ABSTRACT
The genes for chlorate reduction in six bacterial strains were analyzed in order to gain insight into the metabolism. A newly isolated chlorate-reducing bacterium (Shewanella algae ACDC) and three previously isolated strains (Ideonella dechloratans, Pseudomonas sp. strain PK, and Dechloromarinus chlorophilus NSS) were genome sequenced and compared to published sequences (Alicycliphilus denitrificans BC plasmid pALIDE01 and Pseudomonas chloritidismutans AW-1). De novo assembly of genomes failed to join regions adjacent to genes involved in chlorate reduction, suggesting the presence of repeat regions. Using a bioinformatics approach and finishing PCRs to connect fragmented contigs, we discovered that chlorate reduction genes are flanked by insertion sequences, forming composite transposons in all four newly sequenced strains. These insertion sequences delineate regions with the potential to move horizontally and define a set of genes that may be important for chlorate reduction. In addition to core metabolic components, we have highlighted several such genes through comparative analysis and visualization. Phylogenetic analysis places chlorate reductase within a functionally diverse clade of type II dimethyl sulfoxide (DMSO) reductases, part of a larger family of enzymes with reactivity toward chlorate. Nucleotide-level forensics of regions surrounding chlorite dismutase (cld), as well as its phylogenetic clustering in a betaproteobacterial Cld clade, indicate that cld has been mobilized at least once from a perchlorate reducer to build chlorate respiration.
IMPORTANCE
Genome sequencing has identified, for the first time, chlorate reduction composite transposons. These transposons are constructed with flanking insertion sequences that differ in type and orientation between organisms, indicating that this mobile element has formed multiple times and is important for dissemination. Apart from core metabolic enzymes, very little is known about the genetic factors involved in chlorate reduction. Comparative analysis has identified several genes that may also be important, but the relative absence of accessory genes suggests that this mobile metabolism relies on host systems for electron transport, regulation, and cofactor synthesis. Phylogenetic analysis of Cld and ClrA provides support for the hypothesis that chlorate reduction was built multiple times from type II dimethyl sulfoxide (DMSO) reductases and cld. In at least one case, cld has been coopted from a perchlorate reduction island for this purpose. This work is a significant step toward understanding the genetics and evolution of chlorate reduction.
Introduction
Perchlorate (ClO4−) and chlorate (ClO3−) have natural and anthropogenic sources. While recent evidence suggests that these compounds are formed in the atmosphere (1, 2), contamination of drinking water is often a result of human activity. Chlorate has been used as an herbicide and defoliant and as a bleaching agent in the paper industry; perchlorate is a solid oxidant found in flares, explosives, and propellants (3). Bacterial remediation of contaminated water is a viable treatment option, which has spurred both applied (4) and basic (5) science research. Perchlorate and chlorate are respired by dissimilatory perchlorate-reducing bacteria (PRB) and chlorate-reducing bacteria (CRB), respectively, almost all of which are Proteobacteria (6), with a few exceptions (7, 8) (see Fig. S1 in the supplemental material). While all PRB isolated are also chlorate reducers, the reverse is not true. The distinction is at least partly a result of the specificity of the terminal reductase; the perchlorate reductase (PcrAB) can reduce perchlorate and chlorate (9), while the chlorate reductase (ClrABC) can reduce only the latter (10, 11). Chlorite is an obligate intermediate in both pathways and is detoxified by the chlorite dismutase (Cld), which produces chloride and molecular oxygen that is respired. The chlorate reductases of Ideonella dechloratans, Pseudomonas chloritidismutans AW-1, and Pseudomonas sp. strain PDA have been purified as soluble heterotrimers (α1β1γ1) (10–12). ClrABC in I. dechloratans and that in PDA are probably periplasmic, and while fractionation experiments support a cytoplasmic ClrABC in AW-1, a twin-arginine signal motif is predicted (13), suggesting periplasmic localization. By comparison to structurally characterized enzymes EbdABC (14) and NarGHI (15), the α subunit is predicted to contain a bis(molybdopterin guanine dinucleotide)-molybdenum cofactor and a [4Fe-4S] cluster coordinated by one histidine and three cysteines (10). The β subunit is predicted to contain four Fe-S clusters that form an electron transfer pathway between a cytochrome b in the γ subunit (16) and the Fe-S cluster in the α subunit. The δ subunit is homologous to NarJ and most likely participates in proper insertion of the molybdenum cofactor but is not part of the active enzyme (17, 18).
To date, three CRB (Ideonella dechloratans, P. chloritidismutans AW-1, and Alicycliphilus denitrificans BC) have had their genes for chlorate reduction sequenced. As part of our continuing effort to understand the genomics of chloroxyanion respiration by bacteria, genome sequences were completed for four CRB: Ideonella dechloratans, Pseudomonas sp. strain PK, Dechloromarinus chlorophilus NSS, and the newly isolated Shewanella algae ACDC. However, after de novo assembly, the genes for chlorate reduction were found on small contigs, with no information about neighboring regions. Short reads from next-generation sequencing (NGS) technologies often do not unambiguously connect regions surrounding repeats, and as a result, assemblers produce many contigs instead of contiguous finished genomes. A fragmented genome may not be a research impediment if the genes of interest are on a large contig. This was the case in our recent comparative analysis of genes for perchlorate reduction, in which conserved synteny and evidence of horizontal gene transfer led to the identification of a perchlorate reduction genomic island (PRI) that contained metabolic, regulatory, and electron transport chain components (19).
Using a bioinformatics approach, contigs containing genes for chlorate reduction (clrABDC and cld) were extended, connected to neighbors, and confirmed with finishing PCRs. In all four newly sequenced chlorate-reducing bacteria, these genes are flanked by insertion sequences, forming composite transposons. Insertion sequences are small, autonomous, mobile genetic elements that encode a transposase and include accessory genes enclosed by terminal repeat sequences (20). Insertion sequences are widely recognized as drivers of bacterial evolution; they are often present in large numbers and serve as sites for homologous recombination and therefore chromosomal rearrangements, plasmid integration, and deletions (21). As composite transposons, they construct and disseminate novel metabolic pathways, including those involved in the catabolic degradation of substituted aromatics and xenobiotics (22, 23). The identification of chlorate reduction composite transposons is an important step toward understanding the formation and horizontal transfer of this metabolism.
RESULTS
After de novo assembly, genome sequences of PK, NSS, and ACDC contained cld on a contig with a gene cluster encoding a dimethyl sulfoxide (DMSO) reductase family type II enzyme. This was inferred to be the chlorate reductase based on its proximity to cld and phylogenetic clustering with other putative chlorate reductases (10, 11). Since no genetic system is available, the physiological chlorate reductase remains to be validated experimentally in all CRB. For newly sequenced chlorate reducers, the larger genomic context of the chlorate reductase was obscured by fragmented assembly of NGS reads, which produced clrABDC and cld on contigs of less than 16 kb. To resolve neighboring genes, assemblies of I. dechloratans, PK, ACDC, and NSS were improved by mapping reads to the ends of contigs, extending them computationally (24), and predicting links to other contigs (see Materials and Methods).
Shewanella algae ACDC and Dechloromarinus chlorophilus NSS.
