ABSTRACT
Treatment of multidrug-resistant enterococci has become a challenging clinical problem in hospitals around the world due to the lack of reliable therapeutic options. Daptomycin (DAP), a cell membrane-targeting cationic antimicrobial lipopeptide, is the only antibiotic with in vitro bactericidal activity against vancomycin-resistant enterococci (VRE). However, the clinical use of DAP against VRE is threatened by emergence of resistance during therapy, but the mechanisms leading to DAP resistance are not fully understood. The mechanism of action of DAP involves interactions with the cell membrane in a calcium-dependent manner, mainly at the level of the bacterial septum. Previously, we demonstrated that development of DAP resistance in vancomycin-resistant Enterococcus faecalis is associated with mutations in genes encoding proteins with two main functions, (i) control of the cell envelope stress response to antibiotics and antimicrobial peptides (LiaFSR system) and (ii) cell membrane phospholipid metabolism (glycerophosphoryl diester phosphodiesterase and cardiolipin synthase). In this work, we show that these VRE can resist DAP-elicited cell membrane damage by diverting the antibiotic away from its principal target (division septum) to other distinct cell membrane regions. DAP septal diversion by DAP-resistant E. faecalis is mediated by initial redistribution of cell membrane cardiolipin-rich microdomains associated with a single amino acid deletion within the transmembrane protein LiaF (a member of a three-component regulatory system [LiaFSR] involved in cell envelope homeostasis). Full expression of DAP resistance requires additional mutations in enzymes (glycerophosphoryl diester phosphodiesterase and cardiolipin synthase) that alter cell membrane phospholipid content. Our findings describe a novel mechanism of bacterial resistance to cationic antimicrobial peptides.
IMPORTANCE The emergence of antibiotic resistance in bacterial pathogens is a threat to public health. Understanding the mechanisms of resistance is of crucial importance to develop new strategies to combat multidrug-resistant microorganisms. Vancomycin-resistant enterococci (VRE) are one of the most recalcitrant hospital-associated pathogens against which new therapies are urgently needed. Daptomycin (DAP) is a calcium-decorated antimicrobial lipopeptide whose target is the bacterial cell membrane. A current paradigm suggests that Gram-positive bacteria become resistant to cationic antimicrobial peptides via an electrostatic repulsion of the antibiotic molecule from a more positively charged cell surface. In this work, we provide evidence that VRE use a novel strategy to avoid DAP-elicited killing. Instead of “repelling” the antibiotic from the cell surface, VRE diverts the antibiotic molecule from the septum and “traps” it in distinct membrane regions. We provide genetic and biochemical bases responsible for the mechanism of resistance and disclose new targets for potential antimicrobial development.
IMPORTANCE
The emergence of antibiotic resistance in bacterial pathogens is a threat to public health. Understanding the mechanisms of resistance is of crucial importance to develop new strategies to combat multidrug-resistant microorganisms. Vancomycin-resistant enterococci (VRE) are one of the most recalcitrant hospital-associated pathogens against which new therapies are urgently needed. Daptomycin (DAP) is a calcium-decorated antimicrobial lipopeptide whose target is the bacterial cell membrane. A current paradigm suggests that Gram-positive bacteria become resistant to cationic antimicrobial peptides via an electrostatic repulsion of the antibiotic molecule from a more positively charged cell surface. In this work, we provide evidence that VRE use a novel strategy to avoid DAP-elicited killing. Instead of “repelling” the antibiotic from the cell surface, VRE diverts the antibiotic molecule from the septum and “traps” it in distinct membrane regions. We provide genetic and biochemical bases responsible for the mechanism of resistance and disclose new targets for potential antimicrobial development.
Introduction
The emergence of antibiotic resistance in health care-associated pathogens is recognized as a major public health threat by the World Health Organization (1). Vancomycin-resistant enterococci (VRE) are prime examples of such multidrug-resistant organisms, against which new therapies are urgently needed (2). Daptomycin (DAP) is a cyclic antimicrobial lipopeptide that has become a key frontline drug against VRE due to its potent in vitro bactericidal activity. However, an important emerging clinical problem is the development of DAP resistance during therapy (3, 4). DAP is heavily calcium complexed, an event which is absolutely required for its antimicrobial effect becoming a de facto cationic antimicrobial peptide (5). Recent data using fluorescence microscopy in a Bacillus subtilis model (6) indicate that the mechanism of action of DAP involves insertion of the antibiotic molecule into the bacterial membrane in a calcium-dependent manner with a strong preference for interactions with the bacterial division septum. Binding of the antibiotic produces membrane distortions that alter the morphology of the cell and recruit proteins essential for cell division. The defects in cell membrane and cell wall synthesis cause damages in cell integrity that lead to cell death (6). Additionally, a crucial step in the mechanism of DAP-mediated disruption of the cell membrane appears to be the ability of the antibiotic to oligomerize within the cell membrane, a phenomenon that seems to be dependent on the presence of the negatively charged phospholipid phosphatidylglycerol (PG) (5).
Although the mechanisms of DAP resistance have not been fully elucidated, a correlation with evolution of a more relatively positive surface charge has been observed in Staphylococcus aureus (7–9). Electrostatic “repulsion” between relatively positively charged cell surfaces and the calcium-DAP molecule complex is believed to prevent initial interaction of DAP with the cell membrane. Our initial genomic studies in a pair of clinical strains of vancomycin-resistant Enterococcus faecalis recovered from the bloodstream of a patient before and after DAP therapy indicated that the mechanism of resistance was associated with mutations in two groups of genes. The first group of genes, liaFSR, encodes a three-component regulatory system that is involved in cell envelope homeostasis (10). Using an allelic replacement strategy, we showed that deletion of isoleucine in position 177 of the putative LiaF (lipid II-interacting-antibiotic protein) was sufficient to decrease DAP susceptibility (although still below the clinical breakpoint) and led to DAP tolerance, abolishing the in vitro bactericidal activity of the antibiotic (11). Supporting the important role of the LiaFSR system in development of DAP resistance, we found that mutations in liaFSR were commonly observed in DAP-resistant isolates of Enterococcus faecium (12, 13). A second group of genes that seem to be involved in DAP resistance in enterococci are ones encoding enzymes catalyzing reactions implicated in cell membrane phospholipid metabolism (glycerophosphoryl diester phosphodiesterase [GdpD] and cardiolipin synthase [Cls]) (12). Indeed, we previously showed that in vivo development of DAP resistance in enterococci is associated with important changes in cell membrane phospholipid composition (14). Moreover, an important decrease in PG plus a surprising increase in negatively charged phospholipid content (but not in positively charged species as previously described in S. aureus [7, 9]) was observed in DAP-resistant derivatives of both E. faecalis and E. faecium (14) isolated from patients after DAP therapy, suggesting that a more positive surface charge was not the main mediator of DAP resistance in enterococci.
In this study, we demonstrate that the emergence of DAP nonsusceptibility in vancomycin-resistant E. faecalis is associated with an initial cell membrane cardiolipin microdomain redistribution caused by an amino acid deletion in LiaF that diverts the antibiotic away from the division septum, a critical and vital structure for cell survival. The mechanism is completed by changes in phospholipid content brought about by alterations in enzymes involved in phospholipid metabolism. Our work offers novel insights into the mechanisms of bacterial resistance against the attack by antimicrobial peptides and provides potential new targets for antimicrobial development.
RESULTS
Daptomycin resistance is associated with exclusion of BDP-DAP from the division septum.
