ABSTRACT
In infection experiments with genetically distinct Mycobacterium tuberculosis complex (MTBC) strains, we identified clade-specific virulence patterns in human primary macrophages and in mice infected by the aerosol route, both reflecting relevant model systems. Exclusively human-adapted M. tuberculosis lineages, also termed clade I, comprising “modern” lineages, such as Beijing and Euro-American Haarlem strains, showed a significantly enhanced capability to grow compared to that of clade II strains, which include “ancient” lineages, such as, e.g., East African Indian or M. africanum strains. However, a simple correlation of inflammatory response profiles with strain virulence was not apparent. Overall, our data reveal three different pathogenic profiles: (i) strains of the Beijing lineage are characterized by low uptake, low cytokine induction, and a high replicative potential, (ii) strains of the Haarlem lineage by high uptake, high cytokine induction, and high growth rates, and (iii) EAI strains by low uptake, low cytokine induction, and a low replicative potential. Our findings have significant implications for our understanding of host-pathogen interaction and factors that modulate the outcomes of infections. Future studies addressing the underlying mechanisms and clinical implications need to take into account the diversity of both the pathogen and the host.
IMPORTANCE
Clinical strains of the Mycobacterium tuberculosis complex (MTBC) are genetically more diverse than previously anticipated. Our analysis of mycobacterial growth characteristics in primary human macrophages and aerogenically infected mice shows that the MTBC genetic differences translate into pathogenic differences in the interaction with the host. Our study reveals for the first time that “TB is not TB,” if put in plain terms. We are convinced that it is very unlikely that a single molecular mechanism may explain the observed effects. Our study refutes the hypothesis that there is a simple correlation between cytokine induction as a single functional parameter of host interaction and mycobacterial virulence. Instead, careful consideration of strain- and lineage-specific characteristics must guide our attempts to decipher what determines the pathological potential and thus the outcomes of infection with MTBC, one of the most important human pathogens.
Introduction
According to the World Health Organization, tuberculosis (TB) is still among the most deadly infectious diseases worldwide (1). When the pathogenesis of TB is considered, it is striking that only approximately 5 to 10% of immunocompetent individuals progress to active TB after infection. For a long time, host (immune status) and environmental factors (e.g., length and intensity of exposure) have been contemplated as major determinants driving the course of infection, while pathogen diversity has been neglected and considered largely unimportant (2).
This paradigm is now changing due to an increasing number of studies demonstrating that the genetic diversity of the pathogens of the Mycobacterium tuberculosis complex (MTBC) (M. tuberculosis, M. africanum, M. canettii, M. microti, M. caprae, M. bovis, and M. pinnipedii) is higher than previously anticipated. This diversity has a substantial impact on the individual virulence and immunogenicity of clinical isolates (reviewed in reference 2). Using state-of-the-art molecular genotyping techniques (e.g., mycobacterial interspersed repetitive units–variable number of tandem repeats [MIRU-VNTR] typing), we demonstrated that the MTBC consists of two clades. One contains exclusively human-adapted M. tuberculosis lineages/genotypes (“modern” lineages, e.g., Beijing and Euro-American Haarlem), and the other is composed of both animal- and human-adapted lineages, e.g., M. africanum West African 1 and 2 or M. bovis (3). The classification of MTBC into six main phylogenetic lineages has been confirmed by high-throughput sequence analyses (4, 5). These analyses also demonstrated that the level of genetic diversity (mainly single nucleotide polymorphisms [SNPs]) in human-adapted MTBC strains is similar to that in animal-adapted strains, potentially reflecting pathogenic differences as a consequence of adaptation to different hosts. Due to reduced purifying selection, most of this diversity is functional (nonsynonymous SNPs) (4). In an investigation using massive parallel sequencing technologies, we could show that even very closely related strains can harbor significant levels of diversity on the whole-genome level, potentially leading to strain-specific virulence determinants (6). This is supported by the fact that the causative difference between the virulent and avirulent variants of the reference strain H37 (H37Rv and H37Ra) has been linked to just three unique SNPs (7).
There is also mounting evidence that this genetic diversity indeed influences the transmissibility and virulence of clinical MTBC isolates and the immune response and clinical picture they evoke (reviewed in reference 2). The first evidence was based on numerous population-based epidemiological studies (including our own in Hamburg, Germany [8]) that have shown that some strains cause large outbreaks while others do not and appear to be attenuated in particular host populations (reviewed in reference 2), thus strongly arguing for the existence of strain-specific pathogenicity traits. A first indication of host-specific adaptation of the pathogen was the finding that MTBC strains of different lineages do not transmit equally in different host populations (9). The amended classification of clinical isolates in different “principal genetic groups” also led to the discovery of clear differences in immune responses (10, 11), rates of progression to disease (12), and disease type (13, 14). In a study in Madagascar, the correlation of the gamma interferon (IFN-γ) responses with the spoligotypes of the infecting clinical strains showed that strains belonging to the “modern” M. tuberculosis lineages, such as Beijing and Central Asian (CAS), induced lower IFN-γ responses in index cases and their household contacts than “ancient” genotypes, like East African-Indian (EAI) strains (15). The findings of that study are in line with a report that suggests that modern strains (clade 1, M. tuberculosis Euro-American superlineage, e.g., Haarlem or X type, Beijing, Delhi-Cas) may have evolved more toward an attenuated inflammatory host response than strains of ancient lineages (clade 2, M. tuberculosis EAI, M. africanum, and M. bovis) (16). These differential pathogenic properties might be related to a more rapid progression to severe disease that has been found in humans and experimental animals infected with strains of the Beijing lineage (10, 17–20). Further indications for specific adaptations to particular hosts were deciphered by analyzing human genetic variants associated with susceptibility to disease. We identified human genetic variants that protect from infection with a particular MTBC genotype (21). Further analyses have shown that there are additional human variants that protect specifically against other MTBC genotypes only (22).
In spite of this clear evidence for the presence of pathogen factors modulating the course of infection, the underlying mechanisms involved in host-pathogen interaction are only poorly understood. It is not clear yet at what stage of the complex process leading to disease successful or virulent strains might have an advantage over attenuated or less-virulent strains. The interaction of the mycobacterium with the macrophage, its primary host cell, leads to a fast activation of cellular signaling pathways, followed by the release of various mediators, such as cytokines or chemokines (23). The differential release of proinflammatory and anti-inflammatory cytokines contributes to the overall cell activation, which may determine whether the pathogen is eradicated or not (18). Therefore, the analysis of the initial host cell response, including intramacrophage growth, may represent a powerful tool to rapidly characterize the virulence of clinical isolates.
To date, studies analyzing the pathogenicity of clinical isolates have mainly been restricted to a very few isolates. The assessment of virulence of particular strains is difficult. In the current study, we analyzed the infection of human primary macrophages with MTBC clade I and clade II strains. MTBC clade I strains showed an enhanced ability to grow in human macrophages and were associated with an increased bacterial replication in the lungs of infected mice compared to MTBC clade II strains. We also demonstrated that a particular MTBC genotype translates into a distinct biological response profile when getting into contact with human immune cells; however, there was no correlation of a particular cytokine release pattern with a particular virulence phenotype as defined by growth characteristics in human macrophages.
RESULTS
MTBC Beijing, Haarlem, and EAI strains differ in their abilities to induce cytokines in human macrophages.