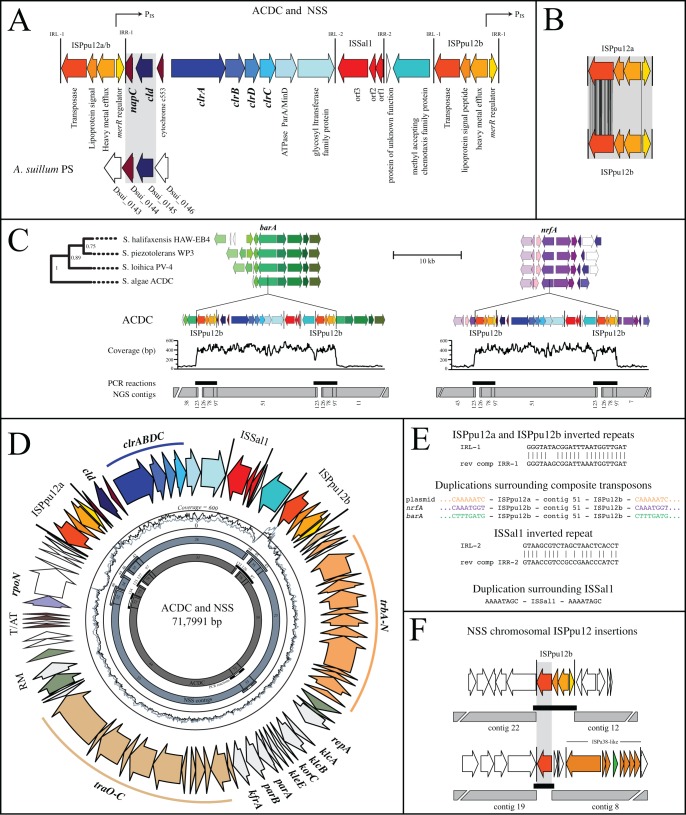
ACDC was isolated from the same marine sediment enrichment from which NSS was previously isolated (6). Details of the physiology of ACDC will be published elsewhere. After improved assembly and annotation (Fig. 1A), two ISPpu12 insertion sequences flanked clrABDC and cld (Fig. 1A). Five copies of this composite transposon were found in ACDC: two chromosomal insertions and one on a multicopy (~3) plasmid (Fig. 1D). The insertion sequences flanking the plasmid-borne copy are isoforms, designated ISPpu12a and ISPpu12b, and contain 68 single nucleotide polymorphisms in the transposase gene, as well as a small deletion of 12 bp in czcD (Fig. 1B). The chromosomal composite transposons contain matching flanking copies of ISPpu12b. The reason for the discrepancy between plasmid and chromosomal insertion sequence isoforms is unknown but was the cause for negligible assembly of this region (Fig. 1C and D). Imperfect 24-bp inverted repeats are located at the boundaries of both ISPpu12 isoforms and are identical to those reported for ISPpu12 in the Pseudomonas putida plasmid pWW0 (Fig. 1E).
FIG 1 .
Comparison of genes involved in chlorate reduction in ACDC and NSS. (A) Structure and annotation of chlorate reduction composite transposons in ACDC and NSS, including synteny with the PRI of A. suillum PS. (B) Locations of single nucleotide polymorphisms (black) in the transposase genes of ISPpu12a and ISPpu12b. (C) Composite transposon insertions in barA and nrfA of ACDC contain two flanking ISPpu12b insertion sequences. (D) Plasmids from ACDC and NSS are nearly identical and contain a chlorate reduction composite transposon flanked by isoforms ISPpu12a and ISPpu12b. T/AT and RM represent toxin/antitoxin and restriction modification systems, respectively. (E) Sequences of inverted repeats and duplications from ISPpu12a/b and ISSal1. (F) ISPpu12b inserted in several chromosomal locations in NSS.
Insertion of the composite transposon into the ACDC chromosome occurred in the open reading frames of nrfA (ammonium-forming nitrite reductase) and barA (hybrid sensor kinase). This finding is supported by conserved synteny surrounding nrfA and barA in closely related species, by the continuous coverage of mapped reads over the insertions, and by PCRs connecting nrfA and barA to the interior of the composite transposon (Fig. 1C). Presumably due to the insertion in nrfA, ACDC reduces nitrate to nitrite but does not reduce nitrite to ammonium (I. C. Clark, unpublished data). Duplications of 8 bp found surrounding composite transposon insertions in nrfA, barA, and the plasmid (Fig. 1E) have been previously observed in other ISPpu12 elements following transposition (25).
Assembly of the NSS genome produced contigs that matched ACDC’s plasmid with 99.9% nucleotide identity (Fig. 1D). The plasmid contains tra and trb gene clusters putatively involved in self-transmissibility. Unlike in ACDC, coverage of the composite transposon in NSS is consistent with it being only plasmid borne. However, NSS does have a full and partial copy of ISPpu12b in its chromosome (Fig. 1F). This is not surprising, considering that ISPpu12 transposes independently, in multiple copies (25), and functions in alpha-, beta-, and gammaproteobacteria (26).
Pseudomonas chloritidismutans AW-1 and Pseudomonas sp. strain PK.
P. chloritidismutans AW-1’s published sequence (GenBank accession no. GQ919187) contains cld and a cytochrome c, which are separated from clrABDC by an insertion sequence. Similar architecture is observed in PK, which contains an insertion sequence in the exact same location (Fig. 2C). However, despite their identical positions, PK’s insertion sequence is ISPst12, while AW-1’s insertion sequence is ISPa16 (Fig. 2C). In both genomes, these insertion sequences have formed tandem duplications of TTAG. Insertion at CTAG has been previously observed for both elements (27). In PK, ISPpu12 insertion sequences again flank the genes for chlorate reduction (Fig. 2A). The ISPpu12 insertion sequences in PK are 99% identical to each other, but one (ISPpu12d) contains a notable 12-bp deletion in czcD. Compared to ACDC and NSS, PK’s ISPpu12 sequences are in the opposite orientation with respect to cld and clrABDC, indicating that the composite transposons found in these bacteria were formed independently. Mapping coverage over this region (Fig. 2C) shows that the genes for chlorate reduction are present in one copy (~chromosomal coverage) but that the ISPpu12d element is present in at least three other locations. We predicted these locations and confirmed them with PCR. These insertions appear to have generated duplications (Fig. 2D). In contrast, duplications at the outer edges of the composite transposon or surrounding individual insertion sequences making up the composite transposon were not found.
FIG 2 .
Comparison of genes involved in chlorate reduction in AW-1 and PK. (A) Structure and annotation of the chlorate reduction composite transposon in PK. (B) Sequences of inverted repeats surrounding ISPpu12, ISPst12, and ISPa16. (C) Conserved synteny and genomic location of chlorate reduction genes in PK and AW-1. (D) Location of three ISPpu12 copies in PK’s chromosome and sequences of duplications created upon insertion.
The left ISPpu12 insertion sequence was absent in the previously published AW-1 sequence, but the chlorate reduction genes are in the same location in the chromosome and in the middle of a conserved region in other Pseudomonas sequences (Fig. 2C). This allowed PCR primers to be designed outward from clrC to a conserved part of ompG in order to finish the rest of AW-1’s sequence in this region. In contrast to PK, which contains two copies of ISPpu12, AW-1 contains only one copy and is therefore not a composite transposon (Fig. 2C).
