Abstract
Soluble concentration gradients play a critical role in controlling tissue formation during embryonic development. The importance of soluble signaling in biology has motivated engineers to design systems that allow precise and quantitative manipulation of gradient formation in vitro. Engineering techniques have increasingly moved to the third dimension in order to provide more physiologically relevant models to study the biological role of gradient formation and to guide strategies for controlling new tissue formation for therapeutic applications. This review provides an overview of efforts to design biomimetic strategies for soluble gradient formation, with a focus on microfluidic techniques and biomaterials approaches for moving gradient generation to the third dimension.
Keywords: biomaterials, diffusion, hydrogels, proteins, tissue engineering
Introduction
Soluble concentration gradients, such as growth factor gradients, play a crucial role during tissue development. Positional information is provided by soluble signals termed “morphogens,” which elicit distinct cell responses dependent on local concentration.[1–7] Morphogen gradient generation is a highly complex and tightly regulated process that is influenced by a variety of cellular and extracellular factors.[1,2,8,9] The unique ability of morphogen gradients to spatially and temporally guide tissue formation has been demonstrated during embryonic development in numerous organisms.[2–6] For example, sonic hedgehog (Shh) concentration gradients help to determine ventral neuronal progenitor identity in the chick dorsal neural tube,[10] Wingless/Int-3 (Wnt-3) gradients regulate melanogenesis in the quail neural tube,[11] and bone morphogenetic protein (BMP) gradients encourage pre-cardiac gene expression in ascidian ventral trunk cells.[12] Thus, natural morphogen gradients represent an elegant mechanism to spatially and temporally control tissue formation.
Our current understanding of the mechanistic details of morphogen concentration gradients is primarily based on the theory of restricted extracellular diffusion. In this model, concentration gradients form due to morphogen transport from the source cells that secrete them to the target cells they act upon. Morphogen transport is spatially guided along specific routes in developing tissues by a variety of factors, including cell organization, the presence of cell surface receptors, and morphogen-binding moieties in the extracellular matrix (ECM).[1–6,8,9] Experiments in chick limb bud and zebrafish models demonstrate the importance of restricted extracellular diffusion in vivo. For example, ECM-associated heparan sulfate proteoglycans (HSPG) modulate Shh signaling range in the chick limb bud.[13] Additionally, retinoic acid was shown to be degraded by Cyp26 immediately upon entering the cell cytoplasm in zebrafish, suggesting that long-range retinoic acid signaling is possible only by extracellular means rather than intracellular transport.[14] Fundamental developmental biology studies such as these have been instrumental in advancing our understanding of the mechanisms that guide soluble gradient formation.
The critical importance of soluble signaling in biology has motivated numerous studies aimed at modeling gradient generation in vitro.[15,16] Many previous studies have applied soluble signaling gradients to cells in two-dimensional (2D) culture, and have been reviewed elsewhere.[17,18] Increasingly, engineers have designed synthetic biomaterials to mimic three-dimensional (3D) biological matrices[19–22] in order to optimize study of cell–cell interactions, mechanical stimuli, and soluble signaling dynamics that are not present in 2D.[20] Synthetic materials provide researchers precise control over parameters such as pore size, crystallinity, molecular weight, copolymer composition and charge, and enable greater mimicry of the in vivo environment.[23–28] In this review we present emerging approaches to control soluble concentration gradients in biomaterials. The particular focus is on biomimetic approaches, which are designed based on principles gleaned from natural tissue development. We specifically focus on approaches that use microfluidics, localized controlled release, and ECM affinity as mechanisms for gradient generation.
Microfluidic Systems for Gradient Generation
Microfluidic systems are ideal tools for developing specifically tuned soluble gradients and characterizing interactions between soluble signals, biomaterials, and cells. In a majority of microfluidic studies, the timing of biomolecule delivery, gradient slope, geometry, and signaling distance may be tuned by adjusting input and output flow rates as well as channel geometry.[29–39] The dimensions of micro-fluidic environments enable one to observe molecule distributions and cell responses via fluorescence and bright field microscopy.[30,31,39–41] Importantly, microfluidic systems may provide particular advantages in the context of biomimicry. The low ratio of cell volume to extracellular fluid or matrix volume in these environments more closely mimics in vivo environments when compared to standard cell culture environments.[42] In addition, the ability to spatially localize morphogen sources and sinks, and the ability to define the region in between, can lead to a direct mimic of the restricted extracellular diffusion mechanism often observed during natural tissue development.
Convective Flow Platforms
Convective flow platforms continuously maintain soluble signal concentrations in source and sink flow channels. This is advantageous because soluble signaling gradients can be maintained for indefinite periods of time.[15,16,31,41,43,44] However, flow platforms can remove cell-secreted factors[31] and flow-induced shear may influence cell behavior in unintended ways.[45,46] In order to address these challenges, convective flow devices can be designed to incorporate a flow barrier (Figure 1) so that biomolecule delivery into the cell culture chamber occurs through diffusion. Examples of barriers include capillary channels[41] and thin polymer films.[31,43,44] In view of the importance of diffusion in natural soluble signaling, these systems may provide a biomimetic mechanism for introducing soluble signals to cells.
Figure 1.
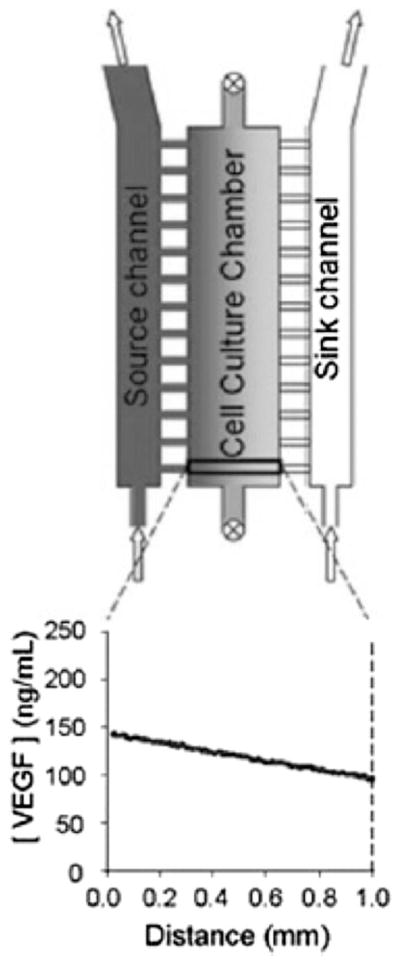
Small molecule gradient generation across a biomaterial using perpendicular capillary extensions from source and sink flow paths.[47]
Though convective flow platforms have mostly focused on presenting soluble signaling molecules to cells cultured in 2D monolayers,[17,18] they have recently been adapted to form soluble gradients in 3D environments. For example, capillary channels were adapted to present a vascular endothelial growth factor (VEGF) gradient to human dermal microvascular endothelial cells (HMDVEC) cultured in a 3D collagen and fibronectin matrix.[47] Results demonstrated that tubulogenesis was more prevalent toward higher concentrations of VEGF. Similar barrier motifs have been demonstrated in other convective flow platforms that generate gradients across 3D hydrogels.[48–50] In future studies, these systems may be promising tools for modeling dynamic changes in morphogen signaling levels via perturbations in source flow rates.