Cytokines are critical effector molecules during the immune response against M. tuberculosis. Thus, we analyzed the cytokine formation of human monocyte-derived macrophages (hMDMs) in response to 13 genetically distinct strains of the MTBC (Table 1) (3). hMDMs were infected at a multiplicity of infection (MOI) of 3:1 with several strains of the clade I group (H37Rv, 4 Beijing and 3 Haarlem strains) and clade II strains (3 EAI, 2 West African 2 [WA2] strains) for 24 h, and the release of tumor necrosis factor alpha (TNF-α), interleukin 12p40 (IL-12p40), IL-1β, RANTES, and IL-6 into the cell culture supernatants was measured by cytometric bead array analysis. In order to take donor-specific response levels into account, data were normalized to the lipopolysaccharide (LPS)-induced responses of the donors. The macrophage responses to distinct isolates were substantially different (Fig. 1): macrophages from all donors analyzed showed only a very weak response to infection with Beijing isolates (4 strains), which is in line with previously published observations (17, 18). In contrast, the infection with Haarlem isolates (3 strains) led to a substantial cytokine response for all 5 mediators analyzed. The response level was increased 5- to 10-fold over the amounts induced by Beijing isolates. Analysis of the culture supernatants after infection with EAI strains showed a very weak response, which resembled the levels observed with Beijing isolates. Analyses of two M. africanum isolates had more ambiguous results: while the WA-2 strain 10514/01 induced a strong release, the response to strain 10517/01 of the same lineage was rather weak. In conclusion, Beijing, Haarlem, and EAI isolates induced a distinct biological response profile: infection with Haarlem isolates led to a strong proinflammatory cytokine response, whereas the response to Beijing and the EAI strains was rather low.
TABLE 1 .
Mycobacterium tuberculosis strains used in this study
| Sample name | Species | Superlineage | Lineage | Note |
|---|---|---|---|---|
| 9679/00 | M. tuberculosis | Clade 1/Euro-American | H37Rv ATCC | Reference strain |
| 1934/03 | M. tuberculosis | Clade 1 | Beijing | |
| 1500/03 | M. tuberculosis | Clade 1 | Beijing | |
| 12594/02 | M. tuberculosis | Clade 1 | Beijing | |
| 49/02 | M. tuberculosis | Clade 1 | Beijing | |
| 4850/03 | M. tuberculosis | Clade 2 | EAI | |
| 1797/03 | M. tuberculosis | Clade 2 | EAI | |
| 947/01 | M. tuberculosis | Clade 2 | EAI | |
| 4130/02 | M. tuberculosis | Clade 1/Euro-American | Haarlem | Nonclustereda |
| 2336/02 | M. tuberculosis | Clade 1/Euro-American | Haarlem | Nonclustereda |
| 9532/03 | M. tuberculosis | Clade 1/Euro-American | Haarlem | Nonclustereda |
| 7761/01 | M. tuberculosis | Clade 1/Euro-American | Haarlem | Clustereda |
| 8921/02 | M. tuberculosis | Clade 1/Euro-American | Haarlem | Clustereda |
| 7679/03 | M. tuberculosis | Clade 1/Euro-American | Haarlem | Clustereda |
| 10514/01 | M. africanum | Clade 2 | West African 2 | |
| 10517/01 | M. africanum | Clade 2 | West African 2 |
a As defined in a longitudinal molecular epidemiological study in Hamburg, Germany (8). Abbreviations: ATCC, American Type Culture Collection; EAI, East African Indian.
FIG 1 .
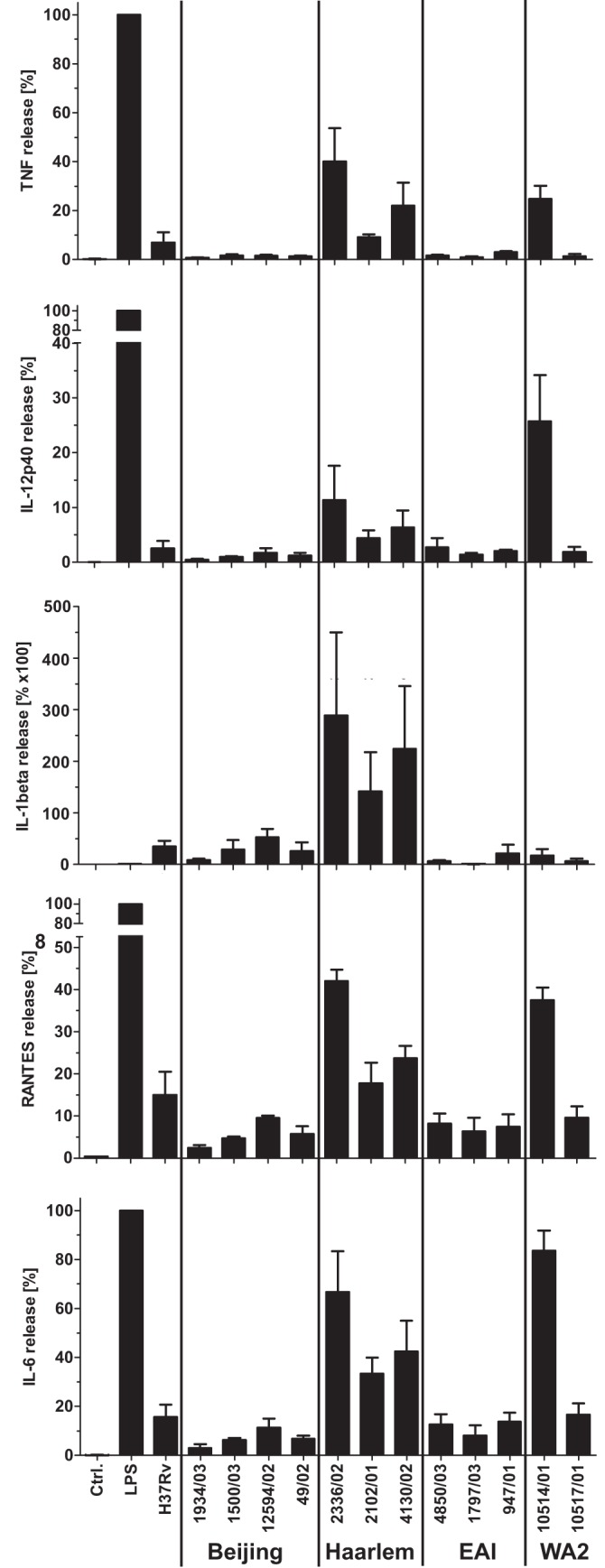
Cytokine release of human macrophages in response to infection with strains of the MTBC. hMDMs were infected with the indicated strains of the MTBC with an MOI of 3:1 for 24 h. The release of TNF, IL12p40, IL-1β, IL-6, and RANTES was measured by cytometric bead array analysis (BD Bioscience). Means ± SE for three independent donors are shown. To adjust for donor-specific response levels, data have been normalized to the lipopolysaccharide (LPS) (10 ng/ml) responses of the donors (=100%).
Uptake of MTBC strains by human macrophages.
