ABSTRACT
Marine bacteria in the Roseobacter and SAR11 lineages successfully exploit the ocean habitat, together accounting for ~40% of bacteria in surface waters, yet have divergent life histories that exemplify patch-adapted versus free-living ecological roles. Here, we use a phylogenetic birth-and-death model to understand how genome content supporting different life history strategies evolved in these related alphaproteobacterial taxa, showing that the streamlined genomes of free-living SAR11 were gradually downsized from a common ancestral genome only slightly larger than the extant members (~2,000 genes), while the larger and variably sized genomes of roseobacters evolved along dynamic pathways from a sizeable common ancestor (~8,000 genes). Genome changes in the SAR11 lineage occurred gradually over ~800 million years, whereas Roseobacter genomes underwent more substantial modifications, including major periods of expansion, over ~260 million years. The timing of the first Roseobacter genome expansion was coincident with the predicted radiation of modern marine eukaryotic phytoplankton of sufficient size to create nutrient-enriched microzones and is consistent with present-day ecological associations between these microbial groups. We suggest that diversification of red-lineage phytoplankton is an important driver of divergent life history strategies among the heterotrophic bacterioplankton taxa that dominate the present-day ocean.
IMPORTANCE
One-half of global primary production occurs in the oceans, and more than half of this is processed by heterotrophic bacterioplankton through the marine microbial food web. The diversity of life history strategies that characterize different bacterioplankton taxa is an important subject, since the locations and mechanisms whereby bacteria interact with seawater organic matter has effects on microbial growth rates, metabolic pathways, and growth efficiencies, and these in turn affect rates of carbon mineralization to the atmosphere and sequestration into the deep sea. Understanding the evolutionary origins of the ecological strategies that underlie biochemical interactions of bacteria with the ocean system, and which scale up to affect globally important biogeochemical processes, will improve understanding of how microbial diversity is maintained and enable useful predictions about microbial response in the future ocean.
Introduction
Heterotrophic marine bacterioplankton taxa have frequently been conceptualized into two ecological categories: those with large genomes, versatile metabolic capabilities, and rapid responses to transient conditions, likened to ecological r-strategists of macroorganisms (1), and those with streamlined genomes, the ability to grow under extremely low substrate concentrations, and the inability to take advantage of enhanced nutrients (2), paralleling the K-strategist paradigm (3–5). Ephemeral patches of organic matter formed at nanometer to millimeter scales through biotic and abiotic processes (6, 7) and harboring nutrient concentrations up to three orders of magnitude higher than bulk seawater (7, 8) are postulated to underlie these divergent strategies, as they provide enriched microhabitats that contrast with the nutrient-poor bulk seawater matrix. While genome sequences have offered insights into differing tactics for obtaining resources in the ocean, evolution of alternate bacterioplankton life history strategies is not yet well understood.
Two phylogenetically related marine taxa, the Roseobacter and SAR11 lineages, exemplify extremes in the free-living to patch-adapted continuum while sharing a common ancestor in the alphaproteobacteria. As two of the most abundant heterotrophic bacterial groups in ocean surface waters (1, 2), the evolutionary paths leading to their divergent ecological strategies have likely influenced where and when fixed carbon is processed in the ocean (9, 10) and what fraction is exported into deep waters (11, 12).
We sought to interpret the evolution of genome properties associated with these marine alphaproteobacterial clades by integrating ancestral gene content reconstruction and patterns of protein-coding sequence evolution. The reconstruction of genome content in ancestral lineages has frequently been modeled using maximum parsimony methods (13–20), but these techniques are not able to model parallel and repeated gene insertions and deletions and are known to underestimate the number of evolutionary events. They also cannot model rate variability among different lineages and gene functions (21). More recently, maximum likelihood approaches have been developed to overcome the disadvantages of parsimony-based reconstructions (21–25). In these likelihood analyses, however, insertion rates are frequently unrealistically assumed to be equal to deletion rates, and no differentiation is made between lateral gene transfer (LGT) and gene duplication.
A recently developed maximum likelihood method implemented in the COUNT software (26, 27) is based on the birth-and-death evolutionary model of multigene families (28). The birth-and-death model assumes that genes are lost, gained, and duplicated independently (29), with constant rates for a fixed family and phylogeny branch, thereby modeling microbial genome evolution in a more realistic way (27, 30). The model is thus described by lineage- and family-specific gene loss and duplication rates, coupled with a lineage-specific family gain process accounting for LGT. Here, we apply this method to Roseobacter and SAR11 clades to address the evolutionary history of their distinct ecological strategies.
RESULTS AND DISCUSSION
Ancestral reconstruction of the marine alphaproteobacterial tree.
Ancestral reconstruction of genome content requires a robust phylogenetic tree describing the evolutionary relationship of the taxa. Using four different phylogenomic approaches which take into account different aspects of heterogeneous evolutionary processes that likely have occurred during the evolution of alphaproteobacterial lineages (P4, RAxML, PhyloBayes, and MrBayes), we obtained a robust phylogenetic position of the marine Roseobacter clade and other major lineages in the alphaproteobacterial tree (Fig. 1; see also Fig. S1 in the supplemental material). However, the SAR11 clade was placed in three alternate evolutionary positions, all of which were supported by extremely high bootstrap values or posterior probabilities within that phylogenomic approach (Fig. 1; see also Fig. S1). Regardless of the specific position in the competing reconstructions, however, the phylogenetic birth-and-death model consistently predicted that the small extant SAR11 genomes (1,300 to 1,500 genes) evolved from a slightly larger common ancestor (~2,000 genes; Fig. 2A; see also Fig. S2A and C), while the large and variable extant Roseobacter genomes (2,000 to 5,000 genes; median, >4,000) evolved from a quite large common ancestor (~8,000 genes; Fig. 2B; see also Fig. S2B and D), a characteristic echoed in the genome size of nonmarine Roseobacter relatives.
FIG 1 .