Pnuematic Valve Systems
Pneumatic valve systems can be used to trigger gradient formation at user-defined times.[51] As an example, Liu et al. seeded populations of mouse embryonic fibroblasts and human hepatocellular carcinoma cells in separate cell culture chambers that were connected by a migration channel. The channel could be opened or closed using a pneumatic valve. When the valve was opened, cell-secreted chemotactic factors from the carcinoma population diffused toward the fibroblast chamber and increased fibroblast motility toward the carcinoma chamber.[39] A separate study by Lii et al. adapted a similar valve system to control diffusion of soluble molecules between neighboring 3D hydrogel matrices. Embryonic stem cells (ESC) in matrigel were localized to a cell culture chamber, and cell tracker dye was encapsulated in a poly(ethylene glycol diacrylate) (PEGDA) hydrogel located in a neighboring chamber. Molecule transfer between the gels was controlled by a pneumatic valve. Upon opening the valve, the cell tracker dye diffused from the PEGDA into the matrigel to stain the ESCs.[38] This precise timing of gradient formation provides a means to create more complex experimental systems and better recreate the signaling dynamics in physiological systems.
Gradients without Pumps or Tubes
Alternative microfluidics platforms have recently been developed to enable gradient generation without the need for extensive flow networks and complex pumping instrumentation. In addition, they are amenable to multiplexing and automation, making them easily accessible methods of gradient generation.[52,53] Huang et al.[40] recently used micropipettes to position juxtaposed matrigel and collagen matrices inside a microfluidic chamber and established sharp boundaries between the matrices using arrays of micron-scale hexagonal posts. Hydrophobic surface chemistry and controlled spacing between the posts prevented the hydrogel precursor solutions from relocating before curing. In addition, the discontinuous barrier defined by the post arrays allowed the neighboring matrigel and collagen matrices to have well-defined points of contact. As a demonstration of molecular transport between the matrices, a population of collagen-encapsulated breast cancer cells secreted chemotactic factors that induced macrophages in the neighboring matrigel to invade the collagen matrix. This strategy may be applicable to numerous hydrogel species and cell co-culture combinations.
Abhyankar et al.[29] developed a gradient generation platform in which the concentration difference between a fluid source and sink results in molecular transport through 3D hydrogel matrices (Figure 2). Controlled gradients of epidermal growth factor (EGF) and short chemoattractant peptides increased neutrophil migration and invasion of rat mammary adenocarcinoma cells into collagen hydrogels, respectively. Gradients in this system can be temporally controlled by repeated replenishment of the source and sink solutions. A recent study generated fetal bovine serum gradients in poly(methyl methacrylate) scaffolds in a similar gradient generation system,[54] suggesting that these systems are amenable to generating gradients inside a variety of materials. In addition, these systems are appropriate for mimicking paracrine movement driven by diffusion.
Figure 2.
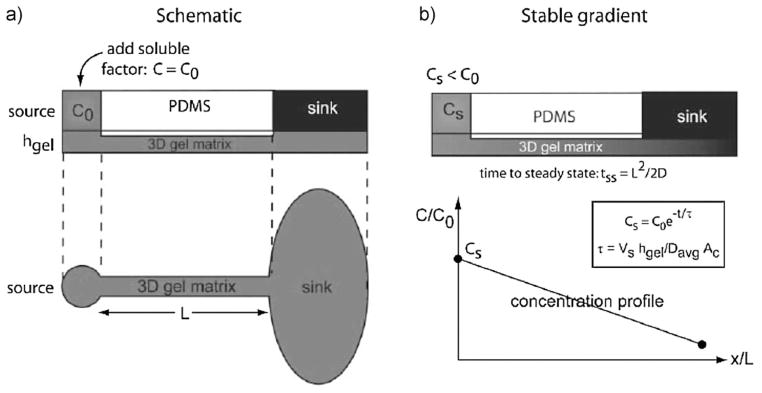
(a) Schematic of polydimethylsiloxane (PDMS) microchannels with defined source and sink regions. (b) Desired concentration profile and calculation of time intervals for replenishing source and sinks.[29]
Passive pumping, or surface-energy driven fluid flow, has been recently developed as a tubeless flow mechanism to transport fluid through microfluidic systems at high flow rates.[52,55] Recently, Khademhosseini and co-workers[56] generated graded distributions of Arg-Gly-Asp-Ser (RGDS) cell adhesion peptide in PEGDA hydrogels by injecting RGDS-PEG molecules into microchannels and allowing evaporation-driven back-flow to imbalance RGDS distribution (Figure 3). Although the study did not strictly aim to generate soluble molecule gradients inside 3D hydrogels, this process could in principle be adjusted to distribute non-cross-linkable molecules inside hydrogel precursor solutions before future curing steps. Notably, this study used passive pumping as a tubeless alternative to a previous gradient generation system that required the use of fluid injection pumps.[32]
Figure 3.
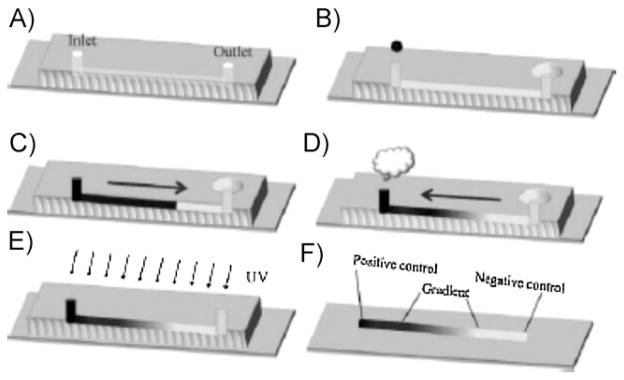
Schematic of passive pumping-aided gradient generation in microfluidic chambers. (A) The microfluidic channel was fashioned in PDMS. (B) The channel was pre-filled with photocrosslinkable hydrogel precursor solution. (C) After the molecule of interest was inserted into the inlet port, it transported down the length of the channel via passive pumping. (D) Evaporation at the inlet port induced a backflow in the system that encouraged concentration gradient formation. (E) Photocrosslinking of the precursor fixes the concentration gradient in place, creating the (F) completed patterned hydrogel. Reproduced with permission from ref.[56] Copyright Wiley-VCH Verlag GmbH & Co. KGaA.
