Abstract
The mechanisms of neuronal differentiation in PC12 cells are still not completely understood. Here, we report that the tumor suppressor PTEN has a profound effect on differentiation by affecting several pathways involved in nerve growth factor (NGF) signaling. When overexpressed in PC12 cells, PTEN (phosphatase and tensin homologue deleted on chromosome ten) blocked neurite outgrowth induced by NGF. In addition, these cells failed to demonstrate the transient mitogenic response to NGF, as well as subsequent growth arrest. Consistent with these observations was a finding that PTEN significantly inhibits NGF-mediated activation of the members of mitogen-activated protein kinase kinase (MEK)/mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K)/AKT signaling pathways, crucial for these processes. While exploring possible mechanisms of PTEN effects on NGF signaling, we discovered a significant down-regulation of both high-affinity (TrkA) and low-affinity (p75) NGF receptors in PTEN-overexpressing clones. Subsequent microarray analysis of several independent clonal isolates revealed a myriad of neuronal genes to be affected by PTEN. All of these changes were validated by quantitative PCR. Of particular interest were the genes for the key enzymes of the dopamine synthesis pathway, receptors for different neurotransmitters, and neuron-specific cytoskeleton proteins, among others. Some, but not all effects could be reproduced by pharmacological inhibitors of PI3K and/or MAPK, suggesting that PTEN may influence some genes by mechanisms independent of these signaling pathways. Our findings may shed new light on the role of this tumor suppressor during normal brain development and suggest a previously uncharacterized mechanism of PTEN action in neuron-like cells.
The PTEN anti-oncogene (phosphatase and tensin homologue deleted on chromosome ten) is among the most frequently mutated genes in high-grade brain tumors, and loss of PTEN is found in a variety of other malignancies (1, 2). Most studies of PTEN function have focused on its lipid-phosphatase activity, which results in inhibition of the phosphoinositide 3-kinase (PI3K) pathway (3-5). PTEN also possesses a protein phosphatase activity (1), but few clear in vivo targets of this function have been identified (6, 7). Inhibition of mitogen-activated protein kinase (MAPK) signaling is another action of PTEN, which at least in part seems to be independent of any effect on PI3K (7, 8). PTEN can influence cellular gene expression (9, 10); many of these changes are reproduced with pharmacological inhibition of the PI3K pathway and are believed to be secondary to cytoplasmic lipid signaling (11). Nonetheless, PTEN is found in the nucleus of most cells (12-14), and a recent study demonstrated that PTEN can bind directly to the p53 anti-oncogene and increase its transcriptional activity independent of lipid-phosphatase signaling (15). Whereas antagonism of the PI3K pathway is clearly a major function of PTEN, these studies indicate that PTEN can have several physiologically significant cellular functions, at least some of which are independent of this mechanism.
Recently, there has been increased interest in the role of PTEN in normal cellular function, particularly neurons. PTEN is present in both developing and adult neurons although expression is quite low during development and seems to be up-regulated at or near birth (14). Loss of PTEN expression is associated with cerebellar dysplasia in human patients (16), and conditional knockout mouse studies have also demonstrated dysplasia and migrational defects (17, 18). PTEN deletion does not seem to prevent neuronal differentiation, however (18). Neurons from animals hemizygous for PTEN also seem to have increased resistance to peroxide toxicity, which is believed to be secondary to increased PI3K signaling and AKT activity in these cells (19). These studies highlight an important role for PTEN in neuronal development and possibly for neuronal survival.
To better understand the role of PTEN function in neurons, we generated stable PC12 rat pheochromocytoma cell lines overexpressing this gene. Nerve growth factor (NGF) normally induces differentiation of PC12 cells into a neuron-like phenotype (20). Although previous studies showed no effect of PTEN loss on neurite outgrowth (14), we found that PTEN overexpression unexpectedly rendered PC12 cells completely resistant to differentiation by NGF. We now report that this phenomenon seems to be due to inhibition of expression of both the high- and low-affinity NGF receptors. Further exploration of this phenomenon revealed that PTEN can induce profound changes in expression of genes in undifferentiated cells that help define the neuronal phenotype. Pharmacological inhibition of PI3K and/or MAPK signaling alone does not recapitulate several of these effects, suggesting that the mechanism of PTEN action on neuronal gene expression may involve other pathways. This finding suggests a mechanism whereby PTEN may influence neuronal differentiation. Preliminary findings have been reported in an abstract at a Society for Neuroscience annual meeting.