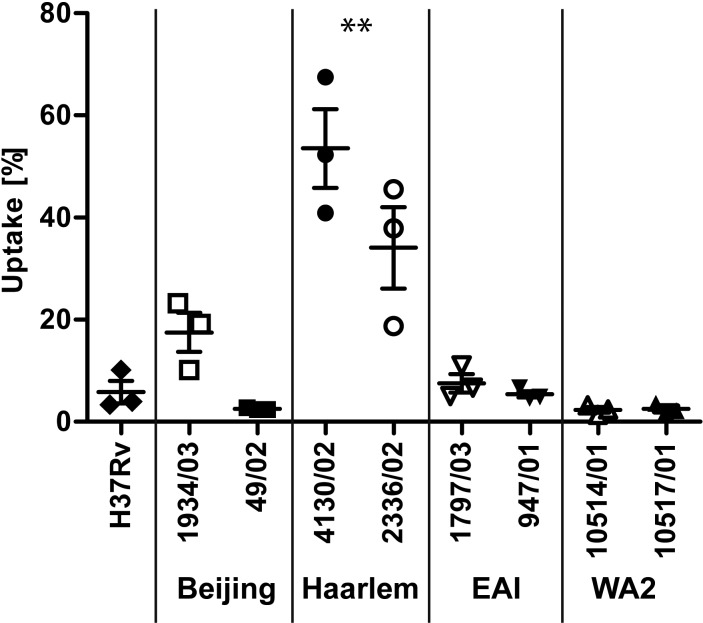
To characterize the interaction of the macrophage with the clinical isolates more closely, we studied the uptake of the strains in human macrophages: 2 × 105 hMDMs were infected with nine different strains of the MTBC: in addition to the reference strain H37Rv, we analyzed two Beijing (1934/03 and 49/02), two Haarlem (4130/02 and 2336/02), two EAI (1797/03 and 947/01), and two WA2 (10514/01 and 10517/01) strains. Cells were infected with an MOI of 0.5 to 1 bacterium per macrophage (Fig. 2) and washed 4 h postinfection (p.i.). The numbers of CFU were determined by serial dilutions of cellular lysates.
FIG 2 .
Differential uptake of MTBC strains by human macrophages. hMDMs were infected with strains of different lineages of the MTBC with an MOI of 1:1 for 4 h. Quantification of viable CFU of the inoculum and 4 h postinfection was conducted by lysis of monolayers, serial dilution, and plating on 7H10 medium. Shown is the uptake (%) of bacteria related to the inoculum of each individual strain (mean ± SE for three independent experiments performed). Strains were grouped into lineages, and the uptake rates were compared by one-way analysis of variance (ANOVA); Tukey multiple-comparison test, **, P < 0.01).
The cells ingested between 2% and 15% of the inoculated bacteria in seven of the nine strains investigated (H37Rv, Beijing, EAI, and WA2). The uptake rates of these seven isolates were not significantly different. Of note, the uptake rates of the M. tuberculosis Haarlem strains 2336/02 and 4130/02 were significantly different from those of the strains of the other lineages: macrophages phagocytosed 34.08% ± 13.8% and 53.56% ± 13.33% (mean ± standard error of the mean [SEM]) of the inoculated bacteria, respectively.
Differential growth of MTBC clade I and clade II strains in human macrophages.
Parallel cultures of MTBC strain-infected macrophages (see above) were cultured for 7 additional days and lysed, and the numbers of CFU were subsequently determined (Fig. 3A): H37Rv and the Beijing and Haarlem isolates showed a substantial growth in hMDMs. Compared to the amount of intracellular bacteria 4 h after infection, 13.3-fold (H37Rv), 11.8- and 16.5-fold (Beijing 49/02 and 1934/03), and 9.0- and 9.4-fold (Haarlem 2336/02 and 4130/02) increases in CFU were observed. In contrast, the fold increase ratios for macrophage cultures infected with clade II strains were consistently low (0.8 and 3.8 for EAI [1797/03 and 947/01] and 0.7 and 3.7 for WA2 10514/01 and 10517/01). These data suggest that there are significant differences in the growth potentials of MTBC clinical isolates in human macrophages (Fig. 3B). They demonstrate that MTBC clade I strains (Beijing, Haarlem, and the H37Rv reference strain) have a better growth capacity in human macrophages than clade II strains.
FIG 3 .
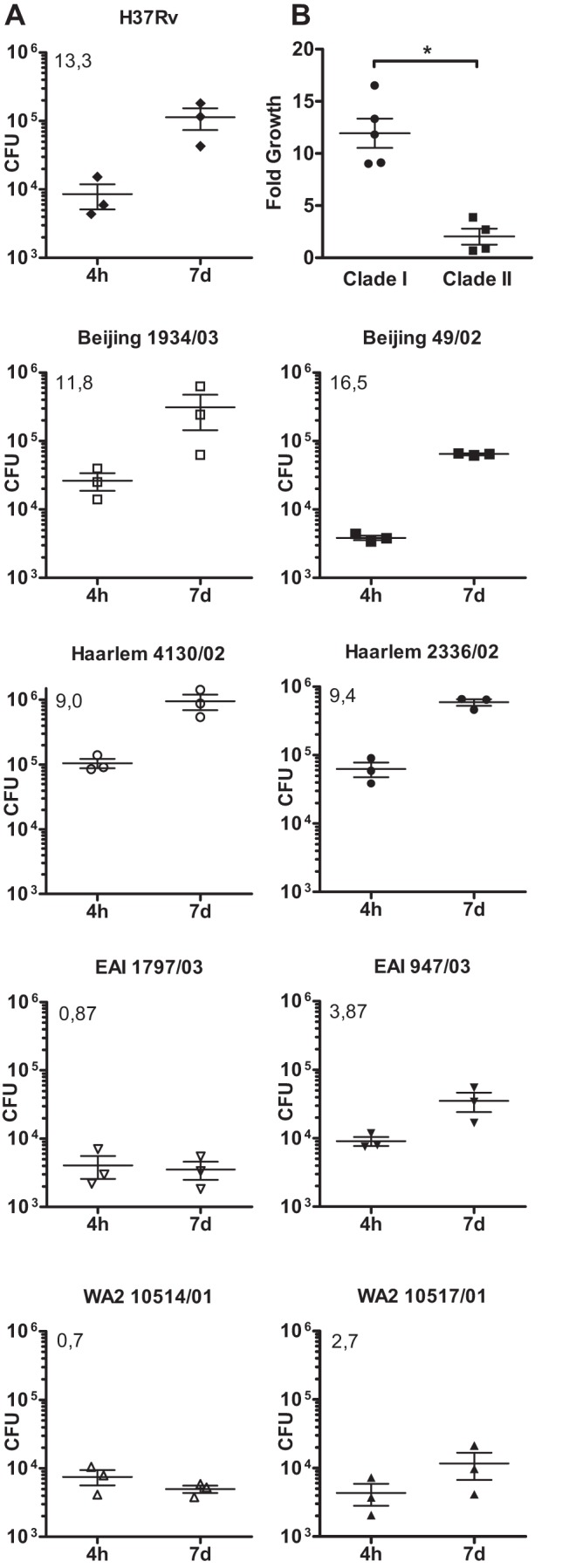
Differential growth of MTBC strains in human macrophages. (A) hMDMs were infected with the indicated strains of different lineages of the MTBC with an MOI of 1:1 for 4 h and 7 days. Quantification of viable CFU at 4 h and 7 days postinfection was conducted by lysis of monolayers, serial dilution, and plating on 7H10 medium. Error bars indicate standard errors of the means from three independent experiments performed with cells from different donors, each consisting of two technical replicates per strain. The mean of the fold increase is shown in the upper left corner of each graph. (B) Fold growth of MTBC clade I and clade II in human macrophages. Shown are the means ± SEM of all clade I and II strains analyzed in panel A (*, P < 0.05, Mann-Whitney U test).
