ABSTRACT
Streptococcus pyogenes (group A Streptococcus [GAS]) causes ~700 million human infections/year, resulting in >500,000 deaths. There is no commercial GAS vaccine available. The GAS surface protein arginine deiminase (ADI) protects mice against a lethal challenge. ADI is an enzyme that converts arginine to citrulline and ammonia. Administration of a GAS vaccine preparation containing wild-type ADI, a protein with inherent enzymatic activity, may present a safety risk. In an approach intended to maximize the vaccine safety of GAS ADI, X-ray crystallography and structural immunogenic epitope mapping were used to inform vaccine design. This study aimed to knock out ADI enzyme activity without disrupting the three-dimensional structure or the recognition of immunogenic epitopes. We determined the crystal structure of ADI at 2.5 Å resolution and used it to select a number of amino acid residues for mutagenesis to alanine (D166, E220, H275, D277, and C401). Each mutant protein displayed abrogated activity, and three of the mutant proteins (those with the D166A, H275A, and D277A mutations) possessed a secondary structure and oligomerization state equivalent to those of the wild type, produced high-titer antisera, and avoided disruption of B-cell epitopes of ADI. In addition, antisera raised against the D166A and D277A mutant proteins bound to the GAS cell surface. The inactivated D166A and D277A mutant ADIs are ideal for inclusion in a GAS vaccine preparation. There is no human ortholog of ADI, and we confirm that despite limited structural similarity in the active-site region to human peptidyl ADI 4 (PAD4), ADI does not functionally mimic PAD4 and antiserum raised against GAS ADI does not recognize human PAD4.
IMPORTANCE
We present an example of structural biology informing human vaccine design. We previously showed that the administration of the enzyme arginine deiminase (ADI) to mice protected the mice against infection with multiple GAS serotypes. In this study, we determined the structure of GAS ADI and used this information to improve the vaccine safety of GAS ADI. Catalytically inactive mutant forms of ADI retained structure, recognition by antisera, and immunogenic epitopes, rendering them ideal for inclusion in GAS vaccine preparations. This example of structural biology informing vaccine design may underpin the formulation of a safe and efficacious GAS vaccine.
Introduction
Group A Streptococcus (GAS) is an exclusively human pathogen that colonizes primarily the upper respiratory tract and the skin. GAS is responsible for common mild infections such as pharyngitis and impetigo and, at a lower frequency, severe invasive conditions, including necrotizing fasciitis and streptococcal toxic shock-like syndrome. Reoccurring GAS infection can elicit nonsuppurative sequelae, including acute rheumatic fever, rheumatic heart disease, and acute poststreptococcal glomerulonephritis (1–3). There is no safe and efficacious commercial GAS vaccine available. GAS vaccinology has focused primarily on the major virulence factor, the surface-exposed M protein. GAS serotypes are designated on the basis of their patterns of M protein expression. M protein has been widely reported to protect against GAS infection (4), and two vaccine formulations based on a subset of M types have reached human clinical trials (5, 6). Despite this progress, there are shortcomings in the targeting of specific M proteins, including the occurrence of many unique serotypes (there are >200 known circulating types of GAS M protein [7]), antigenic variation within the same serotype, differences in the geographical distribution of serotypes (8, 9), and the production of antibodies cross-reactive with human tissue, which can lead to host autoimmune disease (3). In addition to M protein, a number of other GAS surface-localized and secreted antigens have been tested as vaccine candidates in mouse infection models, including fibronectin-binding protein A, R28 protein, protein F1, serum opacity factor (SOF), streptococcal protective antigen, Streptococcus pyogenes cell envelope proteinase (SpyCEP), C5a peptidase, streptococcal hemoprotein receptor, streptococcal pyrogenic exotoxin B (SpeB), streptococcal secreted esterase, streptolysin O (SLO), fibronectin-binding protein 54, streptococcal immunoglobulin-binding protein 35, and trigger factor (4). While all of these antigens show promise, none have progressed past animal-based trials.
