Abstract
A novel influenza A (H1N1) virus has spread rapidly across the globe. Judging its pandemic potential is difficult with limited data, but nevertheless essential to inform appropriate health responses. By analyzing the outbreak in Mexico, early data on international spread, and viral genetic diversity, we make an early assessment of transmissibility and severity. Our estimates suggest that 23,000 (range 6000 to 32,000) individuals had been infected in Mexico by late April, giving an estimated case fatality ratio (CFR) of 0.4% (range: 0.3 to 1.8%) based on confirmed and suspected deaths reported to that time. In a community outbreak in the small community of La Gloria, Veracruz, no deaths were attributed to infection, giving an upper 95% bound on CFR of 0.6%. Thus, although substantial uncertainty remains, clinical severity appears less than that seen in the 1918 influenza pandemic but comparable with that seen in the 1957 pandemic. Clinical attack rates in children in La Gloria were twice that in adults (<15 years of age: 61%; ≥15 years: 29%). Three different epidemiological analyses gave basic reproduction number (R0) estimates in the range of 1.4 to 1.6, whereas a genetic analysis gave a central estimate of 1.2. This range of values is consistent with 14 to 73 generations of human-to-human transmission having occurred in Mexico to late April. Transmissibility is therefore substantially higher than that of seasonal flu, and comparable with lower estimates of R0 obtained from previous influenza pandemics.
On 29 April 2009, the World Health Organization (WHO) announced that the rapid global spread of a strain of influenza A (H1N1) virus detected in the previous week warranted moving the global pandemic alert level to phase 5 (www.who.int/csr/disease/swineflu/). Phase 5 indicates sustained human-to-human transmission of a novel influenza strain of animal origin in one WHO region of the world, and exported cases detected in other regions. In this outbreak, the earliest affected country may have been Mexico, with many cases in other nations associated with travels from that country. There are uncertainties about all aspects of this outbreak, including the virulence, transmissibility, and origin of the virus, and this in turn results in uncertainty in judging the pandemic potential of the virus and when reactive public health responses, such as recommendations to stay at home or to close schools, should be implemented in individual countries. Here we report findings of key early investigations into the outbreak that could aid such policy decisions.
The presence of fatalities [29 confirmed plus 88 suspected deaths in Mexico as of 4 May 2009 (1), 1 confirmed in the United States as of 5 May 2009 (2)] is not necessarily indicative of the virulence of the infection. The interpretation of these statistics depends on the total number of infections, including those with mild infection or who are asymptomatic, which is currently unknown, given the absence of a specific serological test for the new H1N1 influenza strain and associated population-level screening. As of 4 May 2009, 11,356 suspected and 822 laboratory-confirmed cases have been reported in Mexico (1), but these may represent an underestimate of true case numbers as surveillance has understandably focused on severe cases. Furthermore, severe cases in older individuals will be more difficult to identify because of the higher rate of respiratory illness in those over 60 years of age (3), and this could result in an underestimate of overall morbidity. Right censoring of mortality data, which occurs when additional deaths subsequently arise among cases already included in surveillance data, can also bias estimates of the true case fatality ratio (4). Finally, suspected deaths may not all have been caused by infection with the novel virus. These uncertainties necessarily affect any estimate of the case fatality ratio (CFR).