In order to analyze whether the growth of these strains would be similar in their natural habitat, we infected human bronchoalveolar lavage (BAL) cell cultures at a ratio of 3:1 and monitored uptake and intracellular growth of the bacteria. In cells from two independent donors, the clinical isolates Haarlem 2336/02 and Beijing 49/02 multiplied considerably faster in BAL cell cultures than the EAI 1797/03 and WA2 10514/01 strains (Fig. 4). These data demonstrate that clade I strains grow better in human alveolar lung cells than clade II strains.
FIG 4 .
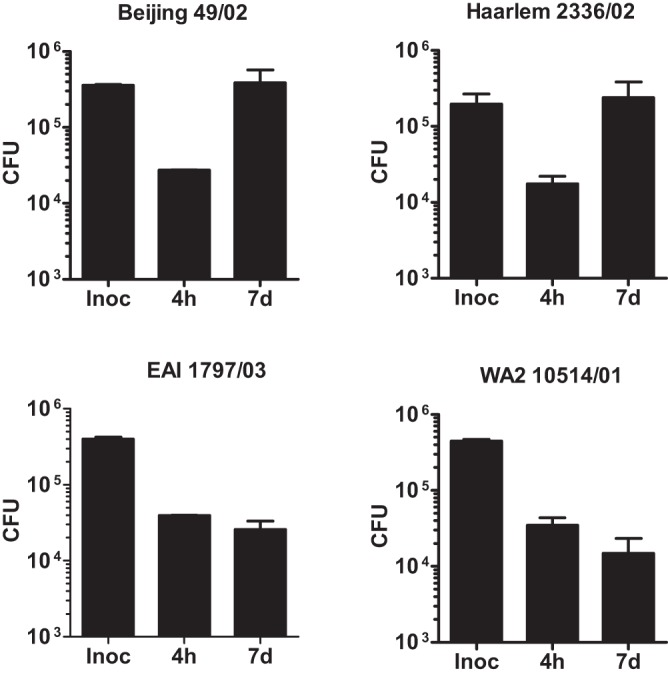
Differential growth of MTBC strains in human bronchoalveolar cells. Cells isolated from bronchoalveolar lavage of healthy donors were infected with the indicated strains of different lineages of the MTBC with an MOI of 1:1 for 4 h and 7 days. Quantification of viable CFU at 4 h and 7 days postinfection was conducted by lysis of monolayers, serial dilution, and plating on 7H10 medium. Error bars indicate standard deviations for two independent experiments performed with cells from different donors, each consisting of two technical replicates per strain.
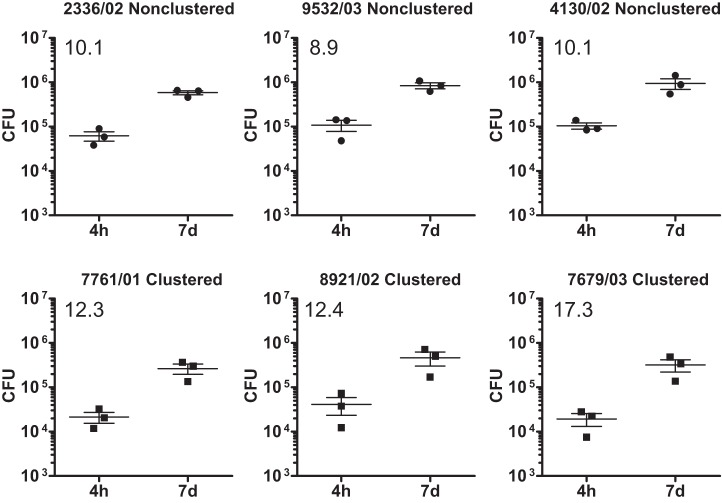
Comparable growth of clustered and nonclustered M. tuberculosis Haarlem strains from Hamburg, Germany.
Strains isolated from TB patients in Hamburg, Germany, belong predominantly to the M. tuberculosis Haarlem genotype. Some of these strains have previously been shown to cause major tuberculosis outbreaks (8), whereas other Haarlem strains are detected only sporadically. In order to analyze whether clinically successful (clustered) Haarlem strains show an enhanced growth in hMDMs, infection experiments were performed with three clustered strains (7761/01, 8921/02, and 7679/03) and three nonclustered strains (2336/02, 9532/03, and 4130/02). All six strains showed a profound growth in cells from three different donors, again demonstrating that clade I strains have a strong capacity to multiply in macrophages derived from healthy European blood donors (Fig. 5). A comparative analysis between clustered and nonclustered isolates showed a slightly superior growth of outbreak isolates; however, this did not reach statistical significance.
FIG 5 .
Growth of different MTBC Haarlem strains in human macrophages. hMDMs were infected with the indicated strains of different lineages of the MTBC with an MOI of 1:1 for 4 h and 7 days. Quantification of viable CFU at 4 h and 7 days postinfection was conducted by lysis of monolayers, serial dilution, and plating on 7H10 medium. Error bars indicate standard errors of the means for three independent experiments performed with cells from different donors, each consisting of two technical replicates per strain. The mean fold increase is shown in the upper left corner of each graph.
Aerosol infection of mice: strains of the EAI and Beijing genotypes differ in CFU development and survival rates.
We extended our study to mouse aerosol infection experiments in order to monitor whether the differential growth characteristics observed in human macrophages would also be seen in the experimental tuberculosis infection in vivo. We infected C57BL/6 (genetically resistant) and DBA/2 (genetically susceptible) mice with H37Rv and two clinical isolates: Beijing 49/02 (clade I) and EAI 1797/03 (clade II). The pulmonary bacterial burden after infection with the Beijing isolate was comparable to that for H37Rv (clade I), which was used as reference strain (Fig. 6A). Increasing bacterial numbers to approximately 107 CFU up to day 21 postinfection were seen for both genotypes, followed by a plateau as shown previously (24). In contrast, the bacterial burden after EAI isolate infection was approximately 100-fold lower than the level of Beijing isolate CFU. Lungs of mice infected with the Beijing isolates also showed higher numbers of cellular infiltrates and larger granulomatous lesions at days 21 and 90 than were seen with EAI strain infection (Fig. 7). These data demonstrate that the EAI strain was less virulent than the Beijing isolate although the mice were infected with twice as many bacteria. To independently investigate the virulences of the different isolates, we also monitored the survival time of susceptible DBA/2 mice (25) after aerosol infection with the strains described above. The median survival time of mice infected with the Beijing genotype was 122 days (Fig. 6B). In contrast, 60% of the mice infected with the EAI strains were still alive on day 450 postinfection. Mice infected with H37Rv succumbed at day 120 or day 200 depending on the infection dose. Mice infected with EAI 1797/03 showed a better survival than mice infected with Beijing 49/02 and also H37Rv. These data have been confirmed in independent experiments with one additional EAI strain and Beijing strain (EAI 4850/03 and Beijing 1934/03) (data not shown). This demonstrates reduced virulence of the clade II EAI strains compared to that of H37Rv and the Beijing isolates, which belong to MTBC clade I.
FIG 6 .
