ABSTRACT
Expression of capsular polysaccharides is a variable trait often associated with more-virulent forms of a bacterial species. For example, typhoid fever is caused by the capsulated Salmonella enterica serovar Typhi, while nontyphoidal Salmonella serovars associated with gastroenteritis are noncapsulated. Here we show that optimization of the immune evasive properties conferred by the virulence-associated (Vi) capsular polysaccharide involved an additional alteration to the cell envelope of S. Typhi, namely inactivation of the fepE gene, encoding the regulator of very-long O-antigen chains. Introduction of the capsule-encoding viaB locus into the nontyphoidal S. enterica serovar Typhimurium reduced complement deposition in vitro and intestinal inflammation in a mouse colitis model. However, both phenotypes were markedly enhanced when the viaB locus was introduced into an S. Typhimurium fepE mutant, which lacks very-long O-antigen chains. Collectively, these data suggest that during the evolution of the S. Typhi lineage, loss of very-long O-antigen chains by pseudogene formation was an adaptation to maximize the anti-inflammatory properties of the Vi capsular polysaccharide.
IMPORTANCE
Genomic comparison illustrates that acquisition of virulence factors by horizontal gene transfer is an important contributor to the evolution of enteric pathogens. Acquisition of complex virulence traits commonly involves horizontal transfer of a large gene cluster, and integration of the gene cluster into the host genome results in the formation of a pathogenicity island. Acquisition of the virulence-associated (Vi) capsular polysaccharide encoded by SPI7 (Salmonella pathogenicity island 7) was accompanied in the human-adapted Salmonella enterica serovar Typhi by inactivation of the fepE gene, encoding the regulator of very-long O-antigen chains. We show that the resulting loss of very-long O-antigen chains was an important mechanism for maximizing immune evasion mediated by the Vi capsular polysaccharide. These data suggest that successful incorporation of a capsular polysaccharide requires changes in the cell envelope of the hosting pathogen.
Introduction
Typhoid fever is a severe systemic infection that presents with fever (1) after an average incubation period of 2 weeks (2). In contrast, gastroenteritis caused by Salmonella enterica serovar Typhimurium is a localized diarrheal disease with an average incubation period of <24 h (3). The swift onset of diarrhea, abdominal pain, and fever during gastroenteritis is explained by the rapid induction of an acute inflammatory response in the intestine, which requires deployment of two type III secretion systems (T3SSs) encoded by Salmonella pathogenicity island 1 (SPI1) and SPI2 (4–6). Although SPI1 and SPI2 are present in S. enterica serovar Typhi (7), the development of intestinal inflammation is slowed markedly during typhoid fever by expression of the virulence-associated (Vi) capsular polysaccharide (8–10). The anti-inflammatory properties of the Vi capsular polysaccharide (11–13) have been proposed to contribute to the long incubation period characteristic of typhoid fever (14, 15).
The biosynthesis of the Vi capsular polysaccharide is encoded by the viaB locus, a 14-kb DNA region located on SPI7, a pathogenicity island present in S. enterica serovar Typhi but absent from Salmonella serovars associated with gastroenteritis (16, 17). The viaB locus contains genes for the regulation (tviA), biosynthesis (tviBCDE), and surface assembly (vexABCDE) of the Vi capsular polysaccharide (18, 19). Expression of the positive regulator TviA is repressed in the intestinal lumen but induced by the two-component system EnvZ/OmpR at an osmolarity encountered in tissue (20, 21). As a result, expression of the Vi capsular polysaccharide is induced when bacteria transit though the intestinal epithelium (22). This regulatory mechanism ensures that S. Typhi is encapsulated by the time it encounters complement. Expression of the Vi capsular polysaccharide inhibits complement deposition (23, 24), because its homopolymeric chains contain approximately 300 residues of (1,4)-2-acetamido-3-O-acetyl-2-deoxy-α-d-galacturonic acid (25), a sugar that does not contain free hydroxyl groups available for ester formation with complement component 3 fragment b (C3b).
Since S. Typhi is strictly human adapted, it is difficult to study the function of its virulence factors in animal models (26). One approach to study the role the Vi capsular polysaccharide plays in vivo has been the introduction of the viaB locus into S. Typhimurium, a natural pathogen of mice (8, 9, 12, 13, 24). However, it is not known whether horizontal transfer of the viaB locus is sufficient to optimally deploy the encoded capsular polysaccharide for immune evasion. Here we show that for the Vi capsular polysaccharide to confer maximal evasion of complement fixation, it is necessary to change the cell envelope of S. Typhimurium, a process that also accompanied the evolution of the S. Typhi lineage.
RESULTS
Very-long O-antigen chains interfere with the function of the Vi capsular polysaccharide in vitro.