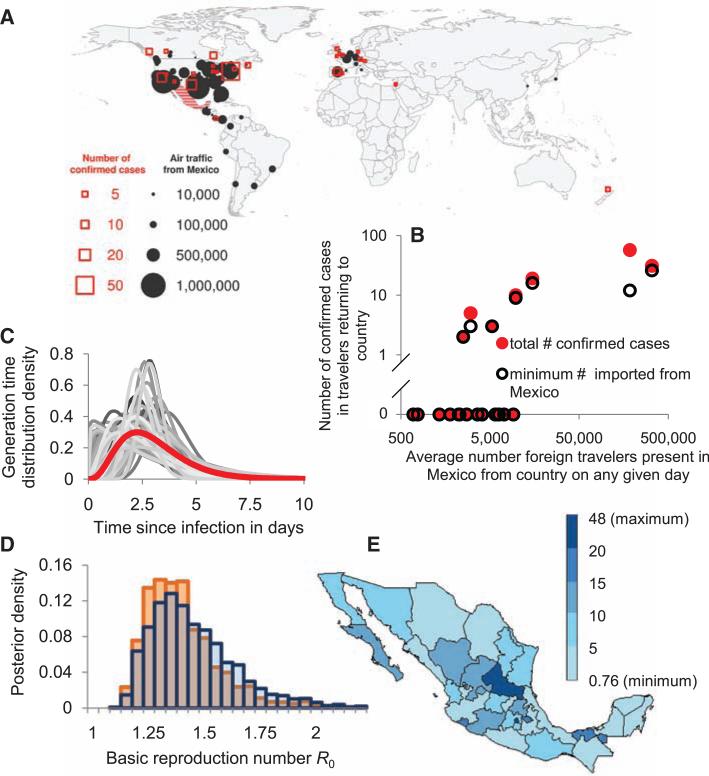
On the basis of international travel patterns, we would expect a proportion of cases of any infection spreading widely in Mexico to be exported by travelers (5). Owing to intense surveillance for influenza-like illness in those returning from Mexico, ascertainment of early cases in newly affected countries was almost certainly more complete and rapid than local surveillance of mild cases in Mexico. Airline passenger flow out of Mexico shows a significant correlation with the frequency of detected confirmed cases worldwide (Spearman correlation coefficient: 0.56, P = 0.004) (Fig. 1, A and B). We thus use data on cases among travelers and backcalculation methods to estimate the total number of people infected in Mexico. Key underlying assumptions in this analysis are that population mixing in Mexico is equally likely between Mexican residents and tourists, and tourists and Mexican residents are at equal risk of infection (despite demographic and other differences). If infections are concentrated away from traveler destinations (Fig. 1E presents the spatial distribution, by state, of cases within Mexico by 5 May), the number of people infected in Mexico will be underestimated, and conversely will be overestimated if the epidemic has disproportionately affected geographical zones visited by travelers. Under the assumption that reporting of infections in travelers was complete, we estimated the number of infections that occurred in Mexico by late April from a model of the interval-censored country case counts, which varied between 18,000 and 32,000 (Table 1), depending on the mean duration of stay of tourists assumed, with perhaps the most credible single value (based on journey duration data) being 23,000. An alternative model that assumed at least one case had been confirmed in every country affected by late April gave lower estimates of the number infected in Mexico, in the range of 6000 to 11,000. However, this model may be viewed as a worst case (from the perspective of resulting CFR estimates), and it fitted the observed number of exported cases in key countries (such as the United States and Canada) substantially worse than did the first model. We used 30 April 2009 as the cut-off date for the data analyzed, but the case data analyzed are subject to delays (clinical onset, testing, and reporting) of up to 1 week, so these estimates may be more representative of infections up to 23 April. The epidemic has subsequently spread further, although the impact of the nonpharmaceutical interventions introduced in Mexico is not yet known.
Fig. 1.
(A). The number of passengers flying out of Mexico by actual destination and the number of confirmed cases as reported on 30 April 2009. (B) The number of cases exported to country j as reported on 30 April 2009 as a function of the estimated average number of foreign travelers in Mexico from country j on any given day in March or April. Black circles: minimal number based on one exposure per epidemiological cluster; filled red circles, total number of confirmed cases. (C) Mean assumed generation time distribution (red) and 100 illustrative draws from the prior distribution, and (D) corresponding posterior distribution of R0 estimates for a stochastic model of an epidemic within Mexico with travelers infected at a rate proportional to the estimated density of travelers per local resident. The two bar charts correspond to a 7-day delay between infection and confirmation (blue) and no delay (orange) in cases among travelers. (E) Number of acute respiratory infection cases per 100,000 inhabitants by state as reported on 5 May 2009 (1), demonstrating spatial distribution of disease within Mexico.
Table 1.