Methods
Plasmids and Cell Lines. A full-length human PTEN cDNA (a gift from A. Yung, M. D. Anderson Cancer Center, Houston) was cloned into pRep7 episomal expression vector (Invitrogen) to generate pRep7.PTEN. pRep7.PTEN or pRep7 (control) were transfected into PC12 cells by using FuGENE6 (Roche Molecular Biochemicals). Four weeks after hygromycin selection (250 μg/ml), individual clones were expanded and analyzed for PTEN expression. Four individual PTEN-overexpressing cell lines (P1-P4), as well as four control lines (C1-C4), were used for the experiments in this study.
Cell Culture and Treatments. PC12 cells were maintained in DMEM supplemented with 10% FBS and antibiotics at 37°C under 5% CO2. PI3K inhibitor LY294002 (21) and MAPK kinase (MEK) 1/2 inhibitor U0126 (22) were from Cell Signaling Technology (Beverly, MA), were used at 10 μM final concentration, and were replenished every 24 h during a 72-h treatment period. Butyrolactone I (Calbiochem) was used at 30 μM (23).
Cell Differentiation and Proliferation Assays. Cells (in 24-well plates, 104 cells per well) were treated with 100 ng/ml NGF (Sigma) in DMEM supplemented with 1% FBS. Cells were incubated for 7 days with NGF replenished every 72 h, and neurite-bearing cells were counted. Only cells with processes equivalent in length to at least two cell diameters were scored as differentiated. For the proliferation assay, cells were treated with 100 ng/ml NGF in DMEM with 1% FBS, and viable cells were counted at different time points by using trypan blue staining. For a baseline, control cells were maintained in the presence of 1% FBS without NGF. No loss of cell viability was detected under this condition. All of the experiments were performed in triplicate.
Western Blotting. Cells were incubated in DMEM with 0.5% FBS for 24 h and followed by 2-h incubation with serum-free DMEM. Cells were treated with 100 ng/ml NGF for 5 and 30 min, washed with ice-cold PBS, and then lysed in lysis buffer (20 mM Tris, pH 8.0/1% Triton X-100/150 mM NaCl/protein and phosphatase inhibitors) (Sigma). Samples were cleared by centrifugation, and equal amounts of proteins (15-30 μg) were denatured in a sample buffer and resolved on a NuPAGE 10% Bistris {[bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane} gel (Invitrogen). Blots were incubated with various primary antibodies (Cell Signaling) followed by secondary antibodies and enhanced chemiluminescent detection (Amersham Pharmacia Biosciences).
Microarray Analysis and Quantitative PCR. For microarray analysis, cells were grown in DMEM supplemented with 10% FBS for 48 h. Total RNA was extracted by using RNeasy kit (Qiagen, Valencia, CA), and RNA quality was assessed by electrophoresis by using the Agilent Bioanalyzer 2100 and spectrophotomeric analysis before cDNA synthesis. All samples were subjected to gene expression analysis with the Affymetrix (Santa Clara, CA) RGU34A high-density oligonucleotide array, which queries 8,000 rat probe sets. The assay was performed at the Microarray Core Facility of the University of Rochester. Three control (C1-C3) and three PTEN-overexpressing (P1-P3) cell lines were processed independently. Data were analyzed by using
microarray analysis suite 5.0 (Affymetrix). Only gene changes with P value < 0.05 were chosen for validation. Real-time quantitative PCR (Q-PCR) was performed by using SYBR Green Master Mix (Applied Biosystems) in an ABI Prism 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's protocols. Before comparative analysis, a validation experiment was performed to determine the relative efficiency of the assay with a primer set for a gene that does not change in the experimental model (GAPDH). This gene was selected based on its behavior in the microarray data set as described (24) and was used as a reference gene for comparative analysis. All reactions were performed in triplicate, and each experiment was replicated at least twice. Data were analyzed by using the sequence detection software (Applied Biosystems).
