs of each sample were prepared in vitro by treating cytoplasmic RNA with oligo(dT) and RNase H. Untreated and treated RNAs were then denatured, electrophoretically separated, and hybridized first to IEX-1 probe (Fig. 3 Upper) and subsequently to GADD45β probe (Fig. 3 Lower). The results may be summarized as follows. (i) Untreated cytoplasmic IEX-1 RNA extracted from mock-infected cells formed a broad band of relatively uniform density reflecting variability in the size of the poly(A) tail (Fig. 3 Upper, lanes 1, 3, 5, 7, and 9). The band present in treated samples corresponds to the poly(A)- RNAs (Fig. 3 Upper, lanes 2, 4, 6, 8, and 10) and migrates faster than untreated RNA, indicating that even the shortest (fastest-migrating) RNA contained some poly(A) sequences. (ii) Untreated cytoplasmic IEX-1 RNA from infected cells formed a similar broad band at 1 h after infection, indicating that the bulk of RNA species was polyadenylated (Fig. 3 Upper, lane 11). In contrast, the majority of RNA forms extracted at 3, 5, and 7 h after infection (Fig. 3 Upper, lanes 13, 15, and 17) migrated very close to the poly(A)- RNAs (Fig. 3 Upper, lanes 14, 16, and 18). We conclude from these data that the bulk of IEX-1 RNA accumulating in the infected cells was deadenylated 1-3 h after infection. (iii) The amounts of GADD45β in mock-infected cells were too low to be quantified (Fig. 3 Lower, lanes 1-10). As predicted from earlier studies, there was a major increase in the GADD45β RNA accumulating in infected cells between 3 and 7 h after infection. The electrophoretic mobility of the untreated GADD45β RNA suggested a relatively uniform distribution of RNA sizes that did not change between 3 and 7 h after infection (Fig. 3 Lower, lanes 11-20). The results indicate that the GADD45β RNA is relatively stable and does not undergo rapid deadenylation characteristic of IEX-1 RNA.
Fig. 3.
Analysis of mRNA deadenylation. HeLa cells were mock-infected (lanes 1-10) or infected with 10 pfu of HSV-1(F) per cell (lanes 11-20), and cytoplasmic RNA was extracted at the indicated hours after infection. Poly(A)- RNA was prepared in vitro by treating the RNA samples with oligo(dT) and RNase H as described in Materials and Methods. Treated (+) and untreated (-) RNA was probed with a 32P-labeled fragment containing the entire coding sequences of IEX-1 or GADD45β.
Progressive 3′-5′ Degradation of c-fos but Not GADD45β mRNA in HSV-1-Infected Cells. In an earlier publication, using real-time PCR analyses based on three sets of primers designed to quantify different portions of the RNAs, we reported that IEX-1 RNA is progressively degraded in a 3′-5′ direction (9). Inasmuch as these results were different from the reports that cellular RNAs are degraded at or near the 5′ terminus after infection with WT virus, it was of interest to determine the pattern of degradation of c-fos RNA as another example of a RNA degraded in a fashion similar to that of IEX-1, and of GADD45β as representative of the second class of RNAs described above.