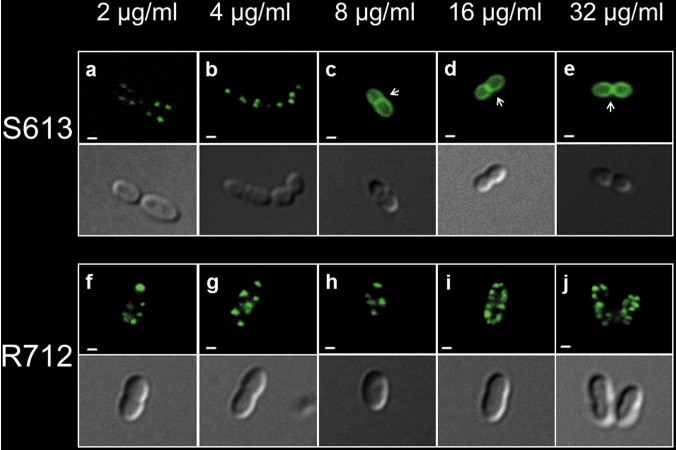
In order to understand the mechanism of enterococcal daptomycin (DAP) resistance, we used a previously described clinically derived pair of E. faecalis strains, a vancomycin-resistant DAP-susceptible (Daps) strain and a DAP-resistant (Dapr) strain recovered from the bloodstream of a patient before and after DAP therapy (S613 and R712, respectively [Table 1]) (12). We initially examined the binding of DAP to the cell membrane of the Daps clinical strain, S613, versus its Dapr variant, R712. We used boron-dipyrromethene (BODIPY)-labeled DAP (BDP-DAP), a fluorescent derivative of DAP that retains antimicrobial activity (MICs of 8 and 32 µg/ml for strain S613 and R712, respectively) (Table 1), to visualize DAP interactions with the cell membrane. Figure 1a and b show that at subinhibitory concentrations, multiple foci of BDP-DAP were bound to the S613 cell membrane and were distributed throughout the cell (“peppering” pattern) but sparing the division septum. At concentrations at and above the MIC, BDP-DAP decorated the E. faecalis S613 cell envelope in a homogeneous pattern, with a clear preference for the division septum (Fig. 1c to e). Indeed, ca. 85% of cells in a microscopic field displayed a linear fluorescence pattern at the septa (108 cells examined, BDP-DAP concentration of 16 µg/ml) (see Fig. S2 in the supplemental material). The binding pattern of BDP-DAP to Dapr strain R712 was similar to that of strain S613 at low concentrations (Fig. 1f and g). Moreover, we were able to observe consistent binding of BDP-DAP to E. faecalis R712 at all concentrations tested (Fig. 1f to j). However, at increasingly higher BDP-DAP concentrations, R712 continued to exhibit the “peppering” pattern. Notably, this pattern excluded the midpoints of the cells where the main septal structures are localized (Fig. 1h to j). Even at 16 and 32 µg/ml, only a small proportion of R712 cells exhibited septal staining with BDP-DAP compared to S613 cells (20% [n = 104] and 49% [n = 110], respectively; P < 0.0001) (Fig. S2). To further confirm that septal exclusion of BDP-DAP from septa is critical in the mechanism of resistance, we quantitated the fluorescence intensity in S613 and R712 cells treated with increasing concentrations of BDP-DAP and normalized to protein contents of the samples. No differences in fluorescent intensities between S613 and R712 cells were observed at any of the BDP-DAP concentrations tested (2 to 32 µg/ml) (all P > 0.05) (Fig. S3), suggesting that the total amount of bound antibiotic was similar in S613 and R712 cells. These data suggest that exclusion of DAP from the main septal structures and diversion of the antibiotic to other sites of the cell membrane mediate, at least in part, DAP resistance in E. faecalis R712.
TABLE 1 .
Daptomycin-susceptible and -resistant Enterococcus faecalis strains and derivatives
| Enterococcus faecalis strain | MIC (µg/ml) of DAP |
MIC (µg/ml) of DAP-BDPb |
Predicted amino acid change |
||||
|---|---|---|---|---|---|---|---|
| MHa | BHIa | TSBa | Clsc | LiaFd | GdpDe | ||
| S613 | 0.5 | 3 | 2 | 8 | None | None | None |
| R712 | 8 | 64 | 64 | 32 | Deletion of Lys at position 61 |
Deletion of Ile at position 177 |
Deletion of Ile at position 170 |
| S613∆liaF177 | 3 | 16 | 16 | 16 | None | Deletion of Ile at position 177 |
None |
| S613∆gdpD170 | 0.75 | 3 | 2 | 8 | None | None | Deletion of Ile at position 170 |
| S613∆liaF177gdpD170 | 3 | 24 | 16 | 16 | None | Deletion of Ile at position 177 |
Deletion of Ile at position 170 |
| S613∆liaF177gdpD170cls61 | 6 | 64 | 64 | 32 | Deletion of Lys at position 61 |
Deletion of Ile at position 177 |
Deletion of Ile at position 170 |
The MIC of daptomycin was determined by the use of Etest (bioMérieux) on the indicated medium.
The MIC of BODIPY-labeled daptomycin (BDP-DAP) was determined by broth microdilution.
Cls, cardiolipin synthase.
LiaF, lipid II-interacting-antibiotic protein.
GdpD, glycerophosphoryl diester phosphodies terase.
FIG 1 .
BODIPY-labeled daptomycin (BDP-DAP) staining of representative E. faecalis S613 (DAP-susceptible) (a to e) and R712 (DAP-resistant) (f to j) cells at increasing concentrations of the fluorescent antibiotic. The images within each panel show bacterial cells captured by fluorescence (top image) and phase-contrast microscopy (bottom image) (bars, 1 µm). (a and b) In E. faecalis S613, binding of the antibiotic in a “peppering” pattern is observed only at subinhibitory concentrations of BDP-DAP. (c to e) At higher BDP-DAP concentrations, saturation of the surface is observed with staining of the division septum (white arrows). (f to j) In E. faecalis R712, the “peppering” staining pattern and binding of the fluorescent antibiotic is observed at all concentrations without visualization of septa. Additional images selected from different microscopic fields are shown in Fig. S2 in the supplemental material.
Diversion of BDP-DAP away from septa is mediated by a mutation in liaF.
Our previous genomic studies comparing E. faecalis S613 to R712 indicated that DAP resistance was associated with the following amino acid changes: (i) deletion of Ile at position 177 of LiaF (LiaF∆177), a member of a three-component regulatory system (LiaFSR) involved in cell envelope homeostasis, (ii) deletion of Ile at position 170 of GdpD (GdpD∆170), a putative glycerophosphoryl diester phosphodiesterase that is likely involved in hydrolyzing deacylated phospholipids (PLs), and (iii) deletion of Lys at position 61 (Cls∆61) of a cardiolipin (CL) synthase (Cls) enzyme. Using an allelic replacement strategy, we then sequentially introduced the resistance-associated alleles belonging to strain R712 into strain S613 and found that LiaF∆177 was a pivotal event in development of DAP resistance (12). Indeed, introduction of the resistant liaF allele alone to Daps S613 (S613∆liaF177) decreased DAP susceptibility (12). Here, we show that S613∆liaF177 resulted in the exclusion of BDP-DAP from septal structures and the “peppering pattern” at higher BDP-DAP concentrations (8 µg/ml) than seen in S613 (4 µg/ml [Fig. 2a]). At 16 µg/ml, homogeneous staining of the cell surface of strain S613∆liaF177 and visualization of septal DAP accumulation in a linear pattern was evident, as seen with S613 at concentrations ≥8 µg/ml (Fig. 2a; see Fig. S4 in the supplemental material).
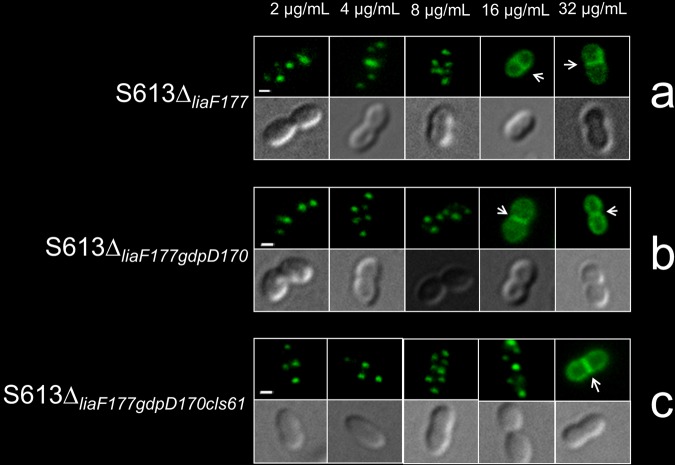
FIG 2 .