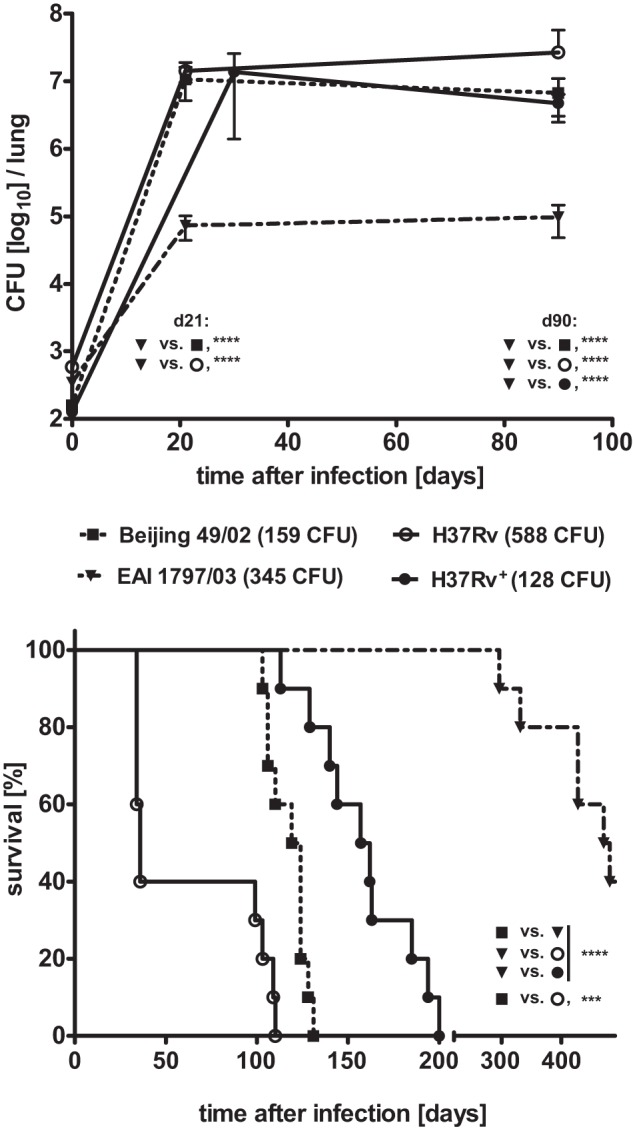
Virulence of selected genotypes of the MTBC in mice. Detection of bacterial replication in the lung (A) or survival time (B) after aerosol infection with an intended infection dose of 200 CFU of representative strains of the EAI (filled triangles, dot-dash line) and Beijing (filled squares, dotted line) genotypes and H37Rv (empty circles, continuous line) as the reference strain. The bacterial burden was analyzed in the lungs of C57BL/6 mice; the survival time after infection with these strains was monitored in DBA/2 mice. Determination of the inoculum size revealed that mice were infected with 588 CFU of the H37Rv strain, which is 1.7× higher than the infection dose of the EAI (345 CFU) and 3.7× higher than the inoculum size of the Beijing (159 CFU) genotype. In order to allow a better comparison of the bacterial replication and survival time between the three genotypes, results from a parallel infection experiment with an inoculum size of 128 CFU of the H37Rv strain are included (filled circles, continuous line). Statistics: for CFU determination, data were log transformed and analyzed by two-way ANOVA with a Bonferroni posthoc test; ****, P < 0.0001; survival, log-rank Mantel Cox test; ****, P < 0.0001; ***, P < 0.0005.
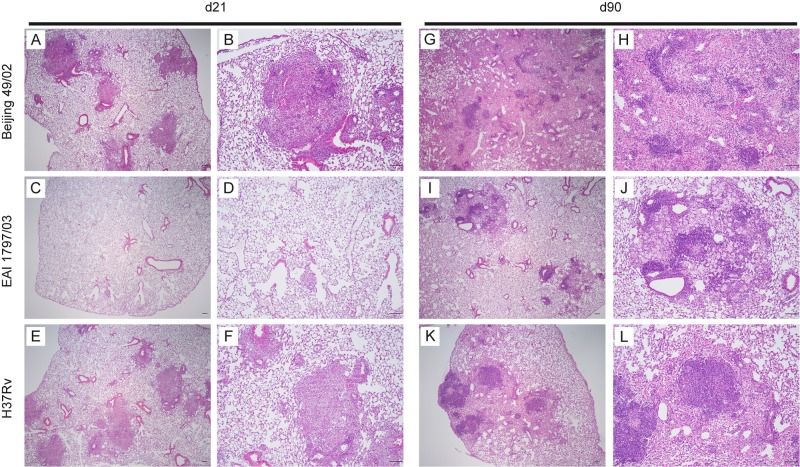
FIG 7 .
Histology of lungs infected with selected genotypes of the MTBC. Representative histopathological findings for the lungs at 21 and 90 days after aerosol infection with isolates of the Beijing (A and B, d21; G and H, d90) or EAI (C and D, d21; I and J, d90) genotype and H37Rv (E and F, d21; K and L, d90) as a reference strain (intended infection dose, 200 CFU). Sections of the lungs were stained with H&E and are shown at low (×40) (A, C, E, G, K, and L) or higher (×100) (B, D, F, H, I, and J) magnification, respectively. Scale bars, 100 µm.
DISCUSSION
The current study identifies lineage-specific differences in the virulence of clinical MTBC isolates in human primary macrophages and in a well-established mouse model of aerosol infection. Based on our data, previous concepts attempting to explain various virulences of clinical isolates, e.g., by a direct relationship between the inflammatory response and strain success, may have been too simplistic. While we confirmed that some strains of the “modern/clade I” MTBC branch induce only a mild inflammatory response, others did not. This seems to be dependent on the lineage they belong to (Haarlem, high; Beijing, low). Even more important, the inflammatory profiles neither correlate with virulence in the experimental setting nor with epidemiological success. Thus, this feature appears to be of limited use for the prediction of virulence characteristics of clinical isolates.
Only a few studies with human macrophages attempted to correlate the release of cytokines induced by M. tuberculosis clinical isolates with the strain lineage: the authors hypothesized that low-inflammatory-response profiles were linked to “modern” MTBC lineages, global expansion, and adaptation to higher population densities (16, 26). While these studies provided the first valuable indications that the early inflammatory profile of clinical isolates may be related to the MTBC phylogeny, their proposal of a general link between weak cytokine induction and strong virulence may have been premature in light of the current study. Portevin et al. (16) reported that the levels of cytokines released by infected human peripheral blood monocyte-derived macrophages were highly variable in response to different strains. Nevertheless, there was a significantly lower immune response to strains belonging to modern lineages than to strains from ancient lineages. This was suggested to provide a selective advantage by favoring more-rapid disease progression and transmission (16). Similar observations were made in other studies specifically for strains of the Beijing lineage that induced low levels of proinflammatory TNF and IL-1β (26).