Results
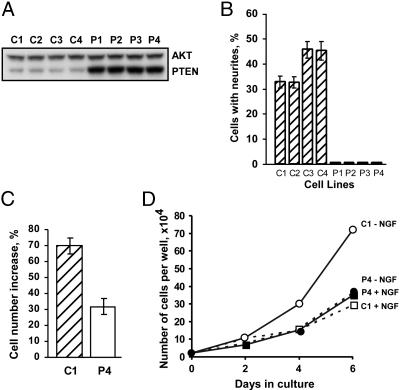
Response of PC12 Cells Overexpressing PTEN to NGF Treatment. To examine the effects of PTEN on differentiated PC12 cells, we generated a series of stable clones overexpressing PTEN (P1-P4) and related controls (C1-C4). As evident from Fig. 1A, in P1-P4 lines PTEN was overexpressed on average 4.2-fold, and its levels were similar among different clonal isolates. We first examined the effects of PTEN on NGF-induced neurite outgrowth. Surprisingly, no processes were observed in any PTEN-overexpressing clones after NGF treatment (Fig. 1B). Higher does of NGF (up to 400 ng/ml) failed to overcome this block (data not shown). In addition to promoting neurite outgrowth, NGF also has a transient mitogenic effect but then induces long-term growth arrest (20, 25). As shown in Fig. 1C, PTEN significantly decreased cell proliferation in a 48-h period during NGF treatment. Although the data are presented only for two cell lines (C1 and P4), this experiment was repeated on other clones as well with similar results. Beyond 48 h, no decrease in cell division was observed whereas control cells exhibited marked inhibition of growth as expected (Fig. 1D). Because growth arrest is an essential event for PC12 differentiation, it is conceivable that the effect of PTEN-overexpressing cells to stop proliferating is responsible for the inability to differentiate. To address this issue, the cells were treated with butyrolactone I, an inhibitor of cdc2 and cdk2 kinases (26). Although this compound blocked cell division without significantly affecting neurite outgrowth of the control cells, the PTEN-overproducing cells did not regain the ability to differentiate (data not shown). This result confirmed that PTEN blocked several cellular responses to NGF independently.
Fig. 1.
Effects of PTEN on NGF responses in PC12 cells. (A) Western blot analysis of PTEN levels in control (subclones C1-C4) and PTEN-overexpressing (subclones P1-P4) cells. Staining for total AKT was included as a loading control. (B) Effect of PTEN on NGF-induced neurite outgrowth in all of the isolated subclones. (C) Inhibition by PTEN of the transient mitogenic effect of NGF. (D) Effect of PTEN on NGF-induced growth arrest. For the experiments in B and C, clones C1 and P4 were selected.
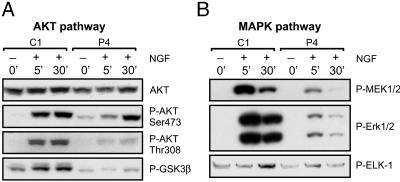
Negative Regulation by PTEN of NGF-Mediated Activation of PI3K/AKT and MEK/MAPK Pathways. PTEN is known to negatively regulate the PI3K/AKT as well as MEK/MAPK pathways (4, 7, 27, 28). Therefore, we investigated the effects of PTEN on these signal transduction cascades in PC12 cells. Serum-starved cells were treated with NGF (100 ng/ml) for the indicated periods, and the activation of the several key members of pathways was then analyzed by Western blotting. As evident from Fig. 2A, PTEN significantly inhibited accumulation of AKT phosphorylated at both Ser-473 and Thr-308 sites after NGF treatment. Consistent with this finding was the observation that phosphorylation of the AKT substrate, glycogen synthase kinase-3β (GSK3β), was inhibited as well. Similarly, PTEN suppressed activation of the MEK/MAPK cascade (Fig. 2B).
Fig. 2.
Negative regulation of NGF signaling by PTEN. PC12 cells (C1 and P4) were treated with NGF, and activation of members of the PI3/AKT (A) and MEK/MAPK (B) signal transduction cascades were analyzed by Western blotting. At all time points, NGF induction of PI3K and MAPK signaling was inhibited by PTEN.