Microfluidics platforms have evolved over time to offer multiple mechanisms for control over soluble signal gradients in cell culture environments. Progress in 3D cell culture within microfluidic systems may result in increasingly complex biomimetic microenvironments, which could lead to a more complete understanding of gradient signaling in biology.
Localized Controlled Release within Biomaterials
While microfluidics platforms are excellent tools for studying the role of soluble signal gradients, these systems are generally designed for in vitro experiments rather than in vivo use or eventual clinical application. An alternative approach to generate gradients for therapeutic strategies involves designing biomaterials that contain biomolecules within localized “depots.” Gradient characteristics can then be controlled through a combination of spatial organization and material properties. These localized release approaches provide a direct mimic of natural morphogen signaling, in which morphogen release from a localized cell source typically occurs. The aim of this section is not to review all biomaterials-based methods for drug delivery, which have been reviewed in detail elsewhere.[57–59] Instead, we focus on approaches in which blended biomaterials are designed to control gradient formation through controlled localization of signaling sources within a material.
Release Depots in Multicomponent Materials
As a demonstration of a biomaterials design that provides strict control over gradient formation, Peret et al. encapsulated protein-loaded poly(lactide-go-glycolide) (PLG) micro-spheres in a PEGDA hydrogel depot via photopolymerization. The depot was submerged in blank PEGDA precursor solution and photopolymerized again, resulting in a continuous gradient-generating hydrogel (Figure 4).[60] Protein gradient magnitude, slope, persistence length, and persistence time could be strictly controlled in this material through a variety of parameters, including protein hydrodynamic radius, microsphere concentration, and hydrogel mesh size. This approach provides an adaptable biomaterials strategy for controlling several aspects of gradient profile generation through simple design, and in principle could be used to incorporate combinations of soluble molecules via mixtures of pre-loaded microspheres.
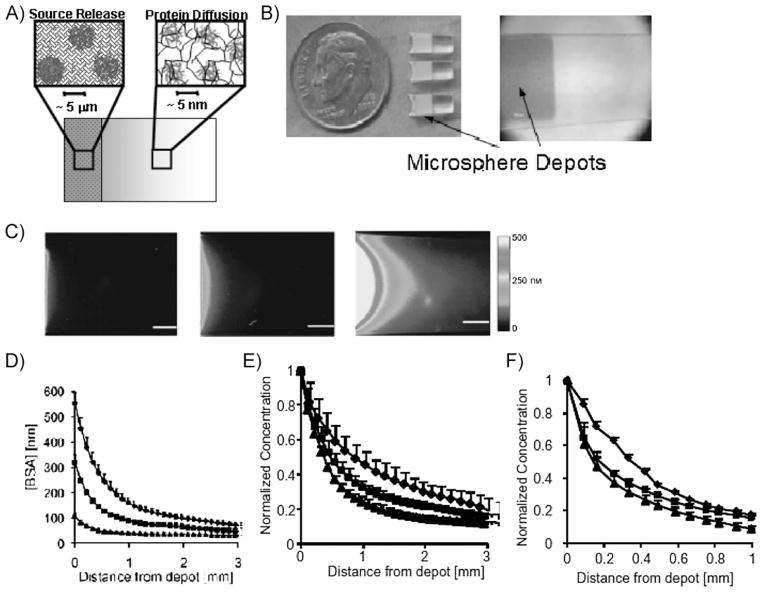
Figure 4.
Generation of controllable protein gradients via localized, sustained release from a microsphere depot. (A) Schematic of the microsphere depot combined with a PEGDA hydrogel. (B) Photographs of the completed multicomponent hydrogel. (C) Fluorescence images of BSA gradient generation (pseudocolored to indicate fluorescence intensity distribution). Left: 10 mg · mL−1 BSA-loaded microspheres. Middle: 30 mg · mL−1 BSA-loaded microspheres. Right: 60 mg · mL−1 BSA-loaded microspheres. (D) Concentration gradients in hydrogels containing 60 mg · mL−1 (◆), 30 mg · mL−1 (■), and 10 mg · mL−1 BSA (▲). (E) Concentration gradients in hydrogels containing lysozyme (◆), ovalbumin (■), and BSA (▲). Lysozyme has the lowest hydrodynamic radius of the three proteins while BSA has the highest. (F) BSA Concentration gradients in hydrogels composed of 5% (◆), 10% (■), and 15% PEGDA (▲). These results demonstrate multiple mechanisms for tailoring gradient characteristics by controlling material properties. Figure portions C–F reproduced with permission from ref.[60] Copyright Wiley-VCH Verlag GmbH & Co. KGaA.
Due to the complexity of the signaling environment during tissue formation, strategies for controlling release for multiple growth factors as well as spatial organization of release are desired. Chen et al.[61] accomplished both of these goals by embedding PLG microspheres in a PLG scaffold, resulting in a scaffold with discrete, spatially located gradients, and distinct release profiles of two growth factors. Specifically, platelet derived growth factor (PDGF)-carrying microspheres were embedded in a VEGF-loaded PLG scaffold, which was delivered to ischemic rat hind limbs. Blood vessel maturation was significantly enhanced in the presence of VEGF and PDGF relative to VEGF alone.[61] In a second study designed to form stable, localized gradients, VEGF and anti-VEGF antibodies were released from distinct regions with in a PLG scaffold to spatially localize VEGF activity. In vivo implantation increased angiogenesis in mouse ischemic hindlimb models while restricting VEGF activity to muscle in close proximity to the scaffold.[62] Thus, these studies offer a method for controlling coordinated gradients of multiple signaling molecules.