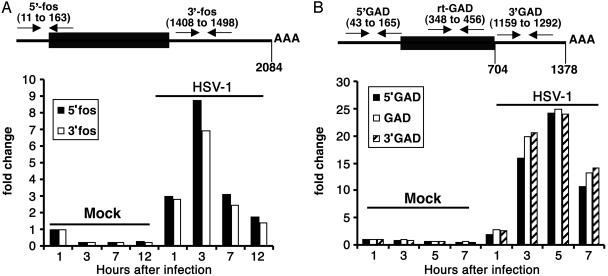
A real-time PCR analysis of c-fos RNA was set up by using two different sets of primers distributed along the RNA sequence. The location of the primers is shown in Fig. 4A Upper, and they measured the 5′ domain (5′ FOS primers) and the 3′ noncoding domain (3′ FOS primers). Replicate cultures of human foreskin fibroblasts were mock-infected or exposed to 10 pfu of HSV-1(F) per cell. The cells were harvested at 1, 3, 7, or 12 h after infection or mock infection. The extracted total RNA was reverse transcribed by using random hexamers and normalized with respect to reverse transcribed 18S rRNA. The amount of c-fos RNA from cells harvested 1 h after mock infection was used as a calibrator for each pair of primers. The results presented in Fig. 4A were consistent with the results of Northern blot hybridization (Fig. 1A), showing that the accumulation of c-fos RNA increased after HSV-1 infection. The c-fos RNA reached peak levels at 3 h after infection and declined thereafter. The comparison of the quantities of c-fos RNA with the two primer sets indicated that portion of the c-fos RNA extracted at different times after infection with WT virus lacked the 3′ sequences measured by the 3′ FOS primers. The loss of 3′ sequences was even more striking in a second experiment (data not shown). In contrast, the amounts of c-fos RNA detected by the two primer sets in mock-infected cells were approximately equal. We conclude from these results that a significant fraction of the c-fos RNA accumulating after the first hour of infection with HSV-1 was truncated at or near the 3′ terminus.
Fig. 4.
Analysis of the 5′ and 3′ regions of c-fos and GADD45β mRNA by real-time PCR. (A Upper) The location of the primers used for real-time PCR along the c-fos RNA sequence. (A Lower) Total RNA was extracted at the indicated hours after mock or HSV-1(F) infection of human foreskin fibroblasts. (B Upper) Location along the GADD45β RNA sequence of the primers used for real-time PCR. (B Lower) Total RNA was extracted at the indicated hours after mock or HSV-1(F) infection of HeLa cells. The amount of RNA accumulating in infected cells was calculated as fold change compared with that of cells harvested 1 h after mock infection.
A second real-time PCR analysis was set up by using three different sets of primers scattered along the GADD45β RNA sequence. The location of the primers is shown in Fig. 4B Upper, and they measured the 5′ domain (5′ GAD primers), the coding domain (rt-GAD primers), and the 3′ noncoding domain (3′ GAD primers). Replicate cultures of HeLa cells were mock-infected or exposed to 10 pfu of HSV-1(F) per cell. The cells were harvested at 1, 3, 5, or 7 h after infection or mock infection. The extracted total RNA was reverse-transcribed by using random hexamers and normalized with respect to reverse-transcribed 18S rRNA. The amount of GADD45β RNA from cells harvested 1 h after mock infection was used as a calibrator for each pair of primers. The results (Fig. 4B) were as follows. (i) As predicted on the basis of the experiments described above, the assays with the three primer sets verified the increase in the total amount of GADD45β RNA after infection. GADD45β RNA reached maximum levels at 5 h after infection and declined after 7 h of infection. The GADD45β RNA present in mock-infected cells stays stable throughout the 7-h duration of the experiment. (ii) In contrast to what is observed with IEX-1 and c-fos RNAs, the quantities of GADD45β RNA detected with the three primer sets in extracts of HSV-1-infected cells were similar. We conclude from these studies that GADD45β was not subject to 3′-5′ degradation after HSV-1 infection.
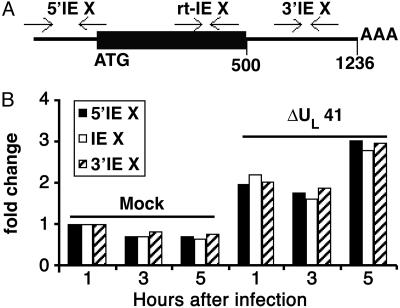
The Progressive 3′-5′ Degradation of IEX-1 RNA Is Mediated by vhs. To determine whether vhs mediates the progressive 3′-5′ degradation of IEX-1 RNA, we compared RNA extracted from ΔUL41-infected cells by using the same sets of IEX-1 primers as described (9). The location of the primers is shown in Fig. 5A, and they measured the accumulated RNAs containing the 5′ domain (5′ IEX primers), the coding domain (rt-IEX primers), and the 3′ noncoding domain (3′ IEX primers). Replicate cultures of HeLa cells were mock-infected or exposed to 10 pfu of ΔUL41-mutant virus per cell. The cells were harvested at 1, 3, or 5 h after infection or mock infection. The real-time PCR was performed as described above. The results are shown in Fig. 5B. Infection of HeLa cells with ΔUL41-mutant virus resulted in a slight up-regulation of IEX-1 RNA. This level of accumulated RNA appeared to be relatively stable throughout the course of infection, and the comparison of the amounts of RNA detected by the three sets of primers showed no evidence of selective 3′-5′ degradation of IEX-1 RNA in ΔUL41-mutant virus-infected cells. These results indicate that the progressive 3′-5′ degradation of IEX-1 RNA was mediated by the UL41 gene product.
