ABSTRACT
Burkholderia pseudomallei causes the potentially fatal disease melioidosis. It is generally accepted that B. pseudomallei is a noncommensal bacterium and that any culture-positive clinical specimen denotes disease requiring treatment. Over a 23-year study of melioidosis cases in Darwin, Australia, just one patient from 707 survivors has developed persistent asymptomatic B. pseudomallei carriage. To better understand the mechanisms behind this unique scenario, we performed whole-genome analysis of two strains isolated 139 months apart. During this period, B. pseudomallei underwent several adaptive changes. Of 23 point mutations, 78% were nonsynonymous and 43% were predicted to be deleterious to gene function, demonstrating a strong propensity for positive selection. Notably, a nonsense mutation inactivated the universal stress response sigma factor RpoS, with pleiotropic implications. The genome underwent substantial reduction, with four deletions in chromosome 2 resulting in the loss of 221 genes. The deleted loci included genes involved in secondary metabolism, environmental survival, and pathogenesis. Of 14 indels, 11 occurred in coding regions and 9 resulted in frameshift mutations that dramatically affected predicted gene products. Disproportionately, four indels affected lipopolysaccharide biosynthesis and modification. Finally, we identified a frameshift mutation in both P314 isolates within wcbR, an important component of the capsular polysaccharide I locus, suggesting virulence attenuation early in infection. Our study illustrates a unique clinical case that contrasts a high-consequence infectious agent with a long-term commensal infection and provides further insights into bacterial evolution within the human host.
IMPORTANCE
Some bacterial pathogens establish long-term infections that are difficult or impossible to eradicate with current treatments. Rapid advances in genome sequencing technologies provide a powerful tool for understanding bacterial persistence within the human host. Burkholderia pseudomallei is considered a highly pathogenic bacterium because infection is commonly fatal. Here, we document within-host evolution of B. pseudomallei in a unique case of human infection with ongoing chronic carriage. Genomic comparison of isolates obtained 139 months (11.5 years) apart showed a strong signal of adaptation within the human host, including inactivation of virulence and immunogenic factors, and deletion of pathways involved in environmental survival. Two global regulatory genes were mutated in the 139-month isolate, indicating extensive regulatory changes favoring bacterial persistence. Our study provides insights into B. pseudomallei pathogenesis and, more broadly, identifies parallel evolutionary mechanisms that underlie chronic persistence of all bacterial pathogens.
Introduction
Burkholderia pseudomallei is a Gram-negative bacterium that causes melioidosis, a potentially fatal infectious disease that is frequently associated with at-risk individuals but can also catastrophically afflict healthy people. Melioidosis is contracted by percutaneous inoculation, inhalation, ingestion, or aspiration following environmental exposure to soil, water, or aerosols containing B. pseudomallei (1). Although Southeast Asia and northern Australia have historically been considered “hot spots” of environmental B. pseudomallei presence, this organism is becoming increasingly recognized as an important cause of morbidity and mortality in other regions (2). Infection with B. pseudomallei can be difficult to treat and requires specific and protracted antibiotic therapy (1).
The clinical picture of melioidosis is multifarious, with presentation and outcome determined by a combination of infecting dose, mode of infection, host risk factors, and yet-to-be-elucidated bacterial virulence determinants (3). Without a prompt diagnosis and access to appropriate antibiotics and state-of-the-art intensive care facilities, the overall mortality rate is ~40%, increasing to >90% in those presenting with septic shock (1). An ongoing prospective study of melioidosis cases in Darwin, Australia (3), has documented 815 culture-confirmed melioidosis cases since October 1989, of which 108 were fatal (13%). Approximately 85% of these patients presented with acute melioidosis following a presumed recent infection, 11% displayed chronic illness (defined as symptoms lasting >2 months), and the remaining 4% had delayed disease activation following latency (3). In all but one case, melioidosis survivors (n = 707) from the Darwin study eventually cleared their initial or relapsed B. pseudomallei infections. This single individual, patient 314 (P314), has remained chronically colonized with B. pseudomallei since being diagnosed with melioidosis in 2000.
Previous longitudinal studies of Burkholderia dolosa, Pseudomonas aeruginosa, and Staphylococcus aureus isolates obtained from chronic infections of patients with cystic fibrosis have identified convergent patterns of evolution that include decreased virulence, increased antimicrobial resistance, and altered metabolic fitness due to similar selective pressures (4–8). Additionally, reductive evolution has been observed in a chronic P. aeruginosa infection (5) and is a common trait among obligate pathogens, including Burkholderia mallei, Rickettsia spp., Chlamydia spp., and Mycobacterium spp. (9–11). These convergent adaptations suggest that some bacterial pathogens undergo key genetic changes in vivo during the transition to chronic disease, resulting in a substantial and sustained niche shift away from being an environmental organism or acute pathogen. In the present study, we have applied next-generation whole-genome sequencing (WGS) technologies to analyze and accurately catalogue genome-wide alterations between the initial isolate (MSHR1043), a 37-month isolate (MSHR1655) and a 139-month isolate (MSHR6686) from P314. Our study of adaptation during chronic persistence substantially adds to the current knowledge of bacterial evolution within the human host.
RESULTS AND DISCUSSION
Assembly and genomic features of the initial P314 isolate, MSHR1043.
The Illumina-454 hybrid assembly of B. pseudomallei MSHR1043 (GenBank accession no. AOGU00000000) is in 44 high-quality contigs totaling 7,221,181 bp and has a G+C content of 68.1%. Alignment of MSHR1043 Illumina reads with MSHR1043 resulted in >99.9% of the reads mapping to this assembly. Further contig joining was not possible with these data because of several large repetitive regions in B. pseudomallei, including 16S RNA, tRNA, Hep_Hag motifs, and variable-number tandem repeat loci. The average size and G+C content of the currently six closed B. pseudomallei genomes (1026b, K96243, MSHR668, 1710b, BPC006, and 1106a) are 7,178,622 bp and 68.2%, respectively. The estimated size and G+C content of MSHR1043 suggest that this strain has not undergone large genetic changes in vivo.
In contrast, 139-month isolate MSHR6686 exhibits four large deletions on chromosome 2, which reduce the size of the genome by 285 kb to ~6.93 Mbp. Thirty-seven-month isolate MSHR1655 is also missing three of these four deleted regions, totaling ~245 kb (see Table S1 in the supplemental material). This observation is consistent with the reductive evolution of bacterial genomes in response to a niche shift toward a more restricted and intimate long-term association with a eukaryotic host (9–11). This phenomenon is similar to that observed in B. mallei, a clonal species within the B. pseudomallei clade that contains a genome smaller than its B. pseudomallei ancestor because of the loss of several loci following its transition from a soil saprophyte to a restricted, primarily equine, niche (9). As expected, assembly of unmapped reads from MSHR6686 did not yield additional contigs, indicating that MSHR6686 has not acquired exogenous DNA. This observation is consistent with previous in vivo evolution studies using closed bacterial genomes that have identified large-scale deletions but not large insertions (5, 12). The lack of MSHR1043 closure prevented comprehensive characterization of structural variation; however, alignment of MSHR1043 contigs against closed B. pseudomallei genomes failed to identify major deletions or novel inversions, suggesting overall colinearity of the genomes. All other mutations (single-nucleotide polymorphisms [SNPs], small [≤15-bp] indels, and deletions) were readily identified.