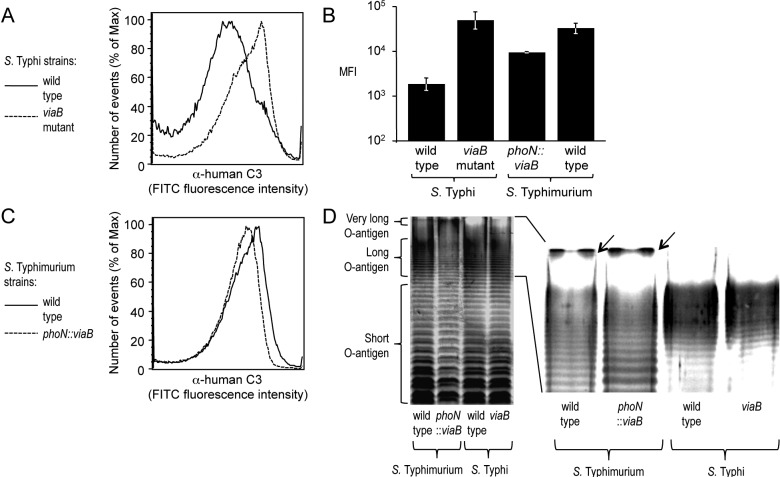
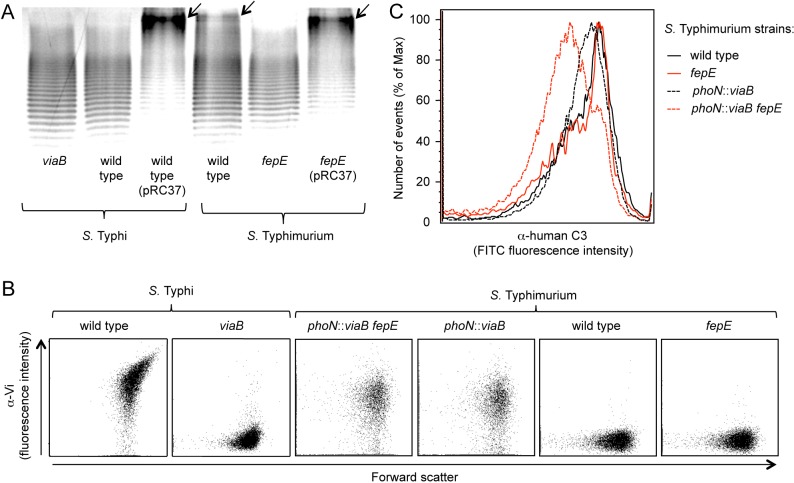
We performed an in vitro assay to determine whether expression of the Vi capsular polysaccharide in S. enterica serovars Typhi and Typhimurium had identical effects on inhibiting complement fixation. Consistent with previous reports (24), analysis of bacterial cells by flow cytometry showed that incubation of a noncapsulated S. Typhi mutant (viaB mutant) in 10% human serum resulted in efficient deposition of C3b on the bacterial surface, while complement deposition was markedly reduced in the capsulated S. Typhi wild-type strain (Ty2) (Fig. 1A and B). Similarly, a capsulated S. Typhimurium strain in which the viaB locus of S. Typhi was inserted chromosomally into the phoN gene (phoN::viaB mutant) deposited less complement on its surface than the noncapsulated S. Typhimurium wild-type strain (IR715) (Fig. 1B and C). However, the magnitude by which expression of the Vi capsular polysaccharide reduced complement fixation was notably larger in S. Typhi than in S. Typhimurium (Fig. 1B).
FIG 1 .
(A to C) Fixation of C3 after incubation of the indicated S. Typhi and S. Typhimurium strains (wild type and mutants) in 10% human serum was detected by flow cytometry using an anti-human C3 (α-human C3) FITC conjugate. The experiments shown in panels A and C were repeated 3 times independently with similar outcomes, and a representative example is shown. The average maximum fluorescence intensity (MFI) values ± standard errors (error bars) determined for these three independent experiments are shown in panel B. (D) Silver-stained SDS-PAGE of LPS preparations from the indicated S. Typhimurium and S. Typhi strains. The positions of short, long, and very-long O-antigen chains are indicated to the left of the gel. A magnification of the region showing long and very-long O-antigen chains is shown on the right, and the presence of very-long O-antigen chains in S. Typhimurium strains is indicated by black arrows.
To investigate possible reasons for differences in the efficacy by which the Vi capsular polysaccharide reduced complement deposition in S. Typhi and S. Typhimurium, we compared expression of lipopolysaccharide (LPS), a surface structure containing O-antigen repeat units that are known to fix complement (27). LPS molecules contain a lipid A anchor and an oligosaccharide core but differ in the number of O-antigen repeat units that extend from the bacterial surface. In S. Typhimurium, O-antigen repeat units are composed of a trisaccharide backbone, consisting of α-d-mannose-(1,4)-α-l-rhamnose-(1,3)-α-d-galactose, and a branching sugar (abequose) that is α-(1,3)-linked to d-mannose in the backbone. Consistent with previous reports, S. Typhimurium exhibited a trimodal distribution in LPS length, including short-LPS species containing between 1 and 15 O-antigen repeat units, long-LPS species carrying between 16 and 35 O-antigen repeat units, and very-long-LPS species with more than 100 O-antigen repeat units (Fig. 1D) (28–30). O-antigen repeat units of S. Typhi carry tyvelose as the branching sugar that is α-(1,3)-linked to the backbone but are otherwise identical to those of S. Typhimurium. Interestingly, in contrast to S. Typhimurium, S. Typhi expressed only short-LPS species and long-LPS species, while very-long-LPS species were absent (Fig. 1D).