Graded Distributions of Signal Carriers
An alternative strategy for forming stable, spatially distinct concentration gradients of multiple biomolecules within 3D biomaterials was demonstrated by Dodla et al. Both step and continuous gradients were generated by inserting agarose hydrogels containing nerve growth factor (NGF)-containing lipid microtubules and immobilized laminin-1 (LN-1) into polysulfone guidance channels.[63] Axonal regeneration for both gradients was similar to nerve tissue grafts, which demonstrates improved functional outcome when compared nerve guidance channels containing either no molecular gradients or a single gradient of NGF or LN-1.[63] In a separate example of this general approach, Wang et al.[64] used a piston-driven gradient generator to suspend a continuous gradient of BMP-2-loaded microspheres and insulin-like growth factor-I (IGF-I)-loaded microspheres in silk fibroin scaffolds. When these scaffolds were seeded with human mesenchymal stem cells (hMSC), markers of osteogenic and chondrogenic differentiation such as calcium deposition and glycosaminoglycan deposition, respectively, increased with increasing BMP-2 concentration. IGF-I enhanced osteogenic and chondrogenic differentiation levels throughout the hydrogel when BMP-2 and IGF-I were presented as opposing gradients. Taken together, these strategies may be useful for recreating the complexities of graded tissue structure such as the discontinuous transition from bone to cartilage in joints.
Immobilized Protein Gradients Inspired by Protein-ECM Affinity and Patterning
The ECM plays a crucial role in gradient formation by directly interacting with soluble proteins through affinity interactions. These affinity interactions are often spatially organized, and can influence how proteins function and interact with cells.[65] For example, in the drosophila testis stem cell niche the leukemia inhibitory factor (LIF) homolog Unpaired (Upd) is secreted by “hub” cells, and Upd-ECM interactions limit its diffusion away from the source such that only stem cells directly adjacent to the hub are able to self-renew in response to Upd signaling.[66] This principal is also demonstrated in the developing Drosophila embryo, where glypicans act as a spatially patterned path for transport of Wingless, a morphogen that specifies the formation of ventral structures.[6,11,67] The glypicans in the ECM bind and stabilize Wingless while spatially defining a transport route through developing tissue.[6] These examples demonstrate the critical role of spatial and affinity-based interactions between morphogens and ECM in natural tissue development and maintenance, and engineering strategies to design biomaterials based on these ideas have begun to emerge.
There have been several examples of strategies for incorporating affinity ligands to sequester soluble molecules in 3D materials.[65,68–78] These affinity-based materials have been used for a variety of biological studies, including sustained growth factor delivery[68,71,75,77,79] and modulation of cell function.[68,69,73,74,79] While studies to date have focused on immobilizing affinity ligands throughout biomaterials, patterning methods discussed below may also be compatible with linking chemistries described for affinity-based materials.[68,69,75]
The nerve guidance channel created by Dodla et al., mentioned previously, contained a continuous gradient of immobilized LN-1, which was initially generated via diffusion of photocrosslinkable LN-1 molecules into the hydrogels. Afterward, the entire material was UV-irradiated to immobilize the LN-1.[63] Luhmann et al.[80] used a piston-driven gradient generator to apply a linear gradient of Ig6-like domain into a fibrin precursor solution. The Ig6-like domain was covalently bound to the fibrin matrix via a Factor-XIIIa mimicking peptide sequence before the entire hydrogel was stabilized by thrombin. The distribution of labeled Ig6-like domain was measured using fluorescence microscopy, and the immobilized gradient maintained a steep slope after 24 h. The examples described above provide strategies that are based on simple diffusion and therefore may be useful for generating gradients using a broad range of bound proteins.
While patterning gradients of immobilized molecules distributions in 3D hydrogels using diffusion is a significant achievement, the complexity of patterns generated using these systems are limited. A possible solution for generating more complex patterns of immobilized molecules in 3D materials has been demonstrated in synthetic hydrogels using several strategies.[81–87] The concept of 3D photo-patterning in PEG hydrogels was first introduced by West and co-workers.[81,83] Hahn et al.,[81] introduced acrylated bioactive groups into PEGDA hydrogels using two-photon microscopy to create complex patterns post-gelation, and subsequent work by Lee et al.[83] demonstrated that cell migration could be guided using this platform. Using a different chemistry for incorporation of photopatterning into PEG hydrogels, DeForest et al.[85] demonstrated that orthogonal “click” chemistry could be used to form complex patterns using two-photon microscopy and that this strategy could be used for 3D cell patterning. In a following study, DeForest et al.[87] demonstrated that complex immobilized gradients could be formed with control over gradient magnitude and slope using the “click” hydrogels. These examples demonstrate how synthetic strategies can be used to create materials with complex, spatially patterns of immobilized ligands, and would be applicable to virtually any biomolecule provided that acrylate or thiol groups could be incorporated.
Challenges and Future Prospects
Triggered Appearance of Soluble Gradients
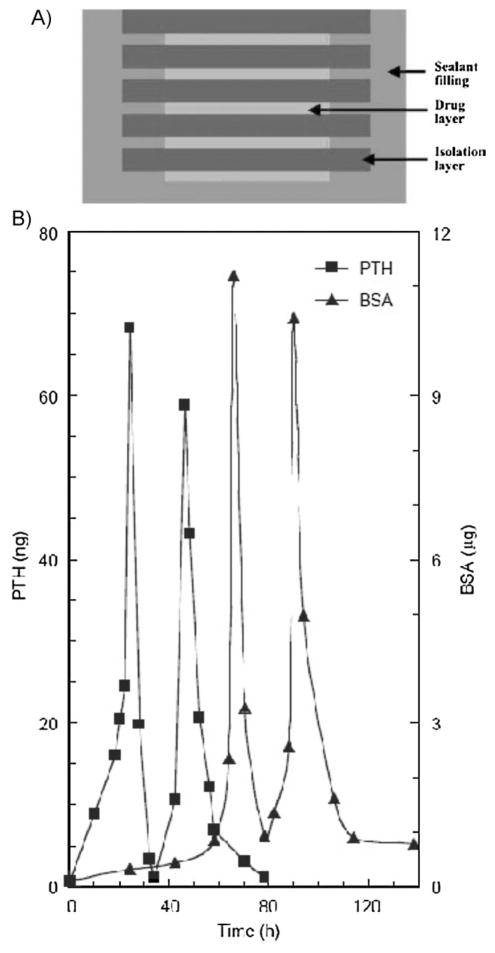
The studies described in the previous sections generally focus on strategies in which release profiles or gradient generation are pre-determined based on initial materials properties. However, tissue formation during both development[1–6,8,9,88] and wound healing[89–91] requires coordination of cell migration, differentiation, and ECM production that is precisely controlled through the timing of distinct growth factor signaling profiles. In view of the importance of both dose and timing during natural tissue development, there is widespread interest in controlling when soluble signaling occurs for tissue engineering strategies. This concept may be extended to control when soluble signal gradients appear in engineered tissues, as well as coordinate the appearance of multiple soluble signal gradients. External stimulation of drug release from materials has been studied for decades, and polymers have been designed to be responsive to pH,[92–94] glucose,[95] temperature,[94,96–100] magnetism,[101–103] ultrasound[104] and electric fields,[105–107] and might be applied to biomolecule delivery for some tissue engineering applications as well. As an advanced example of controlling the timing of soluble signal release, layers of drug-carrying alginate hydrogels were inserted into a chemically stable poly(L-lactic acid) (PLLA) casing along with poly(anhydride) discs.[108] The degradation timing of the polyanhydride layers resulted in a lag time between drug release from alginate hydrogels.[108] Pulsatile release of parathyroid hormone (PTH) and bovine serum albumin (BSA) over time as well as sequential release of drugs encapsulated in separate alginate layers were demonstrated using this technique (Figure 5).[108]
Figure 5.