I. dechloratans and A. denitrificans BC.
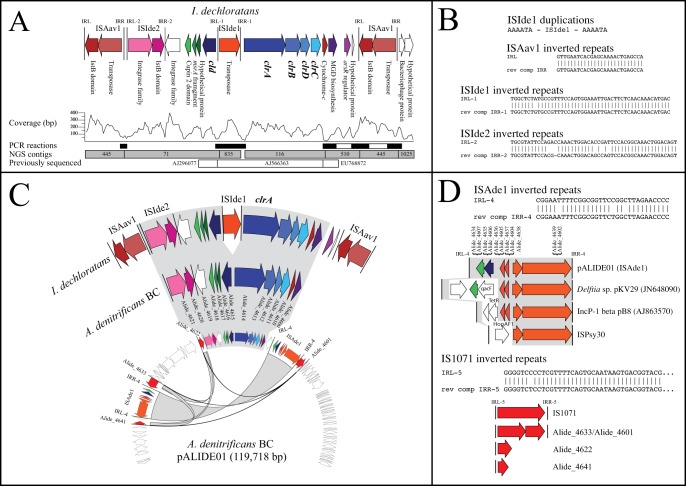
Ideonella dechloratans was one of the first characterized chlorate-reducing bacteria (28), and several fragments of genes putatively involved in chlorate reduction have been sequenced, including cld and a conserved hypothetical gene (GenBank accession no. AJ296077.1) (29), a cytochrome c and molybdopterin-guanine dinucleotide biosynthesis gene (GenBank accession no. EU768872.1) (30), and clrABDC with the insertion sequence ISIde1 (GenBank accession no. AJ566363.1) (10) (Fig. 3A). The genome sequence of I. dechloratans presented here expands previous sequencing efforts, identifying an arsR regulator, a partial methionine sulfoxide reductase gene (msrA), and a cupin 2 domain gene, as well as an insertion sequence that we designate ISIde2 (Fig. 3A).
FIG 3 .
Comparison of genes for chlorate reduction in I. dechloratans and A. denitrificans BC. (A) Structure, annotation, and finishing of the chlorate reduction composite transposon in I. dechloratans. (B) Sequences of inverted repeats and duplications associated with insertion sequences in I. dechloratans. (C) Comparison of syntenic regions in I. dechloratans and A. denitrificans BC pALIDE01. (D) Structure of the ISAde1 transposon containing a cld-like gene and cupin 2 domain gene.
Most interesting, however, is the presence of matching insertion sequences that flank this entire region in I. dechloratans and form a composite transposon (Fig. 3A). The insertion sequences are 99% identical to ISAav1 (27) from Acidovorax citrulli AAC00-1 (GenBank accession no. AF086815), which forms a composite transposon with s-triazine ring cleavage genes, and 99% identical to part of an insertion sequence on Pseudomonas sp. strain ADP’s plasmid pADP-1 (RefSeq accession no. NC_004956), which contains genes for atrazine degradation. The ISAav1 isoform in I. dechloratans has perfect 26-bp inverted repeats (Fig. 3B). Due to the relatively poor quality of the assembled genome (see Table S1 in the supplemental material) and highly variable coverage (Fig. 3A), it was not possible to completely resolve the location(s) of the composite transposon or deduce copy number. However, the presence of high-coverage areas (>200- versus 96-bp average coverage) suggests that it may exist in more than one copy.
Alicycliphilus denitrificans BC was isolated as a benzene-degrading chlorate reducer and has been genome sequenced (31). Genes for chlorate reduction are located on a 119,718-bp plasmid, pALIDE01 (GenBank accession no. CP002450). Apart from the absence of ISIde1, the region within the I. dechloratans composite transposon is remarkably similar to a region on A. denitrificans BC pALIDE01, with just 14 single nucleotide polymorphisms in 12,546 aligned bases. A 6-bp sequence (AAAATA) that was most likely duplicated during transposition of ISIde1 in I. dechloratans is present only once on pALIDE01. The syntenic region (Fig. 3C, gray) includes everything carried within the I. dechloratans composite transposon, except for the ArsR family regulator and a small hypothetical gene.
The A. denitrificans plasmid pALIDE01 shows a history of transposition and recombination in the region surrounding chlorate reduction genes (Fig. 3C). An insertion sequence, 99.8% identical to IS1071 after reconstruction, is fragmented and found in multiple copies (Fig. 3D). The plasmid contains two copies of an ISPsy30-like transposon, which have variable 5′ regions with a conserved resolvase and transposase bound by inverted repeats (Fig. 3D). This transposon, designated ISAde1, carries a toxin/antitoxin module, as well as the cld-like gene and a cupin 2 domain gene. ISAde1 flanks genes for chlorate reduction on pALIDE01 (Fig. 3C), again forming a composite architecture, although the ability of ISPsy30 transposons to mobilize large regions has not been demonstrated.
Phylogenetic analysis of ClrA and Cld.
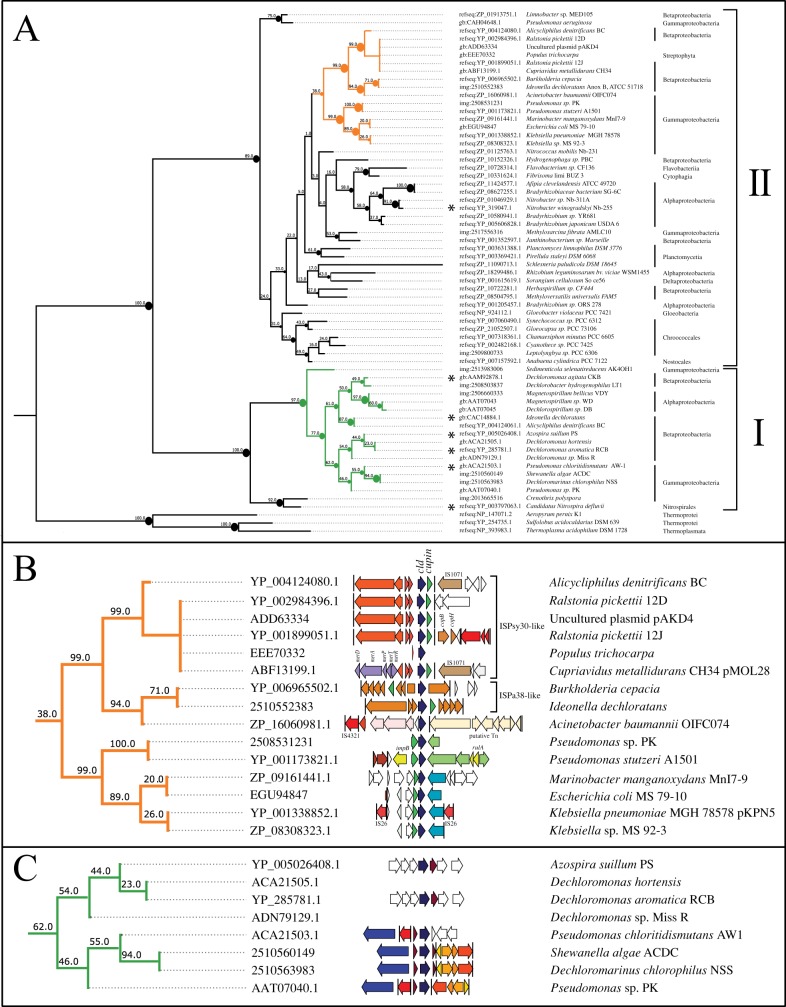
Four new genomes expand the known sequences of ClrA and Cld and provide insight into the evolution of chlorate reduction. Within the larger family of type II DMSO reductases, ClrA resides in a functionally diverse clade that includes ethylbenzene dehydrogenase (EbdA), dimethylsulfide dehydrogenase (DdhA), and selenate reductase (SerA) (Fig. 4A). ClrA proteins from I. dechloratans and A. denitrificans BC group with SerA from Thauera selenatis AX, while ClrA proteins from PK, AW-1, ACDC, and NSS group with DdhA from Rhodovulum sulfidophilum (Fig. 4B). In addition to the canonical αβδγ organization of genes within this clade, two additional genes appear to be conserved in the subclade containing PK, AW-1, ACDC, and NSS. The first gene is annotated as an ATPase and contains MipZ and ParA domains, and the second gene is annotated as a glycosyl transferase (Fig. 4B). The role of these genes in relation to their neighboring reductase is unknown but worthy of further investigation.