Genome-wide point mutations between the initial and 139-month P314 isolates show a strong signal of positive selection.
Twenty-three SNPs between MSHR1043 and MSHR6686 were identified. Nineteen SNPs caused amino acid sequence alterations, one of which resulted in a nonsense mutation. Ten SNPs were predicted to have a disruptive effect on the protein product (Table 1). The strikingly high proportion of nonsynonymous (NS) mutations in MSHR6686 indicates that the in vivo B. pseudomallei population in P314 is under strong positive selection. This phenomenon has been observed in relapse melioidosis (12) and in chronic in vivo infections caused by other free-living bacteria such as P. aeruginosa (5) and B. dolosa (4).
TABLE 1 .
SNP differences between the initial (MSHR1043) and 139-month (MSHR6686) B. pseudomallei isolates from P314
| Location in MSHR1043 |
Nucleotide change |
Amino acid change |
Affected gene in MSHR1043 (K96243) |
SNP effecta | Mutated in MSHR1655? |
Affected protein(s) | Putative protein function(s) |
|---|---|---|---|---|---|---|---|
| seq0001 396179 |
C→T | T194M |
D512_01885 (BPSL0128) |
NS; deleterious | No | NtrX | DNA transcription regulator |
| seq0001 684105 |
C→T | A101A |
D512_03260 (BPSL0430) |
S; neutral | No | Thioesterase | Unknown |
| seq0001 798196 |
C→T | A8T |
D512_03780 (BPSL0530) |
NS; neutral | No | HPr kinase, phosphorylase | Cell adhesion, virulence |
| seq0003 154078 |
A→G | H115R |
D512_07998 (BPSL1362) |
NS; neutral | Yes | PstB | Phosphate uptake |
| seq0003 331699 |
T→C | I33V |
D512_08718 (BPSL1981) |
NS; neutral | Yes | GalU | Sugar, LPS metabolism |
| seq0003 397164 |
A→C | L47R |
D512_09003 (BPSL1924) |
NS; deleterious | No | Hypothetical protein | Unknown |
| seq0003 536977 |
G→A | NAe | NA | NA | No | 99 bp upstream of TetR | Unknown |
| seq0003 678249 |
G→T | R2242L |
D512_10148 (BPSL1712) |
NS; neutral | No | Syringomycin synthetase | Phytopathogenesis |
| seq0003 719972 |
T→C | F84L |
D512_10298 (BPSL1689) |
NS; deleterious | No | Hypothetical protein | Unknown |
| seq0003 720544 |
G→A | G85S |
D512_10303 (BPSL1688) |
NS; deleterious | Yes | Hypothetical protein | Unknown |
| seq0003 943033 |
G→A | Q82stop |
D512_11298 (BPSL1505) |
Nonsense; deleterious |
No | RpoS | Universal stress response |
| seq0003 1040609 |
G→A | G263D |
D512_11778 (BPSL1413) |
NS; deleterious | Yes | Glucose-6-phosphate isomerase |
Glycolysis, gluconeogenesis |
| seq0006 91557 |
T→C | L279P |
D512_14441 (BPSL2409) |
NS; deleterious | Yes | ATP binding protein | Molecular transport |
| seq0007 422966 |
G→A | NA | NA | NA | No | NA | Unknown |
| seq0008 15374 |
G→C | I637M |
D512_17625 (BPSL3016) |
NS; deleterious | Yes | Seca | Membrane transport; secretion |
| seq0017 181448 |
C→T | S97F (S72F)b |
D512_21859 (BPSS0946) |
NS; neutral | Yes | Class A β-lactamase PenA |
β-Lactam resistance |
| seq0020 32593 |
C→A | F99L |
D512_23016 (BPSS1260) |
NS; neutral | No | Hypothetical protein | Unknown |
| seq0028 205054 |
G→T | R458L |
D512_25718 (BPSS1501) |
NS; neutral | Noc | ClpB | Heat shock |
| seq0028 229189 |
T→C | H322R |
D512_25813 (BPSS1520) |
NS; neutral | Yesd | AraC regulator | DNA regulation, virulence |
| seq0028 480279 |
T→C | D54G |
D512_26783 (BPSS1698) |
NS; deleterious | Yes | RsrI | DNA methylation |
| seq0028 553993 |
C→T | A660T |
D512_27123 (BPSS1755) |
NS; deleterious | Yes | RpoD | Primary sigma factor |
| seq0028 1047356 |
G→A | NA | NA | NA | No | NA | Unknown |
| seq0028 1280562 |
G→A | S2S |
D512_30428 (BPSS2321) |
S; neutral | Yes | Hypothetical protein | Unknown |
Resides within the previously described, conserved PenA 70SXXK73 motif (68). S72F causes ~4-fold increased resistance to amoxicillin-clavulanic acid but not to other β-lactams (20, 69).
MSHR1655 possesses a different mutation in this gene (insertion of a repetitive NAP motif at the 3′ end).
MSHR1043 possesses the mutant allele; MSHR1655 and MSHR6686 possess the wild-type allele.
NA, not applicable.
The single nonsense mutation in MSHR6686 occurred in D512_11298, resulting in truncation of the sigma factor protein RpoS (Q82stop). Sigma factors actively recruit RNA polymerase for upregulated transcription in response to certain environmental cues. RpoS, the universal stress response sigma factor, upregulates a large array of genes in response to environmental stimuli, including low pH, oxidative stress, extreme temperatures, and carbon starvation (13, 14). This single mutation therefore has the pleiotropic potential to affect a large portion of the organism’s metabolism. Although the loss of rpoS would be highly disadvantageous for an organism facing a complex natural environment, it potentially removes a metabolic cost and thus can confer selective advantages in the host environment. Identified rpoS mutants in other bacterial species have several selective advantages, including improved nutrient assimilation under nutrient-limiting conditions (15), abolished transcription of certain genes in stationary phase and superinduction of other stationary-phase-induced genes required for host survival (16), increased antibiotic resistance and persistence (17), and biofilm formation (18). Therefore, rpoS abolition in MSHR6686 probably has relatively minor negative consequences in vivo.
MSHR6686 also has a predicted deleterious NS mutation (A660T) in an RpoD protein (encoded by D512_27123). Although RpoD is essential for cellular function, MSHR1043 has genes that encode seven distinct RpoD proteins (D512_07071, D512_11543, D512_10313, D512_09748, D512_30918, D512_27123, and D512_30563). This wide array of RpoD-encoding loci in B. pseudomallei may enable the bacterium to respond and adapt to changing ecological niches, ranging from environmental survival to eukaryotic (e.g., nematode, plant, mammalian) infection. A null mutation in one RpoD locus is unlikely to be lethal, although it is expected to adversely affect certain regulatory pathways. The rpoD mutation, but not the rpoS mutation, is also present in B. pseudomallei MSHR1655 (a previously sequenced strain from P314 isolated 37 months after MSHR1043; GenBank accession no. AAHR00000000), indicating that the rpoD mutation is stable in vivo. It remains unknown how these mutations affect global transcription profiles in P314, and is worthy of further study.