Schematic of pulsed drug release system. (A) The PLLA sealant filling prevents lateral and non-sequential diffusion of drug from the construct. The drug-carrying alginate layers release molecules, and the degradation times of the polyanhydride isolation layers determine lag times between drug release pulses. (B) Pulsed sequential release of PTH and BSA. Reproduced with permission from ref.[108] Reprinted from Biomaterials, 28, X. Liu et al., Pulsatile release of parathyroid hormone from an implantable delivery system, pages 4124–4131. Copyright 2007, with permission from Elsevier.
An alternative to pre-defined release profiles would involve triggering soluble signal release at desired times or in response to specific biological cues. As a proof-of-principle for using a specific biochemical trigger to induce morphogen release, King et al. formed calmodulin-based microspheres that collapse and release their cargo in the presence of the drug trifluoperazine (TFP). TFP caused VEGF-loaded microspheres to collapse to less than 50% of their initial volume, resulting in rapid release of VEGF.[109] The same concept was also recently used for triggered release of BMP-2.[110] One can envision using these “bioresponsive” microspheres and others[111] as localized release depots within biomaterials.
Strategies have also been developed for cell-triggered proteolytic release of soluble biomolecules. For example, covalently bound signal molecules can be released by proteases secreted by cells by incorporating enzymatically degradable peptide tethers.[112,113] Sustained release of protein was also demonstrated for a PEG hydrogel, in which a human neutrophil elastase (HNE)-degradable peptide designed to be susceptible to inflammation was used to cross-link the polymer.[114,115] Since the mechanism for protein release using HNE-degradable PEG hydrogels involved surface, rather than bulk, erosion, release was independent of protein size, and thus could be tuned solely by choice of materials properties, with factors such as peptide concentration or substitutions in the amino acid sequence of the cross-linking peptide being used to tailor release rate.[114,115] This general approach may be amenable to a variety of applications, since there are other available small peptides that are susceptible to proteases.[116] Together, the above strategies may enable soluble gradients to appear at specified times, mimicking those demonstrated during natural tissue formation.
Enhanced Throughput
One of the primary challenges of mimicking natural morphogen gradients is their complexity. For example, the effects of morphogen gradients on tissue formation can be influenced by persistence time, signaling distance, slope, and morphogen potency.[60] This complexity creates a highly complex set of interdependent parameters that need to be optimized, often with little knowledge of ranges that are important physiologically. Further, the design of new materials with degradable matrices[84,113,117–122] and incorporated cells will lead to even more complex sets of conditions that will affect gradient shape, dynamics, and bioactivity. Standard diffusion models may not be appropriate for predicting release rates within these complex materials, let alone their influence on tissue formation. Thus, multiplex testing strategies that can investigate a large number of parameters simultaneously may be important to design gradient systems for in vitro and in vivo use. Early studies indicate that 3D hydrogel arrays formed with both synthetic and natural biomaterials can screen for the effects of adhesion peptides, soluble growth factor concentration, and matrix degradability.[123–126] These strategies and others[38,127–129] may also be applicable to testing combinations of entrapped or tethered soluble signals or loaded microspheres, which could lead to enhanced throughput analysis of soluble gradients.[123–126]
Conclusion
Techniques for generating controlled bimolecular gradients within 3D synthetic environments are just beginning to emerge. To truly exert fine control of soluble molecule concentration gradients in complex tissue engineering systems, consideration should be given not only to spatially controlling the distribution of soluble molecule in a 3D environment, but temporally controlling them as well. Studies in microfluidic systems, controlled release mechanisms and 3D patterning of immobilized molecules are unique toolsets. These technologies may lead toward understanding the origins and temporal progression of morphogen gradients in nature, as well as controlling gradients for tissue engineering applications.
Acknowledgments
The authors gratefully acknowledge the financial support of the NIH (R01HL093282 and Training Grant NIH-5-T32-GM08349), the NSF (CAREER award 0745563) and University of Wisconsin-Madison Graduate Engineering Research Scholars Program.
Biographies

Eric Nguyen received his BS in Biomedical Engineering at Bucknell University in 2009. He became a graduate student at the University of Wisconsin-Madison in 2009 and joined Professor William Murphy’s Bioinspired Materials Laboratory. He is currently funded by the Graduate Engineering Research Scholars fellowship and is a trainee under the National Institutes of Health Biotechnology Training Program. His current research interests include controlling angiogenesis in three-dimensional microenvironments using controlled soluble signaling gradients.

Michael Schwartz received his PhD in chemistry from the University of Wisconsin-Madison in 2003 and spent time as a postdoctoral fellow at the University of California-San Diego and the University of Colorado-Boulder. He joined Professor William Murphy’s lab as an Assistant Scientist in 2010. His research interests include developing engineering strategies for studying complex biological systems and investigating the role of the microenvironment in stem cell and cancer biology.

William Murphy is an Associate Professor of Biomedical Engineering, Pharmacology, and Orthopedics/Rehabilitation at the University of Wisconsin, where he has been since 2004. He received his PhD in Biomedical Engineering from the University of Michigan in 2002, and was a postdoctoral fellow in Chemistry at the University of Chicago from 2002 to 2004. Murphy’s research interests focus on designing “Bioin-spired” materials that mimic and exploit biological systems. Examples of mimicking biology include biomaterials that translate nature’s molecular dynamics into controllable drug delivery. Examples of exploiting biology include biomaterials that bind and regulate specific molecules and stem cells already present in the body to engineer new tissues. He has published over 50 manuscripts and filed 15 patents.