FIG 4 .
Phylogenetic analysis of ClrA and closely related type II DMSO reductases. (A) Location of ClrA proteins within the larger type II DMSO reductase family. Asterisks signify that at least one enzyme in the clade has been shown to reduce chlorate in vitro or through genetic analysis. The black circle represents the ancestral state with proposed chlorate reductase activity. UniProt identifiers of the characterized protein are in parentheses. Conflicting biochemical data are reported in the literature for SerABC from Thauera selenatis (44, 74). (B) Phylogenetics of ClrA and closely related enzymes. Gene neighborhoods reveal two additional conserved genes.
Cld proteins in ACDC, NSS, PK, and AW-1 group phylogenetically with a betaproteobacterial clade of Cld proteins from perchlorate reduction islands (Fig. 5A and C). In ACDC and NSS, cld is surrounded by a napC fragment and a small 3′ portion of pcrD that matches the conserved gene organization of Dechloromonas aromatica RCB and Azospira suillum PS (Fig. 1A) (19). In PK and AW-1, a small 3′ portion of pcrD is present. This is highly suggestive that the direction of horizontal gene transfer was from an RCB/PS-type PRI to a chlorate reduction composite transposon.
FIG 5 .
Phylogenetic analysis of Cld and Cld-like proteins. (A) Respiratory Cld proteins form a monophyletic clade (green) that is distinct from other Cld-like proteins found in many chlorate reducers (orange). Asterisks signify chlorite dismutase activity in vitro or through genetic analysis. Circles visually represent bootstrap support at nodes. (B) Superimposing gene neighborhoods highlights the association of cld-like genes with cupin domain genes and mobile genetic elements in a type II subclade. Cld is shown in blue at the center of the gene alignment, while the cupin 2 protein is green and usually located directly adjacent to Cld. (C) Cld proteins from chlorate-reducing strains ACDC, NSS, PK, and AW-1 group with betaproteobacterial Cld proteins from PS and RCB.
In A. denitrificans BC, I. dechloratans, and PK, an additional cld-like gene was found unassociated with chlorate reduction genes. These encode Cld-like proteins that form a unique type II subclade (Fig. 5A), with exaggerated signatures of horizontal gene transfer (Fig. 5B). In I. dechloratans, the cld-like gene is a passenger on an ISPa38-like transposon, designated ISIde3. ISPa38 transposons contain a cupin 2 gene and are found in the genomes of Aeromonas punctata (pFBAOT6), P. aeruginosa DK2, and NSS. In Burkholderia cepacia, a cld-like gene appears to have replaced a portion of the ISPa38 transposase (Fig. 5B). In A. denitrificans BC, the cld-like gene is colocated with a cupin 2 gene on the plasmid-borne transposon ISAde1. ISAde1 is found in Ralstonia pickettii 12 D and 12 J and in partial form in Cupriavidus metallidurans CH34 (Fig. 5B). The model tree species Populus trichocarpa, which can reduce perchlorate and chlorate (32), contains a cld-like gene that is 99.6% identical to the copy in ISAde1. The contig with the Populus trichocarpa cld also contains a small portion of the antitoxin gene from ISAde1, suggesting that it is bacterial in origin (Fig. 5B). PK contains a cld-like gene and a cupin 2 domain gene that share synteny with a Pseudomonas stutzeri A1501 region comprising phage-associated, UV resistance genes (Fig. 5B).
DISCUSSION
Despite a series of analogous biochemical reactions, genes for chlorate and perchlorate reduction have different genomic architectures and a distinct, yet intertwined, evolutionary history. Composite transposons containing chlorate reduction genes with different flanking insertion sequences (type and orientation) have been identified in five chlorate-reducing bacteria. The transposon ISAde1 that surrounds cld and clr in A. denitrificans BC (Fig. 3C) has not been reported to mobilize genes as a composite and may not be relevant to horizontal transfer of chlorate reduction. However, ISPpu12 and ISAv1 isoforms have been observed in a composite architecture previously, and the former can transpose as a composite (26). Identification of 8-bp direct repeats at the outermost edges of the composite transposon in nrfA, barA, and the plasmid (Fig. 1E) in ACDC strongly suggest that the entire element transposed into these locations. These duplications probably occurred when staggered cuts in the target site were filled in following transposon insertion (20). This, together with the fact that chlorate reduction composite transposons can be plasmid borne, provides a conceivable mechanism by which the metabolism moves horizontally. Indeed, the 99.9% nucleotide identity between plasmids in ACDC and NSS is indicative of recent transfer from one to the other, which is plausible considering that they were isolated from the same enrichment.
The genomic architecture of chlorate reduction genes has implications for the study of this metabolism. In all of the chlorate reducers examined, except for AW-1, which lacks the composite structure, the location and copy number of chlorate reduction genes have the potential to change. The loss of the metabolism by homologous recombination of flanking insertion sequences is possible and has been observed in other composite transposons (22, 23, 33). This is expected at some frequency when cells are grown without chlorate and can dominate in a culture if loss of the metabolism is adventitious. For example, if no regulatory system exists for controlling transcription under chlorate-free conditions, the presence of the composite transposon becomes a metabolic burden and provides a selective advantage to cells that have lost or silenced the metabolism. Composite transposons can also increase in copy number (Fig. 1C) and confound transcriptional studies that assume a single stable locus.
Insertion sequences can activate transcription of neighboring genes by providing full or partial promoters (34). In ACDC, a −35/−10 promoter in ISPpu12 is located upstream of clrABDC. In PK, the promoter is on the opposite strand and could potentially drive cld transcription. It has been suggested that this promoter is active and constitutive in P. putida (25). Insertion sequences are also located between cld and clr in PK, AW-1, and I. dechloratans and could serve to change transcription of surrounding genes. While we cannot rule out the possibility that the repeated localization of insertion sequences between cld and clr is random, it is suggestive either of a physiological role for these insertion sequences or of the historical mechanism of colocalization of cld with clrABDC.
With the exception of A. denitrificans BC, most chlorate reducers are unable to reduce nitrate (AW-1 [35], ASK-1 [36], PDA [37], I. dechloratans [28], and NSS [unpublished data]). I. dechloratans was reported to lose the ability to reduce nitrate after cultivation on chlorate (28), and AW-1 has been reported to regain the ability to denitrify after repeated aerobic subculturing in the presence of nitrate (38). We have identified insertions in the nitrite reductase (nrfA) in ACDC and a putative nitrate-responsive histidine kinase (narX) in PK but have yet to find a genetic basis for NSS’s inability to reduce nitrate, given that it contains genes for a complete denitrification pathway (nar, nir, nor, and nos). The ability of the nitrate reductase NarGHI to reduce chlorate to toxic chlorite has been widely reported (39–41). Early experiments exploited this to isolate chlorate-resistant cells with mutations in parts of the nitrate reduction pathway. This identified the nitrate reductase, as well as genes for Mo cofactor biosynthesis, molybdate transport, and nitrate regulation (42). Given this, it is not surprising that parts of nitrate reduction pathways are inactivated in some chlorate reducers, but more work is needed to fully appreciate the interplay between these metabolisms.