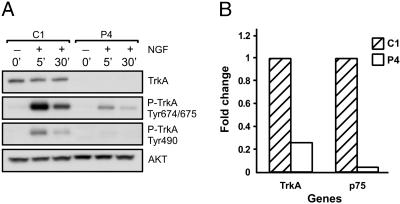
Negative Effect of PTEN on TrkA and p75 Levels. To further elucidate the mechanisms of PTEN effects on NGF signaling, we analyzed the early events of this pathway activation, i.e., autophosphorylation of a high-affinity NGF receptor TrkA at two critical sites. Tyr-674/675 residues lie within the catalytic tyrosine kinase domain and are phosphorylated after NGF binding and receptor dimerization. This event is followed by autophosphorylation at Tyr-490, which is required for Shc association and activation of the MAPK cascade (29). Surprisingly, whereas the intensity of the signal corresponding to activated TrkA was significantly decreased in PTEN-overexpressing cells, the total amount of this receptor was reduced as well (Fig. 3A). Q-PCR analysis demonstrated that PTEN inhibited TrkA expression at the mRNA level. (Fig. 3B). In addition, we analyzed the expression of p75, a low-affinity NGF receptor. Interestingly, the mRNA level for this receptor was also significantly suppressed by PTEN (Fig. 3B). These alterations were consistent among several independent clonal isolates (Table 1). Together, these findings demonstrate that the negative effects of PTEN on NGF signaling are primarily due to down-regulation of NGF receptors expression. However, additional mechanisms of suppression of PI3K/AKT pathway mediated by lipid phosphatase activity of PTEN and predicted by the accepted model of PTEN action can also be involved.
Fig. 3.
Negative regulation of NGF receptor expression by PTEN. (A) PC12 cells (C1 and P4) were treated with NGF for the indicated amount of time, and TrkA activation was analyzed by Western blotting by using antibodies specific for Tyr-674/675 and Tyr-490. Total TrkA as well as AKT (loading control) were also included. Note that reduction in phosphorylation correlated directly with overall reduction in total TrkA protein expression. (B) Suppression of TrkA and p75 mRNA levels in P4 cells as determined by Q-PCR.
Table 1. Effects of PTEN on gene expression in PC12 cells.
| Accession no.
|
Fold change
|
||||
|---|---|---|---|---|---|
| Gene name | Microarray | Q-PCR | P value* | Description | |
| Tyrosine hydroxylase | M10244 | −3.9 | −104.2 | 4 × 10−4 | Key dopamine synthesis enzyme |
| GTP cyclohydrolase I | M58364 | −5.1 | −7.1 | 0.025 | Key dopamine synthesis enzyme |
| Trk A | M85214 | −2.5 | −4.4 | 0.035 | High-affinity NGF receptor |
| P75 | X05137 | −4.7 | −18.9 | 0.039 | Low-affinity NGF receptor |
| 5-HT3 receptor | D49395 | −3.0 | −530.2 | 3 × 10−5 | Serotonin receptor |
| GABA(A) receptor β-3 subunit | X15468 | −17.3 | −182.8 | 3 × 10−5 | Part of the GABA receptor complex |
| PACAP receptor | D14909 | −3.6 | −666.7 | 3 × 10−5 | Receptor for pituitary adenylate cyclase-activating polypeptide |
| Bone morphogenetic protein 7 | D29769 | −24.2 | −57.8 | 10−4 | Multiple roles in neural development and plasticity |
| SHIP | U55192 | 0.92 | 81.9 | 7 × 10−3 | Src homology 2 domain-containing inositol 5′-phosphatase |
| Fez1 | U63740 | −60.56 | −380.7 | 2 × 10−4 | Regulation of axonal outgrowth |
| SNAP-25 | AB003991 | −13.2 | −38.5 | 6 × 10−5 | Regulation of exocytosis in neurons |
| SNAP-23 | AF052596 | 7.37 | 2.9 | 0.019 | Regulation of exocytosis, non-neuronal homologue of SNAP-25 |
P values for each gene were determined by two-tail t test comparing all four control (C1-C4) and PTEN (P1-P4) cell lines.
