ABSTRACT
Human pathogens frequently use protein mimicry to manipulate host cells in order to promote their survival. Here we show that the opportunistic pathogen Pseudomonas aeruginosa synthesizes a structural homolog of the human α2-macroglobulin, a large-spectrum protease inhibitor and important player of innate immunity. Small-angle X-ray scattering analysis demonstrated that the fold of P. aeruginosa MagD (PA4489) is similar to that of the human macroglobulin and undergoes a conformational modification upon binding of human neutrophil elastase. MagD synthesis is under the control of a general virulence regulatory pathway including the inner membrane sensor RetS and the RNA-binding protein RsmA, and MagD undergoes cleavage from a 165-kDa to a 100-kDa form in all clinical isolates tested. Fractionation and immunoprecipitation experiments showed that MagD is translocated to the bacterial periplasm and resides within the inner membrane in a complex with three other molecular partners, MagA, MagB, and MagF, all of them encoded by the same six-gene genetic element. Inactivation of the whole 10-kb operon on the PAO1 genome resulted in mislocalization of uncleaved, in trans-provided MagD as well as its rapid degradation. Thus, pathogenic bacteria have acquired a homolog of human macroglobulin that plays roles in host-pathogen interactions potentially through recognition of host proteases and/or antimicrobial peptides; it is thus essential for bacterial defense.
IMPORTANCE
The pathogenesis of Pseudomonas aeruginosa is multifactorial and relies on surface-associated and secreted proteins with different toxic activities. Here we show that the bacterium synthesizes a 160-kDa structural homolog of the human large-spectrum protease inhibitor α2-macroglobulin. The bacterial protein is localized in the periplasm and is associated with the inner membrane through the formation of a multimolecular complex. Its synthesis is coregulated at the posttranscriptional level with other virulence determinants, suggesting that it has a role in bacterial pathogenicity and/or in defense against the host immune system. Thus, this new P. aeruginosa macromolecular complex may represent a future target for antibacterial developments.
Introduction
The α2-macroglobulin (A2M) is a highly conserved large-spectrum protease inhibitor present in plasma that plays essential roles in innate immunity in humans and other metazoans. The main function of human A2M is to entrap target proteinases, which may be of endo- or exogenous origins (1). A2M is a glycosylated protein composed of four 1,451-amino-acid subunits and several conserved domains (Fig. 1) (2, 3). Recognition and cleavage of the “bait region” of A2M by target proteases involves the formation of a covalent bond between the two molecules through exposure of a preconcealed, conserved cysteine-glutamine thioester bond (CXEQ region) (4–6). This “Venus flytrap” mechanism (7) involves significant conformational changes of the protein and also leads to exposure of the receptor binding domain required for binding of the A2M-protease complex to a cell surface receptor identified as the low-density lipoprotein receptor-related protein (LRP) (2, 8, 9). In addition to transporting proteases, A2M transports a variety of growth factors, cytokines, and hormones (10). The binding of A2M to LRP results in the clearance of A2M and its cargos through the endocytic degradation pathways (1).
FIG 1 .
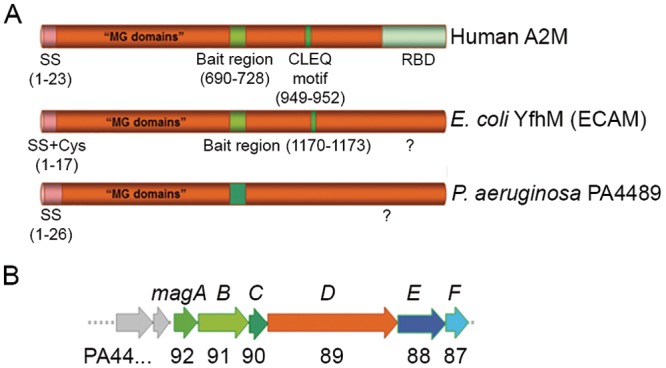
P. aeruginosa PA4489 shares conserved domains with A2M. (A) Representation of human A2M protein, with its conserved domains, compared with those of the E. coli protein (ECAM, YfhM) and the product of the P. aeruginosa PA4489 gene, renamed MagD. Conserved Cys residues are indicated within the lipobox sequence as well as the CLEQ motif, forming the thioester bond. Note the absence of both Cys residues in MagD. SS, signal peptide sequence; MG, macroglobulin; RBD, receptor-binding domain. (B) Genetic organization of the PA4489 gene within the operon of six genes, all predicted to encode proteins of unknown function (http://www.pseudomonas.com). The operon consisting of PA4492 to PA4487 was named the magABCDEF operon and contains genes magA to magF, with PA4489 encoding the P. aeruginosa A2M homolog.
A2M belongs to a family of proteins that includes the C3 complement molecule; these proteins share six conserved macroglobulin (MG) domains and a thioester domain (TED) characterized by the CXEQ sequence (9, 11). Although the C3 molecule is composed of two polypeptide chains, its activation pathway includes proteolytic cleavage and conformational changes that are similar to the one observed for A2Ms, observations that have recently been highlighted by the determination of the crystal structure of A2M and its comparison with the high-resolution structure of C3 (12, 13).
The availability of hundreds of bacterial genomes for bioinformatic analysis allowed the identification of A2M homologs in different bacterial clades, including proteobacteria. Notably, based on an uneven phylogenetic distribution, Budd and coworkers (14) suggested that macroglobulin genes could have been acquired directly from metazoan hosts as colonization and/or defense factors.
Predicted bacterial MG (bMG) proteins can be classified into two subfamilies according to conserved protein domains and the genetic environment of bMG genes. In the majority of bacteria, the MG-encoding gene is adjacent to that encoding penicillin-binding protein 1C (PBP 1C), a membrane-associated molecule involved in cell wall biogenesis. This first class of bMGs displays a common conserved signal peptide and an overlapping lipobox sequence at the Cys residue in the N-terminal region, suggesting that its members are exported into the bacterial periplasm and anchored to the membrane. Furthermore, this class of proteins possesses a conserved bait region and the CLEQ motif required for thioester formation (14) (Fig. 1). The second class of bMGs is encoded within six-gene operons, all encoding proteins of unknown function. In this case, the predicted MG homolog harbors the signal peptide but no Cys residues throughout the whole sequence, which suggests a distinct mechanism involved in localization and substrate capture. A study concerning the first class of bMGs performed on Escherichia coli A2M (ECAM) showed that the protein is capable of binding human neutrophil elastase and undergoes proteolytic cleavage upon target recognition (15). Small-angle X-ray scattering (SAXS) and negative-stain electron microscopy studies revealed that ECAM displays an elongated fold which is reminiscent of that of human complement C3 and undergoes significant conformational changes upon protease binding (16).
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen of humans that provokes acute and chronic infections. Due to its resistance to a majority of clinically employed antibiotics, P. aeruginosa is considered one of the most concerning infectious agents frequently associated with nosocomial infections. P. aeruginosa is capable of either growing as planktonic, mobile bacteria or residing in sessile biofilm communities (18–20). These two ways of life are linked to differential levels of expression of numerous virulence genes, some of which are under the control of the membrane-associated sensors RetS, LadS, and GacS, among others (20, 21). One of the targets of this general regulatory pathway is an operon of six genes, one of which is predicted to encode a homolog of human A2M.
In this work, we structurally and functionally characterized the P. aeruginosa A2M homolog and showed its localization to the bacterial periplasm as a 167-kDa molecule which is cleaved in vivo into a 100-kDa form. The protein is expressed in the majority of P. aeruginosa clinical strains, and its expression is regulated by the RetS/GacS pathway and the RNA-binding protein RsmA. SAXS studies of the recombinant protein demonstrated that it displays an elongated fold that is reminiscent of those of its eukaryotic A2M and C3 counterparts. Despite the fact that P. aeruginosa A2M lacks a conserved β-cysteinyl-glutamyl thioester-harboring region, it is still capable of undergoing a conformational modification upon reacting with neutrophil elastase in vitro. P. aeruginosa A2M, renamed MagD, is associated with the inner bacterial membrane and forms a complex with at least three proteins encoded within the same six-gene operon. These results suggest that macroglobulin-like proteins in pathogenic bacteria may mimic their human homolog by trapping different molecules that are noxious for bacterial survival.