Contributor Information
Eric H. Nguyen, Department of Biomedical Engineering, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Avenue, Madison, Wisconsin 53705, USA
Michael P. Schwartz, Department of Biomedical Engineering, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Avenue, Madison, Wisconsin 53705, USA
William L. Murphy, Email: wlmurphy@wisc.edu, Department of Biomedical Engineering, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Avenue, Madison, Wisconsin 53705, USA, Fax: (608) 265 9239. Department of Pharmacology, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Avenue, Madison, Wisconsin 53705, USA. Department of Orthopedics & Rehabilitation, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Avenue, Madison, Wisconsin 53705, USA
References
- 1.Ashe HL, Briscoe J. Development. 2006;133:385. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- 2.Gurdon JB, Bourillot PY. Nature. 2001;413:797. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence PA. Nat Cell Biol. 2001;3:E151. doi: 10.1038/35083096. [DOI] [PubMed] [Google Scholar]
- 4.Wartlick O, Kicheva A, Gonzalez-Gaitan M. Cold Spring Harb Perspect Biol. 2009;1:a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan D, Lin X. Cold Spring Harb Perspect Biol. 2009;1:93. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JC, Hagemann A, Saka Y, Williams PH. Philos Trans R Soc Lond B. 2008;363:1387. doi: 10.1098/rstb.2007.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akeson A, Herman A, Wiginton D, Greenberg J. Microvasc Res. 2010;80:65. doi: 10.1016/j.mvr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swartz MA. Curr Opin Biotechnol. 2003;14:547. doi: 10.1016/j.copbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Nelson CM. BBA-Mol Cell Res. 2009;1793:903. [Google Scholar]
- 10.Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dongkyun K, Jinsoo S, Jin EJ. Cell Biol Int. 2010;34:763. doi: 10.1042/CBI20090133. [DOI] [PubMed] [Google Scholar]
- 12.Christiaen L, Stolfi A, Levine M. Dev Biol. 2010;340:179. doi: 10.1016/j.ydbio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Saha K, Schaffer DV. Development. 2006;133:889. doi: 10.1242/dev.02254. [DOI] [PubMed] [Google Scholar]
- 14.White RJ, Nie Q, Lander AD, Schilling TF. PLoS Biol. 2007;5:2522. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keenan TM, Folch A. Lab Chip. 2008;8:34. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung BG, Choo J. Electrophoresis. 2010;31:3014. doi: 10.1002/elps.201000137. [DOI] [PubMed] [Google Scholar]
- 17.Meyvantsson I, Beebe DJ. Annu Rev Anal Chem. 2008;1:423. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 18.Gupta K, Kim DH, Ellison D, Smith C, Kundu A, Tuan J, Suh KY, Levchenko A. Lab Chip. 2010;10:2019. doi: 10.1039/c004689b. [DOI] [PubMed] [Google Scholar]
- 19.Lutolf MP. Integr Biol. 2009;1:235. doi: 10.1039/b902243k. [DOI] [PubMed] [Google Scholar]
- 20.Griffith LG, Swartz MA. Nat Rev Mol Cell Biol. 2006;7:211. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 21.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 22.Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SH, Shin H. Adv Drug Deliv Rev. 2007;59:339. doi: 10.1016/j.addr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Lin CC, Anseth KS. Pharm Res. 2009;26:631. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuttelman CR, Rice MA, Rydholm AE, Salinas CN, Shah DN, Anseth KS. Prog Polym Sci. 2008;33:167. doi: 10.1016/j.progpolymsci.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anseth KS, Svaldi DC, Laurencin CT, Langer R. Photopolymerization. Vol. 673. American Chemical Society; Washington: 1997. p. 189. [Google Scholar]
- 27.Anseth KS, Bowman CN, Brannon-Peppas L. Biomaterials. 1996;17:1647. doi: 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- 28.Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Adv Mater. 2010;22:3484. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abhyankar VV, Toepke MW, Cortesio CL, Lokuta MA, Huttenlocher A, Beebe DJ. Lab Chip. 2008;8:1507. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Lokuta MA, Huttenlocher A, Beebe DJ. Lab Chip. 2009;9:1797. doi: 10.1039/b901613a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T, Pinelis M, Maharbiz MM. Biomed Microdevices. 2009;11:65. doi: 10.1007/s10544-008-9210-7. [DOI] [PubMed] [Google Scholar]
- 32.Du YN, Hancock MJ, He JK, Villa-Uribe JL, Wang B, Cropek DM, Khademhosseini A. Biomaterials. 2010;31:2686. doi: 10.1016/j.biomaterials.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie FQ, Yamada M, Kobayashi J, Yamato M, Kikuchi A, Okano T. Biomaterials. 2007;28:4017. doi: 10.1016/j.biomaterials.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 34.Campbell K, Groisman A. Lab Chip. 2007;7:264. doi: 10.1039/b610011b. [DOI] [PubMed] [Google Scholar]
- 35.Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Anal Chem. 2001;73:1240. [Google Scholar]
- 36.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311. [Google Scholar]
- 37.Park JY, Hwang CM, Lee SH, Lee SH. Lab Chip. 2007;7:1673. doi: 10.1039/b710777c. [DOI] [PubMed] [Google Scholar]
- 38.Lii J, Hsu WJ, Parsa H, Das A, Rouse R, Sia SK. Anal Chem. 2008;80:3640. doi: 10.1021/ac8000034. [DOI] [PubMed] [Google Scholar]
- 39.Liu WM, Li L, Wang XM, Ren L, Wang XQ, Wang JC, Tu Q, Huang XW, Wang JY. Lab Chip. 2010;10:1717. doi: 10.1039/c001049a. [DOI] [PubMed] [Google Scholar]
- 40.Huang CP, Lu J, Seon H, Lee AP, Flanagan LA, Kim HY, Putnam AJ, Jeon NL. Lab Chip. 2009;9:1740. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keenan TM, Hsu CH, Folch A. Appl Phys Lett. 2006;89:114103. [Google Scholar]