The molecular mechanisms underpinning B. pseudomallei resistance to clinically administered β-lactam antibiotics are well documented (19–23). An NS SNP (S72F) in the class A β-lactamase gene penA (Table 1) leads to 8-fold increased resistance to amoxicillin-clavulanate in MSHR6686 (24 µg/ml) but no increased resistance to other β-lactams, including ceftazidime (20, 23). This NS SNP is also present in MSHR1655. Both amoxicillin-clavulanate and ceftazidime, which are commonly used to treat B. pseudomallei infections, have been administered on multiple occasions over the period of P314’s infection. However, unlike ceftazidime, amoxicillin-clavulanate has only been administered to treat other organisms complicating P314’s bronchiectasis. Therefore, the PenA S72F mutant has arisen and persisted in the B. pseudomallei population as a consequence of ongoing selective pressures incurred by semiregular amoxicillin-clavulanate treatment for other pathogens. Amoxicillin-clavulanate-resistant B. pseudomallei may potentiate cross-resistance in other pathogens via either recombination of mutant penA into other species or as a side effect of localized enzymatic degradation. The reduced effectiveness of amoxicillin-clavulanate against other pathogens in the airways of P314 remains to be explored.
Many of the remaining NS mutations in MSHR6686 occurred in genes whose functions are not well characterized in B. pseudomallei. Four (21%) NS point mutations were identified in hypothetical proteins (encoded by D512_09003, D512_10298, D512_10303, and D512_23016), three of which were predicted to be detrimental to protein function (Table 1). Other loss-of-function NS SNPs were identified in poorly characterized genes (D512_01885, D512_11778, D512_14441, D512_17625, and D512_26783). Although the functional role of these genes remains enigmatic, our study demonstrates that they are all dispensable in chronic infection and their loss may even be beneficial, either because these loci elicit a strong immunological response or otherwise have evolutionary costs that select for their inactivation.
Genome-wide indels between P314 isolates demonstrate strong selective pressure toward loss-of-function mutations.
A previous study of relapse B. pseudomallei isolate pairs found that only 40% of the indels found occurred in putative coding sequences (12). In contrast, 11/14 (79%) small (≤15-bp) indels (Table 2) identified between MSHR1043 and MSHR6686 occurred in putative coding regions. Seven of these coding sequence indels are also present in MSHR1655. Interestingly, the three intergenic indels occurred exclusively in MSHR1043 and were not observed in MSHR6686, MSHR1655, or closed B. pseudomallei genomes. Nine indels in MSHR6686 caused frameshift mutations that altered peptide length, whereas two were in-frame indels (Table 2). The in-frame indels were predicted to have a neutral impact on protein function, in contrast to the frameshift mutations, which were all detrimental. The overrepresentation of indels in coding regions provides further evidence for positive selection in P314.
TABLE 2 .
Indel differences between the initial (MSHR1043) and 139-month (MSHR6686) B. pseudomallei isolates from P314
| Location in MSHR1043 |
Nucleotide change | Affected gene in MSHR1043 (K96243)e |
Indel effect(s)a | Mutated in MSHR1655? |
Affected protein | Putative function(s) of protein |
|---|---|---|---|---|---|---|
| seq0001 105149 |
CT→C | NAd | NA | Nob | NA | Unknown |
| seq0001 416897 |
CCGCGACGCCGAG→C |
D512_01975 (BPSL0194) |
Deletion (LGVA), codon 20; neutral |
No | Hypothetical protein | Unknown |
| seq0001 1496938 |
GGC→G |
D512_06755 (BPSL1120) |
Frameshift, codon 64; premature stop (381-103 aa); deleterious |
Noc | LPS heptosyltransferase I | Biosynthesis, modification of LPS core |
| seq0003 131174 |
CT→C |
D512_07898 (BPSL1343) |
Frameshift, codon 25; premature stop (270-110 aa); deleterious |
Yes | Hypothetical protein | Unknown |
| seq0003 388954 |
G→GGA |
D512_08953 (BPSL1936) |
Frameshift, codon 100; premature stop (394-110 aa); deleterious |
Yes | OacA | Biosynthesis, modification of LPS O antigen |
| seq0003 537757 |
AC→A | Not assigned (BPSL1804) |
Frameshift, codon 247 in MSHR1043; increased length (399–419 aa); deleterious |
Nob | AmrA | Antibiotic resistance |
| seq0003 721338 |
CGCCGGGGCGG→C |
D512_10308 (BPSL1687) |
Frameshift, codon 85; premature stop (213-179 aa); deleterious |
Yes | Hypothetical protein | Unknown |
| seq0003 1076302 |
GC→G |
D512_11953 (BPSL2093) |
Frameshift, codon 43; premature stop (96-73 aa); deleterious |
Yes | Hypothetical protein | Unknown |
| seq0007 181244 |
C→CCACTCG |
D512_15706 (BPSL2660) |
Insertion (HS), codon 174 (205–207 aa); neutral |
Yes | UreE | Nitrogen fixation |
| seq0007 194989 |
T→TC |
D512_15766 (BPSL2672) |
Frameshift, codon 272; premature stop (637-550 aa); deleterious |
Yes | WbiI | Biosynthesis, modification of LPS O antigen |
| seq0007 196460 |
G→GC |
D512_15771 (BPSL2673) |
Frameshift, codon 121; premature stop (336-141 aa); deleterious |
No | WbiH | Biosynthesis, modification of LPS O antigen |
| seq0020 30723 |
ACGCAT→A | NA | NA | Nob | NA | Unknown |
| seq0024 104413 |
GCATCGA→G | NA | NA | Nob | NA | Unknown |
| seq0028 195058 |
A→AC |
D512_25678 (BPSS1494) |
Frameshift, codon 183; increased length (245-854 aa); deleterious |
Yes | DNA-binding response regulator |
DNA transcription regulation, virulence |
Indel identified in MSHR1043; neither any other previously sequenced B. pseudomallei strains nor the two latter P314 isolates, MSHR1655 and MSHR6686, share these indels.
MSHR1655 has an insertion of a repetitive AHL amino acid motif at codon 278 (total length, 384 amino acids).
NA, not applicable.
MSHR6686 has accumulated four indels affecting lipopolysaccharide (LPS) biosynthesis and modification loci. LPS is an integral component of the outer membrane of Gram-negative bacteria and is a known virulence factor in B. pseudomallei that confers resistance to bactericidal compounds in human serum (24). LPS is also critical for the successful establishment of acute B. pseudomallei infection, enabling the bacterium to escape macrophage killing in vitro (25). LPS is composed of three components: the membrane-associated lipid A, the oligosaccharide-rich core, and the immunogenic outer membrane O antigen. Although LPS is an important virulence determinant, the potent immunogenicity of LPS, particularly its O antigens, appears disadvantageous for long-term persistence. Loss of function of O-antigen biosynthesis and modification is a hallmark of chronic P. aeruginosa infection (5) and may indicate an important strategy employed by B. pseudomallei to evade the immune response.