RESULTS
Expression and regulation of an A2M-like protein in P. aeruginosa.
Bioinformatic analysis indicated that the P. aeruginosa PA4489 gene encodes a homolog of human A2M (http://www.pseudomonas.com) (22). The predicted protein of 167 kDa shares 20% identity and 53% similarity with ECAM (15) and 45% similarity with human A2M (accession no. P01023). P. aeruginosa A2M (from now referred to as MagD) harbors a conserved N-terminal signal peptide that targets proteins across the bacterial inner membrane (IM) and, unlike ECAM and other lipoproteins, does not possess a Cys residue within the signal peptide; it is thus predicted to be present as a soluble molecule in the periplasm. In addition, MagD lacks the conserved CXEQ motif present in the A2M active site that is responsible for the formation of the thioester bond involved in covalent binding of substrates (Fig. 1A). In contrast to that of ECAM, whose gene is adjacent to pbpC, which encodes PBP 1C, the genetic environment of PA4489 predicts an operon with five additional open reading frames that we named magABCDEF (PA4492 to PA4487) (Fig. 1B). Specific antibodies raised against recombinant MagD detected two distinct polypeptides in crude extracts of P. aeruginosa strains, one corresponding to the predicted size of the native MagD protein of approximately 165 kDa (without the predicted signal peptide) and the other a protein of 100 kDa; no labeling was observed in a PAO1 strain with the magD sequence deleted, demonstrating that the two polypeptides are MagD specific (Fig. 2A).
FIG 2 .
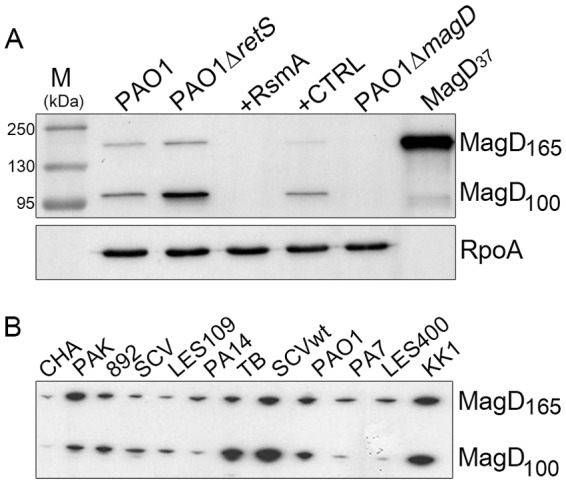
The synthesis of MagD in P. aeruginosa is under the control of RetS and RsmA. (A) Immunoblot analysis of crude P. aeruginosa extracts developed by specific antibodies generated against the recombinant His6-MagD37 protein (7th lane). P. aeruginosa strains were grown to mid-exponential phase, and cells were recovered directly for analysis. Two MagD-specific peptides were identified: a first form corresponding to the entire protein of 165 kDa but probably lacking the predicted signal peptide of 26 amino acids and a second, cleaved form corresponding to a protein of approximately 100 kDa. Note the differences in MagD synthesis in PAO1 ΔretS and PAO1 with RsmA. Lane M, molecular mass markers. (B) Detection of MagD polypeptides in various P. aeruginosa isolates, including cystic fibrosis isolates TB, CHA, SCV, and the Liverpool epidemic strain (LES400). All strains, except CHA, express high levels of MagD, with ratios varying between the 165- and 100-kDa forms. wt, wild type.
Recent studies showed that the leader sequence of PA4492 mRNA is a direct target of RsmA, an RNA-binding protein belonging to the virulence-regulatory cascade that includes two membrane sensors, RetS and LadS, and the two-component system GacS/GacA (20, 21, 23). In order to explore whether the synthesis of MagD is influenced by RetS and RsmA, we performed immunoblotting analysis of two reference strains (PAO1 and PAK) that are deficient for the RetS sensor and of PAO1 overexpressing rsmA. Indeed, the deletion of retS resulted in an increase in the amounts of MagD detected in whole-cell lysates, whereas overproduction of RsmA resulted in a complete turnoff of MagD synthesis (Fig. 2A). MagD was readily detected in several P. aeruginosa strains, mostly clinical isolates obtained from different laboratories (see Table S1 in the supplemental material), with some variations in synthesis levels. The exception was the mucoid clinical isolate CHA, in which the two MagD-specific proteins were barely detected (Fig. 2B). This absence of MagD expression in CHA is due to the perturbation of the same RetS-RsmA regulatory pathway (K. M. Sall, M. G. Casabona, C. Bordi, P. Huber, S. de Bentzman, I. Attrée, and S. Elsen, submitted for publication). Interestingly, in some strains, the amount of the 165-kDa MagD protein was higher than in PAO1, whereas in other strains, the 100-kDa form was more abundant, suggesting that the specific cleavage of the protein may be somehow regulated. Notably, recombinant MagD lacking the first 37 amino acids obtained from expression in E. coli (MagD37) was not cleaved.
MagD shares structural similarities with human macroglobulin, C3, and ECAM.
MagD and eukaryotic A2M belong to the same protein superfamily as components of the complement system, such as factor C3 (15, 16), and share similar domain arrangements (Fig. 1). Previously, we showed that ECAM is an elongated, flexible molecule that undergoes conformational changes upon activation that are potentially reminiscent of those displayed by the other members of the superfamily (16). In order to characterize MagD at a structural level and understand whether it can be activated like its E. coli counterpart, we undertook structural studies of MagD37 by SAXS. Experiments were performed with (i) native protein, (ii) protein treated with methylamine, a small amine used as an activator for ECAM and other C3-like molecules (9, 16), and (iii) MagD incubated with elastase. All samples were purified by gel filtration after the reactions. Data were collected at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France, and are presented in the form log I(s) versus s (nm−1) (Fig. 3A), where I is the measured intensity and s is the scattering angle. The intensity curves for native MagD and the methylamine-treated form (red and blue, respectively, in Fig. 3A) show strikingly similar shapes and are almost perfectly superimposable on the low-angle region, indicating that no major conformational change occurs upon incubation with methylamine. This is corroborated by the radius of gyration (Rg) and maximum particle dimension (Dmax) values calculated from the curves, which were almost the same for native (Rg, 4.57; Dmax, 18.3 nm) and methylamine-treated (Rg, 4.61 nm; Dmax, 17.7 nm) MagD. This result was expected, since MagD does not carry the CXEQ motif that is the hallmark of C3-like molecules and thus in principle should not be activated by a free amine. However, the data collected for MagD after reaction with elastase (green in Fig. 3A) show that the side maximum shifts to slightly higher angles after incubation with the protease and that the Rg and Dmax values are also distinct from that for the native protein (Rg, 5.20 nm; Dmax, 20.3 nm), which suggests that MagD may undergo a conformational modification upon recognition of elastase.
FIG 3 .