Parameter estimates (and 95% confidence intervals) for the cumulative number of influenza A (H1N1) infections in Mexico among Mexican residents by late April 2009, along with corresponding estimates for the case fatality ratio (CFR), the basic reproduction number, and the exponential growth rate, assuming that infections in travelers occurred in Mexico.
| Assumed average duration of stay for visitors arriving by flights | Estimated number of infections among Mexican residents | Case fatality ratio* | Estimated reproduction number R0‡ | Estimated exponential growth rate‡ (doubling time in days‡) | |
|---|---|---|---|---|---|
| Assuming 9 deaths§ | Assuming 101 deaths§ | ||||
| Based on analysis of interval-censored country counts | |||||
| 6 days | 32,000 (26,000, 39,000) | 0.03% (0.02%, 0.03%) | 0.32% (0.26%, 0.39%) | 1.42 | 0.123 (5.6) |
| 9 days (23) | 23,000 (20,000, 280,00) | 0.04% (0.03%, 0.05%) | 0.44% (0.37%, 0.52%) | 1.40 | 0.118 (5.9) |
| 12 days | 180,00 (150,00, 220,00) | 0.05% (0.04%, 0.06%) | 0.55% (0.46%, 0.66%) | 1.39 | 0.114 (6.1) |
| Based on analysis of presence or absence of confirmed disease in countries | |||||
| 6 days | 11,000 (5,000, 27,000) | 0.08% (0.03%, 0.20%) | 0.90% (0.37%, 2.20%) | 1.36 | 0.106 (6.6) |
| 9 days (23) | 7,000 (3,000, 18,000) | 0.12% (0.05%, 0.29%) | 1.36% (0.56%, 3.30%) | 1.33 | 0.098 (7.0) |
| 12 days | 6,000 (2,000, 14,000) | 0.16% (0.07%, 0.39%) | 1.81% (0.74%, 4.40%) | 1.31 | 0.093 (7.4) |
This is a simple estimate of the CFR (4). The numbers of deaths and the data on cases in countries were both obtained as reported on 30 April. Although censoring is important to consider when estimating the CFR (due to the time interval between case report and death), in these data we have both the time between clinical onset and death within Mexico and the time between clinical onset and case confirmation in other countries to consider. Given uncertainty surrounding both of these distributions, we have made the simplifying assumption that these time intervals are similar and thus that the CFR can be estimated by the deaths within Mexico attributed to the novel influenza A (H1N1) divided by the estimated number of infections among Mexican residents based on data reported up to 30 April.
Based on 9 confirmed and 92 suspected deaths (101 total) that were reported by 30 April 2009 (6).
These estimates assume that the estimated cumulative number of influenza A (H1N1) infections in Mexico among Mexican residents relates to the cumulative number of infections up to 23 April 2009 (i.e., 1 week before the data were reported).
On the basis of the 9 confirmed and 92 suspected deaths that were reported by 30 April 2009 (6) and assuming similar times from infection to confirmation and from infection to death, we estimated CFRs in the range of 0.3 to 0.6% from the interval-censored case count model, based on confirmed and suspected deaths combined, or 0.03 to 0.05% for confirmed deaths only. Using the alternative, more pessimistic, country presence/absence model, we estimated CFRs of 0.9 to 1.8% based on suspected and confirmed deaths, and 0.08 to 0.16% from the confirmed deaths alone. These estimates have already changed somewhat as a result of data available after 30 April, but we deliberately report the earlier analysis because it formed part of the evidence base used by WHO to move to phase 5.