Perchlorate, despite having a high redox potential, has a large activation energy that slows inadvertent reduction by metals and enzymes (43). In contrast, chlorate will react abiotically with Fe(II) or Mn(II), and the evolution of an enzyme that overcomes the activation barrier for chlorate was conceivably less difficult than for perchlorate. Many related enzymes, including PcrA (9), NarG (39, 40), and SerA (44), can reduce chlorate in vitro (Fig. 4A; starred clades contain at least one characterized enzyme with chlorate reductase activity). In the case of DdhA, evolution to oxidize a hydrophobic substrate may have precluded turnover of chlorate (45). The most parsimonious hypothesis consistent with these phylogenetic data is that enzymes in this clade evolved from an ancestral enzyme capable of chlorate reduction. If this is correct, the key step in the evolution of chlorate reduction was the presence of a chlorite detoxification system.
Type I chlorite dismutases (Cld proteins) involved in respiratory perchlorate and chlorate reduction are monophyletic (Fig. 5A) (46) but, based on incongruence between protein and species trees, are predicted to have undergone horizontal gene transfer (46, 47). We provide evidence that cld found in four chlorate reduction composite transposons originated from a PRI, based on the transfer of small PRI-specific regions surrounding cld, including part of pcrD (ACDC, NSS, PK, and AW-1) (Fig. 5C) and in some cases a napC homolog (ACDC and NSS) (Fig. 1A). Given that a PRI cld has been coopted for chlorate reduction, why has an active type II chlorite dismutase (46) (Fig. 5A) yet to be associated with a respiratory metabolism? One barrier is the lack of a signal sequence and therefore cytoplasmic localization, which would not protect cells from chlorite produced periplasmically. Another difference is structural: the type II Cld from Nitrobacter winogradskyi is dimeric (48) compared to the tetrameric (49, 50) or pentameric/hexameric (51, 52) type I Cld proteins, but the implications of this difference are unknown. It is also possible that the diversity of respiratory Cld proteins has not been fully surveyed.
In addition to type I Cld proteins, several chlorate reducers contain Cld-like proteins located elsewhere on the chromosome that form a subclade within the type II lineage (Fig. 5A, orange). This subclade contains a set of cld-like genes that are part of transposons (ISPa38-like, ISPsy30-like, and IS26 composite), or associated with phage-related genes, and are therefore presumably highly mobile (Fig. 5B). In all but Acinetobacter baumannii OIFC074, a cupin 2 domain gene is collocated with cld; this occurs in multiple configurations and on different transposons (Fig. 5B). Colocalization of type I cld and cupin genes also occurs in some chlorate reduction composite transposons (Fig. 3A) and PRIs, and as a result, we hypothesize a functional connection between these genes.
Comparative analysis of composite transposons identified the core metabolic genes and several additional genes that may be important for chlorate reduction. These are relatively scarce compared to those in perchlorate reduction islands, suggesting that this mobile metabolism often relies on endogenous systems for electron transport, cofactor biosynthesis, and regulation. At the least, a connection to the host’s electron transport chain must be established. By analogy to the closely related enzymes SerABC (53) and DdhABC (54), and as previously proposed (55), it is possible that ClrABC connects to a quinol oxidase via a cytochrome c. ACDC, NSS, PK, and AW-1 all contain a cytochrome c next to cld that is a possible candidate. However, in I. dechloratans, the cytochrome c in the chlorate reduction composite transposon does not donate electrons to ClrABC in vitro, suggesting that it does not provide the necessary link to the electron transport chain (30). Another cytochrome c in I. dechloratans has been shown to donate electrons to ClrABC in vitro (56), and we identified its sequence (see Fig. S2 in the supplemental material) based on the fragment reported in the literature (55). A quick search (57) for similar proteins gave NirM, the electron donor for nitrite reductase NirS (58), as a top hit.
Regulation of chlorate reduction with respect to other electron acceptors is not well understood. Lack of regulatory genes in the composite transposons of ACDC, NSS, PK, AW-1, and BC implies that regulation may be absent or controlled by a chromosomal system. In I. dechloratans, an ArsR family regulator was identified that may be involved in the observed increase of cld or clrA transcription under anaerobic conditions in the presence of chlorate (59). Although this family of regulators is usually associated with sensing metals, evidence exists for nonmetal signals (60).
The discovery of multiple, independently formed, chlorate reduction composite transposons advances our understanding of this metabolism at the gene level and supports the idea that chlorate reduction is a highly mobile metabolism. Flanking insertion sequences help define a set of genes that move with clrABDC and cld, and although some of these genes may be passengers that were captured as the composite transposons formed, others are likely to have roles in chlorate reduction. We have identified these genes by comparative analysis and hypothesized functions for several. The next step is to develop a genetic system in a chlorate reducer and make clean deletions to test these predictions.
MATERIALS AND METHODS
Genome sequencing.
Whole-genome shotgun sequencing and initial assembly of four chlorate-reducing bacteria, Ideonella dechloratans, Pseudomonas sp. strain PK, Dechloromarinus chlorophilus NSS, and Shewanella algae ACDC, were completed by Eureka Genomics (Hercules, CA). A summary of this sequencing effort is provided in Table S1, and a summary of the assembly is provided in Table S2, both in the supplemental material. Gene calling and annotation were performed using the IMG/ER server (61). Genomes are publically available through IMG (http://img.jgi.doe.gov/cgi-bin/w/main.cgi). A summary of genes internal to chlorate reduction composite transposons is provided in Table S3. Annotation of the manually assembled plasmid from ACDC and NSS was performed using RAST (62).
Improving draft assemblies.
Bowtie (63) was used to map reads (Bowtie -q -n 3 -l 60 -e 200 –best) to assembled genomes. Multiple hits were not allowed in the Bowtie mapping stage. IMAGE (24) was used to extend assembled contigs when possible, and extensions were blasted against all other contigs to search for potential linkages (BLASTn -word_size 10) (64). It should be noted that not all extensions were valid; some were later determined to be erroneous after finishing PCRs were completed. Linkages were used to create a contig graph surrounding chlorate reduction genes. The graph was further simplified using coverage to understand the relative abundance of duplicated regions, which was important for the identification of composite transposons. This workflow is further detailed in Text S1 and Fig. S3 in the supplemental material. PCR and sequencing were performed to confirm linkages. A list of finishing primers is found in Table S4. To further support predicted in silico linkages, reads were remapped to contig junctions and coverage was determined with samtools mpileup (65).
Gene visualization.
Visualization of genes was performed using python scripts and the GenomeDiagram package (66). Additional modifications to diagrams were performed in Adobe Illustrator. ApE was used for viewing and comparing nucleotide sequences and for manually annotating features. Mapped reads and sequenced PCR products were visualized by overlaying them on assembled contigs. Horizontal bars represent PCR products, with black indicating the portion that was Sanger sequenced. Inverted repeats are shown as black vertical lines at the outer edges of insertion sequences. Syntenic regions of high nucleotide identity are highlighted in gray.
Phylogenetics.