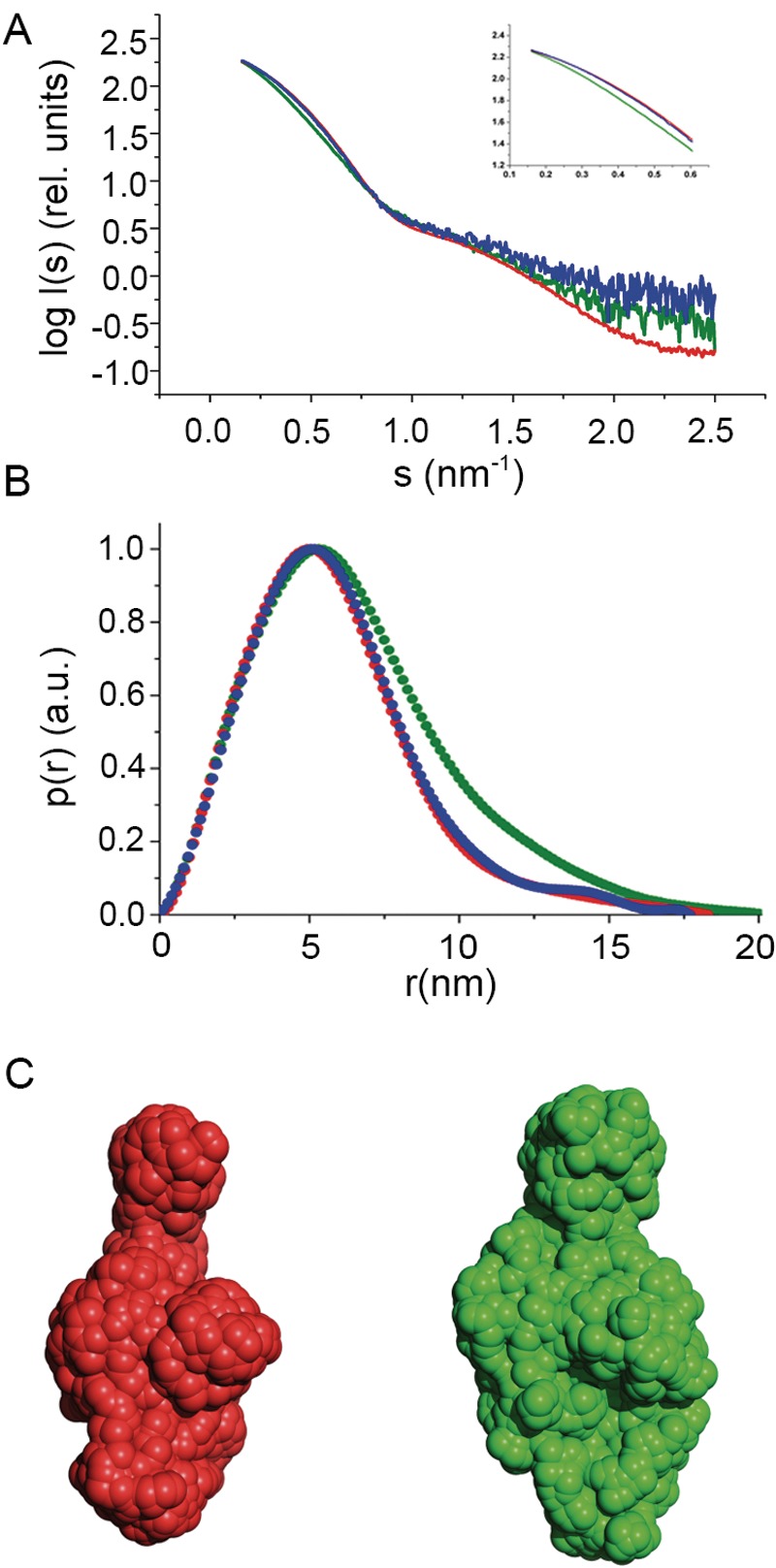
MagD displays minor conformational modifications upon incubation with elastase but not with methylamine. SAXS results for native, methylamine-treated, and elastase-treated MagD. (A) The radially averaged scattered X-ray intensity (I) was plotted as a function of the momentum transfer (s). Scattering patterns for MagD in native form (red), after reaction with methylamine (blue) and elastase (green), were recorded at concentrations of 8.2, 0.3, and 0.6 mg/ml, respectively. (Inset) Detail of differences in curve shapes at lower angles. rel., relative. (B) Distance distributions [p(r) (pair-distance distribution function) in arbitrary units (a.u.)] of native, methylamine-reacted, and elastase-treated MagD forms. All curves were normalized. (C) Ab initio models of MagD generated by SAXS analysis. Each model results from averaging the values from 10 individual models calculated by GASBOR using native (red) and elastase-treated (green) MagD. The envelopes are based on the p(r) functions shown in panel A, and the GNOM files generated were used as input for GASBOR. The models are drawn to scale.
Using the scattering data collected with ID14-3, we initially calculated models of native ECAM using GASBOR with default options. After 10 independent models were generated, values were averaged by DAMAVER (24). Subsequently, a refined averaged model was calculated using GASBOR by employing a fixed core input file calculated by DAMSTART. The envelope of the elastase-reacted form of MagD (Fig. 3A and B) indicates a discrete conformational modification, generating a slightly wider structure. Notably, the native conformation is remarkably similar to that of ECAM (16).
MagD is a periplasmic protein associated preferentially with inner membranes.
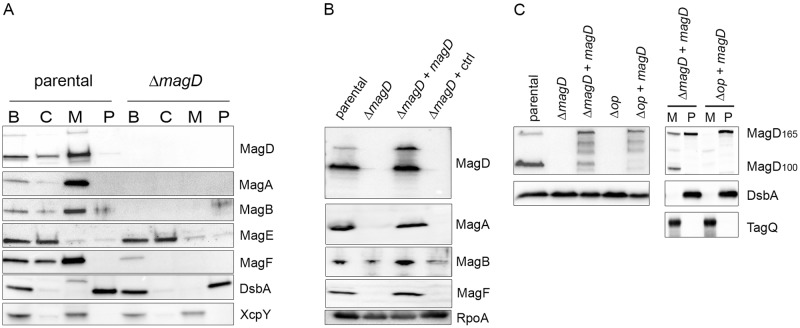
To explore the localization of MagD in P. aeruginosa, we first constructed a fusion protein between the first 64 amino acids of MagD, which encompass its signal peptide, and fluorescent mCherry. The MagD1-64-mCherry-encoding sequence was introduced into the P. aeruginosa PAO1 strain, and the localization of the protein was examined by microscopy both in whole bacteria and in spheroplasts expressing cytoplasmic green fluorescent protein (GFP), as described recently (25). As presented in Fig. 4A, the MagD1-64-mCherry fusion localized to the bacterial periphery without any colocalization with cytoplasmic GFP (upper panel). The mCherry labeling was absent from spheroplast preparations, strongly suggesting that the fusion protein was lost during preparation, as a consequence of being translocated to the periplasmic space. To complete these observations, P. aeruginosa lysates were fractionated into cytoplasmic, membrane, and periplasmic fractions and analyzed by immunoblotting. Interestingly, by this approach, the majority of the native MagD was found associated with bacterial membranes (Fig. 4B, left panel). This was confirmed by high-content mass spectrometry (MS) analysis of P. aeruginosa inner and outer membranes (OM), where MagD peptides were systematically found in the inner membrane preparations (26). Indeed, Western blot analysis performed on membranes separated by centrifugation on discontinuous sucrose gradients confirmed the presence of the two forms of MagD in inner membrane preparations, with a minor fraction found associated with outer membranes (Fig. 4B, right panel). This finding was intriguing since MagD does not possess any predicted transmembrane domains or predicted lipobox sequences that could attach the protein to membrane lipid moieties. We thus postulated that MagD may be associated with the membranes by interacting with other partners.
FIG 4 .
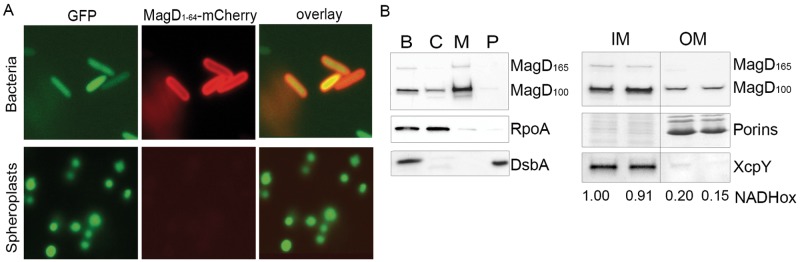
MagD is translocated to the periplasm but appears associated with the inner membrane. (A) The gene encoding MagD1-64-mCherry was expressed under the arabinose-inducible promoter carried within pJN105. The plasmid was introduced into different P. aeruginosa strains expressing cytoplasmic GFP. Spheroplasts obtained by lysozyme treatment and bacteria were observed by fluorescence microscopy using appropriate filters. Note the peripheral labeling in bacteria and the absence of labeling in spheroplasts. (B) Fractionation of PAO1 and immunoblot analyses of MagD. Whole bacterial cells (lane B) were fractionated into the cytoplasm (lane C), total membranes (lane M), and periplasm (lane P) and immunoblotted with anti-MagD antibodies. RpoA and DsbA were used as cytoplasmic and periplasmic markers, respectively. Total membranes were further separated by centrifugation on a sucrose gradient. Two inner membrane (IM) and two outer membrane (OM) fractions were analyzed. The measurement of NADH oxidase activity (NADHox) and immunoblotting with anti-XcpY antibodies were used as IM markers. The OM is characterized by porins (36 kDa) visualized by Coomassie blue staining. MagD is preferentially present in the IM.