Inhibition of Neuronal Gene Expression by PTEN. To more fully explore the effects of PTEN on gene expression in PC12 cells, we performed microarray analysis of mRNA expression profiles in three control (C1-C3) and PTEN-overexpressing (P1-P3) clones. Nearly 8,000 rat genes were simultaneously tested, and >100 genes were found to be significantly affected by PTEN. Approximately two-thirds of these affected genes were inhibited, whereas one-third were induced. Many of the inhibited genes are markers of neuronal phenotype, and several individual gene changes were verified on all four control (C1-C4) and PTEN (P1-P4) clonal isolates by using Q-PCR and, in some instances, Western blotting as well (Table 1). Among the genes significantly down-regulated in PTEN overexpressing cells are the genes for two of the three key enzymes necessary for dopamine synthesis (tyrosine hydroxylase and GTP cyclohydrolase I), neurotransmitter receptors [serotonin, γ-aminobutyric acid (GABA), and pituitary adenylate cyclase-activating polypeptide (PACAP) receptors], as well as a neuron-specific cytoskeleton protein [synaptosome-associated protein (SNAP)-25]. This gene is of particular interest because the nonneuronal homologue, SNAP-23, was simultaneously induced (Table 1). These combined data suggest that PTEN was altering gene expression in PC12 cells toward a less neuronal phenotype.
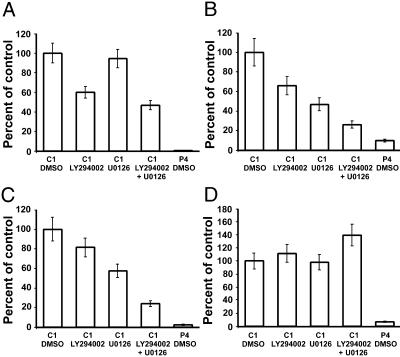
Inhibitors of the PI3K and MAPK Pathways Only Partially Mimic PTEN Effects. In PC12 cells, PTEN seems to oppose at least two signal transduction cascades, PI3K/AKT and MEK/MAPK (Fig. 2). To further understand the contributions of these two pathways to the effects of PTEN on gene expression, control PC12 cells were treated for 72 h with PI3K (LY294002) and MEK (U0126) inhibitors, and expression of selected genes was analyzed by Q-PCR. Interestingly, three different groups of genes were observed (Fig. 4). Group I genes, e.g., tyrosine hydroxylase (Fig. 4A), were down-regulated exclusively by LY294002 compound, and no effect of the MEK inhibitor was observed. Although this result is similar to previously reported effects of PTEN on gene expression, the inhibitory effect was never as profound as PTEN itself. Group II genes, e.g., GTP cyclohydrolase I and p75 (Fig. 4 B and C), were suppressed by both PI3K and MEK inhibitors, and treatment with both compounds simultaneously had an additive effect. Unlike group I genes, the additive effect of PI3K and MAPK inhibition approached the magnitude of the PTEN effect on group II genes. Finally, the genes of group III, e.g., TrkA (Fig. 4D), were not affected by either drug. Together, these results suggest that PTEN possesses several mechanisms by means of which it can modulate gene expression, some of which seem to be independent of known effects on signal transduction.
Fig. 4.
Effects of pharmacological suppression of PI3/AKT and MEK/MAPK on the expression of selected genes. Control (C1) cells were treated with the indicated inhibitors separately or in combination, and the expression was analyzed by using Q-PCR 72 h posttreatment. P4 cells treated with the solvent (DMSO) were also included as a positive control. The selected genes were as follows: tyrosine hydroxylase (A), GTP cyclohydrolase I (B), p75 (C), and TrkA (D). The error bars represent the SE for two to three independent experiments for each gene.
Discussion
In this study, we have examined the effects of PTEN on neuronal phenotype in PC12 cells. To date, the few reported studies of PTEN in neurons have examined the consequences of PTEN deletion on neuronal differentiation (14, 18, 30). These studies have consistently found that PTEN deletions do not prevent neuronal differentiation but can alter neuronal morphology, proliferation, and migration. PTEN is poorly expressed in undifferentiated neurons and PC12 cells, yet expression is substantially up-regulated near birth or after neurotrophin-induced differentiation in culture (14). This finding suggests that PTEN induction mediates one or more functions important for either differentiation and/or adult neuronal function. We thus elected to examine the role of elevated rather than reduced levels of PTEN in undifferentiated cells, to examine the effects of postmitotic PTEN levels on cellular function.