Indel inactivation of the LPS-associated genes wbiI (D512_15766) and oacA (D512_08953) has been reported in P314 isolates (26, 27), and these indels are maintained in MSHR6686. A 1-bp insertion in the highly conserved wbiI gene causes loss of function of an O-antigen biosynthesis pathway, leading to the characteristic “rough” LPS phenotype (caused by modification, reduction, or absence of O-antigen chains) and susceptibility to human serum (26). A coexisting 2-bp insertion in oacA results in an LPS that putatively reacts to B. mallei LPS-specific monoclonal antibody (26). B. mallei lacks OacA (26), which is involved in the modification of O-antigen L-6dTalp residues by acetylation at their O-4 position (26, 27) and may also methylate the O-2 position (26). Inactivation of OacA in MSHR6686 demonstrates a convergent evolutionary strategy with the host-adapted pathogen B. mallei.
We identified two additional genes involved in LPS biosynthesis and modification that appear to be affected by indels in MSHR6686. A 1-bp insertion in the highly conserved wbiH (D512_15771) gene, situated immediately downstream of wbiI, was identified in MSHR6686 but not in MSHR1655 (Table 2). The wbiH mutation potentially results in defunct initiation of O-antigen subunit assembly (28). A 2-bp deletion in D512_06755 (encodes LPS heptosyltransferase I, involved in LPS inner core biosynthesis [29]) caused a frameshift in this protein, resulting in loss of function. Interestingly, the wbiH and D512_06755 indels are not present in MSHR1655, although MSHR1655 possesses a novel indel in D512_06755 that results in the insertion of a repetitive motif (AHL) at codon 278. This indel is also predicted to abolish the function of LPS heptosyltransferase I (Table 2), demonstrating that this locus is under heavy selective pressure to acquire loss-of-function mutations. The continued degradation of LPS biosynthesis and modification pathways indicates that the wbiI and oacA mutants are insufficient for complete abolition of O-antigen immunogenicity, leading to continued downregulation by the bacterium for immune evasion. Alternatively, LPS-associated loci may no longer be under selection in the host environment and are therefore subject to genetic drift. Partial deletion of an O-antigen acetylase wbiA homologue, D512_20407, was also observed in MSHR6686. Although WbiA is required for 2-O-acetylation in B. pseudomallei (27), it is unclear what functional effect the partial deletion of the ostensibly redundant D512_20407 wbiA locus has, if any, on LPS function. Collectively, our study consolidates previous findings that LPS is dispensable in chronic bacterial infections (4, 5).
A 1-bp indel in virG (D512_25678 [or BPSS1494 in K96243]), observed in both MSHR1655 and MSHR6686, resulted in a frameshift that increased protein length from 245 to 854 amino acids (Table 2). VirG is part of a two-component sensor-regulator system for type 6 secretion system cluster 1 (T6SS1) and is essential for the expression of this cluster (30). B. pseudomallei produces six T6SS clusters, although only T6SS1 is necessary for virulence in the hamster and murine models (30, 31). The T6SS1 apparatus is structurally similar to bacteriophage, injecting bacterial effector molecules into the host cell cytosol in a contact-dependent manner (32). On the basis of these prior studies, it is likely that the virG mutation has resulted in the loss of expression of T6SS1, an important virulence factor in B. pseudomallei.
A loss-of-function indel in the periplasmic multidrug efflux lipoprotein AmrA (encoded by BPSL1804 in K96243) was seen in MSHR1043 but not MSHR6686 or MSHR1655. AmrA is part of the aminoglycoside and macrolide resistance operon AmrAB-OprA, which causes the efflux of antibiotics such as gentamicin and erythromycin from the B. pseudomallei cytoplasm (33). Concomitant with defective AmrAB-OprA in MSHR1043, the gentamicin MIC is just 1 µg/ml, whereas the MIC for MSHR6686 is 12 µg/ml. In addition, MSHR1043 fails to grow on Ashdown’s agar, a selective medium for B. pseudomallei that contains 4 µg/ml gentamicin (34). A neutral NS SNP at D512_25813 was also identified in MSHR1043 that was not found in MSHR1655, MSHR6686 (Table 1), or any other publicly available B. pseudomallei genome. The exclusivity of these mutations in MSHR1043 suggests that this strain resides within a lineage distinct from that of MSHR1655 or MSHR6686 or, alternatively, that the MSHR1043 lineage has since become extinct. The coexistence of multiple lineages in vivo following a clonal inoculation event has been documented in isolates from P314 (35), other patients infected with B. pseudomallei (12, 36), and other species (5). WGS of additional midpoint isolates is needed to confirm the evolutionary scenario.
Large deletions have occurred in the 139-month isolate, MSHR6686.
Large genomic deletions are the most visible result of the ongoing process of genome decay that occurs as bacteria adapt to a more restricted niche (5, 9, 12). Reductive evolution involves the loss of nonessential bacterial genes in the host environment, including analogous gene products manufactured by the host that can be exploited by the bacterium (37). Consistent with irreversible host adaption in P314, MSHR6686 has cumulatively lost 285 kb at four distinct loci. The deletions in MSHR6686 have reduced its genome size by 4%, to 6.93 Mbp, resulting in the loss of 221 and 195 genes relative to MSHR1043 and K96243, respectively (see Table S1 in the supplemental material). All of the deletions occur exclusively on chromosome 2, which contains a greater proportion of gene clusters involved in nonessential functions such as secondary metabolism, environmental survival, and pathogenesis than does chromosome 1 (38). Several amino acid metabolism pathways have been deleted from MSHR6686 (see Table S1), indicating that analogous products exist in the host and are being utilized by the bacterium in vivo or that these biosynthesis pathways are redundant in B. pseudomallei. For example, amino acid biosynthesis genes are frequently decayed or lost in obligate intracellular human pathogens such as the Rickettsia spp., Chlamydia spp., and Mycobacterium spp. The loss of these genes therefore probably provides a metabolic advantage in the P314 B. pseudomallei population, enabling a larger proportion of its genome to be committed to central functions such as replication, transcription, and translation (10).
Reductive evolution is also starkly evident in B. mallei, which has a genome that is ~1.5 Mbp smaller than that of its free-living ancestor B. pseudomallei. Interestingly, there is substantial parallel gene loss between MSHR6686 and B. mallei; ~141 kb (49.5%) of the loci absent from MSHR6686 are also absent from B. mallei, spanning (in K96243) BPSS1096 to BPSS1112 and BPSS1123 to BPSS1203 (see Table S1). These deleted loci encode mainly putative secondary metabolic pathways that facilitate bacterial responses to different environmental conditions but not those encountered in the mammalian host. The absence of these loci in both B. mallei and the P314-adapted B. pseudomallei strain provides further insights into the divergence of B. mallei from its B. pseudomallei ancestor (9).