Phylogenetics was performed using a compute cluster running CentOS 5.6 at UC Berkeley’s QB3 Computational Genomics Resource Laboratory. Proteins are labeled in figures with an identification code reflecting their source: uniprot = UniProtID, refseq = NCBI RefSeq ID, gi = NCBI GI number, gb = GenBank locus or accession number, and img = IMG GeneID. The alpha subunit of chlorate reductase (ClrA) from ACDC was used as the query to search IMG and NCBI (nr database) for closely related proteins (BLAST-P). After duplicates were removed, the top 100 sequences were added to prealigned DMSO reductase type II family proteins (Pfam 00384 and 17524 sequences; http://pfam.sanger.ac.uk/, accessed 4 December 2012). CD-HIT (67) was run with a clustering threshold of 0.7, the reduced protein alignment was trimmed with Gblocks (b1 = N/2+1 -b2 = N/2+1 -b3 = N/2 -b4 = 2 -b5 = h, where N is the number of sequences) (68), and the protein phylogeny was reconstructed with FastTree (69) in order to obtain an outgroup basal to the clades of interest. Sequences exterior to this outgroup were discarded, and the remaining sequences were realigned and retrimmed. The best amino acid substitution model was determined with ProtTest (70) to be MTMAM. Subsequent phylogenetic analysis was performed using RAxML with 500 bootstraps (71). The same procedure was followed when building a phylogeny for the subclade containing ClrA. Chlorite dismutase (Cld) sequences were obtained with a PSI-BLAST against NCBI (three iterations, e = 10−3) using ACDC’s type I Cld as the starting query. These sequences were combined with the top 100 hits from a BLAST-P search of both IMG and NCBI (nr) that used the A. denitrificans Cld-like protein as a query. The same workflow as above was used to select an appropriate outgroup and reconstruct the phylogeny with RAxML. 16S rRNA gene sequences from perchlorate- and chlorate-reducing bacterial isolates were obtained from NCBI and newly sequenced genomes. Three nearest-neighbor matches were found and aligned using Ribosomal Database Project tools (72). Phylogenetic analysis was performed with MrBayes 3.2 (73) until the average standard deviation of the split frequencies was less than 0.01. Posterior probabilities were estimated after discarding the first 25% of samples from the cold chain.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
16S rRNA gene-based phylogenetic tree of chlorate- and perchlorate-reducing bacteria. C, chlorate-reducing isolate; P, perchlorate-reducing isolate. Download
Protein sequence of the cytochrome c putatively involved in electron transfer to ClrABC in I. dechloratans. Download
Outline of method used to connect contigs containing clr and cld with neighbors, using ACDC as an example. The steps are as follows. (1) De novo assemble contigs from NGS reads using assembler of choice. (2) Map NGS reads to contigs using short read alignment program (Bowtie, BWA, GEM) and extend contigs with Iterative Mapping and Assembly for Gap Elimination (IMAGE) software. Extended regions are in black. (3) Blast extended regions (black) against all newly extended contigs to create linkages. Build a graph of linkages around genes of interest and superimpose coverage on contigs. Contigs with angled edges are not drawn to scale. (4) When possible, attempt to deconvolute the contig graph using coverage, intuition, and synteny. Repeat elements may connect to many contigs and have higher coverage. Also, a truncated open reading frame (ORF) may contain its N-terminal sequence at the end of one contig and its C-terminal sequence at the beginning of a nearby contig. Synteny in closely related species can be used to support a connection between these contigs. However, they may be separated by repeat elements like insertion sequences. (5) Confirm predictions with PCRs and sequencing of PCR products. Download
Summary of NGS.
Summary of genome assembly and annotation.
Tabular annotation of genes in chlorate reduction composite transposons.
Primers used in this study.
ACKNOWLEDGMENTS
We thank Itai Sharon for insights into bacterial genome finishing.
I.C.C. gratefully acknowledges the support of an NSF Graduate Student Fellowship. Funding for research on perchlorate and chlorate reduction in the laboratory of J.D.C. has been provided through the Energy Biosciences Institute, University of California, Berkeley.
Footnotes
Citation Clark IC, Melnyk RA, Engelbrektson A, Coates JD. 2013. Structure and evolution of chlorate reduction composite transposons. mBio 4(•):e00379-13. doi:10.1128/mBio.00379-13.
REFERENCES
- 1. Dasgupta PK, Martinelango PK, Jackson WA, Anderson TA, Tian K, Tock RW, Rajagopalan S. 2005. The origin of naturally occurring perchlorate: the role of atmospheric processes. Environ. Sci. Technol. 39:1569–1575 DOI: 10.1021/es048612x [DOI] [PubMed] [Google Scholar]
- 2. Balaji Rao BR, Hatzinger PB, Böhlke JK, Sturchio NC, Andraski BJ, Eckardt FD, Jackson W. 2010. Natural chlorate in the environment: application of a new IC-ESI/MS/MS method with a Cl18O3-internal standard. Environ. Sci. Technol. 44:8429–8434 DOI: 10.1021/es1024228 [DOI] [PubMed] [Google Scholar]
- 3. Aziz CE, Hatzinger PB. 2009. Perchlorate sources, source identification and analytical methods, p 55-78 In Stroo HF, Ward CH, In situ bioremediation of perchlorate in groundwater. Springer Verlag, New York, NY [Google Scholar]
- 4. Hatzinger PB. 2005. Perchlorate biodegradation for water treatment. Environ. Sci. Technol. 39:239A–247A DOI: 10.1021/es053280x [DOI] [PubMed] [Google Scholar]
- 5. Coates JD, Achenbach LA. 2004. Microbial perchlorate reduction: rocket-fuelled metabolism. Nat. Rev. Microbiol. 2:569–580 DOI: 10.1038/nrmicro926 [DOI] [PubMed] [Google Scholar]
- 6. Coates JD, Michaelidou U, Bruce RA, O’Connor SM, Crespi JN, Achenbach LA. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balk M, Mehboob F, van Gelder AH, Rijpstra WI, Damsté JS, Stams AJ. 2010. (Per)chlorate reduction by an acetogenic bacterium, Sporomusa sp., isolated from an underground gas storage. Appl. Microbiol. Biotechnol. 88:595–603 DOI: 10.1007/s00253-010-2788-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balk M, van Gelder T, Weelink SA, Stams AJ. 2008. (Per)chlorate reduction by the thermophilic bacterium Moorella perchloratireducens sp. nov., isolated from underground gas storage. Appl. Environ. Microbiol. 74:403–409 DOI: 10.1128/AEM.01743-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kengen SW, Rikken GB, Hagen WR, van Ginkel CG, Stams AJ. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181:6706–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thorell HD, Stenklo K, Karlsson J, Nilsson T. 2003. A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl. Environ. Microbiol. 69:5585–5592 DOI: 10.1128/AEM.69.9.5585-5592.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolterink AF, Schiltz E, Hagedoorn PL, Hagen WR, Kengen SW, Stams AJ. 2003. Characterization of the chlorate reductase from Pseudomonas chloritidismutans. J. Bacteriol. 185:3210–3213 DOI: 10.1128/JB.185.10.3210-3213.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steinberg LM, Trimble JJ, Logan BE. 2005. Enzymes responsible for chlorate reduction by Pseudomonas sp. are different from those used for perchlorate reduction by Azospira sp. FEMS Microbiol. Lett. 247:153–159 DOI: 10.1016/j.femsle.2005.05.003 [DOI] [PubMed] [Google Scholar]