- 42.Paguirigan AL, Beebe DJ. Integr Biol. 2009;1:182. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu HK, Huang B, Zare RN. J Am Chem Soc. 2006;128:4194. doi: 10.1021/ja058530o. [DOI] [PubMed] [Google Scholar]
- 44.Diao JP, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, Shuler ML, Wu MM, DeLisa MP. Lab Chip. 2006;6:381. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- 45.Chisti Y. Crit Rev Biotechnol. 2001;21:67. doi: 10.1080/20013891081692. [DOI] [PubMed] [Google Scholar]
- 46.Tilles AW, Baskaran H, Roy P, Yarmush ML, Toner M. Biotechnol Bioeng. 2001;73:379. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- 47.Shamloo A, Heilshorn SC. Lab Chip. 2010;10:3061. doi: 10.1039/c005069e. [DOI] [PubMed] [Google Scholar]
- 48.Kim MS, Yeon JH, Park JK. Biomed Microdevices. 2007;9:25. doi: 10.1007/s10544-006-9016-4. [DOI] [PubMed] [Google Scholar]
- 49.Mosadegh B, Huang C, Park JW, Shin HS, Chung BG, Hwang SK, Lee KH, Kim HJ, Brody J, Jeon NL. Langmuir. 2007;23:10910. doi: 10.1021/la7026835. [DOI] [PubMed] [Google Scholar]
- 50.Wong AP, Perez-Castillejos R, Love JC, Whitesides GM. Biomaterials. 2008;29:1853. doi: 10.1016/j.biomaterials.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quake SR, Scherer A. Science. 2000;290:1536. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- 52.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe DJ. Lab Chip. 2008;8:717. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 53.Khnouf R, Beebe DJ, Fan ZH. Lab Chip. 2009;9:56. doi: 10.1039/b808034h. [DOI] [PubMed] [Google Scholar]
- 54.Ma L, Zhou C, Lin B, Li W. Biomed Microdevices. 2010;12:753. doi: 10.1007/s10544-010-9429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Resto PJ, Mogen BJ, Berthier E, Williams JC. Lab Chip. 2010;10:23. doi: 10.1039/b917147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He JK, Du YA, Villa-Uribe JL, Hwang CM, Li DC, Khademhosseini A. Adv Funct Mater. 2010;20:131. doi: 10.1002/adfm.200901311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biondi M, Ungaro F, Quaglia F, Netti PA. Adv Drug Deliv Rev. 2008;60:229. doi: 10.1016/j.addr.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 58.Liechty WB, Kryscio DR, Slaughter BV, Peppas NA. Annual Review of Chemical and Biomolecular Engineering. Vol. 1. Annual Review of Chemical and Biomolecular Engineering; Palo Alto: 2010. p. 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothstein SN, Little SR. J Mater Chem. 2011;21:29. [Google Scholar]
- 60.Peret BJ, Murphy WL. Adv Funct Mater. 2008;18:3410. doi: 10.1002/adfm.200800218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen RR, Silva EA, Yuen WW, Mooney DJ. Pharm Res. 2007;24:258. doi: 10.1007/s11095-006-9173-4. [DOI] [PubMed] [Google Scholar]
- 62.Yuen WW, Du NR, Chan CH, Silva EA, Mooney DJ. Proc Natl Acad Sci USA. 2010;107:17933. doi: 10.1073/pnas.1001192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dodla MC, Bellamkonda RV. Biomaterials. 2008;29:33. doi: 10.1016/j.biomaterials.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang XQ, Wenk E, Zhang XH, Meinel L, Vunjak-Novakovic G, Kaplan DL. J Control Release. 2009;134:81. doi: 10.1016/j.jconrel.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schultz GS, Wysocki A. Wound Repair Regen. 2009;17:153. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 66.Yamashita YM, Fuller MT, Jones DL. J Cell Sci. 2005;118:665. doi: 10.1242/jcs.01680. [DOI] [PubMed] [Google Scholar]
- 67.Wilder EL, Perrimon N. Development. 1995;121:477. doi: 10.1242/dev.121.2.477. [DOI] [PubMed] [Google Scholar]
- 68.Benoit DSW, Anseth KS. Acta Biomater. 2005;1:461. doi: 10.1016/j.actbio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 69.Rydholm AE, Held NL, Benoit DSW, Bowman CN, Anseth KS. J Biomed Mater Res. 2008;86A:23. doi: 10.1002/jbm.a.31526. [DOI] [PubMed] [Google Scholar]
- 70.Tsurkan M, Levental K, Freudenberg U, Werner C. Chem Commun. 2010;46:1141. doi: 10.1039/b921616b. [DOI] [PubMed] [Google Scholar]
- 71.Zieris A, Prokoph S, Welzel P, Grimmer M, Levental K, Panyanuwat W, Freudenberg U, Werner C. J Mater Sci Mater Med. 2010;21:915. doi: 10.1007/s10856-009-3913-z. [DOI] [PubMed] [Google Scholar]
- 72.Baldwin A, Kiick K. Biopolymers. 2010;94:128. doi: 10.1002/bip.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freudenberg U, Hermann A, Welzel PB, Stirl K, Schwarz SC, Grimmer M, Zieris A, Panyanuwat W, Zschoche S, Meinhold D, Storch A, Werner C. Biomaterials. 2009;30:5049. doi: 10.1016/j.biomaterials.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Nie T, Akins RE, Kiick KL. Acta Biomater. 2009;5:865. doi: 10.1016/j.actbio.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin CC, Metters AT. Biomacromolecules. 2008;9:789. doi: 10.1021/bm700940w. [DOI] [PubMed] [Google Scholar]
- 76.Spinelli F, Kiick K, Furst E. Biomaterials. 2008;29:1299. doi: 10.1016/j.biomaterials.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai SS, Liu YC, Shu XZ, Prestwich GD. Biomaterials. 2005;26:6054. doi: 10.1016/j.biomaterials.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 78.Yamaguchi N, Chae BS, Zhang L, Kiick KL, Furst EM. Biomacromolecules. 2005;6:1931. doi: 10.1021/bm0500042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wood M, Hunter D, Mackinnon S, Sakiyama-Elbert S. J Biomater Sci Polym Ed. 2010;21:771. doi: 10.1163/156856209X445285. [DOI] [PubMed] [Google Scholar]
- 80.Luhmann T, Hanseler P, Grant B, Hall H. Biomaterials. 2009;30:4503. doi: 10.1016/j.biomaterials.2009.05.041. [DOI] [PubMed] [Google Scholar]