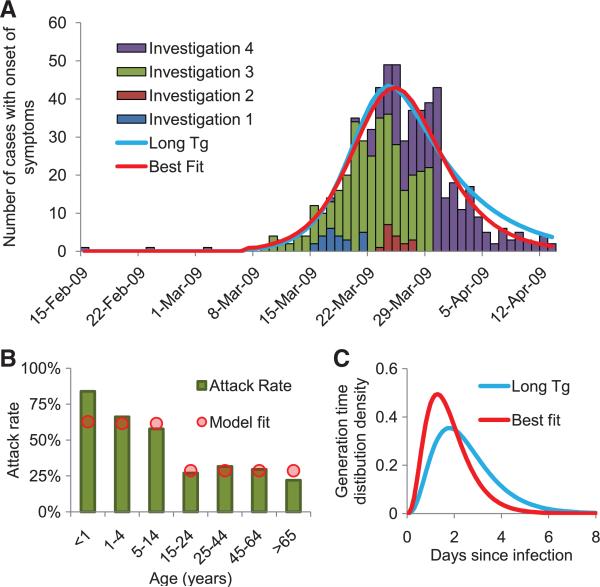
Another source of information on severity comes from the large outbreak of respiratory disease seen in the small, isolated community of La Gloria in Veracruz province, one case of which has been confirmed to have been caused by the novel H1N1 strain. It is possible that other viruses were circulating at the same time as the outbreak, but the overall attack rate is substantially larger than would be expected for a seasonal influenza outbreak. No fatalities among 616 cases have been attributed to infection during the full period of surveillance of that outbreak (Fig. 3A), giving a 95% confidence interval (CI) of 0 to 0.60%.
Fig. 3.
Results of a detailed investigation into an outbreak in the village of La Gloria. (A) The time series of cases based on repeat rounds of investigation into the outbreak, and the best fit of an age-stratified transmission model (see Table 2 for estimates). The graph also shows the best fit of a model where the generation time is constrained to be consistent with earlier estimates for influenza (2.6 days), which does not fit significantly worse than the unconstrained best fit (see Table 2 legend). (B) Observed (bars) and fitted (using best fit, circles) age-specific attack rates; (C) best fit and constrained estimate of the generation time distribution.
Data on the magnitude of the current outbreak in Mexico can also be used to estimate the transmissibility of the virus if the start date of the outbreak is known or can be estimated. Epidemiological investigations into the emergence of the virus in Mexico have focused on the La Gloria outbreak, where the first case in that outbreak is thought to have occurred around 15 February 2009 (Fig. 3A).
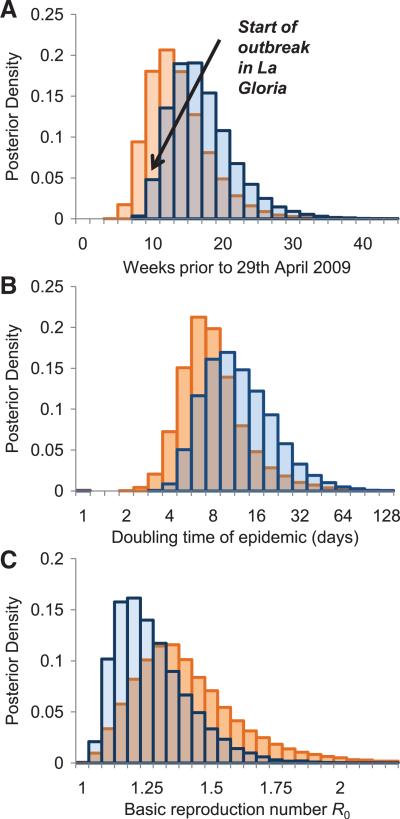
An alternative approach to estimating the start date of the outbreak is to look at the diversity in the genetic sequences of viral samples collected from confirmed cases, assuming that diversity accumulates according to a molecular clock model. Twenty-three complete publicly available hemagglutinin (HA) gene sequences from cases not linked in epidemiological clusters were analyzed with a Bayesian coalescent method that assumes exponential growth of the viral population (7). This yielded an estimate of the time of most recent common ancestor (TMRCA) of 12 January 2009 [95% credible interval (CrI): 3 November 2008 to 2 March 2009]. The genetic model also gave an estimate of the doubling time of the epidemic of 10 days (95% CrI: 4.5 to 37.5 days) (Fig. 2). Assuming exponential growth, the TMRCA is a reasonable estimate of the start of the outbreak, although it is formally an upper bound due to incomplete sampling of the epidemic and the effects of the exponential model prior to distribution. These findings from a population genetic analysis are consistent with the epidemiological investigation of both the start and magnitude of the current epidemic in Mexico. Figure 2 also shows a preliminary version of this analysis based on the first 11 sequences, which gave similar estimates highlighting the power of these methods. [See (8) for further sensitivity analysis and methods.]
Fig. 2.