BODIPY-labeled DAP (BDP-DAP) staining of representative cells from E. faecalis S613 derivatives at increasing concentrations of the fluorescent antibiotic. The images for each BDP-DAP concentration (from 2 to 32 μg/ml) and strain show bacterial cells captured by fluorescence (top image) and phase-contrast (bottom image) microscopy (bars, 1 µm). (a) E. faecalis S613∆liaF177 is a derivative of strain S613 carrying a mutated liaF belonging to E. faecalis R712 (daptomycin resistant). (b) S613∆liaF177gdpD170 is a derivative of S613 carrying both liaF and gdpD alleles from strain R712. (c) S613∆liaF177gdpD170cls61 carries mutated liaF, gdpD, and cls alleles from R712. Introduction of the three mutated alleles prevents BDP-DAP interaction with the septum up to concentrations of 16 µg/ml of BDP-DAP. Additional images selected from different microscopic fields are shown in Fig. S3 in the supplemental material. Arrows indicate visualization of septum in a linear pattern.
We subsequently evaluated the binding pattern of BDP-DAP in an E. faecalis S613 derivative carrying both resistance-associated liaF and gdpD alleles from E. faecalis R712 (S613∆liaF177gdpD170). As shown in Fig. 2b, no difference in the pattern of BDP-DAP staining in strain S613∆liaF177gdpD170 compared to S613∆liaF177 was noted, suggesting that the contribution of the GdpD alteration to diversion of DAP from the septum was minimal. This is consistent with our analysis of S613∆gdpD170, a derivative of S613 carrying GdpD∆170, whose pattern of BDP-DAP binding was identical to that of S613 (data not shown). We next created a derivative of S613 carrying all three of the resistance-associated alleles (S613∆liaF177gdpD170cls61) and found that S613∆liaF177gdpD170cls61 excluded BDP-DAP from septal regions and also displayed the “peppering pattern” at even higher BDP-DAP concentrations (up to 16 µg/ml), similar to R712 (Fig. 2c; see Fig. S4 in the supplemental material). Thus, our findings indicate that DAP resistance involves antibiotic diversion from the septum, initially mediated by a mutation in liaF and ultimately completed by concomitant mutations of other enzymes involved in PL metabolism (i.e., GdpD and Cls). Of note, we were unable to introduce the mutated cls allele alone in S613, S613∆liaF177, or S613∆gdpD170 (after multiple attempts), suggesting that the isolated Lys61 deletion in the putative Cls enzyme may be deleterious unless other changes in cell membrane homeostasis brought about simultaneously by LiaF and GdpD have occurred.
DAP resistance involves redistribution of cardiolipin microdomains away from the division septum.
The hydrophobic fluorescent dye 10-N-nonyl-acridine orange (NAO) has been used previously to visualize CL-enriched domains in Escherichia coli (15, 16) and B. subtilis (17, 18). The association of NAO with CL (but not other PLs) induces a green-to-red shift in its fluorescence emission attributed to the ability of CL (harboring four chains and a small head group) to make microdomains (arrays) within the cell membrane (19, 20). This configuration provides NAO with enough space to form arrays of parallel stacks between the CL arrays (15, 19). As shown in Fig. 3a, visualization of Daps E. faecalis S613 stained with NAO (500 nm) revealed CL-rich domains during early stationary phase of growth. These microdomains were visualized as intense linear staining at the site of the bacterial septum and as polar spots (Fig. 3a), in agreement with previous data generated in E. coli and B. subtilis (15–18). The NAO-stained domains were also visualized under red fluorescence (see Fig. S5 in the supplemental material), indicating that the microdomains consist of CL arrays. Additionally, NAO (500 nm) did not affect the growth of E. faecalis S613. Important changes in CL domain distribution were observed in E. faecalis R712 compared to S613 (Fig. 3b), featuring markedly reduced linear NAO fluorescence at sites of cell division (i.e., linear septal fluorescence was observed in only 37% of R712 cells [n = 65], compared to 82% of S613 cells [n = 56]; P < 0.001). Instead, we observed intense spots of NAO fluorescence (without a linear pattern) with some staining localized at potential sites of cell division (Fig. 3b and Fig. S6); these observations are consistent with the notion that CL domain redistribution is a major mediator of DAP resistance. Using transmission electron microscopy (TEM), we previously showed that R712 (Dapr) harbors a plethora of aberrant septal structures (12). Here, also using TEM, we were able to determine that separation of individual R712 cells was infrequently observed in R712, compared to S613. Indeed, R712 had a significantly higher number of multicell aggregates (>4 cells) than parental S613 (57 versus 16 in 100 cells; P < 0.0001) (Fig. S7). This is consistent with alterations in the ability of these enterococcal cells to fully divide and separate, in association with DAP resistance. Thus, our data suggest that redistribution of CL arrays severely affects cell division machinery. In agreement with this hypothesis, anionic PLs have been proposed to mediate the organization of cell division proteins at specific sites of the cell membrane (CM) (21).
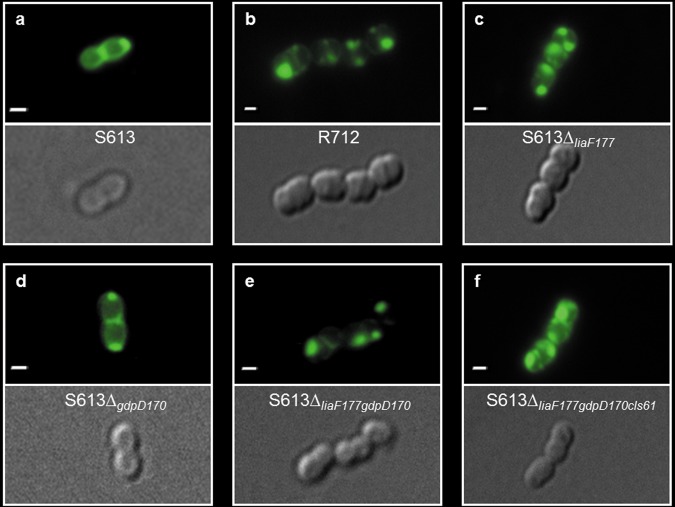
FIG 3 .
Staining of representative cells from E. faecalis S613 (DAP susceptible), R712 (DAP resistant), and S613 derivatives with 10-N-nonyl-acridine orange (NAO) at 500 nM revealing cardiolipin-rich domains in the cell membrane. The top images in panels show bacterial cells captured with a fluorescent microscope (bars, 1 µm). The bottom images in panels are phase-contrast images of the same bacterial cells. (a and d) In E. faecalis S613 and S613∆gdpD170, CL-enriched domains are visualized at the poles and division septa. (b) CL-enriched domain redistribution is observed in DAP-resistant E. faecalis R712. (c) Introduction of a mutated liaF allele from strain R712 into strain S613 was sufficient to alter the distribution of CL domains. (e and f) Introduction of mutated gdpD and cls alleles to S613∆liaF177 did not produce additional alterations in the distribution of CL-rich domains. Additional images selected from different microscopic fields are shown in Fig. S5 in the supplemental material.
Figure 3c and Fig. S6 in the supplemental material show that LiaF∆177 was sufficient to alter the distribution of CL microdomains, since NAO staining of strain S613∆liaF177 was similar to that of strain R712, with most cells lacking clear polar spots and exhibiting altered staining at the sites of cell division. Consistent with the lack of effect of the gdpD mutation on DAP resistance (see above), we were not able to find any substantial changes in CL microdomain distribution in strain S613∆gdpD170 versus strain S613 (Fig. 3d and Fig. S6). Moreover, the pattern of NAO staining of S613∆liaF177gdpD170 and S613∆liaF177gdpD170cls61 was similar to that of S613∆liaF177 (Fig. 3e and f), indicating that the single LiaF∆177 deletion was largely responsible for CL-enriched microdomain redistribution, thus establishing a link between the LiaFSR system and changes in cell membrane PL distribution. Of interest, the LiaFSR system of B. subtilis has been shown to mediate cell envelope responses to antimicrobial peptide attack (22). In order to quantitate the fluorescence differences between septa and nonseptal NAO-stained domains in DAP-resistant R712, we calculated the relative ratio of fluorescent intensity between these sites. The relative ratio of fluorescent intensity between NAO-stained domains versus septa was about 3:1, suggesting that CL-enriched domains are concentrated away from the septum.