The fepE open reading frame encodes the length regulator of very-long O-antigen chains (28) but is interrupted in the S. Typhi genome by a stop codon (7). To test whether this mutation in fepE is responsible for the lack of LPS species with very-long O-antigen chains in S. Typhi, we introduced a plasmid carrying the cloned S. Typhimurium fepE gene (pRC37). In contrast to the wild-type S. Typhi, an S. Typhi strain carrying a plasmid encoding the S. Typhimurium fepE gene (pRC37) produced LPS species with very-long O-antigen chains (Fig. 2A). These data suggested that conversion of fepE into a pseudogene was responsible for the inability of S. Typhi to produce very-long O-antigen chains, which confirmed previous results (31).
FIG 2 .
(A) Silver-stained SDS-PAGE of LPS preparations from the indicated S. Typhimurium and S. Typhi strains (wild type and mutants). Plasmid pRC37 carries the cloned S. Typhimurium fepE gene. The positions of very-long O-antigen chains are indicated by black arrows. (B) Vi capsular polysaccharide expression was detected by flow cytometry. Cells of the indicated S. Typhi and S. Typhimurium strains were labeled with rabbit anti-Vi serum/goat anti-rabbit FITC conjugate (α-Vi on the y axis), and fluorescence intensities were determined for 10,000 particles. Each experiment was repeated 3 times independently with similar outcomes, and a representative example is shown. (C) Fixation of C3 after incubation of the indicated S. Typhi and S. Typhimurium strains in 10% human serum was detected by flow cytometry using an anti-human C3 FITC conjugate. The experiment was repeated 3 times independently with similar outcomes, and a representative example is shown.
We reasoned that very-long O-antigen chains containing an estimated 100 copies of a repeat unit composed of a trisaccharide backbone (28) might rival in length the homopolymeric chains of the Vi capsular polysaccharide comprising approximately 300 sugar residues (25). Therefore, we wanted to test whether the reduced efficacy by which the Vi capsular polysaccharide diminished complement deposition in S. Typhimurium was due to the presence of very-long O-antigen chains. A mutation in fepE was introduced into the wild-type S. Typhimurium and an S. Typhimurium phoN::viaB mutant. Analysis by flow cytometry revealed similar expression levels of the Vi capsular polysaccharide in S. Typhi (Ty2), the S. Typhimurium fepE phoN::viaB mutant (RC60), and the S. Typhimurium phoN::viaB mutant (TH170), while capsule expression was absent in the wild-type S. Typhimurium (IR715), the S. Typhimurium fepE mutant (RC31), and the S. Typhi viaB mutant (STY2) (Fig. 2B). Expression of very-long O-antigen chains was abrogated in the fepE mutant (Fig. 2A). Furthermore, expression of very-long O-antigen chains could be restored in the fepE mutant by introducing the fepE gene cloned on a plasmid (pRC37). Remarkably, complement fixation was markedly reduced in the S. Typhimurium fepE phoN::viaB mutant compared to the S. Typhimurium phoN::viaB mutant (Fig. 2C). These data suggested that very-long O-antigen chains impaired the ability of the Vi capsular polysaccharide to prevent complement deposition on the surface of S. Typhimurium.
Lack of very-long O-antigen chains enhances capsule-mediated suppression of colitis.
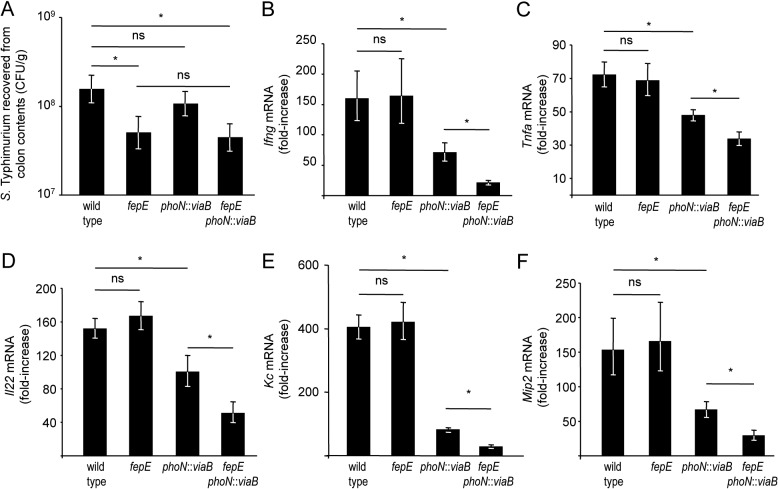
We used the mouse colitis model (5) to investigate the biological relevance of our observations. In this model, mice are preconditioned by treatment with streptomycin, which disrupts the resident microbiota. Subsequent inoculation with S. Typhimurium results in acute cecal inflammation, which is an animal model for human gastroenteritis (reviewed in reference 26). Groups of streptomycin-pretreated mice were either mock infected or inoculated with the wild-type S. Typhimurium or a fepE mutant, phoN::viaB mutant, or fepE phoN::viaB mutant, and the cecum and colon contents were collected 72 h after infection. While the wild-type S. Typhimurium and phoN::viaB mutant were recovered in similar numbers from cecal contents, a small but significant (P < 0.05) reduction in bacterial numbers was observed for strains lacking very-long O-antigen chains (i.e., the fepE mutant and fepE phoN::viaB mutant) (Fig. 3A). These data were consistent with our previous observation that very-long O-antigen chains are required for optimal survival of S. Typhimurium in the lumen of the inflamed gut (32).