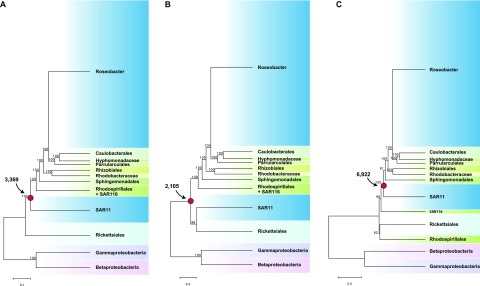
Model-based phylogenomic trees of alphaproteobacteria based on a concatenation of 60 orthologous protein sequences using the P4 Bayesian software with the NDCH and NDRH models (A), the RAxML software (B), and the PhyloBayes software with the CAT model (C). A Bayesian phylogeny using MrBayes with or without the covarion model had the same branching order as the RAxML tree. The node representing the most recent common ancestor (MRCA) of the Roseobacter and SAR11 lineages is indicated with a red dot, and the predicted gene number for the MRCA is indicated. For clarity, only the deep branches connecting the major lineages and their statistical support values are shown. The complete trees are shown in Fig. S1 in the supplemental material.
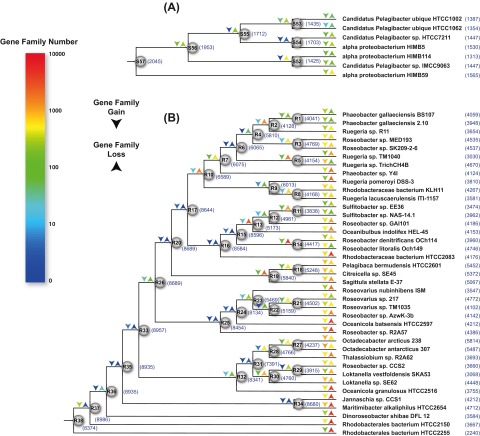
FIG 2 .
Ancestral genome content reconstruction using the COUNT software. The reconstruction is based on the P4-based alphaproteobacterial tree (see Fig. S1 in the supplemental material), but only the parts of the results involving marine SAR11 (A) and Roseobacter (B) are shown. The log-scale color coding represents numbers of reconstructed gain and loss events of each lineage. Numbers in parentheses are predicted gene numbers for ancestral nodes and observed gene numbers for extant lineages. The genome expansion on the Roseobacter branch leading to R37 was statistically significant based on reconstruction of randomized genome content in 100 bootstrapped replicates (see Table S3).
Dynamics of genome content.
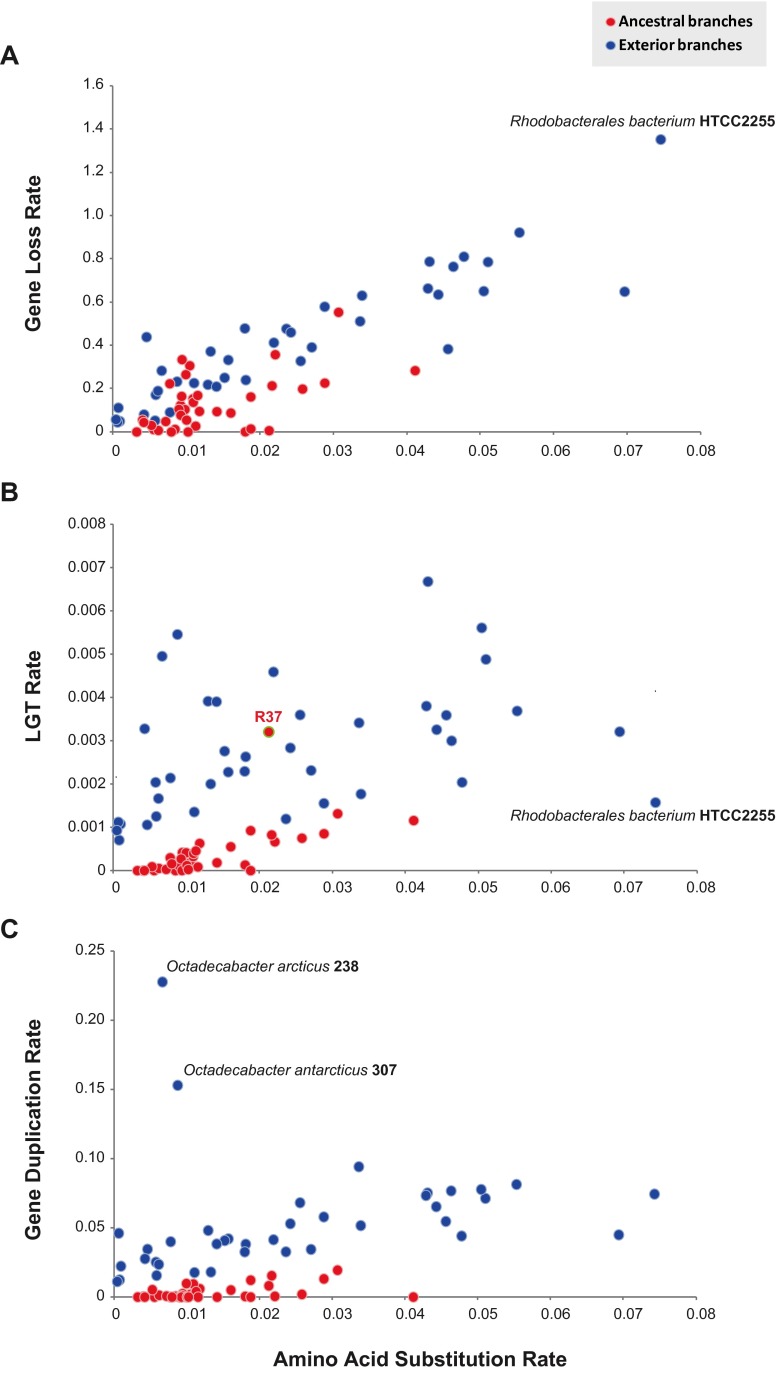
The phylogenetic birth-and-death model imposed on the phylogenomic trees shows a steady trend toward streamlined genomes in the SAR11 lineage, with no abrupt changes in gene family content since the common ancestor (Fig. 2A; see also Fig. S2A and C in the supplemental material). The Roseobacter lineage exhibits a more complicated evolutionary path to net genome reduction, however (Fig. 2B; see also Fig. S2B and D), with the Roseobacter ancestor experiencing an early surge in gene content (leading to the R37 node in Fig. 2B; see also Fig. S2B and D). The model suggests that this surge occurred exclusively through gain of new families rather than expansion of existing ones (see Table S1). The calculated rate of gene loss compared to the amino acid substitution rate for Roseobacter branches varies depending on the underlying phylogenetic tree reconstruction (14 deletions per amino acid substitution for P4, 5 for RAxML, 8 for PhyloBayes), but all three predict that genes were lost at a constant rate for both ancestral and exterior branches (Fig. 3A; see also Fig. S3A and D and Table S2A). For LGT, however, calculated rates are significantly lower for ancestral than for exterior branches (Fig. 3B; see Fig. S3B and E and Table S2A), with the LGT rate following a molecular clock only for the ancestral branches (averaging 0.036, 0.015, or 0.024 gene family acquisitions per amino acid substitution, depending on tree construction; R2 > 0.69 and P < 0.001 in all cases). The notable exception is the ancestral branch leading to Roseobacter node R37, showing a significantly higher LGT rate than any other ancestral branch (see Table S2B), in agreement with the significant surge of genome content on that branch predicted by the birth-and-death model (Fig. 2B; see also Fig. S2B and D) with bootstrapped genome content data sets (see Table S3). For gene duplication, calculated rates are low in the majority of Roseobacter branches (Fig. 3C; see also Fig. S3C and F) and do not follow a molecular clock (P > 0.05) (see Table S2A). The branch leading to the Arctic strain Octadecabacter arcticus 238 is an outlier (P < 0.001; see Table S2B), regardless of whether the observed expansion of insertion sequence families are included or not (see Fig. S4), suggesting that gene duplication rates may be enhanced in polar roseobacters.
FIG 3 .