Fig. 5.
Involvement of UL41 in the progressive 3′-5′ degradation of IEX-1 mRNA. (A) Location along the IEX-1 RNA sequence of the three sets of primers used for real-time PCR. (B) Total RNA was extracted at the indicated hours after mock or ΔUL41-mutant virus infection of HeLa cells.
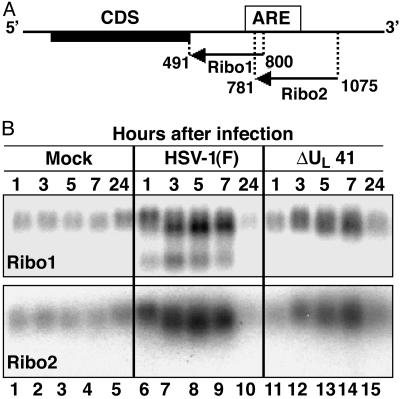
IEX-1 mRNA Is Cleaved Within the ARE in the 3′ UTR. To map in more details the cleavage site in the 3′ UTR of IEX-1 mRNA upon HSV-1 infection, HeLa cells were mock-infected or exposed to 10 pfu of HSV-1(F) or ΔUL41-mutant virus per cell and cytoplasmic RNAs were purified at 1, 3, 5, 7, and 24 h after infection. Two RNA probes antisense to the IEX-1 3′ UTR were generated by transcription in vitro as described in Materials and Methods and used as probes in Northern blot hybridizations. The results as well as the location of the RNA probes are shown in Fig. 6. Specifically, both riboprobes 1 and 2 detected full-length RNAs, but only riboprobe 1 detected the fast-migrating truncated RNA species reported earlier (9) (compare lanes 6-10 of Fig. 6B Lower vs. Upper). These results indicate that the fastest-migrating form of IEX-1 RNA lacked the region of the 3′ UTR downstream of riboprobe 1. Moreover, the results show that the truncation of the RNA was vhs-dependent inasmuch as only the full-length, intact IEX-1 RNA was detected by riboprobe 1 in ΔUL41-mutant virus-infected cells (compare lanes 11-15 with lanes 6-10 of Fig. 6B Upper).
Fig. 6.
Mapping of the vhs-dependent endonucleolytic cleavage site in IEX-1 mRNA. (A) Location along the IEX-1 RNA sequence of Ribo1 and Ribo2, two riboprobes antisense to the 3′ UTR, used for Northern blot analysis. (B) HeLa cells were mock-infected or infected with 10 pfu of HSV-1(F) or ΔUL41-mutant virus per cell. Cytoplasmic RNA was purified from cells harvested at the indicated times after mock infection (lanes 1 and 5), infection with HSV-1(F) (lanes 6-10), or ΔUL41 mutant (lanes 11-15).
It should be noted that the riboprobe 2 was complementary to a region of the IEX-1 3′ UTR-containing ARE, and therefore the endonucleolytic cleavage that generated the fast-migrating forms of IEX-1 RNA occurred in this domain.