Our data suggest that elevated PTEN levels in undifferentiated PC12 cells inhibited expression of several genes that define neuronal phenotype. In particular, both NGF receptors (TrkA and p75) were significantly inhibited, which explains resistance of PTEN-overexpressing cells to NGF treatment. Similarly, levels of tyrosine hydroxylase and GTP cyclohydrolase I, which are two of the three key enzymes in dopamine synthesis, were also reduced. It was particularly striking that a neuron-specific protein, SNAP-25, was significantly inhibited whereas its nonneuronal homologue SNAP-23 was up-regulated. Previous studies have demonstrated that low levels of PTEN expression in undifferentiated cells are necessary to promote migration and tissue development, yet loss of PTEN does not prevent differentiation (18). Although our cell lines averaged 4.2-fold overexpression of PTEN compared with endogenous levels in undifferentiated cells, this difference is comparable with the level of PTEN found in cells differentiated with NGF (14). Although our data may also be specific only to PC12 cells, this result nonetheless raises the possibility that timing and level of PTEN expression in undifferentiated cells must be tightly regulated to promote normal migration and organ development yet permit differentiation as well.
Our original hypothesis predicted that expression of PTEN in undifferentiated cells at postmitotic levels would inhibit the initiation of differentiation by opposing PI3K signaling. Increased PI3K signaling is a hallmark of neurotrophin receptor activation so inhibition of this pathway by PTEN would be expected to block second messengers, which are important for proper differentiation. In fact, it has previously been shown that the exposure of PC12 cells to the PI3K inhibitor wortmannin or expression of a dominant-negative PI3K subunit inhibited neurite outgrowth (31). We were surprised to discover, however, that the consequences of PTEN overexpression are far more complex and cannot be explained by mere suppression of the PI3K pathway. This point is best illustrated by comparison of PTEN effects with those of drug inhibitors of specific second messenger pathways. It seems that some changes induced by PTEN can be replicated by blocking PI3K/AKT and/or MEK/MAPK signaling. However, PTEN effects on other genes either cannot be recapitulated at all by using the specific inhibitors of these pathways or the degree of suppression is not as profound as in PTEN-overexpressing cells. This suggests that other, yet unknown mechanisms may mediate effects of PTEN on gene expression in these cells. Alternatively, some of these differences represent acute vs. chronic changes because the pharmacological inhibitors were applied for only 72 h. Although we believe that those changes not seen with drug inhibitors are due to other mechanisms, it is possible that more prolonged inhibition of PI3K/AKT and/or MEK/MAPK pathways would eventually reproduce all of our findings.
PTEN has several known functions independent of lipid signaling. These effects may provide clues as to possible novel mechanisms of PTEN action on gene expression in our system. For example, the C124S PTEN mutant, which lacks both lipid and protein phosphatase activity, suppressed glioblastoma U87-MG invasion with equal efficiencies as wild-type protein (32). In another study, this mutant inhibited SHEP neuroblastoma cell proliferation (33), a finding that cannot be explained by the current model of PTEN signaling.
Negative regulation of PI3K/AKT cascade by PTEN is generally believed to be due to localization of this protein at the plasma membrane where early events of lipid signaling occur. However, PTEN is also found in the nucleus of most cells. In fact, the loss of nuclear but not cytoplasmic PTEN seems to correlate with the progression of certain tumors, such as thyroid carcinomas (12), endocrine pancreatic tumors (13), esophageal squamous cell carcinoma (34), and melanoma (35). Freeman et al. (15) have shown that PTEN physically associates with and activates the p53 anti-oncogene. Because p53 is a transcription factor, direct binding of PTEN to p53 to potentiate its activity raises the possibility of interactions with other nuclear transcription factors. It seems, therefore, that PTEN may have functions that are independent from its lipid phosphatase activity at the plasma membrane.
Our study suggests that premature induction of PTEN expression has a profound negative effect on development of the neuronal phenotype in PC12 cells. This finding may shed new light on the role of this tumor suppressor in the normal development of neural tissue. It remains unclear whether PTEN levels fluctuate during neural development so it is unclear whether PTEN physiologically regulates differentiation. Nonetheless, our study highlights a previously uncharacterized mechanism of action of PTEN at postmitotic, physiological levels in undifferentiated cells.
Acknowledgments
We thank Charles Mobbs for the gift of PC12 cells and helpful discussions. This work was supported by awards from the Ellison Foundation for Medical Research and the New York Academy of Medicine (to M.G.K.).
A preliminary report of this work was presented at the 32nd Annual Meeting of the Society for Neuroscience, Orlando, FL, November 2-7, 2002 (abstr. 631.18).