Analysis of gene loss rate (A), lateral gene transfer rate (B), and gene duplication rate (C) versus amino acid substitution rate on the Roseobacter branches of the alphaproteobacterial phylogeny constructed using P4. For the exterior Roseobacter branches, LGT rate calculations were highly variable and did not exhibit a clock-like pattern (R2 = 0.14; P = 0.02).
One basal Roseobacter lineage (represented by strain HTCC2255) diverged at node R38 and escaped the early surge, evolving directly toward a highly reduced genome of only 2,240 genes (Fig. 2B; see also Fig. S2B and D in the supplemental material), while in the remaining clades, a trend toward gradual genome reduction followed the rapid early innovation (Fig. 2B; see also Fig. S2B and D). At the tips of the phylogeny, Roseobacter lineages show either gradual genome downsizing or expansion (Fig. 2B; see also Fig. S2B and D). Thus, two time periods of substantial evolutionary change in Roseobacter genomes are predicted: one occurring early in their history and manifested as genome expansion via LGT along the branch leading to node R37, and the second occurring more recently along the branches leading to extant members. A flux of gene family content is also observed in some SAR11 leaf lineages but is of considerably smaller magnitude than that observed at the tips of the Roseobacter phylogeny (Fig. 2; see also Fig. S2).
Biased gene acquisition in roseobacters and SAR11s.
Characterization of gene families based on clusters of orthologous groups (COGs) (31) indicated that putative biological functions gained during the evolution of the SAR11 and Roseobacter lineages were significantly different (chi-square test, P < 0.001). Along the SAR11 branches, acquired families were biased toward cell wall biogenesis (55 families of lipopolysaccharide, cell wall, and polysaccharide synthesis proteins) and pilus synthesis (15 families of assembly proteins). Along the Roseobacter branches, a greater proportion of acquired families were involved in gene regulation (450 families of transcriptional regulators, DNA binding proteins, and sigma factors) and replication/recombination/repair (431 families of transposases, endonucleases, recombinases, methylases, and mismatch repair enzymes) (Fig. 4). During later innovation in the Roseobacter lineage, lateral gene acquisition was biased toward gene regulation (41 families of transcriptional regulators and DNA binding proteins) and defense mechanisms (12 families of antibiotic synthesis and export proteins and multidrug efflux pumps) (Fig. 4), potentially equipping the cells to better compete in microbial communities associated with enriched patches (32). This nonrandom collection of SAR11 and Roseobacter gene functions gained through LGT is indicative of adaptive evolution.
FIG 4 .

Gene families gained per branch in Roseobacter versus SAR11 lineages (left) and in Roseobacter ancestral nodes R37 versus R1 to R36 (right). Letters represent COG categories. Asterisks indicate significant differences in proportions based on Xipe analysis (64) (P < 0.01). The horizontal axis indicates the number of families gained per branch for each COG class. Cell motility families gained in SAR11 represent pilus formation genes.
Pattern of gene loss in Roseobacter strain HTCC2255.
Not all extant Roseobacter lineages have evolved toward genome content suggestive of patch-associated life histories, however. The birth-and-death model predicts that the HTCC2255 lineage lost >5,000 gene families since divergence at the clade ancestor, including those conserved in a majority of roseobacters and involved in motility, chemotaxis, secondary metabolite synthesis and metabolism, signal transduction, and various regulatory functions, making the genetic composition of HTCC2255 and the Roseobacter clade ancestor (node R38 in Fig. 2B; see also Fig. S2B and D in the supplemental material) significantly different (Fig. S5 chi-square test, P < 0.001). In this case, the pattern of gene family loss is suggestive of relaxation of purifying selection on gene families not necessary for a small, free-living bacterioplankter, and in fact the functional profile of the HTCC2255 genome is more similar to that of SAR11 than other roseobacters (Fig. 5).