Protein partners of MagD include three proteins of the mag operon.
In order to identify MagD partners, we set up coimmunoprecipitation (co-IP) experiments using whole bacterial lysates coupled with proteomic analyses. Anti-MagD IPs were performed in parallel with extracts of PAO1 ΔretS (parental) and PAO1 ΔretS ΔmagD (the ΔmagD mutant). Protein contents of inputs and eluates were analyzed by SDS-PAGE, followed by silver nitrate staining (Fig. 5A) and Western blotting using anti-MagD antibodies (Fig. 5B). Anti-MagD antibodies immunoprecipitated both MagD polypeptides (165 and 100 kDa) and additional proteins that were made visible by silver nitrate staining and were absent from control experiments (PAO1 ΔretS ΔmagD). Identification and comparison of proteins from IP eluates uncovered a number of potentially interesting partners that were strongly enriched in the parental strain compared to the ΔmagD strain (Table 1; see also Table S2 in the supplemental material). Notably, MagA (PA4492), MagB (PA4491), and MagF (PA4487), encoded within the same operon as MagD, were among the most enriched binding partners identified and were further investigated (Table 1).
FIG 5 .
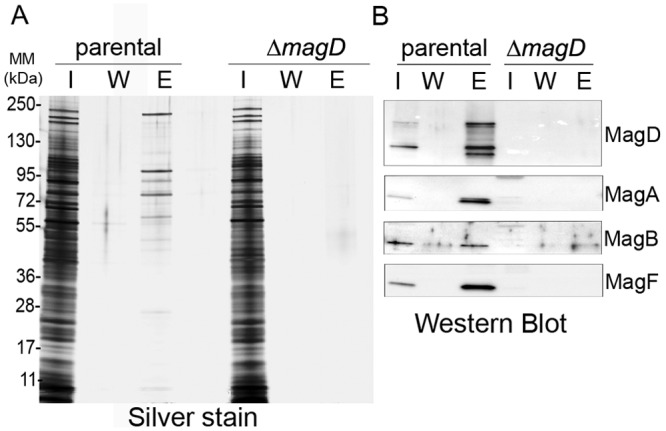
Immunoprecipitation coupled with LC-MS/MS analysis identifies potential MagD partners, including three proteins encoded by the mag operon. (A) Silver-stained SDS-polyacrylamide gel showing the steps of IP experiments. Immunopurified anti-MagD antibodies were used with parental (PAO1 ΔretS) and ΔmagD strains. Twenty microliters of the input (lanes I), washing fraction (lanes W), and eluate (lanes E) were analyzed by SDS-PAGE and revealed by silver staining. Molecular masses (MM) are in kDa. (B) Immunoanalysis of the input, washing fraction, and eluate by anti-MagD polyclonal antibodies showing the presence of MagD only in the eluate of the parental strain. Following LC-MS/MS analysis (Table 1), the same fractions were analyzed for the presence of three proteins encoded within the operon, namely, MagA, MagB, and MagF.
TABLE 1 .
Identification of MagD partners as revealed by immunoprecipitation coupled with mass spectrometry analysesa
| Locus tagb |
Gene name |
Product name | Mean SC of PAO1 ΔretS |
Mean SC of PAO1 ΔretS ΔmagD |
Enrichment |
|---|---|---|---|---|---|
| PA4491 | magB | MagB | 86.5 | 2.5 | 34.6 |
| PA2462 | DUF637 (possible hemagglutinin) |
63 | 2 | 31.5 | |
| PA4487 | magF | MagF | 34.5 | 1.5 | 23 |
| PA3068 | gdhB | NAD-dependent glutamate dehydrogenase |
30 | 0.5 | 60 |
| PA4492 | magA | MagA | 19 | 0 | wt only |
| PA0077 | tssM1 | TssM1 | 19 | 0 | wt only |
| PA5304 | dadA |
d-Amino acid dehydrogenase, small subunit |
18 | 0 | wt only |
| PA2953 | Electron transfer flavoprotein ubiquinone oxidoreductase |
16.5 | 1 | 16.5 |
Proteins immunoprecipitated from the parental and ΔmagD strains with anti-MagD antibodies were analyzed using shotgun proteomics. Proteins identified as having at least 10 spectral counts (SC) in two biological replicates, either only in the parent or enriched at least 10 times (based on SC) in the parent over the ΔmagD strain, are listed. Proteins encoded by the same operon as MagD are boldface.
From http://www.pseudomonas.com. The whole list of identified proteins is provided in the supplemental material.
Proteins encoded by the mag operon display several interesting features (Table 2). First, five of them (with the exception of MagB) possess signal peptides and are predicted to be directed to the periplasm (27). Second, MagB harbors one predicted hydrophobic helix (amino acids 26 to 48). Third, the first and the last genes of the operon encode proteins MagA and MagF, which share 105 out of 269 amino acids. All are annotated in the Pseudomonas genome database (http://www.pseudomonas.com) as being of unknown function. We overexpressed several Mag proteins, raised specific antibodies, and then used them to confirm IP-tandem MS (MS/MS) data. Indeed, MagA, MagB, and MagF were specifically coeluted with MagD, while no protein was eluted in washing fractions (Fig. 5B). Interestingly, while these three MagD partners were readily detected in crude extracts of the parental strain, no signal was obtained from the strain lacking MagD, suggesting the costabilization of the proteins of the presumably multicomponent macroglobulin complex.
TABLE 2.
Characteristics of MagA to -F proteins
| Locus taga |
Protein name |
COGb | pI | Molecular mass (Da) |
Signal peptidec |
|---|---|---|---|---|---|
| PA4492 | MagA | COG4676 | 5.78 | 29,297 | GLPA25LAE |
| PA4491 | MagBd | COG4685 | 5.12 | 64,215 | |
| PA4490 | MagC | COG3234 | 9.90 | 24,001 | GVARG21EPA |
| PA4489 | MagD (A2M) | COG2373 | 5.48 | 167,429 | AAVQA26EDT |
| PA4488 | MagE | COG5445 | 8.82 | 61,710 | LLLRA20AEA |
| PA4487 | MagF | COG4676 | 5.12 | 28,145 | LVAWA19DNPV |
The Pseudomonas genome database is at http://www.pseudomonas.com/.
COG, conserved ortholog group.
The SignalP database is at http://www.cbs.dtu.dk/services/SignalP/. Numbers after amino acids indicate the position in the protein.
The residues of the transmembrane domain (TMH) were amino acids 26 to 48.
Mag complex formation is required for macroglobulin stability, cleavage, and membrane anchoring.
To investigate the localization and stability of MagA, MagB, and MagF in the parental strain PAO1 ΔretS and its MagD-deficient isogenic mutant, we performed immunoblotting analyses of different cellular fractions. The ΔretS strain was used here in order to enhance the detection of Mag proteins. Notably, the three proteins that were identified as MagD partners by IP-MS analysis were found preferentially associated with bacterial membranes, while they were absent from the MagD-deficient strain (Fig. 6A). Importantly, the product of the fifth gene of the operon, encoding MagE (PA4488), was not affected by the magD deletion and was found preferentially in cytoplasmic fractions, further supporting the idea that the magD deletion had no polar effect on downstream genes. To ascertain this observation of costabilization of complex partners, we provided magD in trans from the chromosomally inserted plasmid pSW196 (28) and induced its expression by arabinose. Clearly, MagD synthesis restored the presence of the three other Mag proteins in the ΔretS ΔmagD/pSW196-magD strain (Fig. 6B). Here, we noticed that the level of MagD compared to those of other Mag proteins encoded by the chromosome was important, since high concentrations of arabinose used for induction resulted in degradation of the protein (more details below). Therefore, four proteins, including P. aeruginosa A2M MagD, stabilize each other within the P. aeruginosa envelope by creating a multimolecular complex (Fig. 7).