FIG 3 .
Streptomycin-pretreated mice were infected with the indicated S. Typhimurium strains, and the cecum and colon contents were collected 72 h after infection. (A) Recovery of S. Typhimurium from colon contents. Values are geometric means of CFU per gram colon contents ± standard errors (error bars). (B to F) Transcript levels of Ifng (B), Tnfa (C), Il22 (D), Kc (E), and Mip2 (F) in the cecal mucosa were determined by quantitative real-time PCR. Values are geometric means ± standard errors of fold increases over mRNA levels in mock-infected animals. Values that are statistically significant (P < 0.05) are indicated by a bar and asterisk. Values that are not statistically significant (ns) are indicated.
To assess how the presence of very-long O-antigen chains influences inflammatory responses in the cecal mucosa, we determined mRNA levels of inflammatory markers, including gamma interferon (IFN-γ) encoded by the Infg gene, tumor necrosis factor alpha (TNF-α) encoded by the Tnfa gene, interleukin-22 (IL-22) encoded by the Il22 gene, keratinocyte-derived cytokine (KC) encoded by the Kc gene, and macrophage-inducible protein 2 (MIP-2) encoded by the Mip2 gene, by quantitative real-time PCR. Although the wild-type S. Typhimurium was recovered in significantly greater numbers from colon contents than the fepE mutant (Fig. 3A), both strains elicited similar levels of Ifng, Tnfa, Il22, Kc, and Mip2 expression in the cecal mucosa (Fig. 3B to F). Thus, the presence of very-long O-antigen chains did not alter expression levels of inflammatory markers elicited by S. Typhimurium in the mouse colitis model. Introduction of the S. Typhi viaB locus into the S. Typhimurium genome (phoN::viaB mutant) significantly (P < 0.05) reduced mRNA levels of Ifng, Tnfa, Il22, Kc, and Mip2 compared to those elicited by infection with the wild-type S. Typhimurium (Fig. 3B to F). These data were consistent with our previous observation that expression of the Vi capsular polysaccharide in S. Typhimurium reduces intestinal inflammatory responses in the mouse colitis model (9). Remarkably, introduction of the viaB locus into the fepE mutant (fepE phoN::viaB mutant) significantly (P < 0.05) reduced mRNA levels of Ifng, Tnfa, Il22, Kc, and Mip2 compared to those elicited by infection with the phoN::viaB mutant (Fig. 3B to F). Thus, in the absence of very-long O-antigen chains, the viaB locus suppressed expression of inflammatory markers to a significantly (P < 0.05) greater extent than in their presence. Reduced expression of inflammatory markers was independent of the bacterial burden, because the fepE mutant and the fepE phoN::viaB mutant were recovered in similar numbers (Fig. 3A), whereas the former elicited significantly (P < 0.05) higher mRNA levels of Ifng, Tnfa, Il22, Kc, and Mip2 than the latter (Fig. 3B to F).
We next performed a blinded analysis of histopathological changes observed in the cecal mucosa 72 h after infection to determine the biological consequences of expressing very-long O-antigen chains and/or the Vi capsular polysaccharide. Ceca from mice infected with the wild-type S. Typhimurium or a fepE mutant were devoid of any contents and had severe gross pathological changes, characterized by reduced size with thickening of the cecal wall. Histopathological evaluation revealed epithelial erosion, neutrophil infiltration in the mucosa, and edema in the submucosa. In contrast, ceca from mock-infected mice did not show gross pathological changes or overt histopathology (Fig. 4). Introduction of the S. Typhi viaB locus into the S. Typhimurium genome (phoN::viaB mutant) resulted in a small but significant (P < 0.05) reduction in the severity of histopathological changes compared to mice infected with the wild-type S. Typhimurium or a fepE mutant. Remarkably, infection with a fepE phoN::viaB mutant resulted in a significant (P < 0.05) reduction in the severity of histopathological changes compared to the phoN::viaB mutant. Overall, the results from this histopathological analysis (Fig. 4) substantiated results from expression analysis of inflammatory markers in the cecal mucosa (Fig. 3B to F) and supported the concept that optimal suppression of intestinal inflammation by the viaB locus requires an absence of very-long O-antigen chains.