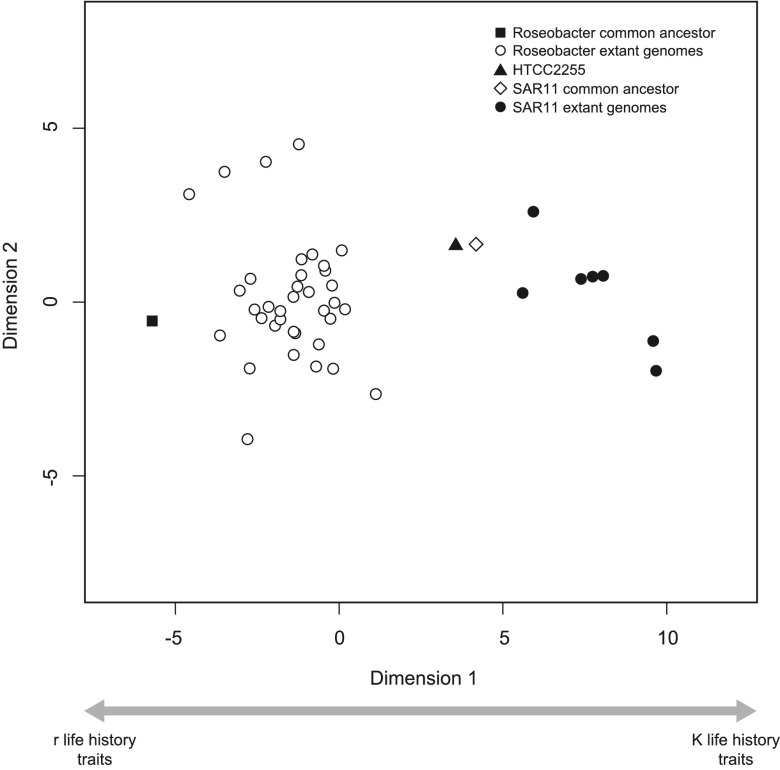
FIG 5 .
High-throughput multidimensional scaling (HiT-MDS) plot of the genetic composition of the Roseobacter and SAR11 extant lineages and predicted composition of their respective common ancestors. The genetic composition was determined by mapping the gene families to COG functional categories. COG classes significantly negatively correlated with dimension 1 and hypothesized to include traits associated with r-selected life histories are G (carbohydrate transport and metabolism), I (lipid transport and metabolism), K (transcription), N (cell motility), P (inorganic ion transport and metabolism), Q (secondary-metabolite biosynthesis, transport, and catabolism), T (signal transduction mechanisms), and V (defense mechanisms). COG classes significantly positively correlated with dimension 1 and hypothesized to include traits associated with K-selected life histories are C (energy production and conversion), D (cell cycle control, cell division, and chromosome partitioning), F (nucleotide transport and metabolism), H (coenzyme transport and metabolism), J (translation, ribosomal structure, and biogenesis), M (cell wall/membrane/envelope biogenesis), O (posttranslational modification and protein turnover, chaperones), and U (intracellular trafficking, secretion, and vesicular transport). No significant correlation was found between dimension 1 and COG classes E (amino acid transport and metabolism) or L (replication, recombination, and repair).
An evolutionary timeline.
The timing of diversification of the lineages was inferred using a maximum likelihood method based on a relaxed molecular clock calibrated by the fossil record (33). This approach dates the occurrence of the common ancestor of SAR11 at 826 (±21) million years ago (mya) (Fig. 6). The prediction from the phylogenetic birth-and-death model that extant SAR11 genomes have been streamlined by only 25 to 30% from their common ancestor emphasizes the importance of the SAR11 position on the alphaproteobacterial tree to the genome streamlining theory (2, 9). If the SAR11 lineage clusters with Rickettsiales at the base of the alphaproteobacterial tree (Fig. 1B), the most recent common ancestor (MRCA) (Fig. 1B) of the SAR11 and Roseobacter lineages is predicted to have had only ~2,100 genes, suggesting only a trivial reduction to the SAR11 ancestor. If the SAR11 lineage branched off either before (Fig. 1A) or after (Fig. 1C) the marine SAR116 lineage (represented by the “Candidatus Puniceispirillum marinum” IMCC1322 genome), the MRCA genome is predicted to contain either ~3,300 or ~6,900 genes (Fig. 1A and C), with the latter most strongly supporting the hypothesis that genomic and metabolic streamlining is the primary evolutionary process influencing the content of extant SAR11 genomes.
FIG 6 .
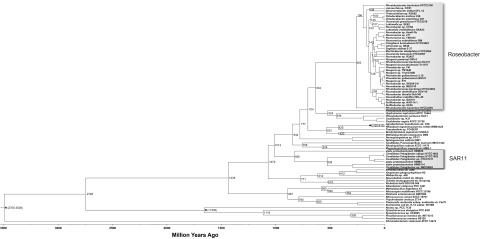
A chronogram of alphaproteobacteria. Nodes with fossil record corrections are indicated with an asterisk. The tree branching order was constructed using RAxML version 7.3.0 software with a data partition model determined using PartitionFinder, and molecular dating was performed using the r8s software.
The timeline of the Roseobacter lineage indicates that their common ancestor (node R38 in Fig. 2B; see Fig. S2B and D in the supplemental material) occurred more recently, at 260 (±7) mya (Fig. 6). In comparison to SAR11, extant Roseobacter genomes exhibited a greater net genome reduction from the common ancestor (50 to 70%) within a considerably shorter evolutionary time frame. Molecular dating of the R37 node at 196 (±7) mya (Fig. 6) places the timing of the first episode of Roseobacter diversification concurrent with the Mesozoic radiation of the eukaryotic red-lineage phytoplankton (dinoflagellates, coccolithophorids, and diatoms), predicted as early as 250 mya (34). Because the cyanobacteria and green algae that dominated the early ocean were not much larger than bacteria (34) and probably of insufficient size to be detected by bacterial chemosensory mechanisms (35), the radiation of larger phytoplankton groups likely offered new habitats for heterotrophic bacterioplankton, particularly for lineages with large genomes encoding chemotaxis, motility, defense, and other functions beneficial for locating and tracking nutrient-enriched microzones (7, 36). Indeed, members of the Roseobacter lineage in the contemporary ocean frequently occur in association with red-lineage phytoplankton cells (37, 38).
Conclusion.