Abbreviations: PTEN, phosphatase and tensin homologue deleted on chromosome ten; PI3K, phosphoinositide 3-kinase; MAPK, mitogen-activated protein kinase; NGF, nerve growth factor; MEK, MAPK kinase; Q-PCR, quantitative PCR; SNAP, synaptosome-associated protein.
References
- 1.Li, J., Yen, C., Liaw, D., Podsypanina, K., Bose, S., Wang, S. I., Puc, J., Miliaresis, C., Rodgers, L., McCombie, R., et al. (1997) Science 275, 1943-1947. [DOI] [PubMed] [Google Scholar]
- 2.Knobbe, C. B., Merlo, A. & Reifenberger, G. (2002) Neuro-oncol. 4, 196-211. [PMC free article] [PubMed] [Google Scholar]
- 3.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375-13378. [DOI] [PubMed] [Google Scholar]
- 4.Stambolic, V., Suzuki, A., de la Pompa, J. L., Brothers, G. M., Mirtsos, C., Sasaki, T., Ruland, J., Penninger, J. M., Siderovski, D. P. & Mak, T. W. (1998) Cell 95, 29-39. [DOI] [PubMed] [Google Scholar]
- 5.Myers, M. P., Pass, I., Batty, I. H., Van der Kaay, J., Stolarov, J. P., Hemmings, B. A., Wigler, M. H., Downes, C. P. & Tonks, N. K. (1998) Proc. Natl. Acad. Sci. USA 95, 13513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura, M., Gu, J., Matsumoto, K., Aota, S., Parsons, R. & Yamada, K. M. (1998) Science 280, 1614-1617. [DOI] [PubMed] [Google Scholar]
- 7.Gu, J., Tamura, M. & Yamada, K. M. (1998) J. Cell Biol. 143, 1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng, L. P., Brown, J. L., Baker, K. M., Ostrowski, M. C. & Eng, C. (2002) Hum. Mol. Genet. 11, 1687-1696. [DOI] [PubMed] [Google Scholar]
- 9.Hong, T. M., Yang, P. C., Peck, K., Chen, J. J., Yang, S. C., Chen, Y. C. & Wu, C. W. (2000) Am. J. Respir. Cell Mol. Biol. 23, 355-363. [DOI] [PubMed] [Google Scholar]
- 10.Matsushima-Nishiu, M., Unoki, M., Ono, K., Tsunoda, T., Minaguchi, T., Kuramoto, H., Nishida, M., Satoh, T., Tanaka, T. & Nakamura, Y. (2001) Cancer Res. 61, 3741-3749. [PubMed] [Google Scholar]
- 11.Stolarov, J., Chang, K., Reiner, A., Rodgers, L., Hannon, G. J., Wigler, M. H. & Mittal, V. (2001) Proc. Natl. Acad. Sci. USA 98, 13043-13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimm, O., Perren, A., Weng, L. P., Marsh, D. J., Yeh, J. J., Ziebold, U., Gil, E., Hinze, R., Delbridge, L., Lees, J. A., et al. (2000) Am. J. Pathol. 156, 1693-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perren, A., Komminoth, P., Saremaslani, P., Matter, C., Feurer, S., Lees, J. A., Heitz, P. U. & Eng, C. (2000) Am. J. Pathol. 157, 1097-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachyankar, M. B., Sultana, N., Schonhoff, C. M., Mitra, P., Poluha, W., Lambert, S., Quesenberry, P. J., Litofsky, N. S., Recht, L. D., Nabi, R., et al. (2000) J. Neurosci. 20, 1404-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman, D. J., Li, A. G., Wei, G., Li, H. H., Kertesz, N., Lesche, R., Whale, A. D., Martinez-Diaz, H., Rozengurt, N., Cardiff, R. D., et al. (2003) Cancer Cell 3, 117-130. [DOI] [PubMed] [Google Scholar]
- 16.Liaw, D., Marsh, D. J., Li, J., Dahia, P. L., Wang, S. I., Zheng, Z., Bose, S., Call, K. M., Tsou, H. C., Peacocke, M., et al. (1997) Nat. Genet. 16, 64-67. [DOI] [PubMed] [Google Scholar]