Cls and GdpD mediate changes in phospholipid content associated with DAP resistance.
In order to quantitatively assess the effects of the above gene mutations on PL content, we determined PL content of the cell membranes of E. faecalis S613 and derivatives, as described previously (Table 2). In our earlier work, we showed that E. faecalis strain S613 contains a large cadre of cell membrane PL species, including several amino-containing, positively charged species, as well as the negatively charged PLs such as PG, CL, phosphatidic acid, and glycerolphospho-diglycodiacylglycerol (GP-DPDAG) (14). As shown in Table 2, LiaF∆177, which reduces the DAP susceptibility of strain S613 and is linked to changes in CL-enriched domain distribution, did not affect actual PL content of S613 cell membranes. Similarly, no significant changes in cell membrane PL content were identified in the double mutant S613∆liaF177gdpD170 compared to S613, despite the fact that DAP MICs increased (Table 2). However, when the liaF mutation was accompanied by changes in both gdpD and cls (i.e., the triple mutant, S613∆liaF177gdpD170cls61), significant alterations in cell membrane PL content were observed versus S613 including, most notably, the following: (i) a decrease in PG (ca. 40%) and CL; (ii) a compensatory increase in other negatively charged PL species; and (iii) an increase in overall amino-containing PLs, with substantially more “flipping” of lysyl-phosphatidylglycerol (L-PG) to the outer leaflet of the CM as seen by significant changes in the amount of amino-containing PLs in the inner versus outer CM leaflet (Table 2). These findings suggest that Cls∆61 (in the background of LiaF∆177 and GdpD∆170) is responsible for additional changes in cell membrane PL composition and asymmetry that augment and fully complete the DAP resistance phenotype in E. faecalis. Moreover, in order to support our findings that CL-enriched microdomain redistribution is the main mediator of DAP resistance and is not related to global changes in membrane structure, we stained E. faecalis S613 and derivatives with the vital nonspecific stain FM 4-64 (23, 24) which has been used to visualize a variety of bacterial membranes, including E. coli and B. subtilis. FM 4-64 is a non-specific lipophilic fluorescent dye (23, 24). As shown in Fig. S8 in the supplemental material, the membranes of S613 cells stained homogeneously with FM 4-64, and the relative intensity of FM 4-64 fluorescence at the septal membranes was double that of other regions of the cells, as described previously for B. subtilis (18, 24). The FM 4-64 staining patterns of strains S613ΔliaF177 and S613ΔliaF177gdpD170 (which exhibited changes in CL-enriched microdomain redistribution) were similar to that of S613, indicating that CL redistribution was not associated with global structural changes of the cell membrane. Conversely, heterogeneous staining of FM 4-64 was observed in R712 (wild-type DAP-resistant strain) and S613ΔliaF177gdpD170cls61 (which exhibits changes in PL content). Indeed, intense nonseptal fluorescence spots were observed in the latter strains (Fig. S8), suggesting that changes in phospholipid content (but not distribution) can affect the overall membrane structure.
TABLE 2 .
Cell membrane phospholipid composition and asymmetry of Enterococcus faecalis S613 and derivatives
| Strain | Cell membrane PL composition (mean % ± SD)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Positively charged PLb |
Total ACPc | Negatively charged PLd |
||||||
| I-LPG | O-LPG | PG | CL | GP-DGDAG | PA | NegPL | ||
| S613 | 6.81 ± 2.66 | 6.52 ± 1.09 | 19.51 ± 4.99 | 20.76 ± 0.11 | 44.0 ± 3.99 | 5.07 ± 1.05 | 4.13 ± 0.01 | 6.54 ± 0.06 |
| S613∆liaF177gdpD170cls61 | 5.48 ± 1.09 | 10.24 ± 1.45 | 27.14 ± 2.62 | 13.22 ± 1.65 | 30.85 ± 1.30 | 9.58 ± 2.05 | 9.27 ± 0.10 | 9.94 ± 0.33 |
| S613∆liaF177gdpD170 | 5.58 ± 2.44 | 7.36 ± 0.62 | 22.63 ± 2.96 | 17.95 ± 0.86 | 39.42 ± 3.26 | 6.77 ± 0.10 | 5.52 ± 0.40 | 7.71 ± 0.85 |
| S613∆gdpD170 | 6.34 ± 0.79 | 7.56 ± 0.42 | 21.63 ± 0.07 | 18.09 ± 0.81 | 40.11 ± 1.88 | 6.35 ± 0.77 | 5.40 ± 1.03 | 8.42 ± 1.79 |
| S613∆liaF177 | 6.09 ± 0.25 | 6.83 ± 2.19 | 19.90 ± 1.64 | 19.95 ± 2.11 | 40.74 ± 2.38 | 6.96 ± 1.12 | 4.72 ± 0.50 | 7.72 ± 1.23 |
Values that are statistically significantly different (P < 0.05) from the values for strain S613 are shown in boldface type. PL, phospholipid.
LPG, lysyl-phosphatidylglycerol; I-LPG, LPG in the inner leaflet of the cell membrane; O-LPG, LPG in the outer leaflet of the cell membrane. The amount of phospholipid flipping is determined by the difference between LPG content in the inner and outer leaflets of the cell membrane.
ACP, amino-containing phospholipid. The amount of total ACP includes O-LPG, I-LPG, and two additional ACPs whose identity has not been established.
Major anionic (negatively charged) phospholipid species isolated from the membrane of E. faecalis strains. PG, phosphatidylglycerol; CL, cardiolipin; GP-DGDAG, glycerolphospho-diglycodiacylglycerol; PA, phosphatidic acid; NegPL, a negatively charged phospholipid species whose structure has not been determined.
DISCUSSION
Cationic antimicrobial peptides are important effectors of the innate host immune response system against bacterial infections (25, 26), and such peptides are currently used as antibiotics in clinical practice (e.g., DAP). Understanding the molecular mechanisms of cationic antimicrobial peptide action, as well as resistance, is likely to offer new insights into bacterial cell membrane homeostasis and thus the possibility of discovering new targets for antimicrobial development. Using BDP-DAP, Pogliano et al. (6) provided novel insights into the mechanism of DAP action in B. subtilis. Indeed, interaction of DAP with the cell membrane and localization of BDP-DAP to the division septum appeared to be a crucial event in the mechanism of bacterial killing by the antibiotic. Moreover, the same authors provided compelling evidence that proteins involved in cell division are recruited to areas where DAP interacts with the cell membrane, a phenomenon that is likely to affect cell division, as previously reported (12, 27). Another important aspect of DAP’s mechanism of action appears to be its oligomerization (which seems to occur in the outer leaflet of the cell membrane), a phenomenon that is dependent on the presence of the phospholipid PG (6, 28, 29). Using lipid vesicles and bacterial membranes from B. subtilis, Muraih et al. (5, 29) showed that DAP molecules interact with one another to form oligomers on liposomal and bacterial membranes. More importantly, DAP’s ability to oligomerize is driven by stoichiometric interactions with PG (5, 29). Previous data originating from studies performed with pairs of clinical strains of DAP-susceptible and DAP-resistant S. aureus and laboratory isolates of B. subtilis (7, 22, 28) suggested that the mechanism of DAP and antimicrobial peptide resistance involves “repulsion” of the antibiotic molecule from the cell surface by changing the overall electrical charge of the cell envelope (becoming more positively charged) and preventing the binding of the antibiotic molecule with its principal target, the cell membrane. In S. aureus, several avenues have been postulated to contribute to increased cell surface charge, including an increased amount of positively charged amino-containing phospholipids in the outer leaflet of the cell membrane (e.g., positively charged L-PG) mediated by the MprF enzyme (9) and enhanced alanylation of lipoteichoic acids by the Dlt proteins (30, 31). Nonetheless, non-DAP-susceptible strains that do not appear to have changes in cell surface charge have also been reported (32).