MSHR6686 has also shed genes encoding putative lipoprotein capsule biosynthesis and modification, chemotaxis, motility and quorum sensing, fatty acid biosynthesis, DNA methylation, thiamine biosynthesis, antibiotic resistance, and virulence factor biosynthesis (see Table S1 in the supplemental material). Many of these loci are not well characterized in B. pseudomallei, and several do not yet have known functions. However, two key loci deleted from MSHR6686 have previously been functionally characterized. The first is an efflux pump encoded by the BpeEF-OprC operon (D512_20002 to D512_20022) that causes the efflux of chloramphenicol and trimethoprim (39); the MSHR6686 mutation has likely rendered this isolate susceptible to these compounds. Trimethoprim-sulfamethoxazole is an antibiotic frequently used in melioidosis treatment, although it has not been routinely administered to P314 because of the patient’s intolerance to this drug. Other putative antibiotic resistance genes, including the BpeGH-OprD operon (D512_22681 to D512_22696) (40) and a metallo-β-lactamase gene, D512_31289, were also deleted, although their substrates and functions remain elusive. Efflux pump mutations have also been observed in a long-term infection with P. aeruginosa, which, like B. pseudomallei, is naturally resistant to many antibiotics (5). Characterization of these antibiotic-resistant loci may provide novel therapeutic options for P314.
The second previously characterized locus abolished in MSHR6686 is the type III secretion 3 (Bsa TTSS3) virulence cluster. TTSS apparatus are common in Gram-negative pathogens and have phenotypic features in common with bacterial flagella. TTSSs allow pathogens to directly inject eukaryotic host cells with virulence proteins that are functionally similar to host proteins yet subvert the host cell solely for the benefit of the bacterium (41). B. pseudomallei produces three TTSS apparatus. Unlike TTSS1 and TTSS2, Bsa TTSS3 is essential for virulence in the hamster model (42), facilitating invasion of nonphagocytic cells and subsequent vacuolar escape (43). Although many of the genes encoding TTSS3 effector molecules (e.g., bopA, bopE, bapA, and bapC) are not mutated in MSHR6686, structural components that form the critical “injectisome” of Bsa TTSS3 are deleted (see Table S1 in the supplemental material). Previous work has shown that the deletion of individual TTSS3 effector molecules may not reduce virulence; however, deletion of the highly conserved gene sctU3 (spaS; D512_25888), which encodes a protein essential to the TTSS3 secretory pathway and which is also deleted from MSHR6686, significantly attenuates virulence (42). TTSS apparatus show signs of convergent evolution, being downregulated in vivo in other pathogenic species (5, 44); a notable exception is B. mallei, which has a fully functional Bsa TTSS3 locus that contributes to its virulence (45). Collectively, our results suggest that TTSS3 is important during initial infection and maintenance of virulence capacity but is disadvantageous for long-term commensal B. pseudomallei infection.
Large deletions have been reported in free-living bacterial species isolated from chronic infections. Comparison of P. aeruginosa isolates obtained 96 months apart from a cystic fibrosis patient revealed a large 188-kb deletion in the latter isolate, with a concomitant loss of ~139 genes (5). Similarly, a study of initial and relapse melioidosis isolate pairs revealed a 330-kb deletion between two B. pseudomallei isolates (1258a and 1258b) obtained 6 months apart (12). Surprisingly, this deletion affected genes BPSS1249 to BPSS1483 (232 genes, 10% of the replicon) located on chromosome 2 yet did not overlap any deleted loci in MSHR6686 (see Table S1 in the supplemental material). There was also no overlap in SNPs or indels, with the exception of BPSS0291 (D512_20007 in MSHR1043), which encodes a lipase-like protein that was deleted from MSHR6686 and contained a frameshift in 354e, a lung isolate obtained 75 months after the original isolate (354a) (12). In general, there was a better correlation between mutations observed in MSHR6686 and those from chronic infections with other pathogenic species (e.g., P. aeruginosa [5] and Burkholderia cenocepacia [44]) than from relapsed B. pseudomallei infections (12). Relapse melioidosis usually results from inadequate duration of or poor patient compliance with the prescribed therapy, and is defined as a recurrent clinical illness after the date for completion of the prescribed treatment for the initial infection (3, 46, 47). Thus, intact virulence capabilities are a necessity. The basis for these two different disease pathologies suggests fundamental selection differences and evolutionary trajectories.
Altered growth rate and morphological differences between MSHR1043 and MSHR6686.
We observed that MSHR6686 takes approximately 1 to 2 days longer to grow than MSHR1043 when plated on both rich and selective media, including chocolate agar, Luria-Bertani agar, and Ashdown’s agar (without gentamicin) (Fig. 1). Growth rate and morphology differences have also been observed in other chronically infecting pathogens, including S. aureus and P. aeruginosa (7, 48). Intriguingly, B. mallei can be differentiated from wild-type B. pseudomallei on the basis of a slower growth rate on both minimal and rich media (49). These parallels suggest that the large deletions observed in MSHR6686, rather than SNPs or indels accumulated in this strain, are responsible for slower growth.
FIG 1 .
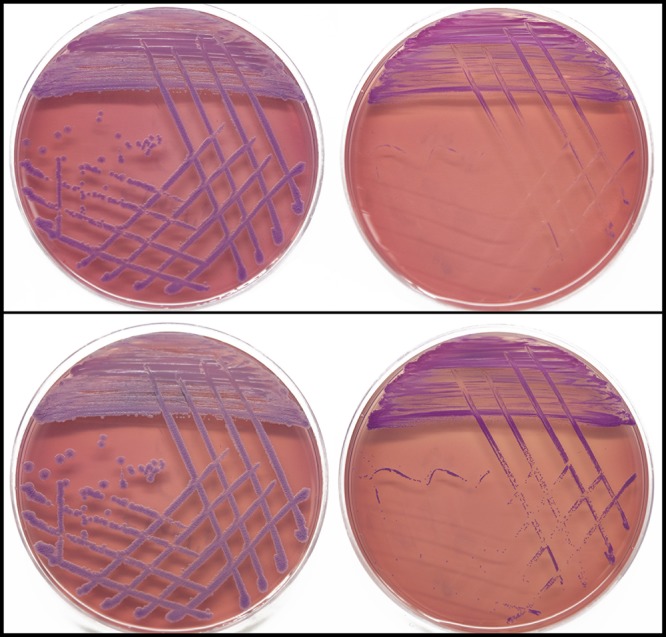
Growth rate and morphology differences between P314 B. pseudomallei isolates. The initial isolate, MSHR1043, grows well after 48 and 72 h of incubation on selective Ashdown’s agar (without gentamicin) (top and bottom left, respectively). In contrast, the 139-month isolate, MSHR6686, exhibits a substantially slower growth rate and morphological differences that make it difficult to recognize as B. pseudomallei (48 and 72 h of growth, top and bottom right, respectively). The growth rate difference is likely due to considerable genetic loss affecting 221 genes on chromosome 2.
Cause of avirulence in the initial P314 strain, MSHR1043.