Although it is a simplified representation of the evolutionary paths taken by heterotrophic marine bacterioplankton (9, 10), the free-living versus patch-adapted dichotomy is nonetheless useful to explore implications of disparate life history strategies of marine bacteria (3, 5). The comparative evolutionary history of the Roseobacter and SAR11 lineages points to the emergence of large eukaryotic phytoplankton as an important event driving divergence of patch-adapted from free-living bacterioplankton, the former of which are implicated in enhancing export flux of organic matter to deeper waters via aggregation (11, 12), and the latter is linked to intensive remineralization of upper ocean fixed carbon through the microbial loop. A future ocean shaped by rising greenhouse gas emissions is consistently predicted to favor picophytoplankton over diatoms, dinoflagellates, and coccolithophorids (39–42) and therefore may favor free-living over patch-adapted bacterioplankton. Subsequent effects on ocean heterotrophy mediated through alterations in the rates and efficiencies (43) of bacterial assimilation of distinct classes of organic compounds (44) could intensify future changes to the oceanic carbon cycle.
MATERIALS AND METHODS
Since resolving the evolutionary position of the SAR11 clade on the alphaproteobacterial tree has proven to be difficult (20, 45), we used multiple evolutionary models to account for the potential heterogeneity in phylogenetic reconstruction and studied the genome evolution of the marine Roseobacter and SAR11 clades in the context of this controversy. Phylogenetic reconstruction used a concatenation of 60 conserved orthologous proteins in 65 alphaproteobacterial genomes (39 and 7 representatives of marine Roseobacter and SAR11 clades, along with additional related lineages; see Fig. S1 in the supplemental material) and 8 outgroup species associated with gammaproteobacteria and betaproteobacteria. Phylogenetic models and software included a maximum likelihood method using a data partition model in the RAxML version 7.3.0 software (46) and a Bayesian method using a data partition model with and without the covarion model in MrBayes version 3.1.2 (47). The partition model involves estimating independent evolutionary models for different genes or subsets of genes, which are implemented in the PartitionFinder software (48). Two alternate partition schemes were chosen depending on the statistical evaluation method. The covarion model takes into account the variation of substitution rate at a site across time (49, 50). Both RAxML and MrBayes are implemented in parallel versions, making them computationally efficient for this large data set on a high-performance computing cluster. Nevertheless, these phylogenetic methods are unable to model other inherent heterogeneities of this data set, including a substantial variation of amino acid composition across sites and across lineages. We thus employed the PhyloBayes and P4 Bayesian software, which are computationally expensive but designed to account for these two aspects.
As there exists a substantial variation in nucleotide G+C content among alphaproteobacterial lineages (~<30% to 70%), and it is known that amino acid composition is affected by the G+C bias (51), the concatenated protein sequence was recoded into the following six Dayhoff groups to reduce this bias (52): cysteine; alanine, serine, threonine, proline, glycine; asparagine, aspartic acid, glutamic acid, glutamine; histidine, arginine, lysine; methionine, isoleucine, leucine, valine; phenylalanine, tyrosine, tryptophan. After recoding was complete, we used a Bayesian method with the CAT model in the PhyloBayes version 3.2e software (53) and a Bayesian method with the node-discrete composition heterogeneity (NDCH) and the node-discrete rate heterogeneity (NDRH) models in the P4 software (54). The CAT model integrates heterogeneity of amino acid composition across sites of a protein alignment (55). The NDCH model allows heterogeneity of amino acid composition across different branches, and the NDRH model allows different rate matrices on different branches (54). All models were used with a Gamma distribution of rate variation among sites.
To study the evolution of life history strategies, we compiled a comprehensive data set of 44,064 orthologous gene families covering the 65 alphaproteobacterial genomes. Gene families were identified using the OrthoMCL software (56). To reconstruct ancestral gene family sizes, we adapted a recently developed pipeline that is suitable for the analysis of such large data sets (27, 57–60), as implemented in the COUNT software package (26). The reconstruction is based on numerical phylogenetic patterns formed by the gene copy numbers across extant genomes in homologous families. Ancestral family sizes are inferred in COUNT by assuming a probabilistic framework involving a phylogenetic birth-and-death model (28) along a rooted phylogeny. In particular, the model is described by lineage- and family-specific gene loss and duplication rates, coupled with a family gain process accounting for arrival by LGT. In contrast to gene-species tree reconciliation methods (61, 62), phylogenetic birth-and-death methods gain expediency by ignoring sequence information and infer ancestral events affecting family sizes by using solely the copy number information. Both larger (27) and smaller (30) ancestral genomes than extant genomes have been predicted with the birth-and-death model when investigating archaeal and virus evolution. In order to infer confidence intervals of the predicted number of gene families in the ancestral nodes, we repeated the procedure with 100 bootstrap data sets generated by randomly sampling gene families (with repetition).
For draft Roseobacter genomes, a regression analysis (see Fig. S6 in the supplemental material) for universal single-copy genes (63) indicated that completeness ranged from 90 to 100%, with a median of 98%. The molecular dating was performed using penalized likelihood based on a relaxed clock model implemented in r8s software version 1.71 (33). Molecular dating requires a phylogenomic tree with fossil calibrations, and thus a few cyanobacterial branches with time constraints were included. This phylogenomic tree was constructed using RAxML version 7.3.0 (46) with an optimized data partition of a concatenation of 61 conserved single-copy orthologous protein sequences. Details of the computational methods can be found in the Text S1 in the supplemental materials. The orthologous sequences, the partitioned amino acid sequences, the gene families that are gained on the Roseobacter and SAR11 branches, and the gene families that are lost on the HTCC2255 branch are available upon request.