- 13. Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. 10.1186/1471-2105-6-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kloer DP, Hagel C, Heider J, Schulz GE. 2006. Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 14:1377–1388 DOI: 10.1016/j.str.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 15. Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NC. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10:681–687 DOI: 10.1038/nsb969 [DOI] [PubMed] [Google Scholar]
- 16. Karlsson J, Nilsson T. 2005. The C subunit of Ideonella dechloratans chlorate reductase: expression, purification, refolding, and heme reconstitution. Protein Expr. Purif. 41:306–312 DOI: 10.1016/j.pep.2005.02.002 [DOI] [PubMed] [Google Scholar]
- 17. Blasco F, Dos Santos P, Magalon A, Frixon C, Guigliarelli B, Santini CL, Giordano G. 1998. NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol. Microbiol. 28:435–447 [DOI] [PubMed] [Google Scholar]
- 18. Vergnes A, Pommier J, Toci R, Blasco F, Giordano G, Magalon A. 2006. NarJ chaperone binds on two distinct sites of the aponitrate reductase of Escherichia coli to coordinate molybdenum cofactor insertion and assembly. J. Biol. Chem. 281:2170–2176 DOI: 10.1074/jbc.M505902200 [DOI] [PubMed] [Google Scholar]
- 19. Melnyk RA, Engelbrektson A, Clark IC, Carlson HK, Byrne-Bailey K, Coates JD. 2011. Identification of a perchlorate reduction genomic island with novel regulatory and metabolic genes. Appl. Environ. Microbiol. 77:7401–7404 DOI: 10.1128/AEM.05758-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Habermann P, Starlinger P. 1982. Bidirectional deletions associated with IS4. Mol. Gen. Genet. 185:216–222 DOI: 10.1007/BF00330790 [DOI] [PubMed] [Google Scholar]
- 22. Slater JH, Weightman AJ, Hall BG. 1985. Dehalogenase genes of Pseudomonas putida PP3 on chromosomally located transposable elements. Mol. Biol. Evol. 2:557–567 [DOI] [PubMed] [Google Scholar]
- 23. Nakatsu C, Ng J, Singh R, Straus N, Wyndham C. 1991. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc. Natl. Acad. Sci. USA 88:8312–8316 DOI: 10.1073/pnas.88.19.8312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsai IJ, Otto TD, Berriman M. 2010. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 11:R41. 10.1186/gb-2010-11-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams PA, Jones RM, Shaw LE. 2002. A third transposable element, ISPpu12, from the toluene-xylene catabolic plasmid pWW0 of Pseudomonas putida mt-2. J. Bacteriol. 184:6572–6580 DOI: 10.1128/JB.184.23.6572-6580.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weightman AJ, Topping AW, Hill KE, Lee LL, Sakai K, Slater JH, Thomas AW. 2002. Transposition of DEH, a broad-host-range transposon flanked by ISPpu12, in Pseudomonas putida is associated with genomic rearrangements and dehalogenase gene silencing. J. Bacteriol. 184:6581–6591 DOI: 10.1128/JB.184.23.6581-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malmqvist Å, Welander T, Moore E, Ternström A, Molin G, Stenström IM. 1994. Ideonella dechloratans gen. nov., sp. nov., a new bacterium capable of growing anaerobically with chlorate as an electron acceptor. Syst. Appl. Microbiol. 17:58–64 DOI: 10.1016/S0723-2020(11)80032-9 [DOI] [Google Scholar]
- 29. Thorell HD, Karlsson J, Portelius E, Nilsson T. 2002. Cloning, characterisation, and expression of a novel gene encoding chlorite dismutase from Ideonella dechloratans. Biochim. Biophys. Acta 1577:445–451 DOI: 10.1016/S0167-4781(02)00446-3 [DOI] [PubMed] [Google Scholar]
- 30. Bohlin J, Bäcklund AS, Gustavsson N, Wahlberg S, Nilsson T. 2010. Characterization of a cytochrome c gene located at the gene cluster for chlorate respiration in Ideonella dechloratans. Microbiol. Res. 165:450–457 DOI: 10.1016/j.micres.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 31. Oosterkamp MJ, Veuskens T, Plugge CM, Langenhoff AA, Gerritse J, Van Berkel WJ, Pieper DH, Junca H, Goodwin LA, Daligault HE, Bruce DC, Detter JC, Tapia R, Han CS, Land ML, Hauser LJ, Smidt H, Stams AJ. 2011. Genome sequences of Alicycliphilus denitrificans strains BC and K601T. J. Bacteriol. 193:5028–5029 DOI: 10.1128/JB.00365-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Aken B, Schnoor JL. 2002. Evidence of perchlorate (ClO4-) reduction in plant tissues (poplar tree) using radio-labeled 36ClO4. Environ. Sci. Technol. 36:2783–2788 DOI: 10.1021/es020560t [DOI] [PubMed] [Google Scholar]
- 33. Meulien P, Downing RG, Broda P. 1981. Excision of the 40kb segment of the TOL plasmid from Pseudomonas putida mt-2 involves direct repeats. Mol. Gen. Genet. 184:97–101 DOI: 10.1007/BF00271202 [DOI] [PubMed] [Google Scholar]
- 34. Charlier D, Piette J, Glansdorff N. 1982. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res. 10:5935–5948 DOI: 10.1093/nar/10.19.5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolterink AF, Jonker AB, Kengen SW, Stams AJ. 2002. Pseudomonas chloritidismutans sp. nov., A non-denitrifying, chlorate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 52:2183–2190 DOI: 10.1099/ijs.0.02102-0 [DOI] [PubMed] [Google Scholar]
- 36. Wolterink A, Kim S, Muusse M, Kim IS, Roholl PJ, van Ginkel CG, Stams AJ, Kengen SW. 2005. Dechloromonas hortensis sp. nov. and strain ASK-1, two novel (per)chlorate-reducing bacteria, and taxonomic description of strain GR-1. Int. J. Syst. Evol. Microbiol. 55:2063–2068 [DOI] [PubMed] [Google Scholar]
- 37. Xu J, Trimble JJ, Steinberg L, Logan BE. 2004. Chlorate and nitrate reduction pathways are separately induced in the perchlorate-respiring bacterium Dechlorosoma sp. KJ and the chlorate-respiring bacterium Pseudomonas sp. PDA. Water Res. 38:673–680 DOI: 10.1016/j.watres.2003.10.017 [DOI] [PubMed] [Google Scholar]
- 38. Cladera AM, García-Valdés E, Lalucat J. 2006. Genotype versus phenotype in the circumscription of bacterial species: the case of Pseudomonas stutzeri and Pseudomonas chloritidismutans. Arch. Microbiol. 184:353–361 DOI: 10.1007/s00203-005-0052-x [DOI] [PubMed] [Google Scholar]
- 39. Afshar S, Johnson E, de Vries S, Schröder I. 2001. Properties of a thermostable nitrate reductase from the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Bacteriol. 183:5491–5495 DOI: 10.1128/JB.183.19.5491-5495.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshimatsu K, Sakurai T, Fujiwara T. 2000. Purification and characterization of dissimilatory nitrate reductase from a denitrifying halophilic archaeon, Haloarcula marismortui. FEBS Lett. 470:216–220 DOI: 10.1016/S0014-5793(00)01321-1 [DOI] [PubMed] [Google Scholar]
- 41. Bell LC, Richardson DJ, Ferguson SJ. 2001. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. The periplasmic enzyme catalyzes the first step in aerobic denitrification. FEBS Lett. 265:85–87 [DOI] [PubMed] [Google Scholar]
- 42. Stewart V. 1988. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol. Rev. 52:190–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urbansky ET. 1998. Perchlorate chemistry: implications for analysis and remediation. Bioremediat. J. 2:81–85 [Google Scholar]