While our data confirm that Beijing strains induce low cytokine levels and grow better in human macrophages, there is no general correlation of low cytokine induction with high virulence, since Euro-American Haarlem strains induced high levels of cytokines in all donors investigated, associated with a high virulence in human primary macrophages. Enhanced cytokine and chemokine formation induced by Haarlem isolates in human macrophages compared to results with strains of the Beijing lineage was also recently observed by others (27). The limited value of proinflammatory cytokines as a sole predictive parameter for replication and virulence is further supported by our observation that the cytokine profile in response to EAI strains resembles the very low cytokine levels induced by Beijing isolates. However, the EAI strains only have a very limited replication rate in human macrophages. A direct correlation between cytokine induction and growth is jeopardized by the results for the two M. africanum WA2 isolates investigated. Infection with WA2 strain 10514/01 induced a strong host cell response, indicated by the secretion of TNF-α, IL-6, IL-12 p40, and RANTES. In contrast, macrophages infected with WA2 strain 10517/01 hardly produced any cytokines. Of note, neither strain showed profound growth in macrophages compared to results for Beijing and Haarlem isolates. These data hold also true for a larger test panel of Haarlem strains that comprise attenuated (not involved in outbreaks; nonclustered) and highly successful strains (involved in outbreaks; clustered) determined in a longitudinal population-based epidemiological study in Hamburg, Germany (8). Cytokine induction was similarly high in clustered and nonclustered strains (data not shown), while growth in the human macrophage model was enhanced in successful strains, although this did not reach statistical significance. Taken together, it is likely that mechanisms independent from the capacity for cytokine production may better correlate with mycobacterial growth in macrophages. In order to identify these processes, we are currently performing detailed gene expression profiling experiments in macrophages infected with different lineages of MTBC.
Findings of the current study also suggest that uptake into human monocytic cells may be involved in strain success. Compared to that for all other strains investigated, the uptake of the Haarlem isolates was 3- to 5-fold enhanced in macrophages differentiated from healthy European blood donors. Currently, the molecular mechanisms for this enhanced uptake and for successful growth remain elusive.
Considering the full panel of strains analyzed, there is evidence for at least three different pathogenic profiles: (i) strains of the Beijing lineage are characterized by low uptake, low cytokine induction, and a high replicative potential, (ii) strains of the Haarlem lineage by high uptake, high cytokine induction, and high growth rates, and (iii) EAI strains by low uptake, low cytokine induction, and low replicative potential. Similar to the EAI isolates, the M. africanum West African strains showed a low-virulence phenotype in human macrophages but were characterized to induce a different cytokine induction profile.
Overall, the data presented here support previous suggestions that modern/clade I and ancient/clade II have clear pathogenic differences that are nonetheless independent from cytokine induction profiles. We extended previous findings for murine bone marrow-derived macrophages (28) to human monocyte-derived macrophages, alveolar macrophages, and aerosol mouse infection models. An important question remains to be considered: how can the specific growth characteristics of clade I and clade II strains observed in independent experimental systems be explained?
A potential explanation is based on the detailed analysis of the phylogenetic tree obtained by MIRU-VNTR analysis (3). All strains belonging to the clade I carry the M. tuberculosis-specific TbD1 deletion, which was identified in a seminal study by Brosch et al. (29). Based on the presence or absence of TbD1, M. tuberculosis strains can be divided into “ancestral” and “modern” strains, the latter comprising representatives of major epidemics like the Beijing, Haarlem, and African M. tuberculosis isolates. In molecular terms, TbD1 stands for a DNA fragment of 2,153 bp which encodes two proteins: the transmembrane transport protein mmpL6 (Rv1557) and the putative membrane protein mmpS6. In TbD1-deficient strains, mmpL6 is truncated and mmpS6 is absent. Thus, based on the current study, the deletion of these two proteins is associated with enhanced growth in human macrophages. The comparative analysis of clade I and II strains as shown here represents a first functional correlation that the deletion of these two proteins indeed confers some selective advantage to “modern” M. tuberculosis strains.
When the data are seen from a host cell perspective, it needs to be considered that our experiments were performed with macrophages from healthy blood donors from Northern Europe. The observation that all “modern” or clade I strains were characterized by massive growth, whereas “ancient,” clade II strains were not or showed only a very limited growth potential in human macrophages, may indicated that Caucasian donors may have developed mechanisms that limit the replication of ancient TB strains. It is intriguing to speculate on what would happen if our study were performed with cells from donors originating from West or East Africa. If macrophages from African blood donors were to show an enhanced “in vitro susceptibility” to clade II strains, this would indicate that specific host factors, which differ between ethnic groups with distinct genetic backgrounds, may also impact the host’s ability to control a particular strain. This is particularly interesting since it was recently shown in a study in Ghana that the development of tuberculosis as determined by X-ray data and clinical parameters does not differ between patients infected with a clade II M. africanum or a clade I African M. tuberculosis isolate (22, 30).
It is a very common theme in microbiology that bacteria exert different virulence characteristics due to the expression of distinct virulence factors. At the species and subspecies levels, these factors are often encoded on virulence plasmids and pathogenicity islands, mechanisms which to our knowledge do not contribute to differences in mycobacterial virulence. At the strain level, it has become apparent that genomically diverse isolates of Streptococcus mutans, for example, exhibit considerable phenotypic heterogeneity (31), and genetic diversity in Salmonella enterica serovar Enteritidis has recently been linked to genetic polymorphisms in known Salmonella virulence pathways (32). The current study demonstrates that the genetic diversity of MTBC strains also translates into pathogenic consequences. The identities of the individual factors which are functionally responsible for the differential behaviors of the MTBC complex strains are currently not known. Thus, a thorough structural analysis of these isolates is needed to identify and functionally characterize these virulence factors. In conclusion, strain- and lineage-specific characteristics must no longer be ignored when attempting to understand what determines the transmissibility and the pathogenic potential and thus the outcome of infection with MTBC
MATERIALS AND METHODS
Ethics statement.
All experiments performed with human cells were approved by the Ethics Committee of the University of Lübeck, Lübeck, Germany (permits 07/125, 10/215, and 12/116). The animal experiments performed were approved by the Ethics Committee for Animal Experiments of the Ministry for Environment, Nature, Protection and Agriculture of the State of Schleswig-Holstein (Kommission für Tierversuche/Ethik-Kommission des Landes Schleswig-Holstein), Kiel, Germany, permit V312-72241.123-3 (112-11/10) (“Untersuchung des immunmodulatorischen Potentials von epidemiologisch relevanten klinischen Stämmen des M.tb-Komplexes”/“Study on the immunomodulatory potential of epidemiologically relevant clinical strains of the M.tb-complex”).
Bacteria.
Bacterial strains were initially cultured from clinical samples on Löwenstein/Jensen (L/J) medium at the National Reference Center for Mycobacteria in Borstel, Germany. The MTBC clinical isolates used in this study were handled to minimize in vitro passaging. All cultures used in this study were derived from frozen stocks prepared after a single in vitro passage of original archived samples. The virulent lab strain H37Rv, however, has undergone countless rounds of in vitro passaging. All strains were further characterized by various genotyping methods and susceptibility testing as described elsewhere (3). The unweighted-pair group method using average linkages (UPGMA) tree of the strains used is depicted in Fig. S1 in the supplemental material. Homogenous bacterial suspensions were prepared from L/J cultures in 10 ml 7H9 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.05% Tween 80, and 0.2% glycerol and incubated in 30-ml square medium bottles (Nalgene) at 37°C without shaking (preculture). Growth to mid-log phase was monitored by measuring the optical density at 600 nm (OD600) every second day (Bio-Tek Synergy). Precultures were transferred into a roller bottle system (Corning) and incubated at 37°C/5 rpm. Twenty milliliters of fresh medium was gradually added to a final volume of 100 ml (main culture). Mid-log-phase bacterial suspensions (OD600, 0.2 to 0.4) were stored in 1-ml aliquots at −80°C for further investigations. Numbers of CFU were determined for each strain investigated. Furthermore, sterility controls (Ziehl-Neelsen staining, blood agar, and LB medium) were performed for all precultures and main cultures.