SUPPLEMENTAL MATERIAL
Supplemental methods
Three alternate well-supported phylogenomic trees of alphaproteobacteria based on a concatenation of 60 orthologous protein sequences. (A) A Bayesian tree using the P4 software. This fifty-percent majority-rule consensus of 1,000 trees was sampled from the posterior distribution of a Metropolis-coupled Markov chain Monte Carlo (MCMCMC) with the NDCH model of 14 across-tree composition vectors and the NDRH model of 3 across-tree rate matrix vectors with Dayhoff-recoded data and with a gamma model accounting for among-site rate variation (GTR + G + NDCH + NDRH). A chi-square test using a posterior predictive distribution rejected the homogeneous model, which does not take into account the variation in amino acid composition across the branches of the tree. The number of vectors in the NDCH and NDRH models was determined by Bayes factors. The scale bar indicates the number of substitutions per site. The value near each internal branch is the posterior probability for that branch. (B) A maximum likelihood phylogeny using the RAxML version 7.3.0 software with a data partition model determined using PartitionFinder. Values at the nodes show the number of times the clade defined by that node appeared in the 100 bootstrapped data sets using RAxML. A Bayesian phylogeny using MrBayes with the same partition scheme and with or without the covarion model had the same branching order as the RAxML tree. (C) A Bayesian phylogeny using the PhyloBayes version 3.2e software based on Dayhoff-recoded protein sequences with the CAT model, a gamma model for among-site rate variation, and Poisson substitution matrix (Poisson + G + CAT). The value near each internal branch is the posterior probability for that branch. Trees are rooted using species from betaproteobacteria and gammaproteobacteria. Download
Ancestral genome content reconstruction using the COUNT software. The reconstruction is based on the RAxML-based (see Fig. S1B) and PhyloBayes-based (see Fig. S1C) alphaproteobacterial tree, but only the parts of the results involving marine SAR11 (A, C) and Roseobacter (B, D) are shown. The log-scale color coding represents numbers of reconstructed gain and loss events of each lineage. Numbers in parentheses are predicted gene numbers for ancestral nodes and observed gene numbers for extant lineages. The genome expansion on the Roseobacter branch leading to R37 was statistically significant based on reconstruction of randomized genome content in 100 bootstrapped replicates (see Table S3). Download
Analysis of gene loss rate (A, D), lateral gene transfer rate (B, E), and gene duplication rate (C, F) versus amino acid substitution rate on the Roseobacter branches of the alphaproteobacterial phylogeny constructed using RAxML (see Fig. S1B) and PhyloBayes (see Fig. S1C). For the exterior Roseobacter branches, LGT rate calculations were highly variable and did not exhibit a clock-like pattern (R2 = 0.13; P = 0.02); this is the case for both reconstructions. Download
Gene duplication rate versus amino acid substitution rate on the Roseobacter branches of the alphaproteobacterial phylogeny constructed by P4 (A), RAxML (B), and PhyloBayes (C) after 667 transposase families were removed. The transposases were identified using RPS-BLAST against the COG database, using the best BLASTP hit to the NCBI RefSeq database and using the ISsaga database. Download
Genetic composition in the common ancestor of the Roseobacter clade (R38) versus the basal Roseobacter lineage Rhodobacterales bacterium HTCC2255. Letters represent COG categories. Asterisks indicate significant differences using Xipe resampling analysis. The horizontal axis indicates the percentage that each COG category represents in the genome. Download
Regression model for Roseobacter genome coverage estimation based on genome statistics for 39 cultured Roseobacter strains. The x axis is the ratio of the number of conserved single-copy genes universally present in Roseobacter species to the number of predicted protein-encoding genes; the y axis is the number of nucleotides sequenced. The data were fit to a power regression model (R2 = 0.94). Download
Gene number (predicted for ancestral nodes) and predicted number of gene families gained, lost, expanded, and contracted in the Roseobacter and SAR11 clade genomes. The ancestral reconstruction is based on an alphaproteobacterial tree estimated using P4 (A), RAxML (B), and PhyloBayes (C). Note that the gain of a gene family or expansion of an existing gene family could be a result of either LGT or gene duplication. The relative contribution of these two mechanisms is presented in Fig. 3 and Fig. S3.
Statistical analyses of gene loss, LGT, and gene duplication rates for Roseobacter branches. (A) Analysis of covariance results of F tests for ancestral compared to exterior branches; (B) deleted t residual analysis from regressions of gene loss rate, LGT rate, and gene duplication rate versus amino acid substitution rate, conducted separately for ancestral and exterior branches.
Gene count estimates and confidence intervals of ancestral Roseobacter and SAR11 nodes for three ancestral reconstructions of the alphaproteobacterial tree.
ACKNOWLEDGMENTS
We thank Peter Foster, David Posada, Miguel Fonseca, and Rob Lanfear for advice on phylogenetic analyses, Camille English for assistance with graphics, Shalabh Sharma, Brian Hopkinson, and Yongjie Huang for bioinformatics assistance, Roman Stocker for discussions, and the Georgia Advanced Computing Resource Center at the University of Georgia for computational resources.
This research was funded by grants from the Gordon and Betty Moore Foundation, the National Science Foundation (MCB-0702125), and the Natural Sciences and Engineering Research Council of Canada.
H L. conceived of the study; H.L. and M.C. designed bioinformatics protocols; H.L. and M.A.M. analyzed the data; H.L., A.L.H., and M.A.M. interpreted the data; H.L. and M.A.M. wrote the paper.
Footnotes
Citation Luo H, Csürös M, Hughes AL, Moran MA. 2013. Evolution of divergent life history strategies in marine alphaproteobacteria. mBio 4(4):e00373-13. doi:10.1128/mBio.00373-13.