- 17.Backman, S. A., Stambolic, V., Suzuki, A., Haight, J., Elia, A., Pretorius, J., Tsao, M. S., Shannon, P., Bolon, B., Ivy, G. O. & Mak, T. W. (2001) Nat. Genet. 29, 396-403. [DOI] [PubMed] [Google Scholar]
- 18.Marino, S., Krimpenfort, P., Leung, C., van der Korput, H. A., Trapman, J., Camenisch, I., Berns, A. & Brandner, S. (2002) Development (Cambridge, U.K.) 129, 3513-3522. [DOI] [PubMed] [Google Scholar]
- 19.Li, L., Liu, F., Salmonsen, R. A., Turner, T. K., Litofsky, N. S., Di Cristofano, A., Pandolfi, P. P., Jones, S. N., Recht, L. D. & Ross, A. H. (2002) Mol. Cell. Neurosci. 20, 21-29. [DOI] [PubMed] [Google Scholar]
- 20.Greene, L. A. & Tischler, A. S. (1976) Proc. Natl. Acad. Sci. USA 73, 2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlahos, C. J., Matter, W. F., Hui, K. Y. & Brown, R. F. (1994) J. Biol. Chem. 269, 5241-5248. [PubMed] [Google Scholar]
- 22.Favata, M. F., Horiuchi, K. Y., Manos, E. J., Daulerio, A. J., Stradley, D. A., Feeser, W. S., Van Dyk, D. E., Pitts, W. J., Earl, R. A., Hobbs, F., et al. (1998) J. Biol. Chem. 273, 18623-18632. [DOI] [PubMed] [Google Scholar]
- 23.Bang, O. S., Park, E. K., Yang, S. I., Lee, S. R., Franke, T. F. & Kang, S. S. (2001) J. Cell Sci. 114, 81-88. [DOI] [PubMed] [Google Scholar]
- 24.Brooks, A. I., Stein, C. S., Hughes, S. M., Heth, J., McCray, P. M., Jr., Sauter, S. L., Johnston, J. C., Cory-Slechta, D. A., Federoff, H. J. & Davidson, B. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6216-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, D. R. & Stephens, R. M. (1994) J. Neurobiol. 25, 1404-1417. [DOI] [PubMed] [Google Scholar]
- 26.Dahia, P. M., Gimm, O., Chi, H., Marsh, D. J., Reynolds, P. R. & Eng, C. (2000) J. Med. Genet. 37, 715-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng, L. P., Smith, W. M., Brown, J. L. & Eng, C. (2001) Hum. Mol. Genet. 10, 605-616. [DOI] [PubMed] [Google Scholar]
- 28.Li, J., Simpson, L., Takahashi, M., Miliaresis, C., Myers, M. P., Tonks, N. & Parsons, R. (1998) Cancer Res. 58, 5667-5672. [PubMed] [Google Scholar]
- 29.Segal, R. A. & Greenberg, M. E. (1996) Annu. Rev. Neurosci. 19, 463-489. [DOI] [PubMed] [Google Scholar]
- 30.Kwon, C. H., Zhu, X., Zhang, J., Knoop, L. L., Tharp, R., Smeyne, R. J., Eberhart, C. G., Burger, P. C. & Baker, S. J. (2001) Nat. Genet. 29, 404-411. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, T. R., Blader, I. J., Hammonds-Odie, L. P., Burga, C. R., Cooke, F., Hawkins, P. T., Wolf, A. G., Heldman, K. A. & Theibert, A. B. (1996) J. Cell Sci. 109, 289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, H., Cheville, J. C., Pan, Y., Roche, P. C., Schmidt, L. J. & Tindall, D. J. (2001) J. Biol. Chem. 276, 38830-38836. [DOI] [PubMed] [Google Scholar]
- 33.van Golen, C. M., Schwab, T. S., Ignatoski, K. M., Ethier, S. P. & Feldman, E. L. (2001) Cell Growth Differ. 12, 371-378. [PubMed] [Google Scholar]
- 34.Tachibana, M., Shibakita, M., Ohno, S., Kinugasa, S., Yoshimura, H., Ueda, S., Fujii, T., Rahman, M. A., Dhar, D. K. & Nagasue, N. (2002) Cancer 94, 1955-1960. [DOI] [PubMed] [Google Scholar]
- 35.Whiteman, D. C., Zhou, X. P., Cummings, M. C., Pavey, S., Hayward, N. K. & Eng, C. (2002) Int. J. Cancer 99, 63-67. [DOI] [PubMed] [Google Scholar]