- 44. Martinez-Espinosa RM, Dridge EJ, Bonete MJ, Butt JN, Butler CS, Sargent F, Richardson DJ. 2007. Look on the positive side! The orientation, identification and bioenergetics of archaeal membrane-bound nitrate reductases. FEMS Microbiol. Lett. 276:129–139 DOI: 10.1111/j.1574-6968.2007.00887.x [DOI] [PubMed] [Google Scholar]
- 45. Hanlon SP, Toh TH, Solomon PS, Holt RA, McEwan AG. 2004. Dimethylsulfide: acceptor oxidoreductase from Rhodobacter sulfidophilus. Eur. J. Biochem. 239:391–396 [DOI] [PubMed] [Google Scholar]
- 46. Maixner F, Wagner M, Lücker S, Pelletier E, Schmitz-Esser S, Hace K, Spieck E, Konrat R, Le Paslier D, Daims H. 2008. Environmental genomics reveals a functional chlorite dismutase in the nitrite oxidizing bacterium “Candidatus Nitrospira defluvii.” Environ. Microbiol. 10:3043–3056 DOI: 10.1111/j.1462-2920.2008.01646.x [DOI] [PubMed] [Google Scholar]
- 47. Bender KS, Rice MR, Fugate WH, Coates JD, Achenbach LA. 2004. Metabolic primers for detection of (per)chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Appl. Environ. Microbiol. 70:5651–5658 DOI: 10.1128/AEM.70.9.5651-5658.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mlynek G, Sjöblom B, Kostan J, Füreder S, Maixner F, Gysel K, Furtmüller PG, Obinger C, Wagner M, Daims H, Djinović-Carugo K. 2011. Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi. J. Bacteriol. 193:2408–2417 DOI: 10.1128/JB.01262-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goblirsch BR, Streit BR, Dubois JL, Wilmot CM. 2010. Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 15:879–888 DOI: 10.1007/s00775-010-0651-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mehboob F, Wolterink AF, Vermeulen AJ, Jiang B, Hagedoorn PL, Stams AJ, Kengen SW. 2009. Purification and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol. Lett. 293:115–121 DOI: 10.1111/j.1574-6968.2009.01517.x [DOI] [PubMed] [Google Scholar]
- 51. De Geus DC, Thomassen EA, Hagedoorn PL, Pannu NS, van Duijn E, Abrahams JP. 2009. Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J. Mol. Biol. 387:192–206 DOI: 10.1016/j.jmb.2009.01.036 [DOI] [PubMed] [Google Scholar]
- 52. Kostan J, Sjöblom B, Maixner F, Mlynek G, Furtmüller PG, Obinger C, Wagner M, Daims H, Djinović-Carugo K. 2010. Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium Candidatus Nitrospira defluvii identification of a catalytically important amino acid residue. J. Struct. Biol. 172:331–342 DOI: 10.1016/j.jsb.2010.06.014 [DOI] [PubMed] [Google Scholar]
- 53. Lowe EC, Bydder S, Hartshorne RS, Tape HL, Dridge EJ, Debieux CM, Paszkiewicz K, Singleton I, Lewis RJ, Santini JM, Richardson DJ, Butler CS. 2010. Quinol-cytochrome c oxidoreductase and cytochrome C4 mediate electron transfer during selenate respiration in Thauera selenatis. J. Biol. Chem. 285:18433–18442 DOI: 10.1074/jbc.M110.115873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McDevitt CA, Hugenholtz P, Hanson GR, McEwan AG. 2002. Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 44:1575–1587 DOI: 10.1046/j.1365-2958.2002.02978.x [DOI] [PubMed] [Google Scholar]
- 55. Bäcklund AS, Bohlin J, Gustavsson N, Nilsson T. 2009. Periplasmic c cytochromes and chlorate reduction in Ideonella dechloratans. Appl. Environ. Microbiol. 75:2439–2445 DOI: 10.1128/AEM.01325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bäcklund AS, Nilsson T. 2011. Purification and characterization of a soluble cytochrome c capable of delivering electrons to chlorate reductase in Ideonella dechloratans. FEMS Microbiol. Lett. 321:115–120 DOI: 10.1111/j.1574-6968.2011.02321.x [DOI] [PubMed] [Google Scholar]
- 57. Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hasegawa N, Arai H, Igarashi Y. 2001. Two c-type cytochromes, NirM and NirC, encoded in the nir gene cluster of Pseudomonas aeruginosa act as electron donors for nitrite reductase. Biochem. Biophys. Res. Commun. 288:1223–1230 DOI: 10.1006/bbrc.2001.5919 [DOI] [PubMed] [Google Scholar]
- 59. Lindqvist MH, Johansson N, Nilsson T, Rova M. 2012. Expression of chlorite dismutase and chlorate reductase in the presence of oxygen and/or chlorate as the terminal electron acceptor in Ideonella dechloratans. Appl. Environ. Microbiol. 78:4380–4385 DOI: 10.1128/AEM.07303-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gao CH, Yang M, He ZG. 2012. Characterization of a novel ArsR-like regulator encoded by Rv2034 in Mycobacterium tuberculosis. PLoS One 7:e36255. 10.1371/journal.pone.0036255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2011. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 DOI: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 65. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079 DOI: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pritchard L, White JA, Birch PR, Toth IK. 2006. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics 22:616–617 DOI: 10.1093/bioinformatics/btk021 [DOI] [PubMed] [Google Scholar]
- 67. Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152 DOI: 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 DOI: 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 69. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 DOI: 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]
- 71. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 DOI: 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 72. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huelsenbeck JP, Ronquist F. 2001. MrBayes: bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 DOI: 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 74. Schröder I, Rech S, Krafft T, Macy JM. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 272:23765–23768 DOI: 10.1074/jbc.272.38.23765 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
16S rRNA gene-based phylogenetic tree of chlorate- and perchlorate-reducing bacteria. C, chlorate-reducing isolate; P, perchlorate-reducing isolate. Download
Protein sequence of the cytochrome c putatively involved in electron transfer to ClrABC in I. dechloratans. Download
Outline of method used to connect contigs containing clr and cld with neighbors, using ACDC as an example. The steps are as follows. (1) De novo assemble contigs from NGS reads using assembler of choice. (2) Map NGS reads to contigs using short read alignment program (Bowtie, BWA, GEM) and extend contigs with Iterative Mapping and Assembly for Gap Elimination (IMAGE) software. Extended regions are in black. (3) Blast extended regions (black) against all newly extended contigs to create linkages. Build a graph of linkages around genes of interest and superimpose coverage on contigs. Contigs with angled edges are not drawn to scale. (4) When possible, attempt to deconvolute the contig graph using coverage, intuition, and synteny. Repeat elements may connect to many contigs and have higher coverage. Also, a truncated open reading frame (ORF) may contain its N-terminal sequence at the end of one contig and its C-terminal sequence at the beginning of a nearby contig. Synteny in closely related species can be used to support a connection between these contigs. However, they may be separated by repeat elements like insertion sequences. (5) Confirm predictions with PCRs and sequencing of PCR products. Download
Summary of NGS.
Summary of genome assembly and annotation.
Tabular annotation of genes in chlorate reduction composite transposons.
Primers used in this study.