MSHR1043 is avirulent in the intraperitoneal murine model, even at a high infecting dose (A. Tuanyok, unpublished data). We were therefore interested in identifying the potential molecular basis of virulence attenuation in MSHR1043. The Prokaryotic Genome Automatic Annotation Pipeline used for MSHR1043 annotation identified 13 frameshift indels in this strain, of which 4 also occurred in MSHR6686 and MSHR1655 (Table 3). Only one of these indels causes loss-of-function of an essential virulence factor, WcbR, a fatty acid synthase within the 35-kb capsular polysaccharide I (CPS-I) cluster (50). Intact CPS-I prevents phagocytosis by host immune cells (51) and is functional in B. mallei (52). In vitro knockouts of wcbR have a demonstrated reduction in CPS-I production and expression (50). The 1-bp indel in wcbR has persisted throughout P314’s infection (see Figure S1 in the supplemental material), indicating that it occurred prior to MSHR1043 isolation and has favorable consequences in vivo. We speculate that this CPS-I mutation was responsible for virulence attenuation early in P314’s infection and was a critical step in the progression to chronic-carriage disease.
TABLE 3 .
Frameshift indels in P314 isolates MSHR1043, MSHR1655, and MSHR6686 not found in closed or draft genome-sequenced B. pseudomallei genomes
| Protein | Gene (K96243) | Contig | Start | End |
|---|---|---|---|---|
| Type I polyketide synthase WcbR | BPSL2789 | seq0007 | 325993 | 333551 |
| Bbp50 integrase | NAa | seq0028 | 553262 | 553702 |
| Integrase | NA | seq0028 | 987262 | 988081 |
| Aromatic amino acid aminotransferase | BPSS2200 | seq0028 | 1122114 | 1123340 |
NA, not applicable.
In conclusion, we have characterized genome-wide changes occurring in B. pseudomallei during an ongoing chronic-carriage infection of the human respiratory tract. The mutational pattern demonstrates a predominantly positive selection signal that conveys benefits for host adaptation and commensalism. The slow growth rate of chronically infecting bacteria, their exquisite ability to evade killing by antibiotics and the immune system, and the inherent heterogeneity of chronic in vivo populations make eradication on the basis of current treatments difficult, if not impossible. P314’s infection represents a hitherto undescribed B. pseudomallei scenario in the global melioidosis literature that we have termed “chronic carriage” to denote a long-term, persistent human infection that has become, paradoxically, asymptomatic. Although asymptomatic carriage of B. pseudomallei has been suspected in goats (53), it has been conventionally believed that the organism is not carried asymptomatically by humans and that any culture-positive clinical specimen necessarily represents disease requiring therapy (54). Protracted carriage of B. pseudomallei has also been observed in some cystic fibrosis patients (55, 56), suggesting that B. pseudomallei commensalism may be more common than previously thought.
MATERIALS AND METHODS
Ethics statement.
This study has been approved by the Human Research Ethics Committee of the Northern Territory Department of Health and Menzies School of Health Research, approval number HREC 02/38 (Clinical and Epidemiological Features of Melioidosis). Written informed consent was provided by the study participant.
Clinical history of P314 and bacterial isolates used in this study.
P314 is a white female with bilateral non-cystic fibrosis bronchiectasis but no other melioidosis risk factors who originally presented with a chronic cough productive of green sputum in July 2000 when 61 years old. P314 had previously been treated for Mycobacterium avium complex and P. aeruginosa pulmonary infections and had prior surgery for chronic sinusitis. A chest X-ray showed patchy pulmonary infiltrates associated with bilateral basal bronchiectasis, and a computed tomography scan showed extensive cystic bronchiectasis of the left lower lobe. B. pseudomallei was cultured from sputum (MSHR1043) and nose and throat swabs at that time (57). P314’s symptoms initially improved after 14 days of intravenous ceftazidime, but her sputum remained B. pseudomallei positive. P314 was subsequently given intravenous meropenem for a week, followed by 3 weeks of intravenous ceftazidime. Because of a severe trimethoprim-sulfamethoxazole allergy, P314 was administered doxycycline for the oral “eradication phase” of therapy, which is standard for melioidosis treatment completion (1).
P314 has subsequently had numerous courses of intravenous ceftazidime and meropenem, trials of nebulized ceftazidime, and prolonged courses of oral doxycycline and amoxicillin-clavulanate to treat periods of increased cough and sputum production. In hindsight, these exacerbations and even the initial melioidosis presentation may not have reflected disease due to B. pseudomallei infection but rather standard bacterial infective exacerbations of bronchiectasis. In August 2003, 37 months after MSHR1043 was isolated, P314 underwent surgical lobectomy of the lower left lung lobe. B. pseudomallei was cultured from this lung tissue (MSHR1655). Despite excellent recovery, P314 continued to have respiratory exacerbations that were persistently culture positive for B. pseudomallei. In recent years, P314’s respiratory symptoms have improved, although a productive cough with yellow or green sputum persists. MSHR6686 was isolated in January 2012 (139 months [11.5 years] after MSHR1043) during a routine visit for bronchiectasis management. P314 has since not been administered antibiotics with activity against B. pseudomallei and has reported being the least symptomatic since her initial melioidosis diagnosis. P314’s sputum remained B. pseudomallei positive as of January 2013.
MSHR1043 and MSHR6686 were isolated from P314’s sputum samples by direct plating on chocolate agar. Cultures were screened against the TTS1 assay (58) to confirm their identity as B. pseudomallei prior to WGS.
Antimicrobial testing.
Etests (bioMérieux, Baulkham Hills, NSW, Australia) were used to determine the MICs of ceftazidime, gentamicin, and amoxicillin-clavulanate for P314’s isolates. Etesting was carried out according to the manufacturer’s instructions.
WGS and hybrid assembly.
High-quality and high-yield genomic DNA was obtained from B. pseudomallei cultures of MSHR1043 and MSHR6686 as previously described (59). The strains underwent paired-end WGS (500-bp fragments with ~800× and ~100× coverage, respectively) using the Illumina Genome Analyzer IIx platform (Illumina Inc., San Diego, CA). MSHR1043 was also sequenced using the 454 Genome Sequencer FLX instrument (454 Life Sciences, Branford, CT). Mira 3.4.0 (60) was used to perform de novo hybrid 454-Illumina assembly of MSHR1043. MSHR1043 Illumina reads were reduced to ~80× coverage by quality filtering with FASTX-Toolkit v0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/) prior to MIRA assembly. 454 and Illumina reads were quality filtered by MIRA as part of the hybrid assembly process.
The raw hybrid MSHR1043 assembly was subjected to manual contig assembly to improve draft quality. Gap5 v1.2.11 (61) was used to check and, where possible, stitch MSHR1043 MIRA-generated contigs. A second hybrid assembly was performed using the MIRA-assembled contigs as a scaffold for unfiltered Illumina reads in Velvet v1.1.04 (62). Regions of synteny between MSHR1043 and publicly available B. pseudomallei genomes were then identified using progressiveMAUVE (v2.3.1) alignment (63), BLAST (64), and the SynMap module of CoGe (http://genomevolution.org/CoGe/SynMap.pl). These tools enabled stitching of additional contigs in nonrepetitive, conserved loci. B. pseudomallei MSHR1655, which was isolated from P314 37 months after MSHR1043, was used as a reference. All contig stitches were rigorously validated by aligning unfiltered Illumina reads with the MSHR1043 reference using Burrows-Wheeler Aligner (BWA) v0.5.9 (65) with manual verification of correct read mapping and pairing over the putative contig joins. Variant calling was performed using the Genome Analysis Toolkit (GATK) v2.1-11 (66). All SNPs and indels were corrected, and incorrectly assembled contigs were manually separated. This process was iterated until no SNPs, indels, or misassemblies were identified in MSHR1043 with BWA and the GATK. To verify the length of MSHR1043, filtered Illumina reads were realigned with the reference with subsequent assembly of unaligned reads using MIRA. Lastly, contigs <800 bp in size were discarded.