FIG 6 .
The membrane-localized MagA-MagB-MagD-MagF complex is required for macroglobulin localization and stability. PAO1 ΔretS (parent) (A) and PAO1 ΔretS ΔmagD (ΔmagD mutant) (B) bacteria were fractionated into the cytoplasm (lanes C), total membranes (lanes M), and periplasm (lanes P) and developed by antibodies directed against MagA, MagB, MagF, and MagD. Antibodies against DsbA and XcpY were used for periplasmic and membrane controls, respectively. Three Mag proteins (A, B, and F) cofractionate in the membrane fraction with MagD in the parental strain and are absent in the ΔmagD mutant. (B) Total extracts of PAO1 ΔretS (lane parental), PAO1 ΔretS ΔmagD (lane ΔmagD), PAO1 ΔretS ΔmagD in which MagD was provided in trans by pSW196 (lane ΔmagD + magD), and the control strain with an empty vector, pSW196 (lane ΔmagD + ctrl), were examined for the presence of Mag proteins by Western blotting. Note that the expression of magD in trans hinders the degradation of MagA, MagB, and MagF. (C) PAO1 and its isogenic Δop mutant, from which the whole mag operon was deleted, in the presence or absence of magD provided in trans, were analyzed for the two MagD forms. DsbA antibodies were used as a loading control. In the right panel, membrane (lanes M) and periplasmic (lanes P) fractions were prepared and analyzed. DsbA was used as a marker for the periplasm, and TagQ was used as a marker for total membranes. In the Δop strain, in trans-supplied MagD is absent from membranes and present only as the 16- kDa form of MagD in the periplasm.
FIG 7 .
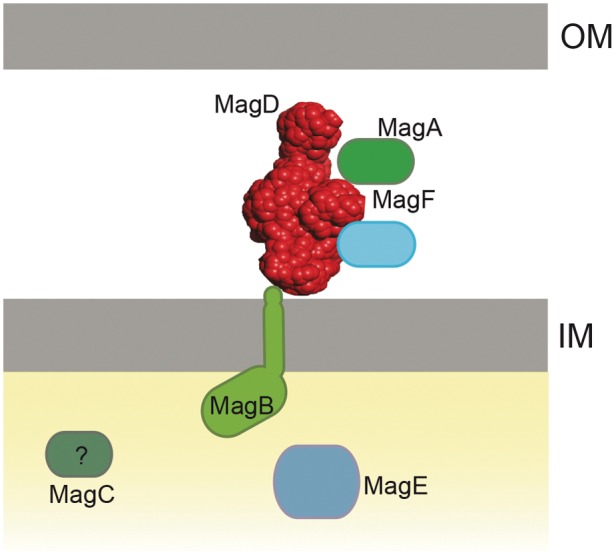
Schematic representation of the P. aeruginosa Mag complex localized to the bacterial envelope. MagD is structurally reminiscent of C3 and A2M molecules. It is translocated across the inner membrane to the periplasm and is associated with the inner membrane, probably through interactions with MagB, which harbors one transmembrane domain in its C terminus. The orientation of the MagB in the inner membrane was chosen arbitrarily. MagA and MagF, encoded by the same operon, are also partners of the membrane-associated Mag complex.
In order to investigate the role of proteins encoded by the mag operon with regard to MagD, we deleted the whole 10-kb operon from the PAO1 chromosome and then provided magD in trans, inducing its expression by arabinose. In this operon-deleted genetic background (the Δop strain), we observed an evident, rapid degradation of MagD provided in trans (Fig. 6C, left panel). Interestingly however, when MagD could be detected, it appeared only as the noncleaved 165-kDa form (Fig. 6C, right panel). Furthermore, this noncleaved form was found preferentially in the bacterial periplasm and was absent from bacterial membranes. Thus, the Mag proteins encoded by the operon are essential for the correct localization of MagD by anchoring it to the inner membrane and ensuring its proteolytic cleavage into the 100-kDa form.
DISCUSSION
In this work, we characterized a novel protein complex that resides within the P. aeruginosa envelope whose main component, MagD, structurally mimics MG molecules of innate immunity, such as A2M and C3. MagD is encoded within an operon harboring six genes (magA, magB, magC, magD, magE, magF) and is a 167-kDa protein with several MG domains in its N-terminal region (14, 22; http://www.pseudomonas.com). Despite the fact that MagD is homologous to human A2M and ECAM (9, 16, 29), it lacks the two essential cysteine residues. The first cysteine, which belongs to the lipobox region, is thought to anchor the protein to the membrane. The second cysteine within the CLEQ motif, conserved in E. coli and human A2M, participates in the formation of the thioester bond required for covalent binding of substrate proteases. Despite these differences, recombinant MagD (produced in E. coli) displays a notable structural resemblance in its native form to both ECAM and human C3 (9, 13, 16), as determined by SAXS experiments. However, incubation of MagD with methylamine, which targets the nonexposed thioester bond in proteins of the macroglobulin family (9, 16), did not engender the measurable conformational modification seen in the case of ECAM or C3, an expected difference due to the absence of the catalytic Cys within the MagD sequence. Interestingly, however, incubation of MagD with elastase generated a minor conformational modification, suggesting that the protease could recognize and bind to the bait region of the macroglobulin. This indicates that despite the lack of the classical CXEQ motif, one of the functions of MagD may involve host protease recognition, but details of the precise mechanism still await clarification.
The C3 complement molecule, as well as other A2Ms, undergoes proteolytic cleavage upon activation (12, 30). Interestingly, MagD appears to be cleaved from a 165-kDa to a 100-kDa form in all clinical isolates tested. Unfortunately, to date, we have been unable to determine the exact site and role of this cleavage in MagD activation, and its function is only speculative. It seems that the 65-kDa fragment of the cleaved protein is rapidly degraded, since only the 100-kDa polypeptide could be detected by both immunodetection and mass spectrometry analysis. Furthermore, the cleavage required the formation of a multimolecular complex in the bacterial periplasm.
Despite the absence of the conserved cysteine in the lipobox region and no predicted transmembrane domain, MagD associates with the bacterial IM through interactions with MagB. MagB is encoded within the same operon, fractionates to the IM, and harbors one predicted transmembrane helix. We found, by performing IP experiments followed by shotgun proteomics, that in addition to MagD and MagB, MagA and MagF are involved in complex formation. This was corroborated by the fact that both MagA and MagF were found in IM fractions. Moreover, in the absence of MagD (∆magD strain), MagA, MagB, and MagF were degraded. However, MagE, encoded by the fifth gene of the operon, was equally well expressed and stable in wild-type and magD-deleted strains. To our surprise, MagE, despite possessing the signal peptide, which should allow its transfer across the inner membrane, was found only in the bacterial cytoplasm. We cannot exclude the possibility that the predicted signal peptide is not functional or that a specific external signal is required for protein translocation. It is also possible that cytoplasmic MagE interacts with the cytoplasmic domain of MagB and links the Mag complex with other cytoplasmic components. MagD partners may participate in proper localization of the complex and its processing/activation. Indeed, when MagD is found in bacteria without its partners (the Δop strain), it is still translocated into the periplasmic space, but it does not anchor to the membrane, it is not correctly processed, and it degrades rapidly.
Finally, one additional protein is encoded by the mag operon, MagC, which was never identified by mass spectrometry analysis of either IP eluates or IM and OM fractions. MagC was also absent from the recent analyses of the periplasmic compartment (31). This may suggest that MagC is also cytoplasmic; its participation in complex formation requires further investigation. The current view of the Mag complex in the P. aeruginosa envelope is depicted in Fig. 7.