REFERENCES
- 1. Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP, Henriksen JR, King GM, Belas R, Fuqua C, Brinkac L, Lewis M, Johri S, Weaver B, Pai G, Eisen JA, Rahe E, Sheldon WM, Ye W, Miller TR, Carlton J, Rasko DA, Paulsen IT, Ren Q, Daugherty SC, Deboy RT, Dodson RJ, Durkin AS, Madupu R, Nelson WC, Sullivan SA, Rosovitz MJ, Haft DH, Selengut J, Ward N. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910–913 [DOI] [PubMed] [Google Scholar]
- 2. Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245 [DOI] [PubMed] [Google Scholar]
- 3. Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, Ferriera S, Lapidus A, Anderson I, Kyrpides N, Munk AC, Detter C, Han CS, Brown MV, Robb FT, Kjelleberg S, Cavicchioli R. 2009. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:15527–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polz MF, Hunt DE, Preheim SP, Weinreich DM. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2009–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, Johnson J, Montgomery R, Ferriera S, Beeson K, Williamson SJ, Tovchigrechko A, Allen AE, Zeigler LA, Sutton G, Eisenstadt E, Rogers YH, Friedman R, Frazier M, Venter JC. 2010. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468:60–66 [DOI] [PubMed] [Google Scholar]
- 6. Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5:966–966 [DOI] [PubMed] [Google Scholar]
- 7. Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. U. S. A. 105:4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackburn N, Fenchel T. 1999. Influence of bacteria, diffusion and shear on micro-scale nutrient patches, and implications for bacterial chemotaxis. Mar. Ecol. Prog. Ser. 189:1–7 [Google Scholar]
- 9. Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS. 2012. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3(5):e00252-12. 10.1128/mBio.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morris JJ, Lenski RE, Zinser ER. 2012. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3(2):e00036-12. 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gärdes A, Iversen MH, Grossart HP, Passow U, Ullrich MS. 2011. Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5:436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lapoussière A, Michel C, Starr M, Gosselin M, Poulin M. 2011. Role of free-living and particle-attached bacteria in the recycling and export of organic material in the Hudson Bay system. J. Mar. Syst. 88:434–445 [Google Scholar]
- 13. Boussau B, Karlberg EO, Frank AC, Legault BA, Andersson SG. 2004. Computational inference of scenarios for α-proteobacterial genome evolution. Proc. Natl. Acad. Sci. U. S. A. 101:9722–9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dagan T, Martin W. 2007. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl. Acad. Sci. U. S. A. 104:870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hao W, Golding GB. 2004. Patterns of bacterial gene movement. Mol. Biol. Evol. 21:1294–1307 [DOI] [PubMed] [Google Scholar]
- 16. Kunin V, Ouzounis CA. 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13:1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mirkin BG, Fenner TI, Galperin MY, Koonin EV. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 3:2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Snel B, Bork P, Huynen MA. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 12:17–25 [DOI] [PubMed] [Google Scholar]
- 20. Viklund J, Ettema TJ, Andersson SG. 2012. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol. Biol. Evol. 29:599–615 [DOI] [PubMed] [Google Scholar]
- 21. Hao W, Golding GB. 2006. The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res. 16:636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cohen O, Rubinstein ND, Stern A, Gophna U, Pupko T. 2008. A likelihood framework to analyse phyletic patterns. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:3903–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Didelot X, Darling A, Falush D. 2009. Inferring genomic flux in bacteria. Genome Res. 19:306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marri PR, Hao W, Golding GB. 2007. The role of laterally transferred genes in adaptive evolution. BMC Evol. Biol. 7:S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marri PR, Hao W, Golding GB. 2006. Gene gain and gene loss in streptococcus: is it driven by habitat? Mol. Biol. Evol. 23:2379–2391 [DOI] [PubMed] [Google Scholar]
- 26. Csűros M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912 [DOI] [PubMed] [Google Scholar]
- 27. Csűros M, Miklós I. 2009. Streamlining and large ancestral genomes in Achaea inferred with a phylogenetic birth-and-death model. Mol. Biol. Evol. 26:2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39:121–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nye TM. 2009. Modelling the evolution of multi-gene families. Stat. Methods Med. Res. 18:487–504 [DOI] [PubMed] [Google Scholar]
- 30. Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6:223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 32. Slightom RN, Buchan A. 2009. Surface colonization by marine Roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75:6027–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanderson MJ. 2003. R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19:301–302 [DOI] [PubMed] [Google Scholar]
- 34. Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJ. 2004. The evolution of modern eukaryotic phytoplankton. Science 305:354–360 [DOI] [PubMed] [Google Scholar]
- 35. Jackson GA. 1987. Simulating chemosensory responses of marine microorganisms. Limnol. Oceanogr. 32:1253–1266 [Google Scholar]
- 36. Miller TR, Belas R. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648–1659 [DOI] [PubMed] [Google Scholar]
- 37. González JM, Simó R, Massana R, Covert JS, Casamayor EO, Pedrós-Alió C, Moran MA. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jasti S, Sieracki ME, Poulton NJ, Giewat MW, Rooney-Varga JN. 2005. Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71:3483–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marinov I, Doney SC, Lima ID. 2010. Response of ocean phytoplankton community structure to climate change over the 21st century: partitioning the effects of nutrients, temperature and light. Biogeosciences 7:3941–3959 [Google Scholar]
- 40. Morán XAG, López-Urrutia A, Calvo-Díaz A, Li WKW. 2010. Increasing importance of small phytoplankton in a warmer ocean. Glob. Change Biol. 16:1137–1144 [Google Scholar]
- 41. Riebesell U, Körtzinger A, Oschlies A. 2009. Sensitivities of marine carbon fluxes to ocean change. Proc. Natl. Acad. Sci. U. S. A. 106:20602–20609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taylor GT, Muller-Karger FE, Thunell RC, Scranton MI, Astor Y, Varela R, Ghinaglia LT, Lorenzoni L, Fanning KA, Hameed S, Doherty O. 2012. Ecosystem responses in the southern Caribbean sea to global climate change. Proc. Natl. Acad. Sci. U. S. A. 109:19315–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alonso-Sáez L, Gasol JM. 2007. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73:3528–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson CE, Carlson CA. 2012. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ. Microbiol. 14:1500–1516 [DOI] [PubMed] [Google Scholar]
- 45. Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappé MS, Giovannoni SJ. 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 47. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 48. Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29:1695–1701 [DOI] [PubMed] [Google Scholar]