SNP, indel, and deletion detection and characterization.
BWA was used to align Illumina reads for MSHR6686 against the MSHR1043 reference genome on the basis of default parameters for paired-end Illumina data. The GATK and SAMtools v0.1.18 (67) were subsequently used to call SNPs and small indels (≤15 bp) following the removal of duplicate reads with Picard MarkDuplicates v1.6 (http://picard.sourceforge.net) and Smith-Waterman realignment of the bam file around regions with a high mismatch rate according to the GATK. SNPs and indels were restricted to haploid calls (using the ploidy filter) and filtered for quality using the GATK with the following parameters for SNPs and indels: clusterSize, 3; clusterWindowSize, 10; MLEAF, <0.95; QD, <10.0; MQ, <30; FS, >20; QUAL, <30; DP, <(average genome coverage)/4 or (average genome coverage) × 3. Larger deletions were detected in MSHR6686 by using the coverageBed utility of BEDTools v2.15.0 (68) on the basis of a 1-kb window size. To detect the acquisition of exogenous DNA in MSHR6686, Illumina reads for MSHR6686 were aligned against MSHR1043 and assembly was performed with unaligned reads using MIRA. All mutations were visually verified on the basis of Illumina read mapping in Tablet v1.12.08.29 (69). Two indels in seq0003 388954 (affecting oacA) and seq0007 194989 (affecting wbiI) have been previously characterized by dideoxy chain termination sequencing (26) and were confidently identified by our pipeline. Annotation of variants was performed by using SnpEff v3.1 (70) with manual BLAST verification against the NCBI Microbes genome database. Functional characterization of SNPs was carried out using PROVEAN v1.1 (71).
Nucleotide sequence accession number.
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under accession no. AOGU00000000. The version described in this paper is the first version, AOGU01000000.
SUPPLEMENTAL MATERIAL
Genes deleted from the 139-month B. pseudomallei isolate, MSHR6686, relative to MSHR1043. Purple shading, loci also absent from B. mallei; yellow shading, Bsa TTSS loci absent from MSHR6686. The 37-month isolate, MSHR1655, retains genes in deleted region 4 but, like MSHR6686, is missing regions 1, 2, and 3
WcbR alignment comparing P314 Burkholderia pseudomallei isolates with wild-type B. pseudomallei strains.
ACKNOWLEDGMENTS
This work was supported by project grants from the Australian National Health and Medical Research Council, by the U.S. Department of Homeland Security Science and Technology Directorate (HSHQDC-10-C-00139 and HSHQDC-10-C-00135), and by NIH-NIAID (U01 AI-075568).
We thank Nicolette Janke, Meagan Seymour, Heidie O’Neill, Joshua Stone, Stephanie Warrington, and Jeffrey Foster of Northern Arizona University; Vanessa Theobald, Glenda Harrington, and Patiyan Andersson of the Menzies School of Health Research; and John Gillece, Remy Hilsabeck, and James Schupp of the Translational Genomics Research Institute for laboratory and technical assistance. We acknowledge the Edgewood Chemical Biological Center for 454 sequencing of MSHR1043.
Footnotes
Citation Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, Keim P, Pearson T, Currie BJ. 2013. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 4(4):e00388-13. doi:10.1128/mBio.00388-13.
REFERENCES
- 1. Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N. Engl. J. Med. 367:1035–1044 [DOI] [PubMed] [Google Scholar]
- 2. Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl. Trop. Dis. 4:e900. 10.1371/journal.pntd.0000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR, Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. 2011. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet. 43:1275–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 190:7910–7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. 2011. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One 6:e24301. 10.1371/journal.pone.0024301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goerke C, Wolz C. 2010. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int. J. Med. Microbiol. 300:520–525 [DOI] [PubMed] [Google Scholar]
- 9. Losada L, Ronning CM, DeShazer D, Woods D, Fedorova N, Kim HS, Shabalina SA, Pearson TR, Brinkac L, Tan P, Nandi T, Crabtree J, Badger J, Beckstrom-Sternberg S, Saqib M, Schutzer SE, Keim P, Nierman WC. 2010. Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements. Genome Biol. Evol. 2:102–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andersson SG, Kurland CG. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263–268 [DOI] [PubMed] [Google Scholar]
- 11. Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011 [DOI] [PubMed] [Google Scholar]
- 12. Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One 7:e36507. 10.1371/journal.pone.0036507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65:189–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subsin B, Thomas MS, Katzenmeier G, Shaw JG, Tungpradabkul S, Kunakorn M. 2003. Role of the stationary growth phase sigma factor RpoS of Burkholderia pseudomallei in response to physiological stress conditions. J. Bacteriol. 185:7008–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 16. Farewell A, Kvint K, Nyström T. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039–1051 [DOI] [PubMed] [Google Scholar]
- 17. Hong SH, Wang X, O’Connor HF, Benedik MJ, Wood TK. 2012. Bacterial persistence increases as environmental fitness decreases. J. Microb. Biotechnol. 5:509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heydorn A, Ersbøll B, Kato J, Hentzer M, Parsek MR, Tolker-Nielsen T, Givskov M, Molin S. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chantratita N, Rholl DA, Sim B, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Thanwisai A, Chua HH, Ooi WF, Holden MT, Day NP, Tan P, Schweizer HP, Peacock SJ. 2011. Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 108:17165–17170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tribuddharat C, Moore RA, Baker P, Woods DE. 2003. Burkholderia pseudomallei class A beta-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob. Agents Chemother. 47:2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sam IC, See KH, Puthucheary SD. 2009. Variations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomallei. J. Clin. Microbiol. 47:1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarovich DS, Price EP, Von Schulze AT, Cook JM, Mayo M, Watson LM, Richardson L, Seymour ML, Tuanyok A, Engelthaler DM, Pearson T, Peacock SJ, Currie BJ, Keim P, Wagner DM. 2012. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One 7:e30789. 10.1371/journal.pone.0030789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA, Schweizer HP. 2011. Molecular investigations of PenA-mediated β-lactam resistance in Burkholderia pseudomallei. Front. Microbiol. 2:139. 10.3389/fmicb.2011.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeShazer D, Brett PJ, Woods DE. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081–1100 [DOI] [PubMed] [Google Scholar]