FIG 4 .
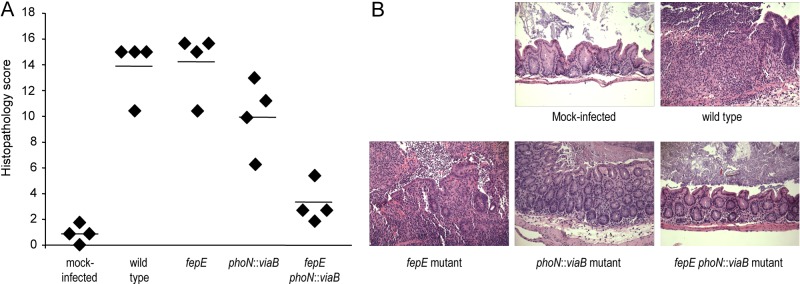
Streptomycin-pretreated mice were either mock infected or infected with the indicated S. Typhimurium strains, and the cecum was collected 72 h after infection. (A) Combined histopathology score of pathological changes observed in sections from the cecum. Each symbol represents the combined histopathology score for an individual animal. The average for each group of mice is indicated by a short line. (B) Representative images of histopathological changes.
DISCUSSION
The fact that the vast majority of the 2,587 known serovars of S. enterica are zoonotic and associated with a localized, self-limiting gastroenteritis in immune-competent individuals (33) suggests that human-restricted specialists associated with systemic febrile illnesses, such as S. enterica serovar Typhi or Paratyphi A, evolved from ancestral zoonotic organisms that caused gastroenteritis (34). While the S. enterica species is estimated to be 40 to 63 million years old (35), S. Typhi represents a clonal lineage that emerged only recently, between 10,000 and 150,000 years ago (36, 37). After the S. Typhi lineage had passed through approximately 75% of its evolutionary history, it exchanged some 23% of its genome by horizontal gene transfer with the S. Paratyphi A lineage, presumably during coexistence in a shared human reservoir (38). Subsequent to this large-scale genetic exchange, which might mark the origin of typhoid and paratyphoid fever, the lineages of S. Typhi and S. Paratyphi A became isolated again, and both of their genomes subsequently accumulated pseudogenes at an accelerated rate (39).
Many pseudogenes present in S. Typhi encode functions required for the gastrointestinal lifestyle of S. Typhimurium. For example, the genome of S. Typhi strain CT18 contains pseudogenes in 7 of its 11 chaperone/usher-type fimbrial operons (40), which encode adhesins required by S. Typhimurium to colonize the intestinal lumen (41). Furthermore, the S. Typhi genome carries pseudogenes in operons functioning in anaerobic respiration (ttrS, dmsA, dmsB, narV, and narW) (7, 42), and these functions are required by S. Typhimurium to outgrow obligate anaerobic bacteria in the lumen of an acutely inflamed gut during gastroenteritis (43, 44). While these genes are likely maintained in S. Typhimurium because they aid in intestinal growth and transmission during gastroenteritis (45), they can be seen as dispensable for the extraintestinal lifestyle of S. Typhi, a pathogen that spreads by means of water, milk, and food products contaminated by individuals with chronic gallbladder carriage (46). Thus, pseudogene formation in S. Typhi is commonly viewed as a process leading to random losses of genetic functions that are inherited from an ancestral organism associated with gastroenteritis but that are not required for causing typhoid fever in humans.
Our results suggest that, surprisingly, the formation of one pseudogene, namely, a fepE allele interrupted by a stop codon, resulted in a gain of function during the evolution of the S. Typhi lineage. Specifically, we show that inactivation of fepE resulted in an enhanced functionality of the Vi capsular polysaccharide. The Vi capsular polysaccharide is an important virulence factor of S. Typhi (reviewed in reference 47) and has been developed into a vaccine against typhoid fever (48). Expression of the Vi capsular polysaccharide inhibits complement deposition (23, 24), a process that was more efficient in strains lacking a functional fepE gene. One consequence of complement activation is the formation of a membrane attack complex on the bacterial surface that leads to lysis unless serum resistance mechanisms are deployed. One study reports that the Vi capsular antigen is not required for serum resistance of S. Typhi (31), but in this study, bacteria were grown in Luria-Bertani (LB) broth, a condition that represses expression of the Vi capsular polysaccharide (20, 21). In several other studies, expression of the Vi capsular polysaccharide was shown to increase serum resistance of S. Typhi (23, 24, 49), which is consistent with its role in preventing complement deposition (Fig. 1A).