(A) Starting from publicly available HA viral sequences, a posterior distribution of the estimated TMRCA was derived using a Bayesian coalescent model, which assumes exponential population growth (coded in BEAST 1.4), with the date of the first known human case highlighted. Details of the BEAST analysis and parameter estimates are presented in (8). Posterior distribution of the doubling time of the epidemic (B) and of R0 (C). The bar charts show the results obtained from the first 11 sequences available on 2 May 2009 (orange) and from an updated analysis with 23 epidemiologically unlinked sequences available on 7 May 2009 (blue). The differences in estimates arise due to some sequences in the smaller sample being from epidemiological clusters, highlighting the importance of careful sampling.
The reproduction number, defined as the number of cases one case generates on average over the course of their infectious period, is a key measure of transmissibility and can be estimated in a number of ways from the data currently available.
First, by assuming exponential growth, the growth rate of the epidemic (r) can be inferred from estimates of the current cumulative number of infections (Yf) and estimated start date and size for the outbreak (t0 and Y0, respectively). The basic reproduction number (R0) can be estimated from the exponential growth rate if one also assumes that the generation time distribution for the new H1N1 strain is similar to that of other strains of seasonal and pandemic viruses (9, 10) [Table 1 and (8)]. Using the date of 15 February as the first case of the La Gloria outbreak (8) gives reproduction number estimates of between 1.31 and 1.42, depending on which variant of the geographical backcalculation model is used. Extending a more sophisticated Bayesian estimation method (11) that allows for stochastic variability intrinsic to epidemic dynamics and parameter uncertainty gave similar but slightly higher estimates for R0 with wider ranges: posterior median = 1.40; 95% CrI: 1.15 to 1.90 (Fig. 1C).
Second, by assuming a prior distribution on the generation time distribution informed by previous estimates of influenza, the Bayesian coalescent population genetic analysis yielded a second set of estimates for R0: posterior median = 1.22; 95% CrI: 1.05 to 1.60 (Fig. 2C).
Third, R0 can also be estimated from analysis of the dynamics of the epidemic within defined settings. Detailed data collected by the Mexican authorities investigating the La Gloria outbreak indicate that 616 individuals from a resident population of 1575 had acute respiratory infection between 15 February and 14 April 2009 (Fig. 3A). Data on the age distribution of cases and the dates of disease onset were used in our analysis. Figure 3B shows that the clinical attack rate varied markedly as a function of age, with 61% of individuals under 15 years old affected, dropping to 29% of people over that age. The corresponding relative risk is 2.13, with a 95% CI of 1.89 to 2.39. Based on all confirmed cases in Mexico as reported on 5 May 2009 (1), the corresponding relative risk is 1.52 (95% C I: 1.33 to 1.73). The overall community attack rates seen in La Gloria are comparable to (or higher than) those seen in previous pandemics (12).
Fitting alternative epidemic models to the La Gloria data (8) demonstrated that a model with heterogeneous mixing by age plus age-dependent susceptibility to infection was required to adequately fit the data with plausible parameter estimates. The resulting maximum-likelihood estimate of R0 was 1.58 with a 95% CI of 1.34 to 2.04 (Table 2). This analysis also provided the only independent estimate of the mean generation time, Tg (1.91 days; 95% CI: 1.30 to 2.71 days) (Table 2), shorter than earlier estimates for influenza (9, 10), though not significantly so. It is biologically plausible that R0 and Tg could be correlated, because both are linked to the underlying replicative fitness of the virus. More data are needed. Owing to parameter identifiability issues, it was not possible to estimate age-dependent infectiousness, as well as age-dependent mixing, from these data. Although these estimates are informative, it should be emphasized that some uncertainties remain regarding the denominator population and that a range of other models may fit the data as well as the model choice shown here. Household data would be particularly useful in reducing remaining uncertainty.
Table 2.