It was previously shown that the leader sequence of the PA4492 (magA) mRNA is a direct target of the RNA-binding protein RsmA (23). This suggests that the mag operon is part of a regulon comprising several operons, all under the control of the same regulatory cascade. Indeed, RsmA binds and downregulates the expression of a battery of genes involved in the formation and/or maintenance of the P. aeruginosa biofilm, including psl and pel operons necessary for exopolysaccharide synthesis, as well as the type VI secretion system (T6SS) (23). RsmA activity is counterbalanced by two small regulatory RNAs, RsmY and RsmZ, whose expression is notably under the control of three IM sensors, LadS, RetS, and GacS (20, 21, 23). The fact that MagD synthesis is upregulated under the conditions that favor biofilm formation, e.g., during chronic infections, is intriguing. Indeed, under these conditions, P. aeruginosa downregulates the majority of so-called aggressive virulence factors, such as the type III secretion system (T3SS) machinery, which injects toxins into the host cell (32). T3SS toxins are major players in the defense against the host immune system, notably in phagocytosis by macrophages and neutrophils (32–36). The absence of the active T3SS, which inactivates immune cells, may render the bacterium more vulnerable to external, damaging molecules, such as proteases and antimicrobial peptides released by these cells. Under these conditions, the overexpression of the protease inhibitor may be essential for bacterial survival. Indeed, recognition of elastase by the recombinant protein suggests that MagD may play a function that emulates the process undergone by human A2M, which traps and inactivates external proteases.
There are several instances where pathogenic bacteria develop mechanisms of resistance to metazoan immune molecules. For example, many bacterial species, including P. aeruginosa, harbor the periplasmic serine protease inhibitor ecotin, which protects bacteria against attack by neutrophils (37). Another example is a conserved bacterial protein, Ivy, capable of binding and inhibiting host lysozyme (38, 39). Macroglobulin-like molecules are encoded in diverse bacterial species; however, only a few possess the complex genetic environment similar to that of P. aeruginosa (14). In the majority of bacteria, the macroglobulin-encoding gene is located in the vicinity of the one coding for PBP 1C (14), an enzyme involved in peptidoglycan synthesis (40), suggesting that bacterial MGs may play a protective role, notably during bacterial division and cell wall synthesis.
The IP-MS/MS experiment allowed the identification of, in addition to three Mag proteins, a few putative partners, notably TssM1, which was recovered with 19 spectral counts uniquely from the parental strain. Intriguingly, TssM1 is a conserved inner membrane protein of the type VI secretion system (H1-T6SS) machinery, which plays a crucial role in bacterium-bacterium interactions (41–43). P. aeruginosa H1-T6SS-encoding genes, situated in the Hcp secretion island I (HSI-I) cluster, are coregulated with the mag operon by the same regulatory cascade, with RsmA targeting several H1-T6SS mRNAs (23). Interestingly, T6SSs inject proteins within the periplasm of the neighboring species, thus attacking directly the target peptidoglycan (41, 44). We cannot exclude the possibility that the macroglobulin complex identified in this work plays a protective role against a bacterial aggressor.
Initial evidence for a potential role of mag genes in bacterial virulence came from work of Potvin et al. (45). Using high-throughput screening of P. aeruginosa mutants attenuated in a chronic rat infection model, these authors identified three mutants with independent transposon insertions in the PA4491 (magB), PA4489 (magD), and PA4488 (magE) genes (45).
Further phenotypic characterization of these mutants showed either that they were affected in motility (swarming) or that they were attenuated for virulence in an alternative model host, Drosophila melanogaster. This suggests that the Mag complex, identified and characterized here, may play a key role in virulence and/or bacterial defense in a number of pathogens and thus represents a novel potential target for future antibacterial development.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa and E. coli were grown at 37°C with agitation in Luria-Bertani (LB) broth supplemented with antibiotics when needed. Antibiotics used were ampicillin (100 µg/ml), tetracycline (10 µg/ml), gentamycin (25 µg/ml), and kanamycin (25 µg/ml) for E. coli and carbenicillin (300 µg/ml), tetracycline (100 µg/ml), and gentamycin (200 µg/ml) for P. aeruginosa. P. aeruginosa strains were cultured on Pseudomonas isolation agar (Difco) plates. For analysis, overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.1 and cultivated further to an OD600 of 1.0 to 1.5. Bacterial cells were harvested by centrifugation and immediately treated for further experiments or frozen at −20°C. P. aeruginosa strains were obtained from different laboratories as indicated in Table S1 in the supplemental material.
Genetic constructions.
P. aeruginosa strains with the magD sequence deleted were constructed as follows. The magD sequence corresponding to codons 37 to TGA was obtained by PCR and cloned into the SmaI-digested vector pEX100T (46). Then, the internal sequence was deleted by PstI digestion and religation of the vector. The genetic construct used to delete the whole magABCDEF operon was obtained by splicing by overlap extension (SOE) PCR and cloned into pEX100T. The mutants were created by homologous recombination. pEX100T vectors carrying deletions were introduced into the desired P. aeruginosa strain using pRK2013 as a helper plasmid (46, 47). Mutants were created in the PAO1 and PAO1 ΔretS backgrounds for magD deletion and only in PAO1 for the whole-operon deletion (Δop). The sequence encoding the first 64 amino acids of MagD fused to the mCherry protein was obtained by PCR amplification and was cloned into pJN-mCherry (24). For overexpression in E. coli, the magD gene lacking its first 111 nucleotides was PCR amplified, sequenced, and cloned as an EcoRI-HindIII fragment into the pETDuet-1 vector (Novagen). The overexpressed protein lacks the first 37 amino acids and harbors a hexahistidine sequence at the N terminus. The magA, -B, -C, -E, and -F genes were synthesized using E. coli codon usage and cloned into pUC57 by GenScript. The sequences harbor NdeI and BamHI restriction sites at their 5′ and 3′ ends, respectively, and are made so as to encode proteins without a signal peptide. For magB, the sequence encoding a putative transmembrane helix was also excluded. All genes were cloned into pET15b (Novagen) for overexpression. For complementation, the magD gene was synthetized by Proteogenix and cloned into a mini-CTX derivative, pSW196 (28), harboring the arabinose-inducible promoter pBAD to drive magD expression. The rsmA gene was PCR amplified and cloned, after being sequenced, into the pIApX2 vector. As a control, pIApX2-GFP (48) was introduced in the same P. aeruginosa strain and cultivated under the same conditions. The list of primers used for amplifications are given in Table S1 in the supplemental material.
Protein expression and antibody production.
The plasmids were introduced into E. coli BL21(DE3) Star (Invitrogen), and protein production was induced by IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) in 500 ml of LB broth for 3 h at 37°C. The His6-MagD37 protein was obtained in the soluble fraction, while the other Mag proteins were obtained in inclusion bodies. No His6-MagC protein could be obtained. Proteins were purified by Ni2+ affinity chromatography using standard protocols (Novagen) and an Äkta purifier (GE Healthcare). Rabbit polyclonal serum raised against His6-MagD37 was obtained from Covalab. Antibodies against other Mag proteins were obtained by immunization of mice by AgroBio.
Small-angle scattering experiments.
SAXS measurements were recorded at the ID14-3 beamline of the European Synchrotron Radiation Facility (Grenoble, France). Prior to data collection, a scattering curve of bovine serum albumin reference solution (5.4 mg/ml) was recorded. Experiments were performed at concentrations of 2.3 and 8.2 mg/ml for native His6-MagD37, 0.3 mg/ml for the methylamine-activated form, and 0.6 mg/ml for elastase-incubated His6-MagD37. Between measurements, scattering from a buffer sample was recorded, and these data were subsequently subtracted from the respective sample curves. No radiation damage was observed during the 10-s exposure frames, and all data were recorded at 25°C. Data were treated by following default parameters of the PRIMUS software package (49). The radius of gyration (Rg) and the forward scattering value I(0) were estimated using the Guinier approximation (50). Both parameters, as well the maximum particle dimension, Dmax, were calculated by the GNOM software (51). Ab initio models of His6-MagD37 were generated using GASBOR (24). A final-average model was generated from 10 independent models using DAMAVER through their pairwise superposition (52).
Bacteria and spheroplast preparation for MagD1-64-mCherry analysis.