Our data indicate that vancomycin-resistant E. faecalis, unlike S. aureus and B. subtilis, utilizes a distinct mechanism to overcome the attack by calcium-decorated DAP. Using BDP-DAP, we evaluated the accumulation of the antibiotic in the cell envelope of a pair of clinical strains of E. faecalis, a DAP-susceptible strain (S613) and a DAP-resistant strain (R712). To our surprise, BDP-DAP was not repelled from the cell surface of DAP-resistant E. faecalis R712 (even at low concentrations), indicating that the mechanism of resistance is not dependent on repulsion of cationic calcium-DAP complexes from the cell envelope as previously proposed (7–9, 28). Furthermore, we provide evidence that the amount of bound BDP-DAP is similar between R712 versus S613 at all concentrations tested, suggesting that the amount of DAP binding is not the critical determinant of resistance but the pattern of binding to vital structures (e.g., division septum). Our data also suggest that the mechanism of resistance relies on diversion of the antimicrobial peptide from the septum and provide further evidence that interactions of DAP with the septal structures are crucial for its antimicrobial activity. Our results also indicate that saturation of the division septum by the antimicrobial peptide molecule is concentration dependent and occurs only at concentrations of the antibiotic at or above the MIC, as has been shown previously (6), consistent with clinical observations that higher doses than those approved for staphylococci may have clinical benefit against enterococci (33–35).
In order to understand the biochemical basis for DAP septal diversion, we developed a NAO staining-based technique of enterococcal cells. NAO interacts with CL-enriched microdomains in the cell membrane that can be visualized by fluorescence microscopy. Using NAO staining, it has been shown in E. coli and B. subtilis that CL-enriched domains are localized at the division septum and cell poles (15–18). Moreover, CL and other anionic PLs have been shown to be important for the division machinery (21). NAO staining of E. faecalis S613 (DAP susceptible) revealed CL-enriched domains localized at the septum and poles. However, a striking change in CL-enriched microdomain distribution was observed in the DAP-resistant derivative R712 in which the CL-enriched arrays were redistributed away from the division septum. This phenomenon was confirmed by calculating the relative fluorescence intensities between septa and NAO-stained domains, strongly suggesting that diversion of DAP from the septum is likely to be mediated by changes in CL localization (and possibly other anionic PLs). Moreover, we were able to show that a single substitution in LiaF (deletion of Ile at position 177 of the putative protein) was sufficient to affect the distribution of CL domains without altering cell membrane PL composition. Taken together, our data suggest that the LiaFSR system, which is predicted to regulate the cell envelope response to antibiotics and antimicrobial peptides, is likely to mediate changes in the cell membrane by altering PL distribution (but not content). It is also tempting to speculate that changes in localization of PLs may also affect the distribution of lipoteichoic acids and transmembrane proteins, including those involved in cell wall homeostasis, affecting cell division. Indeed, CL has been shown to affect the function of transmembrane transporters (36, 37). Furthermore, using the vital stain FM 4-64, we also provide evidence that CL-enriched microdomain distribution is not associated with global structural changes in the cell membrane.
Although PL redistribution was important as an initial step toward resistance, it was not sufficient to confer clinical resistance. Indeed, introduction of substitutions in enzymes involved in PL metabolism (GdpD and Cls) were necessary to confer high levels of resistance to DAP. Interestingly, introduction of Cls∆61 into E. faecalis S613 harboring the mutated liaF and gdpD alleles produced a striking change in PL content and DAP resistance similar to that of E. faecalis R712, suggesting that the biochemical changes in cell membrane composition brought about by this mutation increased the ability of the enterococcal cell to resist the attack by the antimicrobial peptide. Of note, we recently showed that a different substitution in Cls (R218Q) (38), which is also associated with DAP resistance, increased the catalytic activity of the enzyme, suggesting that changes in Cls are likely to confer a “gain-in-function” phenotype, as has been previously shown with MprF. It is also important to note that Cls∆61 could be introduced only when the LiaF and GdpD substitutions were also present, suggesting that enzymatic changes in Cls may be compatible only under specific cell membrane conditions and that changes in enzyme activity may be deleterious unless compensatory events also occur.
On the basis of our results, a cogent model for the mechanism of DAP resistance has emerged. In DAP-susceptible E. faecalis, cell membrane architecture is preserved, with PG and CL constituting the main PL species. In the absence of activation of the LiaFSR system (wild-type LiaF), CL-enriched domains are localized at sites of cell membrane curvature, namely the septum and poles (similar to what has been shown in E. coli and B. subtilis). In the presence of low concentrations of positively charged calcium-bound DAP (sub-MIC), the antibiotic binds to the cell membrane, initially sparing the septum. When the concentration of DAP increases to or above the MIC, the antibiotic accumulates at the septum, producing alterations in cell membrane architecture and impairment of cell division, leading to cell death. In DAP-resistant E. faecalis, activation of the LiaSR two-component regulatory system results in redistribution of CL-enriched microarrays away from the septum. Subsequent changes in PL synthesis enzymes (such as GdpD and Cls) alter the PL composition which, among other effects, drastically reduces PG content in the cell membrane. Due to these changes, calcium-complexed DAP cannot effectively bind to the septum and is diverted to cell membrane sites that, while deficient in PG, are rich in other negatively charged PLs. Consequently, DAP is diverted to those sites but cannot oligomerize therein, failing to fully distort cell membrane architecture and homeostasis. Using this sophisticated strategy, enterococci are able to survive the DAP attack. Our findings suggest a new paradigm in the mechanism of resistance to antimicrobial peptides; future studies should concentrate on dissecting the LiaFSR response in enterococci with the intention of using this information to develop novel antimicrobial agents that may impair the cell membrane response to daptomycin and other antibiotics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and their phenotypes are listed in Table 1. Enterococcus faecalis S613 is a vancomycin-resistant clinical bloodstream isolate recovered from a patient with recurrent bacteremia in 2005 (3). E. faecalis R712 is a derivative of S613, recovered from blood from the same patient after ca. 14 days of daptomycin (DAP) and amikacin therapy (3). Strains S613∆liaF177 and S613∆liaF177gdpD170 are laboratory derivatives of S613 into which the liaF and gdpD alleles from strain R712 were introduced and have been described previously (12). Strain S613∆liaF177gdpD170cls61, generated here, harbors allelic replacements of liaF, gdpD. and cls (see below). Unless otherwise noted, enterococci were cultured in brain heart infusion (BHI) broth at 37°C with gentle agitation. DAP and boron-dipyrromethene (BODIPY) fluorescence (FL)-labeled DAP (BDP-DAP) were provided by Cubist Pharmaceuticals (Lexington, MA). DAP MICs were determined on Mueller-Hinton (MH) and BHI agars by Etest (bioMérieux). DAP MICs in trypticase soy broth (TSB) were determined by broth microdilution supplemented with Ca2+ (50 mg/liter). Results were interpreted based on the breakpoints recommended by the Clinical and Laboratory Standards Institute (CLSI) (39) on MH agar. BDP-DAP MICs were determined in Luria-Bertani (LB) broth (supplemented with Ca2+ [50 mg/liter]) by microdilution with increasing concentrations of the fluorescent antibiotic.
Mutant construction and genetic manipulations.
In order to create an E. faecalis S613 derivative harboring the mutated liaF, gdpD, and cls alleles from E. faecalis R712, we used the p-chloro-phenylalanine (p-Chl-Phe) sensitivity counterselection system (PheS*) (40) with plasmid pHOU3 (which carries the chloramphenicol acetyltransferase gene conferring resistance to chloramphenicol) as described previously (12). Briefly, primers 5′-ATAAGAATGCGGCCGCGATGCAGCTGTTGCACGTTT (forward) and 5′-GGAATTCCGCGTATCCCGCCAATAGCC (reverse) ~500 bp upstream and downstream, respectively, of the codon encoding position 61 of Cls were used for PCR amplifications using the genomic DNA of strain R712 as the template. PCR products were cloned into pHOU3 using NotI and EcoRI. The plasmid constructs were electroporated into E. faecalis CK111 and subsequently delivered to E. faecalis S613∆liaF177gdpD170 by conjugation (12, 41). First, recombination integrants were selected on plates containing chloramphenicol (15 µg/ml) and vancomycin (32 µg/ml). In order to obtain the desired replacement, first-event integrants were subsequently grown on p-chloro-phenylalanine, and colonies were tested by replica plating in the presence and absence of chloramphenicol (12). Chloramphenicol-susceptible mutants were characterized by pulsed-field gel electrophoresis, and the open reading frames of the three genes (liaF, gdpD, and cls) were sequenced in their entirety in both directions. Growth curves of the mutant (S613∆liaF177gdpD170cls61) were performed in BHI and compared to the growth curves of strains S613, S613∆liaF177, S613∆liaF177gdpD177 and R712 as shown in Fig. S1 in the supplemental material.