Epidemiological parameters estimated by fitting an age-stratified mathematical model to the outbreak in the village of La Gloria (Fig. 3). For sensitivity analysis and model selection, we tested several reduced model variants. None fitted significantly worse, but several produced implausible estimates of the generation time. The respective best-fit values are as follows: 1. No asymptomatics and no misreporting, no assortative mixing: psymp = 1, θ ≡ 0, R0 = 1.37, Tg = 1.39, and ρchild = 2.80. 2. No asymptomatics and no misreporting: psymp = 1, R0 = 1.41, Tg = 1.53 days, θ = 0.31, and ρchild = 2.22. 3. No assortative mixing: same as variant 1. 4, model with long generation time consistent with previous estimates from influenza (also shown in Fig. 3), Tg ≡ 2.60 days, R0 = 1.97, ρchild = 2.52, θ = 0.51, and psymp = 0.72. (The symbol “≡” denotes parameters defined to take fixed values.)
| Best estimate | 95% confidence interval | Description | |
|---|---|---|---|
| R 0 | 1.58 | 1.34–2.04 | Basic reproduction number |
| T g | 1.91 | 1.30–2.71 | Mean generation time (days) |
| p symp | 86% | 69–100% | Proportion of cases that are symptomatic and ascertained |
| ρ child | 2.06 | 1.60–3.31 | Susceptibility of children relative to adults |
| θ | 0.50 | 0.00–0.72 | Assortativity of mixing between children and adults (0 = random, 1 = fully assortative) |
| Assumed value | |||
| f L | 1/3 | Assumed | Fraction of the generation time that is latent (uninfectious) |
| ϕ child | 1 | Assumed | Infectiousness of children relative to adults |
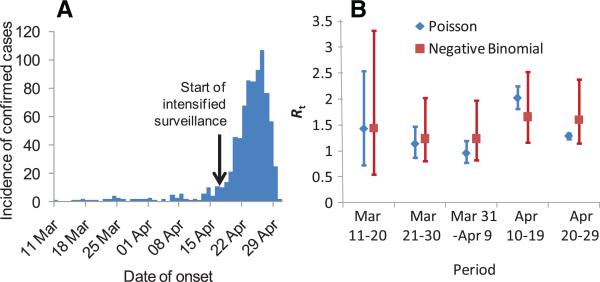
Fourth, the time-dependent reproduction number (Rt) can be estimated from the time series of reported disease onsets among confirmed cases in Mexico (Fig. 4). These data are subject to much uncertainty because of marked changes in surveil-lance over the reporting interval, plus the non-specificity of symptoms that are similar to existing and perhaps simultaneously circulating strains of influenza. However, we developed methods for analyzing such data (8) that account for substantial underreporting, with a change in the underreporting rate from 17 April when surveillance within Mexico was intensified. The average value of Rt estimated for Mexico up until the end of April was 1.37 (95% CrI: 1.24 to 1.59) for a model with Poisson case counts, and 1.47 (95% CrI: 1.21 to 1.88) for a perhaps more plausible negative binomial model allowing daily case counts to be overdispersed (8).
Fig. 4.
(A) Time course of the Mexican epidemic with (B) the posterior estimates (median and 95% CrI) of the reproduction number over time obtained under Poisson and negative binomial models from the analysis of confirmed cases. The estimate ofthe negativebinomial dispersion parameter k is for a low-to-moderate overdispersion, but this is enough to greatly increase the uncertainty in R(t).
Given estimates of R0 and the current epidemic size x, we can estimate the number of generations Nt of transmission of the virus among humans that is necessary to explain the current epidemic. Assuming a simple branching process with reproduction number R0, the mean number of generations of transmission is given by Nt = ln(x/x0)/ln(R0), assuming the epidemic was started by x0 humans being infected from animal sources. Assuming x0 = 1 gives estimates of Nt between 14 and 73. But even if we assume that 5% of cases were infected directly from animal sources, we obtain an estimated 5 to 22 generations of transmission, indicating sustained human-to-human transmission in Mexico.
All of the R0 estimates are comparable with, but perhaps on the low end of, R0 estimates obtained from analysis of previous pandemics [1.4 to 2.0 for 1918, 1957, and 1968 (9, 13–15)].