GFP-expressing bacteria and spheroplasts were prepared as described previously (25). Briefly, overnight cultures were diluted to an OD600 of 0.15 and incubated up to mid-log phase at 37°C with shaking. Cells were harvested, and spheroplasts were created as described previously (25, 31). Induction was carried out by the addition of 0.25% arabinose for 1.5 h. In order to visualize intact bacteria, 1 ml of culture was harvested by a rapid centrifugation step, resuspended in 100 µl fresh LB broth, and immobilized in 1% agarose. Observations were carried out with a Zeiss Axiovert operating system.
Fractionation of P. aeruginosa.
Fractionation of bacterial cells was performed using exponentially grown cultures (OD600 = 1). The pellet, equivalent to 2.109 bacteria, was resuspended in 1 ml buffer A (10 mM Tris-HCl, 200 mM MgCl2, pH 8) in the presence of protease inhibitor cocktail (Complete, Roche) and 0.5 mg/ml lysozyme and incubated for 30 min at 4°C with gentle agitation. The periplasmic fraction was recovered after centrifugation at 8,000 × g and 4°C for 15 min. After one wash, the pellet, resuspended in 10 mM Tris-HCl, 10 mM MgCl2, pH 8, was disrupted by sonication. Unbroken bacteria were eliminated by centrifugation at 8,000 × g for 15 min. Then, the supernatant was ultracentrifuged at 200,000 × g and 4°C for 45 min (TLA120 Beckman rotor) to obtain the cytosolic fraction (supernatant) and the total membrane fraction. All fractions were resuspended in 4× SDS-PAGE loading buffer and incubated for 5 min at 100°C before SDS-PAGE or Western blotting. E. coli RNA polymerase (RpoA) and disulfide oxidoreductase (DsbA) were used as internal markers for cytosolic and periplasmic fractions, respectively.
Inner and outer membrane separation.
Inner and outer membranes of PAO1 ΔretS were obtained as described previously with minor modifications (25). Briefly, overnight cultures of PAO1 ΔretS were diluted to an OD600 of 0.15 and cultured to the mid-log phase of growth. At this point, cells were harvested by centrifugation and resuspended in 25 ml buffer A (10 mM Tris-HCl, 20% sucrose, 10 µg/ml DNase, 10 µg/ml RNase, pH 7.4). Cells were broken using a Microfluidizer at 15,000 lb/in2, and the total membrane fraction was obtained by a centrifugation step at 200,000 × g for 1 h. This sample was resuspended in 500 µl of 20% sucrose and loaded on top of a discontinuous sucrose gradient composed of eight layers of sucrose, the volumes of which were (from bottom to top) 55% (1.4 ml), 50% (1.5 ml), 45% (1.5 ml), 42.5% (1.3 ml), 40% (1.5 ml), 37.5% (1.3 ml), 35% (1.5 ml), and 30% (1.0 ml). Centrifugation at 90,000 × g was carried out for 72 h, and 500-µl fractions were collected from the top. Fractions were characterized by SDS-PAGE, immunoblotting, and NADH oxidase activity. Porins were used as outer membrane markers. NADH oxidase activity and XcpY antibodies were used as inner membrane markers (53, 54).
Immunoblotting analysis.
Western blotting analyses were done on a Hybond LFP-polyvinylidene difluoride (PVDF) transfer membrane (GE Healthcare) after electrotransfer in Laemmli buffer containing 20% ethanol. The membranes were blocked with 5% nonfat dry milk before incubation with primary antibodies overnight at 4°C. Dilutions of polyclonal antibodies were anti-MagD at 1:40,000, anti-MagA, -MagB, -MagE, and -MagF at 1:1,000, anti-RpoA (NeoClone) at 1:10,000, anti-DsbA at 1:10,000 (obtained from R. Voulhoux, CNRS, Marseille, France), anti-XcpY at 1:2,000 (54), and anti-TagQ at 1:10,000 (25). The secondary horseradish peroxidase (HRP)-conjugated antibodies against rabbit or mouse were obtained from Sigma and used at a dilution of 1:50,000. Detection was performed with a Luminata Western HRP substrate kit (Millipore).
Immunoprecipitation.
Total extracts from PAO1 ΔretS or PAO1 ΔretS ΔmagD cultures (equivalent to 30 OD600 units) resuspended in buffer A (10 mM Tris-HCl, pH 8, 20% sucrose) and protease inhibitor cocktail (Complete, Roche) were obtained using a Microfluidizer. Unbroken bacteria were eliminated by centrifugation at 8,000 × g for 10 min prior to use. Protein A magnetic beads (Dynabeads immunoprecipitation kit protein A; Invitrogen) were incubated with immunopurified anti-MagD antibodies for 30 min at room temperature in antibody binding buffer (Invitrogen kit). The covalent cross-link was realized using 5 mM BS3 (Bis-sulfosuccinimidyl-suberate). After washes with wash buffer (Invitrogen kit), the anti-MagD-coupled beads were added to total extracts of PAO1 ΔretS or PAO1 ΔretS ΔmagD and the incubation was carried out for 2 h at room temperature in buffer A containing 100 mM NaCl. The magnetic beads were washed three times, and the elution was performed for 10 min at 70°C with reagents provided by the manufacturer.
Proteomic analyses.
Protein digestion and nano-liquid chromatography (LC)–MS/MS analyses were done as previously described (25, 26). The details are given in Table S2 in the supplemental material. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (55) with the data set identifier PXD000189.
SUPPLEMENTAL MATERIAL
Proteomic analyses. Download
P. aeruginosa strains and oligonucleotides.
Proteins found to be enriched in the parental strain compared to the ΔmagD strain by spectral counting.
ACKNOWLEDGMENTS
We thank Michel Ragno (BCI, iRTSV) for help with protein purification and antibody characterization, Frank Gabel (IBS) for help with SAXS data collection, the European Synchrotron Radiation Facility (ESRF), for access to the BIOSAXS beamline (ID14-3), and Stéphanie Bouillot for help with microscopy (BCI, iRTSV). We thank Marie-Odile Fauvarque (BGE, iRTSV) for interest in this work as well as all team members for stimulating discussions. We thank Romé Voulhoux for the anti-XcpY and anti-DsbA antibodies.
This work was supported by grants from INSERM and the CNRS. A.D. and I.A. received financial support from the Rhone-Alpes FINOVI Foundation. M.G.C. is a Ph.D. student financed by the French Cystic Fibrosis Foundation (VLM).
M.R.-G., M.G.C., D.N., F.C., and Y.C. performed the experiments. M.R.-G., Y.C., S.E., A.D., and I.A. contributed reagents and analyzed the data. A.D. and I.A. designed the project and wrote the manuscript.
Footnotes
Citation Robert-Genthon M, Casabona MG, Neves D, Couté Y, Cicéron F, Elsen S, Dessen A, Attrée I. 2013. Unique features of a Pseudomonas aeruginosa α2-macroglobulin homolog. mBio 4(4):e00309-13. doi:10.1128/mBio.00309-13.