Discussion
In this report, we identified two classes of cellular RNAs, differing with respect to stability and accumulation of the translation products. The first class, exemplified by IEX-1, IκBα, and c-fos RNAs are subjected to rapid degradation. In the course of these studies on individual RNAs, we have identified deadenylation, RNA decay by a 3′-5′ pathway, and endonucleolytic cleavage. A characteristic of the degradation process is that the partially degraded RNAs consisting of the 5′ portions of the truncations persisted and were readily detected in the cytoplasm of infected cells. None of these processes were detected in cells infected with the ΔUL41-mutant virus. Translation products of these RNAs were at best detected for a very short time after infection with WT virus. Failure of the translation product to accumulate in infected cells may also be attributed to the transport of unspliced RNA to the cytoplasm enabled and mediated by the infected cell no. 27 (ICP27).
The second class of RNAs, exemplified by GADD45β and TTP RNAs, showed no evidence of accumulation or persistence of deadenylated RNAs or 3′-5′-degraded intermediates. The unavoidable conclusion derived from comparisons of the two classes of RNAs is that the turnover of GADD45β and TTP RNAs differs from that of IEX-1, IκBα, or c-fos RNAs in HSV-1-infected cells. Consistent with the hypothesis that these RNAs retain their integrity longer is the evidence that GADD45β shown in Fig. 2 and TTP (data not shown) proteins readily accumulate in infected cells. One obvious conclusion of the results presented in this report is that the degradation of RNAs mediated by the UL41 gene product is not uniform.
The results presented in this report raise four questions. The first is the basis of the discrimination of the two classes of RNAs. As indicated in the Introduction, the structure of IEX-1 mRNA raised the possibility that the mode of degradation of the RNA was related to the presence of ARE within the 3′ UTR. Consistent with this hypothesis, the two other ARE-containing RNAs, IκBα and c-fos, were degraded in a similar fashion, whereas GADD45β and TTP RNAs lacking the ARE in their 3′ UTR were not subjected to similar degradation. One prediction arising from these studies is that vhs targets in a specific fashion the degradation of ARE-containing RNAs.
The second question relates to the persistence of truncated, partially degraded RNAs in HSV-1-infected cells. The problem stems from the observation that in uninfected cells these ARE-containing RNAs are rapidly degraded and products of 3′-5′ degradation do not accumulate. For example, Mukherjee et al. (13) had to use synthetic RNA oligonucleotides that contained three consecutive phosphothioate derivatives to identify in vitro mRNA turnover intermediates. A central question is why the partially degraded RNAs persist in the cytoplasm of HSV-1-infected cells. We envision at least two possibilities. The first is that the vhs protein while mediating the degradation of these RNAs also alters the sequence of events leading to the final disposal of the truncated remnants. An alternative explanation is that vhs activates inducible pathways that sequester the RNAs (e.g., stress granules) that prevent both expression and degradation of the RNAs (21). Preliminary studies (A.E. and B.R., unpublished data) support at least in part this hypothesis. An earlier study (22) reported that vhs degrades in vitro a short β-globin RNA substrate in specific size bands, suggesting a possible vhs endonucleolytic activity. Furthermore, Zelus et al. reported that vhs RNase activity has some preference for A- or U-rich regions as compared with C-rich regions.
The third question concerns the targeting by HSV-1 of specific RNAs for rapid destruction. One possible explanation is that ARE-containing RNAs frequently encode regulatory proteins acting in the cells that produce them or on other cells (e.g., cytokines) (10, 23). It could be expected that large viruses such as HSV-1 would evolve mechanisms that would block the synthesis or lead to degradation of cellular proteins whose actions may abrogate viral replication or enhance the immune response against the virus. Although GADD45β and TTP are also host responses to stress, their function may not be deleterious to the virus. Interestingly, TTP is an AU-rich binding protein that promotes decay of ARE-containing mRNAs (24).
Last, as noted in the Introduction, a key observation made early in the course of studies of viral replication is the decreased incorporation of amino acids into cellular proteins. The discrepancy between the selective expression of cellular proteins by RNAs induced after infection noted in this study and the overall pattern of decrease in cellular protein synthesis in WT virus-infected cells but not in ΔUL41-infected cells remains unresolved. Among numerous explanations that could account for this discrepancy is that the cellular proteins expressed after infection are a minority, invisible in the background of cellular proteins whose synthesis is shut off. This hypothesis is not readily tenable in that it implies that ARE-containing RNAs constitute the majority of RNAs made in uninfected cells. An alternative explanation is that vhs protein has two distinct functions, that of mediating selective degradation of some RNAs and that of selective suppression of translation of other RNAs. Heretical as it may seem, this hypothesis requires further investigation.