A second consequence of complement deposition and activation of the alternative pathway is the production of anaphylatoxins (C3a and C5a). Anaphylatoxins are potent enhancers of cytokine responses elicited by stimulating the Toll-like receptor 4 (TLR4)/MD2/CD14 receptor complex with LPS (reviewed in reference 50). Thus, suppression of complement activation by the Vi capsular polysaccharide might explain why expression of this virulence factor diminishes the induction of TLR4/MD2/CD14-dependent proinflammatory responses (10–13). Expression of the Vi capsular polysaccharide in S. Typhimurium attenuates intestinal inflammation elicited in bovine ligated ileal loops (8) and in the mouse colitis model (9). Interestingly, suppression of intestinal inflammation by the Vi capsular polysaccharide was significantly enhanced when production of very-long O-antigen chains was abrogated by inactivation of the fepE gene. These data suggest that conversion of fepE into a pseudogene enhanced the ability of S. Typhi to suppress or delay intestinal inflammation using the Vi capsular polysaccharide.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains used in this study are presented in Table 1. A plasmid carrying the S. enterica serovar Typhimurium fepE gene cloned into the low-copy-number vector pWSK129 has been described previously (32). Bacterial cultures were routinely incubated with aeration at 37°C in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter) or on LB agar plates (15 g agar per liter) unless capsule expression was desired. To induce expression of the Vi capsular polysaccharide, bacteria were grown in broth containing 10 g tryptone and 5 g yeast extract per liter. The following antibiotics were added as necessary at appropriate concentrations: chloramphenicol (Cm), 0.03 mg/ml; carbenicillin, 0.1 mg/ml; kanamycin (Kan), 0.05 mg/ml; and nalidixic acid, 0.05 mg/ml.
TABLE 1 .
Bacterial strains used in this study
| Bacterial strain |
Description or relevant phenotype or genotype |
Reference |
|---|---|---|
|
S. Typhimurium strains |
||
| IR715 | ATCC 14028; Nalr derivative | 53 |
| RC31 | IR715 fepE::pGP704 | 32 |
| TH170 | IR715 phoN::viaB | 9 |
| RC60 | IR715 phoN::viaB fepE::pGP704 | This study |
|
S. Typhi strains |
||
| Ty2 | Wild-type strain; ATCC 700931 | |
| STY2 | Ty2 viaB::Kan | 10 |
Construction of mutants.
The fepE::pGP704 mutation was transduced from S. Typhimurium RC31 into the S. Typhimurium phoN::viaB mutant TH170 using transduction with phage P22 HT int-105 to yield strain RC60.
Animal experiments.
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, Davis, and performed according to Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines. Groups (n = 4) of female mice (C57BL/6J mice from Jackson Laboratory) aged 6 to 8 weeks were inoculated intragastrically with 20 mg streptomycin in a volume of 0.1 ml 24 h prior to intragastric inoculation with either 0.1 ml sterile LB broth or 1 × 109 CFU of the indicated S. Typhimurium strains. Colon contents and cecal tissues were harvested at 72 or 120 h after infection, homogenized in phosphate-buffered saline (PBS), serially diluted, and plated on LB agar plates containing the appropriate antibiotics to enumerate CFU.
RNA extraction and quantitative real-time PCR.
Cecal tissue was homogenized in a minibeadbeater (BioSpec Products), and RNA was extracted by the TRI reagent method (Molecular Research Center) as described previously (51). Reverse transcription was performed using TaqMan reagent (Applied Biosystems), and 2 µl of converted cDNA was used with a 250 nM concentration of primers listed in Table 2 and SYBR green (Applied Biosystems) for real-time PCR in the ViiA 7 system (Life Technologies). Data were analyzed using the comparative threshold cycle method. Transcription levels of Ifng, Tnfa, IL22, Kc, and Mip2 genes were normalized to Gapdh mRNA (encoding glyceraldehyde-3-phosphate dehydrogenase [GAPDH]).
TABLE 2 .
Nucleotide primers for quantitative real-time PCR
| Target gene | Nucleotide sequence |
|---|---|
| Gapdh | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ |
| 5′-AGGTCGGTGTGAACGGATTTG-3′ | |
| Ifng | 5′-TCAAGTGGCATAGATGTGGAAGAA-3′ |
| 5′-TGGCTCTGCAGGATTTTCATG-3′ | |
| Tnfa | 5′-TTGGGTCTTGTTCACTCCACGG-3′ |
| 5′-CCTCTTTCAGGTCACTTTGGTAGG-3′ | |
| IL22 | 5′-GGCCAGCCTTGCAGATAACA-3′ |
| 5′-GCTGATGTGACAGGAGCTGA-3′ | |
| Kc | 5′-TGCACCCAAACCGAAGTCAT-3′ |
| 5′-TTGTCAGAAGCCAGCGTTCAC-3′ | |
| Mip2 | 5′-AGTGAACTGCGCTGTCAATGC-3′ |
| 5′-AGGCAAACTTTTTGACCGCC-3′ |
Histopathology.