BDP-DAP staining of E. faecalis strains.
BODIPY FL-labeled daptomycin (BDP-DAP) (Cubist Pharmaceuticals) is a fluorescent derivative of daptomycin that retains antimicrobial activity. By using fluorescence microscopy, bacterial cells can be imaged, and the sites of accumulation of BDP-DAP in the cell membrane can be determined as previously described (6, 22, 28). Organisms were grown to exponential phase in LB broth at 37°C with gentle shaking. One hundred microliters of each sample was incubated with BDP-DAP at increasing concentrations (2 µg/ml to 32 µg/ml) in LB (supplemented with Ca2+ [50 mg/liter]) for 10 min in darkness. Excess unincorporated BDP-DAP was removed by washing the cells in LB broth three times, and the cells were immobilized on a coverslip treated with poly-l-lysine (Sigma-Aldrich). Bacterial cells were viewed with an Olympus IX71 microscope equipped with PlanApo N 100× objective. Fluorescence was observed using a standard fluorescein isothiocyanate (FITC) filter set (excitation at 490 nm and emission at 528 nm). Data acquisition was performed by SlideBook 5.0 software and processed using Adobe Photoshop CS5. A minimum of two independent experiments was performed for each strain on different days. In order to estimate the amount of BDP-DAP bound to E. faecalis strain S613 versus strain R712, we quantitated the fluorescence intensity in S613 and R712 cells treated with increasing concentrations of BDP-DAP and normalized to protein contents of the samples. S613 and R712 cells were treated with BDP-DAP as described above, and fluorescence was measured following excitation at 488 nm and emission at 510 nm. Protein extraction from the cells was performed by previously described methods (42). Protein concentrations were estimated using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific) according to the manufacturer’s instructions.
Staining of E. faecalis strains using NAO and FM 4-64.
Visualization of cardiolipin (CL)-rich domains was performed using the fluorescent dye 10-N-nonyl acridine orange (NAO). NAO has been shown to bind preferentially to CL-enriched arrays in the cell membrane, allowing visualization of CL-enriched domains in the cell membrane (19). Association of NAO with CL molecules results in a green-to-red shift in fluorescence (20). For microscopic examination, cells were grown in TSB to exponential phase (A600 of ~0.3). NAO (Molecular Probes) was added to growth medium to a final concentration of 500 nM, which did not inhibit growth of E. faecalis. Samples were allowed to stain for 3.5 h at 37°C in darkness with gentle shaking. After incubation, E. faecalis cells were washed three times with 0.9% saline and immediately immobilized on a coverslip treated with poly-l-lysine (Sigma-Aldrich). Fluorescent images were captured by an Olympus IX71 microscope with a PlanApo N 100× objective. The focus was set under phase-contrast conditions and then shifted to light emission conditions to capture fluorescent images and minimize photobleaching. The exposure time was 500 ms. Green fluorescence from NAO was detected using a standard fluorescein isothiocyanate (FITC) filter (excitation at 490 nm and emission at 528 nm). Image acquisition was performed using the SlideBook 5.0 software package. At least three independent sets of experiments were performed for each strain on different days. Captured images were processed using Adobe Photoshop CS5. In a single experiment, the viability of cells was confirmed by placing coverslips with NAO-stained bacteria on BHI agar and incubating the coverslips at 37°C overnight. All strains grew, indicating that NAO was not toxic to enterococci and that the images were taken while the cells were alive. Additionally, green-to-red fluorescence shift in the images was examined using the Nikon A1 confocal laser microscope equipped with 100× Plan-Apo/1.4-numerical-aperture (NA) oil objective. Fluorescence was viewed using the PicoQuant imaging system. Excitation was at 490 nm and emission was at either 528 or 617 nm at exposure times of 500 ms. Fluorescent areas identified at both wavelengths indicate a green-to-red shift, a property exhibited by NAO when bound to cardiolipin (see Fig. S4 in the supplemental material). Images of individual cells were used to calculate the ratio of NAO fluorescence intensity at the division septum versus nonseptal NAO-stained domains in strain R712 (43, 44). Intensity was quantitated by acquiring pixel-to-area values in the areas of interest (ImageJ software). Three septal areas versus nonseptal domains were averaged prior to calculation of the fluorescence intensity ratio. FM 4-64 (Molecular Probes) was added to a final concentration of 1 µg/ml to a cell culture in TSB that was harvested in stationary growth phase. After incubation at 37°C for 30 min, the cells were fixed on slides coated with a layer of 0.8% agarose gel in water. Fluorescent images were viewed using an Olympus IX71 microscope with a PlanApo N 100× objective. Red fluorescence was detected using a standard tetramethyl rhodamine isothiocyanate (TRITC) filter (excitation at 541 nm and emission at 617 nm). Image acquisition was performed using the SlideBook 5.0 software package. At least two independent experiments were performed on different days for each strain. Images were processed with Adobe Photoshop CS5.
Transmission electron microscopy.
Transmission electron microscopy (TEM) was used to assess ultrastructural characteristics of the pair of E. faecalis strains, strains S613 and R712, using standard methods as previously described (12). Chaining (≥4 cells) was evaluated in both S613 and R712 by counting ≥100 cells of each isolate from different fields.
Cell membrane phospholipid composition and asymmetry.
Phospholipids (PLs) were extracted from enterococcal isolates using a methodology described before (45, 46). Briefly, extraction of PLs was performed on enterococcal isolates grown in TSB for 18 h (late stationary phase). The PLs were separated using two-dimensional thin-layer chromatography (TLC) using Silica Gel 60 F254 high-performance (HP) TLC plates (Merck). A first dimension of chloroform−methanol−25% ammonium hydroxide (65:25:6, by volume) and a second dimension of chloroform-water-methanol-glacial acetic acid-acetone (45:4:8:9:16, by volume) in the vertical and horizontal orientation, respectively, were used for the separation of the PLs (46). The identification of TLC PL spots was made in comparison to control two-dimensional (2D) TLC plates of known PL standards, which were purchased from Avanti Polar Lipids (Alabaster, AL). For quantitative analysis, individual PLs from TLC plates were digested with 0.3 ml of 70% perchloric acid (0.3 ml) at 180°C for 3 h and then quantified spectrophotometrically at A660 (46). Experiments were performed at least three times on separate days. Data were reported as the mean (± standard deviation [SD]).
Outer cell membrane (CM) lysyl-phosphatidylglycerol (LPG) distribution was determined using fluorescamine analysis, since this fluorophore specifically labels only surface-exposed amino-PLs in the outer CM leaflet as described previously (7, 46). These experiments were performed in parallel with the PL extraction and analyses described above. PL spots on the TLC plate were confirmed by exposure to iodine vapors and spraying with CuSO4 (100 mg/ml) containing 8% phosphoric acid (vol/vol) and heated at 180°C (47). LPG in the inner leaflet of the cell membrane (I-LPG) (positively charged), and other amino-containing PLs were visualized by ninhydrin staining.
LC/ESI-MS/MS analysis.