Overall, our transmissibility estimates are consistent with the lowest values used in earlier detailed computer simulations used to study scenarios in pandemic mitigation (16, 17), indicating that the conclusions regarding control policy effectiveness reached by those analyses could be relevant to the current epidemic. However, the key tradeoff remains the balancing of the economic and societal cost of interventions, such as school closure, against the numbers of lives saved through use of such measures. Where substantial antiviral stockpiles are available, a secondary trade-off is the extent to which large-scale prophylaxis is justified, given the potential risks of high-level resistance developing (18–21). At present, estimates of disease severity are insufficiently robust to allow these trade-offs to be properly evaluated, but that uncertainty should diminish rapidly in coming weeks as more data on severe cases in the United States and other countries become available.
As the situation develops, a key issue is to optimize study designs and surveillance protocols to be most informative in estimating some of these unknown factors, thus potentially informing and refining the public health response. Clearly, detailed investigations of transmission in households and schools will be useful, as would be the consistent collection and dissemination of electronic patient records, which could be used to detect cofactors in the severity of infection.
In conclusion, while the emerging data from Mexico and other countries have enabled important insights into the origin, extent, transmissibility, and severity of the unfolding pandemic [including detailed epidemiological analysis of data from the U.S. outbreak recently published (22)], many uncertainties remain and should not be underestimated. The incubation and infectious periods have not yet been reliably ascertained, leaving uncertainty in estimates of the generation time. Much remains to be done to estimate clinical severity of infection, to understand regional variations seen so far (or indeed, whether they exist). As the epidemic spreads further, it is likely that severity will vary from country to country depending on health care resources and the public health measures adopted to mitigate impact. The existence of any cross-immunity (perhaps not mediated via HA-specific antibodies) from past exposure to prior influenza A subtypes is unknown, but the strong age dependence in clinical attack rates seen in La Gloria is intriguing. Cross-immunity would imply that R0 could be higher in fully susceptible populations than estimated here. The future evolution of the transmissibility, antigenicity, virulence, and antiviral resistance profile of this or any influenza virus is difficult to predict. It is also unclear whether this strain will displace existing influenza A subtypes from the human population, as occurred in the past three pandemics. The extent to which seasonal damping of transmission in North America and Europe is responsible for the moderate transmissibility seen to date is uncertain; the progress of transmission in the Southern Hemisphere (which is just entering its influenza season) needs to be carefully monitored in the next few months. To reduce all these uncertainties, it is essential that public health agencies around the world continue to collect high-quality epidemiological data in a focused, resource-efficient manner despite the expected increases in case numbers in coming weeks. Epidemiological analysis and modeling are useful tools for guiding such efforts and interpreting the resulting data.
Note added in proof
We cited two sources (1, 6) for confirmed and suspected deaths in Mexico, reported by 4 May 2009 and 30 April 2009, respectively. These sources are not publicly available at present. However, similar reports are publicly available: The Mexican government Web site (24) gives some data on the 5 May situation report (25) documenting 26 confirmed deaths and 114 suspected deaths (77 without samples for analysis), and Morbidity and Mortality Weekly Report (26) lists 7 confirmed and 77 suspected deaths posted on 30 April. Since this article appeared online, the number of deaths in Mexico up to 23 April has been determined to be 21, resulting in a revised estimate of the CFR of 0.091% (range: 0.066 to 0.35%) (24).