Cecal tissues were formalin fixed, sectioned, stained with hematoxylin and eosin (H&E) and submitted to a veterinary pathologist for blinded scoring using a scale described previously (52). Representative images of tissue sections were taken using an Olympus BX41 microscope.
Analysis of Vi and LPS expression and C3 deposition by flow cytometry.
Detection of Vi and LPS expression by flow cytometry was performed as described previously (9) using the DNA-specific stain propidium iodide, rabbit anti-Vi serum, or anti-O:4 (1:250 dilution; Becton Dickinson) and goat anti-rabbit fluorescein isothiocyanate (FITC) conjugate (1:250 dilution; Jackson ImmunoResearch). Binding of complement component 3 (C3) was determined by flow cytometry as described previously (24) using the DNA-specific stain propidium iodide, human serum (10% dilution; Quidel), and fluorescein isothiocyanate (FITC)-conjugated goat anti-human C3b monoclonal antibody (1:250 dilution; MP Biomedicals). All samples were analyzed on an LSRII instrument (BD).
Statistical analysis.
Relative abundance values of S. Typhimurium strains and fold changes in mRNA levels were converted logarithmically (log10) prior to statistical analysis using a one-tailed parametric test (Student’s t test. A P value of <0.05 was considered to be significant.
ACKNOWLEDGMENT
We acknowledge support by Public Health Service grant AI044170 to A.J.B.
Footnotes
Citation Crawford RW, Wangdi T, Spees AM, Xavier MN, Tsolis RM, Bäumler AJ. 2013. Loss of very-long O-antigen chains optimizes capsule-mediated immune evasion by Salmonella enterica Serovar Typhi. mBio 4(4):e00232-13. doi:10.1128/mBio.00232-13.
REFERENCES
- 1. Nasrallah SM, Nassar VH. 1978. Enteric fever: a clinicopathologic study of 104 cases. Am. J. Gastroenterol. 69:63–69 [PubMed] [Google Scholar]
- 2. Olsen SJ, Bleasdale SC, Magnano AR, Landrigan C, Holland BH, Tauxe RV, Mintz ED, Luby S. 2003. Outbreaks of typhoid fever in the United States, 1960–99. Epidemiol. Infect. 130:13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glynn JR, Palmer SR. 1992. Incubation period, severity of disease, and infecting dose: evidence from a Salmonella outbreak. Am. J. Epidemiol. 136:1369–1377 [DOI] [PubMed] [Google Scholar]
- 4. Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O’Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 8. Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, Lawhon SD, Chu H, Lau T, Bevins CL, Adams LG, Bäumler AJ. 2007. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect. Immun. 75:4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haneda T, Winter SE, Butler BP, Wilson RP, Tükel C, Winter MG, Godinez I, Tsolis RM, Bäumler AJ. 2009. The capsule-encoding viaB locus reduces intestinal inflammation by a Salmonella pathogenicity island 1-independent mechanism. Infect. Immun. 77:2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Bäumler AJ. 2005. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect. Immun. 73:3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirose K, Ezaki T, Miyake M, Li T, Khan AQ, Kawamura Y, Yokoyama H, Takami T. 1997. Survival of Vi-capsulated and Vi-deleted Salmonella typhi strains in cultured macrophage expressing different levels of CD14 antigen. FEMS Microbiol. Lett. 147:259–265 [DOI] [PubMed] [Google Scholar]
- 12. Wilson RP, Raffatellu M, Chessa D, Winter SE, Tükel C, Bäumler AJ. 2008. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell. Microbiol. 10:876–890 [DOI] [PubMed] [Google Scholar]
- 13. Jansen AM, Hall LJ, Clare S, Goulding D, Holt KE, Grant AJ, Mastroeni P, Dougan G, Kingsley RA. 2011. A Salmonella typhimurium-Typhi genomic chimera: a model to study Vi polysaccharide capsule function in vivo. PLoS Pathog. 7:e1002131. 10.1371/journal.ppat.1002131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raffatellu M, Chessa D, Wilson RP, Tükel C, Akçelik M, Bäumler AJ. 2006. Capsule-mediated immune evasion: a new hypothesis explaining aspects of typhoid fever pathogenesis. Infect. Immun. 74:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsolis RM, Young GM, Solnick JV, Bäumler AJ. 2008. From bench to bedside: stealth of enteroinvasive pathogens. Nat. Rev. Microbiol. 6:883–892 [DOI] [PubMed] [Google Scholar]
- 16. Selander RK, Beltran P, Smith NH, Helmuth R, Rubin FA, Kopecko DJ, Ferris K, Tall BD, Cravioto A, Musser JM. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Selander RK, Smith NH, Li J, Beltran P, Ferris KE, Kopecko DJ, Rubin FA. 1992. Molecular evolutionary genetics of the cattle-adapted serovar Salmonella dublin. J. Bacteriol. 174:3587–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Virlogeux I, Waxin H, Ecobichon C, Popoff MY. 1995. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology 141:3039–3047 [DOI] [PubMed] [Google Scholar]