Major lipids separated on the 2D-TLC plate were scraped off, extracted with chloroform, and confirmed by normal-phase liquid chromatography-electrospray ionization-tandem mass spectrometry (LC/ESI-MS/MS) analysis as described before (14). Briefly, normal-phase LC was performed on an Agilent 1200 Quaternary LC system equipped with an Ascentis Silica high-performance liquid chromatography (HPLC) column (5 µm; 250 by 2.1 mm; Sigma-Aldrich, St. Louis, MO). The mobile phases were chloroform-methanol-aqueous ammonium hydroxide (800:195:5, vol/vol) (mobile phase A solvent), chloroform-methanol-water-aqueous ammonium hydroxide (600:340:50:5, vol/vol) (mobile phase B solvent), and chloroform-methanol-water-aqueous ammonium hydroxide (450:450:95:5, vol/vol) (mobile phase C solvent). The elution program consisted of the following: 100% mobile phase A solvent was held isocratically for 2 min and then linearly increased to 100% mobile phase B solvent over 14 min and held at 100% mobile phase B solvent for 11 min. The LC gradient was then changed to 100% mobile phase C solvent over 3 min, held at 100% C for 3 min, finally returned to 100% mobile phase A solvent over 0.5 min, and held at 100% phase A solvent for 5 min. The total LC flow rate was 300 µl/min. To achieve optimum ESI efficiency, a postcolumn splitter was used to divert ~10% of the LC effluent into the mass spectrometer, a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA). Instrumental settings for negative ion electrospray (ESI) and MS/MS analysis of lipid species were as follows: ion spray voltage (IS) = −4,500 V; curtain gas (CUR) = 20 lb/in2; gas source–I (GSI) = 20 lb/in2; declustering potential (DP) = −55 V; and focusing potential (FP) = −150 V. The MS/MS analysis used nitrogen as the collision for the gas. Data analysis was performed using Analyst QS software (Applied Biosystems, Foster City, CA).
SUPPLEMENTAL MATERIAL
Growth curves of Enterococcus faecalis. Growth expressed as optical density (A600) performed in brain heart infusion broth of E. faecalis S613 (▲), R712 (∆), S613ΔliaF177 (■), S613ΔliaF177gdpD170 (♦), and S613ΔliaF177gdpD170cls61 (●). Representative growth curves from at least two independent experiments are shown. Download
Staining of E. faecalis S613 (DAP-susceptible) and R712 (DAP-resistant) strains with BODIPY-labeled DAP (BDP-DAP) at concentrations of 16 µg/ml and 32 µg/ml. The images for a strain at a specific BDP-DAP concentration show representative bacterial cells captured under green fluorescence taken from different fields (bars, 1 µm). Only a small proportion of cells exhibiting fluorescent septal staining with BDP-DAP at 16 and 32 µg/ml was observed in R712 cells compared to S613 cells (20% [n = 104] and 49% [n = 110], respectively; P < 0.0001; two different blinded observers). Download
Fluorescence intensities of DAP-susceptible E. faecalis S613 and DAP-resistant E. faecalis R712. Cells were treated with BDP-DAP, and fluorescence was normalized to cell protein content. Rfu, relative fluorescence units. Download
Staining of E. faecalis S613 derivatives with BODIPY-labeled DAP (BDP-DAP) at concentrations of 16 µg/ml and 32 µg/ml. The images for a strain at a specific BDP-DAP concentration show representative bacterial cells captured under green fluorescence taken from different fields (bars, 1 µm). At 16 µg/ml, there was a statistically significant difference in the percentage of cells exhibiting fluorescent linear septa between strains S613ΔliaF177gdpD170cls61 (32%) and S613ΔliaF177 (57%) and S613ΔliaF177gdpD170 (65%) (P < 0.005). At 32 µg/ml, fluorescent septa were seen in strains S613ΔliaF177gdpD170cls61 (57%), S613ΔliaF177 (69%), and S613ΔliaF177gdpD170 (72%). At least 100 cells were counted for each strain at the indicated concentration by two different blinded observers. Download
10-N-Nonyl acridine orange (NAO) staining of E. faecalis S613 (DAP-susceptible) and R712 (DAP-resistant) strains. The green-to-red shift indicates that the phospholipid domains are likely to contain cardiolipin. (a and b) Excitation was at 490 nm, and emission was at either 528 (a) or 617 (b) nm (bars, 2 µm). (c) Colocalization of green and red fluorescent domains. Download
10-N-Nonyl acridine orange (NAO) staining of representative cells from E. faecalis S613 (DAP susceptible), R712 (DAP resistant), and S613 derivatives. The images for each strain show representative bacterial cells captured with green fluorescence taken from different fields (bars, 1 µm). Fluorescent linear septa were seen in 26% of E. faecalis S613ΔliaF177, 30% of S613ΔliaF177gdpD170, and 28% of S613ΔliaF177gdpD170cls61, respectively (all significantly different [P < 0.001] from the values for strain S613). Linear septa were observed in 81% of S613ΔgdpD170 ([P = 1] from the values for strain S613). At least 50 cells were counted for each strain by two blinded observers. Download
Transmission electron microscopy of E. faecalis daptomycin-susceptible S613 and daptomycin-resistant R712. Red arrows indicate alterations in points of cell contact, leading to infrequent cell separation in strain R712 compared to strain S613 (bars, 2 µm). Download
FM 4-64 staining of representative cells from E. faecalis S613 (DAP susceptible), R712 (DAP resistant), and S613 derivatives. The images for each strain show representative bacterial cells captured with red fluorescence taken from different fields (bars, 1 µm). Homogeneous staining of cell membranes is observed in strains S613ΔliaF177, S613ΔliaF177gdpD170, and S613ΔgdpD170. In contrast, structural alterations of the cell membrane are observed in strains R712 and S613ΔliaF177gdpD170cls61. Download
ACKNOWLEDGMENTS
C.A.A. is supported by National Institutes of Health (NIH-NIAID) grant R01 AI093749. A.S.B. is supported by NIH-NIAID grant R01 AI39108-15, Y.S. is supported by NIH-NIAID grant, R01 AI080714, and B.E.M. is supported by NIH-NIAID grant, R01 AI047923. This work was also supported in part by a John S. Dunn Foundation Collaborative Research Award.
The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to Silvia Munoz-Price and John P. Quinn for providing enterococcal strains S613 and R712. We thank Joe Pogliano, Danielle M. McGrath, Isabel Reyes, I-Hsiu Huang, and Hung Ton-That for technical support. We thank Jared A. Silverman for providing BODIPY FL-labeled daptomycin.
Footnotes
Citation Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, Reyes J, Diaz L, Weinstock GM, Murray BE, Shamoo Y, Dowhan W, Bayer AS, Arias CA. 2013. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4(4):e00281-13. doi:10.1128/mBio.00281-13.