Supplementary Material
Acknowledgments
We thank all those in Mexico and WHO (in particular J. Fitzner, K. Vandemaele and A. Merianos) who helped to collate the data used in this analysis. We also thank A. Borquez for help with translation and data collection and R. Eggo for data collation. We thank R. Anderson, K. Fukada, R. Hatchett, M. Lipsitch, D. Shay and L. Wolfson for useful discussions and comments. We thank the U.S. Centers for Disease Control; the Instituto de Salud Carlos III, Spain; Statens Erum Institut, Denmark; Erasmus MC Rotterdam, Netherlands; University of Regensburg, Germany; and the WHO collaborating centre for Reference and Research on Influenza, Australia, for posting viral sequences on GenBank. The work at Imperial College was funded by the Medical Research Council UK Centre grant. We also acknowledge additional support for individual staff members from the National Institute of General Medical Sciences (NIH) Models of Infectious Disease Agent Study (MIDAS) programme, The Royal Society (C.F., W.P.H., N.C.G., A.R., O.G.P.), Research Councils UK (S.C.), Bill and Melinda Gates Foundation (M.V.K., T.D.H., J.G.), The Wellcome Trust (R.F.B., grant GR082623MA), Biotechnology and Biological Sciences Research Council UK (T.J.), Microsoft Research (W.R.H.), and a studentship from the Medical Research Council (H.E.J.). GenBank accession numbers: GQ117067, FJ973557, FJ966082, FJ966952, FJ966960, FJ966974, FJ966971, FJ969511, GQ117040, FJ985753, GQ117119, FJ982430, GQ117097, GQ117059, GQ117103, GQ117112, CY039527, FJ984364, FJ984397, FJ985763, FJ974021, GQ117056, and FJ966982 for the main analysis and CY039527, FJ966082, FJ966959, FJ966960, FJ966974, FJ969509, FJ969511, FJ966952, FJ966982, FJ971076, and FJ973557 for the preliminary analysis.
References and Notes
- 1.México Dirección General Adjunta de Epidemiología Brote de influenza humana A H1N1 México. 2009 May 4 and 5; [Google Scholar]
- 2.WHO [5 May 2009];Swine Influenza—Update 15. ( www.who.int/csr/don/2009_05_05/en/index.html)
- 3.Glezen WP, Payne AA, Snyder DN, Downs TD. J. Infect. Dis. 1982;146:313. doi: 10.1093/infdis/146.3.313. [DOI] [PubMed] [Google Scholar]
- 4.Ghani AC, et al. Am. J. Epidemiol. 2005;162:479. doi: 10.1093/aje/kwi230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingsworth TD, Ferguson NM, Anderson RM. Nat. Med. 2006;12:497. doi: 10.1038/nm0506-497. [DOI] [PubMed] [Google Scholar]
- 6.Mexico Dirección General Adjunta de Epidemiología Brote de influenza porcina México. 2009 Apr 30; [Google Scholar]
- 7.Drummond AJ, Rambaut A. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Additional details on methods, data, and results are in the Supporting Online Material.
- 9.Ferguson NM, et al. Nature. 2005;437:209. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 10.Wallinga J, Lipsitch M. Proc. Biol. Sci. 2007;274:599. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipsitch M, et al. Science. 2003;300:1966. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost WH, Sydenstricker E. Public Health Rep. 1919;34:491. [Google Scholar]
- 13.Mills CE, Robins JM, Lipsitch M. Nature. 2004;432:904. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gani R, et al. Emerg. Infect. Dis. 2005;11:1355. doi: 10.3201/eid1109.041344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viboud C, et al. Vaccine. 2006;24:6701. doi: 10.1016/j.vaccine.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson NM, et al. Nature. 2006;442:448. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germann TC, Kadau K, Longini IM, Jr., Macken CA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5935. doi: 10.1073/pnas.0601266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong MD, et al. N. Engl. J. Med. 2005;353:2667. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- 19.Lackenby A, et al. Euro Surveill. 2008;13 doi: 10.2807/ese.13.05.08026-en. available at www.eurosurveillance.org/images/dynamic/EE/V13N05/art8026.pdf. [DOI] [PubMed] [Google Scholar]
- 20.Moscona A. N. Engl. J. Med. 2005;353:2633. doi: 10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- 21.Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Emerg. Infect. Dis. 2009;15:155. doi: 10.3201/eid1502.081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novel Swine-Origin Influenza A Virus Investigation Team N. Engl. J. Med. 2009 10.1056/NEJMoa0903810. [Google Scholar]
- 23.Hudman LE, Jackson RH. Geography of Travel and Tourism. Cengage Learning, Thomson Learning; Clifton Park, NY: 2002. p. 4. [Google Scholar]
- 24. http://portal.salud.gob.mx/contenidos/noticias/influenza/estadisticas.html.
- 25. http://portal.salud.gob.mx/sites/salud/descargas/pdf/influenza/presentacion20090505.pdf.
- 26. www.cdc.gov/mmwr/preview/mmwrhtml/mm5817a5.htm.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.