- 19. Wetter M, Goulding D, Pickard D, Kowarik M, Waechter CJ, Dougan G, Wacker M. 2012. Molecular characterization of the viaB locus encoding the biosynthetic machinery for Vi capsule formation in Salmonella Typhi. PLoS One 7:e45609. 10.1371/journal.pone.0045609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Rüssmann H, Bäumler AJ. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol. Microbiol. 74:175–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winter SE, Winter MG, Godinez I, Yang H-J, Russmann H, Andrews-Polymenis HL, Bäumler AJ. 2010. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog. 6:e1001060. 10.1371/journal.ppat.1001060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tran QT, Gomez G, Khare S, Lawhon SD, Raffatellu M, Bäumler AJ, Ajithdoss D, Dhavala S, Adams LG. 2010. The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect. Immun. 78:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Looney RJ, Steigbigel RT. 1986. Role of the Vi antigen of Salmonella typhi in resistance to host defense in vitro. J. Lab. Clin. Med. 108:506–516 [PubMed] [Google Scholar]
- 24. Wilson RP, Winter SE, Spees AM, Winter MG, Nishimori JH, Sanchez JF, Nuccio SP, Crawford RW, Tükel Ç, Bäumler AJ. 2011. The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica serotype Typhi. Infect. Immun. 79:830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heyns K, Kiessling G. 1967. Strukturaufklarung des Vi-antigens aus Citrobacter freundii (E. coli) 5396/3. Carbohydr. Res. 3:340–353 [Google Scholar]
- 26. Tsolis RM, Xavier MN, Santos RL, Bäumler AJ. 2011. How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect. Immun. 79:1806–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joiner KA, Hammer CH, Brown EJ, Cole RJ, Frank MM. 1982. Studies on the mechanism of bacterial resistance to complement-mediated killing. I. Terminal complement components are deposited and released from Salmonella minnesota S218 without causing bacterial death. J. Exp. Med. 155:797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murray GL, Attridge SR, Morona R. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence: identification of FepE as a second Wzz. Mol. Microbiol. 47:1395–1406 [DOI] [PubMed] [Google Scholar]
- 29. Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA. 1992. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J. Bacteriol. 174:5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Batchelor RA, Haraguchi GE, Hull RA, Hull SI. 1991. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 173:5699–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bravo D, Silva C, Carter JA, Hoare A, Alvarez SA, Blondel CJ, Zaldívar M, Valvano MA, Contreras I. 2008. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J. Med. Microbiol. 57:938–946 [DOI] [PubMed] [Google Scholar]
- 32. Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Bäumler AJ. 2012. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog. 8:e1002918. 10.1371/journal.ppat.1002918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PA, Weill FX. 2010. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161:26–29 [DOI] [PubMed] [Google Scholar]
- 34. Bäumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McQuiston JR, Herrera-Leon S, Wertheim BC, Doyle J, Fields PI, Tauxe RV, Logsdon JM., Jr. 2008. Molecular phylogeny of the salmonellae: relationships among Salmonella species and subspecies determined from four housekeeping genes and evidence of lateral gene transfer events. J. Bacteriol. 190:7060–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, Achtman M. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39–45 [DOI] [PubMed] [Google Scholar]
- 37. Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, Achtman M. 2006. Evolutionary history of Salmonella typhi. Science 314:1301–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Didelot X, Achtman M, Parkhill J, Thomson NR, Falush D. 2007. A bimodal pattern of relatedness between the Salmonella Paratyphi A and Typhi genomes: convergence or divergence by homologous recombination? Genome Res. 17:61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40:987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, Bäumler AJ. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng W, Liou SR, Plunkett G, III, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Bäumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3:e00413-12. 10.1128/mBio.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. 2008. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 76:403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stone WJ. 1912. The medical aspect of chronic typhoid infection (typhoid bacillus carriers). Am. J. Med. Sci. 143:544–557 [Google Scholar]
- 47.Wangdi T, Winter SE, Bäumler AJ.Typhoid fever: “you can’t hit what you can’t see.” Gut Microbes. 2012. 3:88–92. [DOI] [PMC free article] [PubMed]
- 48. Robbins JD, Robbins JB. 1984. Reexamination of the protective role of the capsular polysaccharide (Vi antigen) of Salmonella typhi. J. Infect. Dis. 150:436–449 [DOI] [PubMed] [Google Scholar]
- 49. Hashimoto Y, Li N, Yokoyama H, Ezaki T. 1993. Complete nucleotide sequence and molecular characterization of ViaB region encoding Vi antigen in Salmonella typhi. J. Bacteriol. 175:4456–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haas PJ, van Strijp J. 2007. Anaphylatoxins: their role in bacterial infection and inflammation. Immunol. Res. 37:161–175 [DOI] [PubMed] [Google Scholar]
- 51. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339:708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to utilize ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U. S. A. 108:17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stojiljkovic I, Bäumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]