- 49. Galtier N. 2001. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 18:866–873 [DOI] [PubMed] [Google Scholar]
- 50. Tuffley C, Steel M. 1998. Modeling the covarion hypothesis of nucleotide substitution. Math. Biosci. 147:63–91 [DOI] [PubMed] [Google Scholar]
- 51. Foster PG, Hickey DA. 1999. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48:284–290 [DOI] [PubMed] [Google Scholar]
- 52. Hrdy I, Hirt RP, Dolezal P, Bardonová L, Foster PG, Tachezy J, Embley TM. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432:618–622 [DOI] [PubMed] [Google Scholar]
- 53. Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288 [DOI] [PubMed] [Google Scholar]
- 54. Foster PG. 2004. Modeling compositional heterogeneity. Syst. Biol. 53:485–495 [DOI] [PubMed] [Google Scholar]
- 55. Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21:1095–1109 [DOI] [PubMed] [Google Scholar]
- 56. Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 23:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Szöllosi G, Daubin V. 2012. Modeling gene family evolution and reconciling phylogenetic discord, p 29–51 In Anisimova M, Evolutionary genomics, vol 856 Humana Press, New York City, NY. [DOI] [PubMed] [Google Scholar]
- 60. Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol. Direct 7:46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Akerborg O, Sennblad B, Arvestad L, Lagergren J. 2009. Simultaneous Bayesian gene tree reconstruction and reconciliation analysis. Proc. Natl. Acad. Sci. U. S. A. 106:5714–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kamneva OK, Knight SJ, Liberles DA, Ward NL. 2012. Analysis of genome content evolution in PVC bacterial super-phylum: assessment of candidate genes associated with cellular organization and lifestyle. Genome Biol. Evol. 4:1375–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun S, Varaljay V, Vila-Costa M, Westrich JR, Moran MA. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798 [DOI] [PubMed] [Google Scholar]
- 64. Rodriguez-Brito B, Rohwer F, Edwards RA. 2006. An application of statistics to comparative metagenomics. BMC Bioinformatics 7:162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods
Three alternate well-supported phylogenomic trees of alphaproteobacteria based on a concatenation of 60 orthologous protein sequences. (A) A Bayesian tree using the P4 software. This fifty-percent majority-rule consensus of 1,000 trees was sampled from the posterior distribution of a Metropolis-coupled Markov chain Monte Carlo (MCMCMC) with the NDCH model of 14 across-tree composition vectors and the NDRH model of 3 across-tree rate matrix vectors with Dayhoff-recoded data and with a gamma model accounting for among-site rate variation (GTR + G + NDCH + NDRH). A chi-square test using a posterior predictive distribution rejected the homogeneous model, which does not take into account the variation in amino acid composition across the branches of the tree. The number of vectors in the NDCH and NDRH models was determined by Bayes factors. The scale bar indicates the number of substitutions per site. The value near each internal branch is the posterior probability for that branch. (B) A maximum likelihood phylogeny using the RAxML version 7.3.0 software with a data partition model determined using PartitionFinder. Values at the nodes show the number of times the clade defined by that node appeared in the 100 bootstrapped data sets using RAxML. A Bayesian phylogeny using MrBayes with the same partition scheme and with or without the covarion model had the same branching order as the RAxML tree. (C) A Bayesian phylogeny using the PhyloBayes version 3.2e software based on Dayhoff-recoded protein sequences with the CAT model, a gamma model for among-site rate variation, and Poisson substitution matrix (Poisson + G + CAT). The value near each internal branch is the posterior probability for that branch. Trees are rooted using species from betaproteobacteria and gammaproteobacteria. Download
Ancestral genome content reconstruction using the COUNT software. The reconstruction is based on the RAxML-based (see Fig. S1B) and PhyloBayes-based (see Fig. S1C) alphaproteobacterial tree, but only the parts of the results involving marine SAR11 (A, C) and Roseobacter (B, D) are shown. The log-scale color coding represents numbers of reconstructed gain and loss events of each lineage. Numbers in parentheses are predicted gene numbers for ancestral nodes and observed gene numbers for extant lineages. The genome expansion on the Roseobacter branch leading to R37 was statistically significant based on reconstruction of randomized genome content in 100 bootstrapped replicates (see Table S3). Download
Analysis of gene loss rate (A, D), lateral gene transfer rate (B, E), and gene duplication rate (C, F) versus amino acid substitution rate on the Roseobacter branches of the alphaproteobacterial phylogeny constructed using RAxML (see Fig. S1B) and PhyloBayes (see Fig. S1C). For the exterior Roseobacter branches, LGT rate calculations were highly variable and did not exhibit a clock-like pattern (R2 = 0.13; P = 0.02); this is the case for both reconstructions. Download
Gene duplication rate versus amino acid substitution rate on the Roseobacter branches of the alphaproteobacterial phylogeny constructed by P4 (A), RAxML (B), and PhyloBayes (C) after 667 transposase families were removed. The transposases were identified using RPS-BLAST against the COG database, using the best BLASTP hit to the NCBI RefSeq database and using the ISsaga database. Download
Genetic composition in the common ancestor of the Roseobacter clade (R38) versus the basal Roseobacter lineage Rhodobacterales bacterium HTCC2255. Letters represent COG categories. Asterisks indicate significant differences using Xipe resampling analysis. The horizontal axis indicates the percentage that each COG category represents in the genome. Download
Regression model for Roseobacter genome coverage estimation based on genome statistics for 39 cultured Roseobacter strains. The x axis is the ratio of the number of conserved single-copy genes universally present in Roseobacter species to the number of predicted protein-encoding genes; the y axis is the number of nucleotides sequenced. The data were fit to a power regression model (R2 = 0.94). Download
Gene number (predicted for ancestral nodes) and predicted number of gene families gained, lost, expanded, and contracted in the Roseobacter and SAR11 clade genomes. The ancestral reconstruction is based on an alphaproteobacterial tree estimated using P4 (A), RAxML (B), and PhyloBayes (C). Note that the gain of a gene family or expansion of an existing gene family could be a result of either LGT or gene duplication. The relative contribution of these two mechanisms is presented in Fig. 3 and Fig. S3.
Statistical analyses of gene loss, LGT, and gene duplication rates for Roseobacter branches. (A) Analysis of covariance results of F tests for ancestral compared to exterior branches; (B) deleted t residual analysis from regressions of gene loss rate, LGT rate, and gene duplication rate versus amino acid substitution rate, conducted separately for ancestral and exterior branches.
Gene count estimates and confidence intervals of ancestral Roseobacter and SAR11 nodes for three ancestral reconstructions of the alphaproteobacterial tree.