REFERENCES
- 1. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 2. Arias CA, Contreras GA, Murray BE. 2010. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 16:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Munoz-Price LS, Lolans K, Quinn JP. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41:565–566 [DOI] [PubMed] [Google Scholar]
- 4. Sabol K, Patterson JE, Lewis JS, II, Owens A, Cadena J, Jorgensen JH. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 49:1664–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muraih JK, Pearson A, Silverman J, Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta 1808:1154–1160 [DOI] [PubMed] [Google Scholar]
- 6. Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 194:4494–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bayer AS, Schneider T, Sahl HG. 2013. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 1277:139–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2312–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146 [DOI] [PubMed] [Google Scholar]
- 11. Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Arias CA. 2013. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 57:2831–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J. Med. 365:892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob. Agents Chemother. 56:4354–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mishra NN, Bayer AS, Tran TT, Shamoo Y, Mileykovskaya E, Dowhan W, Guan Z, Arias CA. 2012. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 7:e43958. 10.1371/journal.pone.0043958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mileykovskaya E, Dowhan W. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182:1172–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kicia M, Janeczko N, Lewicka J, Hendrich AB. 2012. Comparison of the effects of subinhibitory concentrations of ciprofloxacin and colistin on the morphology of cardiolipin domains in Escherichia coli membranes. J. Med. Microbiol. 61:520–524 [DOI] [PubMed] [Google Scholar]
- 17. Barák I, Muchová K, Wilkinson AJ, O’Toole PJ, Pavlendová N. 2008. Lipid spirals in Bacillus subtilis and their role in cell division. Mol. Microbiol. 68:1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. 2004. Cardiolipin domains in Bacillus subtilis marburg membranes. J. Bacteriol. 186:1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH. 2001. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 507:187–190 [DOI] [PubMed] [Google Scholar]
- 20. Petit JM, Maftah A, Ratinaud MH, Julien R. 1992. 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur. J. Biochem. 209:267–273 [DOI] [PubMed] [Google Scholar]
- 21. Mileykovskaya E, Dowhan W. 2005. Role of membrane lipids in bacterial division-site selection. Curr. Opin. Microbiol. 8:135–142 [DOI] [PubMed] [Google Scholar]
- 22. Hachmann AB, Angert ER, Helmann JD. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fishov I, Woldringh CL. 1999. Visualization of membrane domains in Escherichia coli. Mol. Microbiol. 32:1166–1172 [DOI] [PubMed] [Google Scholar]
- 24. Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710–720 [DOI] [PubMed] [Google Scholar]
- 26. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 27. Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hachmann AB, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55:4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muraih JK, Harris J, Taylor SD, Palmer M. 2012. Characterization of daptomycin oligomerization with perylene excimer fluorescence: stoichiometric binding of phosphatidylglycerol triggers oligomer formation. Biochim. Biophys. Acta 1818:673–678 [DOI] [PubMed] [Google Scholar]
- 30. Bertsche U, Weidenmaier C, Kuehner D, Yang SJ, Baur S, Wanner S, Francois P, Schrenzel J, Yeaman MR, Bayer AS. 2011. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and d-alanylation. Antimicrob. Agents Chemother. 55:3922–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang SJ, Kreiswirth BN, Sakoulas G, Yeaman MR, Xiong YQ, Sawa A, Bayer AS. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mishra NN, Rubio A, Nast CC, Bayer AS. 2012. Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. Int. J. Microbiol. 2012:683450. 10.1155/2012/683450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kullar R, Davis SL, Levine DP, Zhao JJ, Jr, Crank CW, Segreti J, Sakoulas G, Cosgrove SE, Rybak MJ. 2011. High-dose daptomycin for treatment of complicated gram-positive infections: a large, multicenter, retrospective study. Pharmacotherapy 31:527–536 [DOI] [PubMed] [Google Scholar]
- 34. Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall AD, Steed ME, Arias CA, Murray BE, Rybak MJ. 2012. Evaluation of standard- and high-dose daptomycin versus linezolid against vancomycin-resistant Enterococcus isolates in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 56:3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosch JW, Hsu FF, Caparon MG. 2007. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J. Bacteriol. 189:801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vega LA, Caparon MG. 2012. Cationic antimicrobial peptides disrupt the Streptococcus pyogenes ExPortal. Mol. Microbiol. 85:1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davlieva M, Zhang W, Arias CA, Shamoo Y. 2013. Biochemical characterization of cardiolipin synthase mutations associated with daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 57:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. CLSI 2011. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. CLSI Document MS100-S21. CLSI, Wayne, PA [Google Scholar]
- 40. Kristich CJ, Chandler JR, Dunny GM. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Panesso D, Montealegre MC, Rincón S, Mojica MF, Rice LB, Singh KV, Murray BE, Arias CA. 2011. The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BMC Microbiol. 11:20. 10.1186/1471-2180-11-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bogdanov M, Dowhan W. 1995. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J. Biol. Chem. 270:732–739 [DOI] [PubMed] [Google Scholar]
- 43. Lunde CS, Rexer CH, Hartouni SR, Axt S, Benton BM. 2010. Fluorescence microscopy demonstrates enhanced targeting of telavancin to the division septum of Staphylococcus aureus. Antimicrob. Agents Chemother. 54:2198–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pereira PM, Filipe SR, Tomasz A, Pinho MG. 2007. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3627–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dixit BL, Gupta CM. 1998. Role of the actin cytoskeleton in regulating the outer phosphatidylethanolamine levels in yeast plasma membrane. Eur. J. Biochem. 254:202–206 [DOI] [PubMed] [Google Scholar]
- 46. Mukhopadhyay K, Whitmire W, Xiong YQ, Molden J, Jones T, Peschel A, Staubitz P, Adler-Moore J, McNamara PJ, Proctor RA, Yeaman MR, Bayer AS. 2007. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153:1187–1197 [DOI] [PubMed] [Google Scholar]
- 47. Tsai M, Ohniwa RL, Kato Y, Takeshita SL, Ohta T, Saito S, Hayashi H, Morikawa K. 2011. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 11:13. 10.1186/1471-2180-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curves of Enterococcus faecalis. Growth expressed as optical density (A600) performed in brain heart infusion broth of E. faecalis S613 (▲), R712 (∆), S613ΔliaF177 (■), S613ΔliaF177gdpD170 (♦), and S613ΔliaF177gdpD170cls61 (●). Representative growth curves from at least two independent experiments are shown. Download
Staining of E. faecalis S613 (DAP-susceptible) and R712 (DAP-resistant) strains with BODIPY-labeled DAP (BDP-DAP) at concentrations of 16 µg/ml and 32 µg/ml. The images for a strain at a specific BDP-DAP concentration show representative bacterial cells captured under green fluorescence taken from different fields (bars, 1 µm). Only a small proportion of cells exhibiting fluorescent septal staining with BDP-DAP at 16 and 32 µg/ml was observed in R712 cells compared to S613 cells (20% [n = 104] and 49% [n = 110], respectively; P < 0.0001; two different blinded observers). Download
Fluorescence intensities of DAP-susceptible E. faecalis S613 and DAP-resistant E. faecalis R712. Cells were treated with BDP-DAP, and fluorescence was normalized to cell protein content. Rfu, relative fluorescence units. Download
Staining of E. faecalis S613 derivatives with BODIPY-labeled DAP (BDP-DAP) at concentrations of 16 µg/ml and 32 µg/ml. The images for a strain at a specific BDP-DAP concentration show representative bacterial cells captured under green fluorescence taken from different fields (bars, 1 µm). At 16 µg/ml, there was a statistically significant difference in the percentage of cells exhibiting fluorescent linear septa between strains S613ΔliaF177gdpD170cls61 (32%) and S613ΔliaF177 (57%) and S613ΔliaF177gdpD170 (65%) (P < 0.005). At 32 µg/ml, fluorescent septa were seen in strains S613ΔliaF177gdpD170cls61 (57%), S613ΔliaF177 (69%), and S613ΔliaF177gdpD170 (72%). At least 100 cells were counted for each strain at the indicated concentration by two different blinded observers. Download
10-N-Nonyl acridine orange (NAO) staining of E. faecalis S613 (DAP-susceptible) and R712 (DAP-resistant) strains. The green-to-red shift indicates that the phospholipid domains are likely to contain cardiolipin. (a and b) Excitation was at 490 nm, and emission was at either 528 (a) or 617 (b) nm (bars, 2 µm). (c) Colocalization of green and red fluorescent domains. Download
10-N-Nonyl acridine orange (NAO) staining of representative cells from E. faecalis S613 (DAP susceptible), R712 (DAP resistant), and S613 derivatives. The images for each strain show representative bacterial cells captured with green fluorescence taken from different fields (bars, 1 µm). Fluorescent linear septa were seen in 26% of E. faecalis S613ΔliaF177, 30% of S613ΔliaF177gdpD170, and 28% of S613ΔliaF177gdpD170cls61, respectively (all significantly different [P < 0.001] from the values for strain S613). Linear septa were observed in 81% of S613ΔgdpD170 ([P = 1] from the values for strain S613). At least 50 cells were counted for each strain by two blinded observers. Download
Transmission electron microscopy of E. faecalis daptomycin-susceptible S613 and daptomycin-resistant R712. Red arrows indicate alterations in points of cell contact, leading to infrequent cell separation in strain R712 compared to strain S613 (bars, 2 µm). Download
FM 4-64 staining of representative cells from E. faecalis S613 (DAP susceptible), R712 (DAP resistant), and S613 derivatives. The images for each strain show representative bacterial cells captured with red fluorescence taken from different fields (bars, 1 µm). Homogeneous staining of cell membranes is observed in strains S613ΔliaF177, S613ΔliaF177gdpD170, and S613ΔgdpD170. In contrast, structural alterations of the cell membrane are observed in strains R712 and S613ΔliaF177gdpD170cls61. Download