REFERENCES
- 1. Rehman AA, Ahsan H, Khan FH. 2012. α-2-Macroglobulin: a physiological guardian. J. Cell. Physiol. 228:1665–1675 [DOI] [PubMed] [Google Scholar]
- 2. Sottrup-Jensen L, Sand O, Kristensen L, Fey GH. 1989. The alpha-macroglobulin bait region. Sequence diversity and localization of cleavage sites for proteinases in five mammalian alpha-macroglobulins. J. Biol. Chem. 264:15781–15789 [PubMed] [Google Scholar]
- 3. Armstrong PB, Quigley JP. 1999. Alpha2-macroglobulin: an evolutionarily conserved arm of the innate immune system. Dev. Comp. Immunol. 23:375–390 [DOI] [PubMed] [Google Scholar]
- 4. Arakawa H, Nishigai M, Ikai A. 1989. Alpha 2-macroglobulin traps a proteinase in the midregion of its arms. An immunoelectron microscopic study. J. Biol. Chem. 264:2350–2356 [PubMed] [Google Scholar]
- 5. Delain E, Pochon F, Barray M, Van Leuven F. 1992. Ultrastructure of alpha 2-macroglobulins. Electron Microsc. Rev. 5:231–281 [DOI] [PubMed] [Google Scholar]
- 6. Qazi U, Kolodziej SJ, Gettins PG, Stoops JK. 2000. The structure of the C949S mutant human alpha(2)-macroglobulin demonstrates the critical role of the internal thiol esters in its proteinase-entrapping structural transformation. J. Struct. Biol. 131:19–26 [DOI] [PubMed] [Google Scholar]
- 7. Meyer C, Hinrichs W, Hahn U. 2012. Human alpha2-macroglobulin—another variation on the Venus flytrap. Angew. Chem. Int. Ed. Engl. 51:5045–5047 [DOI] [PubMed] [Google Scholar]
- 8. Sottrup-Jensen L, Gliemann J, Van Leuven F. 1986. Domain structure of human alpha 2-macroglobulin. Characterization of a receptor-binding domain obtained by digestion with papain. FEBS Lett. 205:20–24 [DOI] [PubMed] [Google Scholar]
- 9. Doan N, Gettins PG. 2007. Human alpha2-macroglobulin is composed of multiple domains, as predicted by homology with complement component C3. Biochem. J. 407:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feige JJ, Negoescu A, Keramidas M, Souchelnitskiy S, Chambaz EM. 1996. Alpha 2-macroglobulin: a binding protein for transforming growth factor-beta and various cytokines. Horm. Res. 45:227–232 [DOI] [PubMed] [Google Scholar]
- 11. Xiao T, DeCamp DL, Spran SR. 2000. Structure of a rat alpha 1-macroglobulin receptor-binding domain dimer. Protein Sci. 9:1889–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. 2006. Structure of C3b reveals conformational changes that underlie complement activity. Nature 444:213–216 [DOI] [PubMed] [Google Scholar]
- 13. Marrero A, Duquerroy S, Trapani S, Goulas T, Guevara T, Andersen GR, Navaza J, Sottrup-Jensen L, Gomis-Rüth FX. 2012. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew. Chem. Int. Ed. Engl. 51:3340–3344 [DOI] [PubMed] [Google Scholar]
- 14. Budd A, Blandin S, Levashina EA, Gibson TJ. 2004. Bacterial alpha2-macroglobulins: colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 5:R38. 10.1186/gb-2004-5-6-r38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doan N, Gettins PG. 2008. Alpha-macroglobulins are present in some gram-negative bacteria: characterization of the alpha2-macroglobulin from Escherichia coli. J. Biol. Chem. 283:28747–28756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neves D, Estrozi LF, Job V, Gabel F, Schoehn G, Dessen A. 2012. Conformational states of a bacterial α2-macroglobulin resemble those of human complement C3. PLoS One 7:e35384. 10.1371/journal.pone.0035384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 18. Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14:47–70 [PMC free article] [PubMed] [Google Scholar]
- 19. Mikkelsen H, Sivaneson M, Filloux A. 2011. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13:1666–1681 [DOI] [PubMed] [Google Scholar]
- 20. Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 21. Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 103:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72:612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Svergun DI, Petoukhov MV, Koch MH. 2001. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80:2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Casabona MG, Silverman JM, Sall KM, Boyer F, Couté Y, Poirel J, Grunwald D, Mougous JD, Elsen S, Attree I. 2013. An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ. Microbiol. 15:471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Casabona MG, Vandenbrouck Y, Attree I, Couté Y. 7 June 2013. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics 10.1002/pmic.201200565 [DOI] [PubMed] [Google Scholar]
- 27. Lewenza S, Gardy JL, Brinkman FS, Hancock RE. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harpel PC, Hayes MB, Hugli TE. 1979. Heat-induced fragmentation of human alpha 2-macroglobulin. J. Biol. Chem. 254:8669–8678 [PubMed] [Google Scholar]
- 30. Dodds AW, Law SK. 1998. The phylogeny and evolution of the thioester bond-containing proteins C3, C4 and alpha 2-macroglobulin. Immunol. Rev. 166:15–26 [DOI] [PubMed] [Google Scholar]
- 31. Imperi F, Ciccosanti F, Perdomo AB, Tiburzi F, Mancone C, Alonzi T, Ascenzi P, Piacentini M, Visca P, Fimia GM. 2009. Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics 9:1901–1915 [DOI] [PubMed] [Google Scholar]
- 32. Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7:654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dacheux D, Attree I, Schneider C, Toussaint B. 1999. Cell death of human polymorphonuclear neutrophils induced by a Pseudomonas aeruginosa cystic fibrosis isolate requires a functional type III secretion system. Infect. Immun. 67:6164–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dacheux D, Toussaint B, Richard M, Brochier G, Croize J, Attree I. 2000. Pseudomonas aeruginosa cystic fibrosis isolates induce rapid, type III secretion-dependent, but ExoU-independent, oncosis of macrophages and polymorphonuclear neutrophils. Infect. Immun. 68:2916–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matteï PJ, Faudry E, Job V, Izoré T, Attree I, Dessen A. 2011. Membrane targeting and pore formation by the type III secretion system translocon. FEBS J. 278:414–426 [DOI] [PubMed] [Google Scholar]
- 36. Tosi T, Pflug A, Discola KF, Neves D, Dessen A. 2013. Structural basis of eukaryotic cell targeting by type III secretion system (T3SS) effectors. Res. Microbiol. 164:605–619 [DOI] [PubMed] [Google Scholar]
- 37. Eggers CT, Murray IA, Delmar VA, Day AG, Craik CS. 2004. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem. J. 379:107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abergel C, Monchois V, Byrne D, Chenivesse S, Lembo F, Lazzaroni JC, Claverie JM. 2007. Structure and evolution of the ivy protein family, unexpected lysozyme inhibitors in gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:6394–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Monchois V, Abergel C, Sturgis J, Jeudy S, Claverie JM. 2001. Escherichia coli ykfE ORFan gene encodes a potent inhibitor of C-type lysozyme. J. Biol. Chem. 276:18437–18441 [DOI] [PubMed] [Google Scholar]
- 40. Matteï PJ, Neves D, Dessen A. 2010. Bridging cell wall biosynthesis and bacterial morphogenesis. Curr. Opin. Struct. Biol. 20:749–755 [DOI] [PubMed] [Google Scholar]
- 41. Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwarz S, Hood RD, Mougous JD. 2010. What is type VI secretion doing in all those bugs? Trends Microbiol. 18:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basler M, Mekalanos JJ. 2012. Type 6 secretion dynamics within and between bacterial cells. Science 337:815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. 2012. New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol. Microbiol. 88, 921–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, Levesque RC. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5:1294–1308 [DOI] [PubMed] [Google Scholar]
- 46. Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195–1204 [DOI] [PubMed] [Google Scholar]
- 47. Konyecsni WM, Deretic V. 1988. Broad-host-range plasmid and M13 bacteriophage-derived vectors for promoter analysis in Escherichia coli and Pseudomonas aeruginosa. Gene 74:375–386 [DOI] [PubMed] [Google Scholar]
- 48. Thibault J, Faudry E, Ebel C, Attree I, Elsen S. 2009. Anti-activator ExsD forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J. Biol. Chem. 284:15762–15770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36:1277–1282 [Google Scholar]
- 50. Guinier A, Fournet G. 1955. Small angle scattering of X-rays. Wiley, New York, NY. [Google Scholar]
- 51. Svergun DI. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25:495–503 [Google Scholar]
- 52. Volkov VV, Svergun DI. 2003. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36:860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aubert D, MacDonald DK, Valvano MA. 2010. BcsKC is an essential protein for the type VI secretion system activity in Burkholderia cenocepacia that forms an outer membrane complex with BcsLB. J. Biol. Chem. 285:35988–35998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Michel G, Bleves S, Ball G, Lazdunski A, Filloux A. 1998. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology 144:3379–3386 [DOI] [PubMed] [Google Scholar]
- 55. Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H. 2013. The proteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41:D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomic analyses. Download
P. aeruginosa strains and oligonucleotides.
Proteins found to be enriched in the parental strain compared to the ΔmagD strain by spectral counting.