Isolation, differentiation, and cultivation of hMDMs and BAL cells.
Mononuclear cells were isolated from peripheral blood mononuclear cells (PBMC) of healthy volunteers by density gradient centrifugation. Monocytes were separated (purity consistently greater than 92%) by counterflow elutriation. Human monocyte-derived macrophages (hMDMs) were generated in the presence of 10 ng/ml recombinant human macrophage colony-stimulating factor (M-CSF) from highly purified monocytes as described elsewhere (33). For the isolation of bronchoalveolar lavage (BAL) cells, a flexible bronchoscopy was performed according to German guidelines with intravenous and local anesthesia at the physician’s discretion (34). The bronchoscope was wedged into a subsegmental bronchus of the middle lobe, and bronchoalveolar lavage with a total volume of 250 ml sterile saline was performed. Single-cell suspensions from BAL fluid were obtained by passing the BAL fluid through a stainless steel sieve (Teesieb Profi Plus; WMF, Geislingen, Germany) with a mesh aperture of 0.5 mm. BAL cells (4 × 105) (containing approximately 80% alveolar macrophages) were cultured in 500 µl RPMI 1640 with 10% fetal calf serum (FCS) and 4 mM l-glutamine, 10 ng/ml M-CSF, penicillin G (100 U/ml), and amphotericin B (5 µg/ml) in 48-well flat-bottom microtiter plates (Nunc).
Infection experiment analysis of mycobacterial growth.
hMDMs (2 × 105) were cultured in 500 µl RPMI 1640 with 10% FCS and 4 mM l-glutamine in 48-well flat-bottom microtiter plates (Nunc) at 37°C in a humidified atmosphere containing 5% CO2. Macrophages were infected with MTBC strains at the indicated MOI. Four hours postinfection, nonphagocytosed bacteria were removed by washing three times with 0.5 ml Hanks’ balanced salt solution (HBSS) (Invitrogen, Karlsruhe, Germany) at 37°C. After washing and after 3 days of cultivation, 0.5 ml medium was added to the macrophage culture. At day 7, supernatants were completely removed, and macrophage cultures were lysed at 4 h and 7 days postinfection by adding 10 µl 10% saponin solution (Sigma, Steinheim, Germany) in HBSS at 37°C for 15 min. Lysates were serially diluted in sterile water containing 0.05% Tween 80 (Merck, Darmstadt, Germany) and plated twice on 7H10 agar containing 0.5% glycerol (Serva) and 10% heat-inactivated bovine calf serum (BioWest, France). After 3 weeks at 37°C, the CFU were counted.
Quantification of cytokine production.
Cytokine levels of TNF-α, IL-6, IL-1β, IL12p40, and RANTES in supernatants from uninfected and infected macrophage cultures were determined 24 h postinfection (p.i.) by cytometric bead array (CBA) technology using a FACSArray bioanalyzer system (BD Biosciences) according to the manufacturer’s instructions. Data and cytokine concentrations were analyzed using the FCAP Array software program (BD Biosciences).
M. tuberculosis aerosol infection experiments with mice.
Female 6- to 8-week-old specific-pathogen-free mice were maintained in individually ventilated cages (IVC) (Ebeco, Castrop-Rauxel, Germany) under biosafety level III conditions. Mice were aerogenically infected (Glas-Col, Terre-Haute, IN) with approximately 200 CFU of representative isolates of the Beijing and EAI genotypes, as well as the reference strain H37Rv. Inoculum size was confirmed 24 h after infection by determining the bacterial burden in the lung. Bacterial replication in the lungs of C57BL/6 mice was determined. At indicated time points after infection, lungs from five animals per group were aseptically removed, weighed, and homogenized in distilled sterile water containing 0.05% Tween 80. Tenfold serial dilutions of lung homogenates were plated on Middlebrook 7H10 agar supplemented with 10% bovine serum and incubated at 37°C for 21 days. Colonies on plates were enumerated, and results were expressed as log10 CFU per organ. The survival time after infection was monitored in DBA/2 mice. Mice that lost 25% of their original weight during the course of infection were scored as moribund and had to be sacrificed.
Histopathology.
For histopathological analysis, lung lobes were fixed in 4% formalin-phosphate-buffered saline (PBS), set in paraffin blocks, and sectioned (2 to 3 µm). Histology was performed using standard protocols for hematoxylin-eosin (H&E) staining.
SUPPLEMENTAL MATERIAL
UPGMA tree (left panel) based on the copy numbers of 24 MIRU-VNTR loci (right panel, spoligotyping results whereby a repeat region’s presence or absence is indicated by a black box or white box, respectively) of the 16 strains investigated. The tree was calculated using the MIRU-VNTRplus server (http://www.miru-vntrplus.org; distance measure, categorical). Download
ACKNOWLEDGMENTS
This work was supported in part by BMBF grant project 01KI0784, WP d: “Pathogenic variability of clinical M. tb isolates: implications for the modulation of the immune system, infectivity and disease progression” (to S.N. and N.R.), the EU FP7 TB-PAN-NET (FP7-223681) project (S.N.), and DFG SPP1580 priority program Re 1228/5-1 to N.R.
We gratefully acknowledge Silvia Maaß and Martina Ackermann for expert help and technical assistance.
N.R., S.H., K.W., and S.N. conceived and designed the experiments. S.H., K.W., J.B., L.N., M.E., C.H., C.L., and R.D. performed the experiments. N.R., K.W., S.H., and S.N. analyzed the data. N.R., S.H., K.W., C.H., S.E., and S.N. wrote the article.
Footnotes
Citation Reiling N, Homolka S, Walter K, Brandenburg J, Niwinski L, Ernst M, Herzmann C, Lange C, Diel R, Ehlers S, Niemann S. 2013. Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice. mBio 4(4):e00250-13. doi:10.1128/mBio.00250-13.