- 81.Hahn MS, Miller JS, West JL. Adv Mater. 2006;18:2679. [Google Scholar]
- 82.Polizzotti BD, Fairbanks BD, Anseth KS. Biomacromolecules. 2008;9:1084. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 83.Lee SH, Moon JJ, West JL. Biomaterials. 2008;29:2962. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv Mater. 2009;21:5005. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoffmann JC, West JL. Soft Matter. 2010;6:5056. [Google Scholar]
- 87.DeForest CA, Sims EA, Anseth KS. Chem Mater. 2010;22:4783. doi: 10.1021/cm101391y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 89.Gillitzer R, Goebeler M. J Leukoc Biol. 2001;69:513. [PubMed] [Google Scholar]
- 90.Werner S, Grose R. Physiol Rev. 2003;83:835. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 91.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Nature. 2008;453:314. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 92.Lowman AM, Morishita M, Kajita M, Nagai T, Peppas NA. J Pharm Sci. 1999;88:933. doi: 10.1021/js980337n. [DOI] [PubMed] [Google Scholar]
- 93.Gupta KC, Ravi Kumar MNV. Polym Int. 2000;49:141. [Google Scholar]
- 94.Shi J, Liu LH, Sun XM, Cao SK, Mano JF. Macromol Biosci. 2008;8:260. doi: 10.1002/mabi.200700177. [DOI] [PubMed] [Google Scholar]
- 95.Ishihara K, Kobayashi M, Ishimaru N, Shinohara I. Polym J. 1984;16:625. [Google Scholar]
- 96.Bae YH, Okano T, Kim SW. J Polym Sci, Part B Polym Phys. 1990;28:923. [Google Scholar]
- 97.Okano T, Bae YH, Jacobs H, Kim SW. J Control Release. 1990;11:255. [Google Scholar]
- 98.Bae YH, Okano T, Kim SW. J Control Release. 1989;9:271. [Google Scholar]
- 99.Bae YH, Okano T, Hsu R, Kim SW. Makromol Chem Rapid Commun. 1987;8:481. [Google Scholar]
- 100.Alexandridis P, Holzwarth JF, Hatton TA. Macromolecules. 1994;27:2414. [Google Scholar]
- 101.Satarkar NS, Hilt JZ. J Control Release. 2008;130:246. doi: 10.1016/j.jconrel.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 102.Saslawski O, Weingarten C, Benoit JP, Couvreur P. Life Sci. 1988;42:1521. doi: 10.1016/0024-3205(88)90009-4. [DOI] [PubMed] [Google Scholar]
- 103.Lu ZH, Prouty MD, Guo ZH, Golub VO, Kumar C, Lvov YM. Langmuir. 2005;21:2042. doi: 10.1021/la047629q. [DOI] [PubMed] [Google Scholar]
- 104.Kooiman K, Bohmer MR, Emmer M, Vos HJ, Chlon C, Shi WT, Hall CS, de Winter S, Schroen K, Versluis M, de Jong N, van Wamel A. J Control Release. 2009;133:109. doi: 10.1016/j.jconrel.2008.09.085. [DOI] [PubMed] [Google Scholar]
- 105.Kwon IC, Bae YH, Kim SW. Nature. 1991;354:291. doi: 10.1038/354291a0. [DOI] [PubMed] [Google Scholar]
- 106.Yuk SH, Cho SH, Lee HB. Pharm Res. 1992;9:955. doi: 10.1023/a:1015821504229. [DOI] [PubMed] [Google Scholar]
- 107.Whiting CJ, Voice AM, Olmsted PD, McLeish TCB. J Phys Condens Matter. 2001;13:1381. [Google Scholar]
- 108.Liu X, Pettway GJ, McCauley LK, Ma PX. Biomaterials. 2007;28:4124. doi: 10.1016/j.biomaterials.2007.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.King WJ, Mohammed JS, Murphy WL. Soft Matter. 2009;5:2399. [Google Scholar]
- 110.King WJ, Toepke MW, Murphy WL. Acta Biomater. doi: 10.1016/j.actbio.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.King W, Murphy W. Polym Chem. doi: 10.1039/c0py00244e. [DOI] [Google Scholar]
- 112.Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmokel H, Bezuidenhout D, Djonov V, Zilla P, Hubbell JA. FASEB J. 2003;17:2260. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 113.Tokatlian T, Shrum CT, Kadoya WM, Segura T. Biomaterials. 2010;31:8072. doi: 10.1016/j.biomaterials.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aimetti AA, Tibbitt MW, Anseth KS. Biomacromolecules. 2009;10:1484. doi: 10.1021/bm9000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Aimetti AA, Machen AJ, Anseth KS. Biomaterials. 2009;30:6048. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nagase H, Fields GB. Biopolymers. 1996;40:399. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 117.Metters AT, Anseth KS, Bowman CN. Polymer. 2000;41:3993. [Google Scholar]
- 118.Burdick JA, Philpott LM, Anseth KS. J Polym Sci, Part A Polym Chem. 2001;39:683. [Google Scholar]
- 119.Gobin AS, West JL. FASEB J. 2002;16:751. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 120.West JL, Hubbell JA. Macromolecules. 1999;32:241. [Google Scholar]
- 121.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proc Natl Acad Sci USA. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hudalla GA, Eng TS, Murphy WL. Biomacromolecules. 2008;9:842. doi: 10.1021/bm701179s. [DOI] [PubMed] [Google Scholar]
- 123.Lan-Levengood S, Murphy WL. Curr Stem Cell Res Ther. 2010;5:261. doi: 10.2174/157488810791824557. [DOI] [PubMed] [Google Scholar]
- 124.King WJ, Jongpaiboonkit L, Murphy WL. J Biomed Mater Res. 2010;93A:1110. doi: 10.1002/jbm.a.32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jongpaiboonkit L, King WJ, Murphy WL. Tissue Eng A. 2009;15:343. doi: 10.1089/ten.tea.2008.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jongpaiboonkit L, King WJ, Lyons GE, Paguirigan AL, Warrick JW, Beebe DJ, Murphy WL. Biomaterials. 2008;29:3346. doi: 10.1016/j.biomaterials.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Lab Chip. 2005;5:111. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 128.Koh WG, Itle LJ, Pishko MV. Anal Chem. 2003;75:5783. doi: 10.1021/ac034773s. [DOI] [PubMed] [Google Scholar]
- 129.Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, Khademhosseini A. Lab Chip. 2007;7:756. doi: 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]