- 25. Arjcharoen S, Wikraiphat C, Pudla M, Limposuwan K, Woods DE, Sirisinha S, Utaisincharoen P. 2007. Fate of a Burkholderia pseudomallei lipopolysaccharide mutant in the mouse macrophage cell line RAW 264.7: possible role for the O-antigenic polysaccharide moiety of lipopolysaccharide in internalization and intracellular survival. Infect. Immun. 75:4298–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tuanyok A, Stone JK, Mayo M, Kaestli M, Gruendike J, Georgia S, Warrington S, Mullins T, Allender CJ, Wagner DM, Chantratita N, Peacock SJ, Currie BJ, Keim P. 2012. The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PLoS Negl. Trop. Dis. 6:e1453. 10.1371/journal.pntd.0001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brett PJ, Burtnick MN, Heiss C, Azadi P, DeShazer D, Woods DE, Gherardini FC. 2011. Burkholderia thailandensis oacA mutants facilitate the expression of Burkholderia mallei-like O polysaccharides. Infect. Immun. 79:961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ortega X, Hunt TA, Loutet S, Vinion-Dubiel AD, Datta A, Choudhury B, Goldberg JB, Carlson R, Valvano MA. 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187:1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klena JD, Gray SA, Konkel ME. 1998. Cloning, sequencing, and characterization of the lipopolysaccharide biosynthetic enzyme heptosyltransferase I gene (waaC) from Campylobacter jejuni and Campylobacter coli. Gene 222:177–185 [DOI] [PubMed] [Google Scholar]
- 30. Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, Peacock SJ, Prior JL, Atkins TP, Deshazer D. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pilatz S, Breitbach K, Hein N, Fehlhaber B, Schulze J, Brenneke B, Eberl L, Steinmetz I. 2006. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74:3576–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ashdown LR. 1979. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology 11:293–297 [DOI] [PubMed] [Google Scholar]
- 35. Pearson T, U’Ren JM, Schupp JM, Allan GJ, Foster PG, Mayo MJ, Gal D, Choy JL, Daugherty RL, Kachur S, Friedman CL, Leadem B, Georgia S, Hornstra H, Vogler AJ, Wagner DM, Keim P, Currie BJ. 2007. VNTR analysis of selected outbreaks of Burkholderia pseudomallei in Australia. Infect. Genet. Evol. 7:416–423 [DOI] [PubMed] [Google Scholar]
- 36. Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, Dale JL, Ginther JL, Leadem B, Colman RE, Foster JT, Tuanyok A, Wagner DM, Peacock SJ, Pearson T, Keim P. 2010. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog. 6:e1000725. 10.1371/journal.ppat.1000725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wixon J. 2001. Featured organism: reductive evolution in bacteria: Buchnera sp., Rickettsia prowazekii and Mycobacterium leprae. Comp. Funct. Genomics 2:44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar A, Chua KL, Schweizer HP. 2006. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob. Agents Chemother. 50:3460–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar A, Mayo M, Trunck LA, Cheng AC, Currie BJ, Schweizer HP. 2008. Expression of resistance-nodulation-cell-division efflux pumps in commonly used Burkholderia pseudomallei strains and clinical isolates from northern Australia. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl 1):S145–S151 [DOI] [PubMed] [Google Scholar]
- 41. Mota LJ, Cornelis GR. 2005. The bacterial injection kit: type III secretion systems. Ann. Med. 37:234–249 [DOI] [PubMed] [Google Scholar]
- 42. Warawa J, Woods DE. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101–108 [DOI] [PubMed] [Google Scholar]
- 43. Dowling AJ, Wilkinson PA, Holden MT, Quail MA, Bentley SD, Reger J, Waterfield NR, Titball RW, Ffrench-Constant RH. 2010. Genome-wide analysis reveals loci encoding anti-macrophage factors in the human pathogen Burkholderia pseudomallei K96243. PLoS One 5:e15693. 10.1371/journal.pone.0015693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mira NP, Madeira A, Moreira AS, Coutinho CP, Sá-Correia I. 2011. Genomic expression analysis reveals strategies of Burkholderia cenocepacia to adapt to cystic fibrosis patients’ airways and antimicrobial therapy. PLoS One 6:e28831. 10.1371/journal.pone.0028831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ulrich RL, DeShazer D. 2004. Type III secretion: a virulence factor delivery system essential for the pathogenicity of Burkholderia mallei. Infect. Immun. 72:1150–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Limmathurotsakul D, Chaowagul W, Chantratita N, Wuthiekanun V, Biaklang M, Tumapa S, White NJ, Day NP, Peacock SJ. 2008. A simple scoring system to differentiate between relapse and re-infection in patients with recurrent melioidosis. PLoS Negl. Trop. Dis. 2:e327. 10.1371/journal.pntd.0000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Currie BJ, Fisher DA, Anstey NM, Jacups SP. 2000. Melioidosis: acute and chronic disease, relapse and re-activation. Trans. R. Soc. Trop. Med. Hyg. 94:301–304 [DOI] [PubMed] [Google Scholar]
- 48. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:557–562 [DOI] [PubMed] [Google Scholar]
- 49. Redfearn MS, Palleroni NJ, Stanier RY. 1966. A comparative study of Pseudomonas pseudomallei and Bacillus mallei. J. Gen. Microbiol. 43:293–313 [DOI] [PubMed] [Google Scholar]
- 50. Cuccui J, Milne TS, Harmer N, George AJ, Harding SV, Dean RE, Scott AE, Sarkar-Tyson M, Wren BW, Titball RW, Prior JL. 2012. Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect. Immun. 80:1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reckseidler-Zenteno SL, DeVinney R, Woods DE. 2005. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect. Immun. 73:1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. DeShazer D, Waag DM, Fritz DL, Woods DE. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253–269 [DOI] [PubMed] [Google Scholar]
- 53. Thomas AD. 1981. Prevalence of melioidosis in animals in northern Queensland. Aust. Vet. J. 57:146–148 [DOI] [PubMed] [Google Scholar]
- 54. White NJ. 2003. Melioidosis. Lancet 361:1715–1722 [DOI] [PubMed] [Google Scholar]
- 55. Schülin T, Steinmetz I. 2001. Chronic melioidosis in a patient with cystic fibrosis. J. Clin. Microbiol. 39:1676–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O’Carroll MR, Kidd TJ, Coulter C, Smith HV, Rose BR, Harbour C, Bell SC. 2003. Burkholderia pseudomallei: another emerging pathogen in cystic fibrosis. Thorax 58:1087–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mayo M, Kaesti M, Harrington G, Cheng AC, Ward L, Karp D, Jolly P, Godoy D, Spratt BG, Currie BJ. 2011. Burkholderia pseudomallei in unchlorinated domestic bore water, tropical Northern Australia. Emerg. Infect. Dis. 17:1283–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, Wilkins PP. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J. Clin. Microbiol. 44:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Currie BJ, Gal D, Mayo M, Ward L, Godoy D, Spratt BG, LiPuma JJ. 2007. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect. Dis. 7:68. 10.1186/1471-2334-7-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bonfield JK, Whitwham A. 2010. Gap5—editing the billion fragment sequence assembly. Bioinformatics 26:1699–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 65. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup; 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. 2010. Tablet—next generation sequence assembly visualization. Bioinformatics 26:401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes deleted from the 139-month B. pseudomallei isolate, MSHR6686, relative to MSHR1043. Purple shading, loci also absent from B. mallei; yellow shading, Bsa TTSS loci absent from MSHR6686. The 37-month isolate, MSHR1655, retains genes in deleted region 4 but, like MSHR6686, is missing regions 1, 2, and 3
WcbR alignment comparing P314 Burkholderia pseudomallei isolates with wild-type B. pseudomallei strains.