REFERENCES
- 1. WHO 2012. Global tuberculosis report 2012. WHO, Geneva, Switzerland [Google Scholar]
- 2. Gagneux S, Small PM. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328–337 [DOI] [PubMed] [Google Scholar]
- 3. Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160 http://dx.doi.org/doi:10.1371/journal.ppat.1000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6:e311. 10.1371/journal.pbio.0060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niemann S, Köser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, Kokko-Gonzales P, Murray LJ, Rigatti R, Smith VP, Arends FP, Cox HS, Smith G, Archer JA. 2009. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One 4:e7407. 10.1371/journal.pone.0007407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee JS, Krause R, Schreiber J, Mollenkopf HJ, Kowall J, Stein R, Jeon BY, Kwak JY, Song MK, Patron JP, Jorg S, Roh K, Cho SN, Kaufmann SH. 2008. Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe 3:97–103 [DOI] [PubMed] [Google Scholar]
- 8. Diel R, Seidler A, Nienhaus A, Rüsch-Gerdes S, Niemann S. 2005. Occupational risk of tuberculosis transmission in a low incidence area. Respir. Res. 6:35. 10.1186/1465-9921-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. López B, Aguilar D, Orozco H, Burger M, Espitia C, Ritacco V, Barrera L, Kremer K, Hernandez-Pando R, Huygen K, van Soolingen D. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Jong BC, Hill PC, Brookes RH, Gagneux S, Jeffries DJ, Otu JK, Donkor SA, Fox A, McAdam KP, Small PM, Adegbola RA. 2006. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J. Infect. Dis. 193:1279–1286 [DOI] [PubMed] [Google Scholar]
- 12. De Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, Jackson-Sillah DJ, Fox A, Deriemer K, Gagneux S, Borgdorff MW, McAdam KP, Corrah T, Small PM, Adegbola RA. 2008. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 198:1037–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NT, Thuong NT, Stepniewska K, Huyen MN, Bang ND, Loc TH, Gagneux S, van Soolingen D, Kremer K, van der Sande M, Small P, Anh PT, Chinh NT, Quy HT, Duyen NT, Tho DQ, Hieu NT, Torok E, Hien TT, Dung NH, Nhu NT, Duy PM, van Vinh Chau N, Farrar J. 2008. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 4:e1000034 http://dx.doi.org/10.1371/journal.ppat.1000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong Y, Cave MD, Zhang L, Foxman B, Marrs CF, Bates JH, Yang ZH. 2007. Association between Mycobacterium tuberculosis Beijing/W lineage strain infection and extrathoracic tuberculosis: insights from epidemiologic and clinical characterization of the three principal genetic groups of M. tuberculosis clinical isolates. J. Clin. Microbiol. 45:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rakotosamimanana N, Raharimanga V, Andriamandimby SF, Soares JL, Doherty TM, Ratsitorahina M, Ramarokoto H, Zumla A, Huggett J, Rook G, Richard V, Gicquel B, Rasolofo-Razanamparany V. 2010. Variation in gamma interferon responses to different infecting strains of Mycobacterium tuberculosis in acid-fast bacillus smear-positive patients and household contacts in Antananarivo, Madagascar. Clin. Vaccine Immunol. 17:1094–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Portevin D, Gagneux S, Comas I, Young D. 2011. Human macrophage responses to clinical isolates from the mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 7:e1001307 http://dx.doi.org/doi:10.1371/journal.ppat.1001307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., III 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84–87 [DOI] [PubMed] [Google Scholar]
- 18. Manca C, Reed MB, Freeman S, Mathema B, Kreiswirth B, Barry CE, III, Kaplan G. 2004. Differential monocyte activation underlies strain-specific Mycobacterium tuberculosis pathogenesis. Infect. Immun. 72:5511–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanveer M, Hasan Z, Kanji A, Hussain R, Hasan R. 2009. Reduced TNF-alpha and IFN-gamma responses to Central Asian strain 1 and Beijing isolates of Mycobacterium tuberculosis in comparison with H37Rv strain. Trans. R. Soc. Trop. Med. Hyg. 103:581–587 [DOI] [PubMed] [Google Scholar]
- 20. Hanekom M, van der Spuy GD, Streicher E, Ndabambi SL, McEvoy CR, Kidd M, Beyers N, Victor TC, van Helden PD, Warren RM. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 45:1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herb F, Thye T, Niemann S, Browne EN, Chinbuah MA, Gyapong J, Osei I, Owusu-Dabo E, Werz O, Rüsch-Gerdes S, Horstmann RD, Meyer CG. 2008. ALOX5 variants associated with susceptibility to human pulmonary tuberculosis. Hum. Mol. Genet. 17:1052–1060 [DOI] [PubMed] [Google Scholar]
- 22. Intemann CD, Thye T, Niemann S, Browne ENL, Amanua Chinbuah M, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, Helm S, Rüsch-Gerdes S, Horstmann RD, Meyer CG. 2009. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 5:e1000577 http://dx.doi.org/doi:10.1371/journal.ppat.1000577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flynn JL, Chan J. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93–129 [DOI] [PubMed] [Google Scholar]
- 24. Reiling N, Hölscher C, Fehrenbach A, Kröger S, Kirschning CJ, Goyert S, Ehlers S. 2002. Cutting edge: Toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to airborne infection with Mycobacterium tuberculosis. J. Immunol. 169:3480–3484 [DOI] [PubMed] [Google Scholar]
- 25. North RJ, Jung YJ. 2004. Immunity to tuberculosis. Annu. Rev. Immunol. 22:599–623 [DOI] [PubMed] [Google Scholar]
- 26. Sarkar R, Lenders L, Wilkinson KA, Wilkinson RJ, Nicol MP. 2012. Modern lineages of Mycobacterium tuberculosis exhibit lineage-specific patterns of growth and cytokine induction in human monocyte-derived macrophages. PLoS One 7:e43170. 10.1371/journal.pone.0043170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang C, Peyron P, Mestre O, Kaplan G, van Soolingen D, Gao Q, Gicquel B, Neyrolles O. 2010. Innate immune response to Mycobacterium tuberculosis Beijing and other genotypes. PLoS One 5:e13594. 10.1371/journal.pone.0013594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Homolka S, Niemann S, Russell DG, Rohde KH. 2010. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 6:e1000988 http://dx.doi.org/10.1371/journal.ppat.1000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99:3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thye T, Niemann S, Walter K, Homolka S, Intemann CD, Chinbuah MA, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, Rüsch-Gerdes S, Horstmann RD, Ehlers S, Meyer CG. 2011. Variant G57E of mannose binding lectin associated with protection against tuberculosis caused by Mycobacterium africanum but not by M. tuberculosis. PLoS One 6:e20908. 10.1371/journal.pone.0020908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmer SR, Miller JH, Abranches J, Zeng L, Lefebure T, Richards VP, Lemos JA, Stanhope MJ, Burne RA. 2013. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One 8:e61358. 10.1371/journal.pone.0061358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Allard MW, Luo Y, Strain E, Pettengill J, Timme R, Wang C, Li C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW. 2013. On the evolutionary history, population genetics and diversity among isolates of salmonella enteritidis PFGE pattern JEGX01.0004. PLoS One 8:e55254. 10.1371/journal.pone.0055254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reiling N, Blumenthal A, Flad HD, Ernst M, Ehlers S. 2001. Mycobacteria-induced TNF-alpha and IL-10 formation by human macrophages is differentially regulated at the level of mitogen-activated protein kinase activity. J. Immunol. 167:3339–3345 [DOI] [PubMed] [Google Scholar]
- 34. Häussinger K, Ballin A, Becker HD, Bölcskei P, Dierkesmann R, Dittrich I, Frank W, Freitag L, Gottschall R, Guschall WR, Hartmann W, Hauck R, Herth F, Kirsten D, Kohlhäufl M, Kreuzer A, Loddenkemper R, Macha N, Markus A, Stanzel F, Steffen H, Wagner M, Working Party on Recommendations for Quality Standards in Endoscopy of the German Society of Pulmonology (Section Endoscopy) 2004. Recommendations for quality standards in bronchoscopy. Pneumologie 58:344–356 (In German.) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UPGMA tree (left panel) based on the copy numbers of 24 MIRU-VNTR loci (right panel, spoligotyping results whereby a repeat region’s presence or absence is indicated by a black box or white box, respectively) of the 16 strains investigated. The tree was calculated using the MIRU-VNTRplus server (http://www.miru-vntrplus.org; distance measure, categorical). Download