Acknowledgments
We thank Dr. Beatrice Fineschi and the Biological Sciences Collegiate Divisions for making the real-time PCR machine available and Guido Franzoso (University of Chicago) for the gift of the GADD45β-specific mAb. These studies were aided by National Cancer Institute Grants CA78766, CA71933, CA83939, CA87661, and CA88860 (to B.R.) of the U.S. Public Health Service. A.E. is a recipient of a fellowship from the Philippe Foundation.
Abbreviations: ARE, AU-rich element; TTP, tristetraprolin; pfu, plaque-forming units; HSV, herpes simplex virus; vhs, virion host shutoff.
References
- 1.Strom, T. & Frenkel, N. (1987) J. Virol. 61, 2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Read, G. S. & Frenkel, N. (1983) J. Virol. 46, 498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong, A. D. & Frenkel, N. (1987) Proc. Natl. Acad. Sci. USA 84, 1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng, P., Everly, D. N., Jr., & Read, G. S. (2001) J. Virol. 75, 10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karr, B. M. & Read, G. S. (1999) Virology 264, 195-204. [DOI] [PubMed] [Google Scholar]
- 6.Lam, Q., Smibert, C. A., Koop, K. E., Lavery, C., Capone, J. P., Weinheimer, S. P. & Smiley, J. R. (1996) EMBO J. 15, 2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, T. J., Morrison, L. A. & Leib, D. A. (2002) J. Virol. 76, 2054-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taddeo, B., Esclatine, A. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 17031-17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taddeo, B., Esclatine, A., Zhang, W. & Roizman, B. (2003) J. Virol. 77, 6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bevilacqua, A., Ceriani, M. C., Capaccioli, S. & Nicolin, A. (2003) J. Cell. Physiol. 195, 356-372. [DOI] [PubMed] [Google Scholar]
- 11.Bakheet, T., Williams, B. R. & Khabar, K. S. (2003) Nucleic Acids Res. 31, 421-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewer, G. & Ross, J. (1988) Mol. Cell. Biol. 8, 1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee, D., Gao, M., O'Connor, J. P., Raijmakers, R., Pruijn, G., Lutz, C. S. & Wilusz, J. (2002) EMBO J. 21, 165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bresnahan, W. A., Hultman, G. E. & Shenk, T. (2000) J. Virol. 74, 10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ejercito, P. M., Kieff, E. D. & Roizman, B. (1968) J. Gen. Virol. 2, 357-364. [DOI] [PubMed] [Google Scholar]
- 16.Poon, A. P. & Roizman, B. (1997) Virology 229, 98-105. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C. Y., Xu, N. & Shyu, A. B. (1995) Mol. Cell. Biol. 15, 5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taddeo, B., Luo, T. R., Zhang, W. & Roizman, B. (2003) Proc. Natl. Acad. Sci. USA 100, 12408-12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung, P., Ellison, K. S., Verity, R. & Smiley, J. R. (2000) J. Virol. 74, 2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soliman, T. M., Sandri-Goldin, R. M. & Silverstein, S. J. (1997) J. Virol. 71, 9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha, N. & Anderson, P. (2002) Biochem. Soc. Trans. 30, 963-969. [DOI] [PubMed] [Google Scholar]
- 22.Zelus, B. D., Stewart, R. S. & Ross, J. (1996) J. Virol. 70, 2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tebo, J., Der, S., Frevel, M., Khabar, K. S., Williams, B. R. & Hamilton, T. A. (2003) J. Biol. Chem. 278, 12085-12093. [DOI] [PubMed] [Google Scholar]
- 24.Blackshear, P. J. (2002) Biochem. Soc. Trans. 30, 945-952. [DOI] [PubMed] [Google Scholar]