We previously characterized arginine deiminase (ADI) as a GAS vaccine candidate. ADI is localized on the cell surface and produces opsonic antibodies capable of protecting mice against lethal challenges with homologous and heterologous GAS isolates (10). ADI is one of three enzymes in the ADI pathway and converts arginine to citrulline with the concomitant production of ammonia. In GAS, the enzymatic activity of ADI protects cells from low-pH environments (11, 12). Administration of a GAS vaccine preparation containing wild-type ADI, a protein with inherent enzymatic activity, may result in undesirable safety concerns. Some of the other previously reported GAS vaccine antigens possess enzyme activity, including C5a peptidase (13), SLO (14, 15), SpyCEP (15, 16), SOF (17), and SpeB (18). These antigens have been successfully deactivated via truncation or site-directed mutagenesis as a means of improving their safety profile.
Here we used X-ray crystallography and structural immunogenic epitope mapping to inform vaccine safety and design. We determined the crystal structure of GAS ADI at 2.48 Å resolution. A number of individual residues were targeted for site-directed mutagenesis on the basis of their positions in the GAS structure and following the comparison of GAS ADI to other ADI structures in which the active site was known, including those of Mycoplasma arginini (19) and Pseudomonas aeruginosa (20–22). We identify two site-directed mutant forms of ADI, the D166A and D277A mutant proteins, with unaltered antigenic characteristics and an ideal safety profile, as novel GAS vaccine components.
RESULTS
GAS ADI structure and active site.
The structure of GAS ADI was determined at 2.48 Å resolution by molecular replacement by using the structure of M. arginini ADI (19) as the search model (see Table S1 in the supplemental material). The asymmetric unit contains 8 molecules, the Cα atoms of which superpose with root mean square distances (RMSDs) ranging from 0.19 to 0.53 Å. As with other available ADI structures, GAS ADI consists of two domains, one consisting of five ββαβ motifs repeated around a pseudo-5-fold axis that contains the active site. The other is an orthogonal four-helix bundle (Fig. 1A and C). GAS ADI has an analogous structure in ADIs from other species. It superposes with those of M. arginini and P. aeruginosa with RMSDs of 1.31 and 1.56 Å, respectively, with deviations in one loop close to the active site (GAS ADI residues 268 to 274), one loop distal to the active site (residues 338 to 341), and the largest differences found in the four-helix bundle domain (residues 97 to 100 and 129 to 152). The GAS ADI active-site residues (D166, E220, H275, D277, and C401; Fig. 1B) superpose closely with those of M. arginini and P. aeruginosa, and several molecules in the asymmetric unit even contain a region of strong positive density in the active site, coinciding with the citrulline moiety bound to the M. arginini structure (19). This indicates that a molecule, possibly a bound substrate molecule, has been copurified with the protein, but the resulting electron density maps are not distinct enough to model this with certainty.
FIG 1 .
Enzymatic and structural comparisons of wild-type and mutant ADIs. (A) Model of the GAS ADI crystal structure illustrating the pseudo-5-fold axis of the catalytic domain in green, blue, pink, cyan, and red, respectively, with the helical orthogonal bundle in orange and the remaining structural elements in gray. (B) ADI active-site residues targeted for site-directed mutagenesis. (C) Topology diagram of GAS ADI, colored as in panel A. (D) Colorimetric assay for the determination of citrulline. Triplicate data from two independent replicate experiments are presented. Error bars indicate the standard deviations. Wild-type ADI produced a significantly greater amount of citrulline than each mutant ADI, as determined by one-way ANOVA (P < 0.05; indicated by an asterisk). (E) Tryptophan emission fluorescence spectra of wild-type ADI and mutant proteins. Wild-type ADI and the D166A, E220A, H275A, and D277A mutant ADIs exhibited nearly identical emission maxima (347 to 349 nm), whereas that of C401A mutant ADI was red shifted to 330 nm, which is indicative of significantly altered tertiary and/or quaternary structure. (F) UV CD spectra (200 to 260 nm) comparing the secondary structures of mutant ADIs to that of wild-type ADI. (G) ESI-MS spectra of wild-type and mutant ADIs. Wild-type ADI and the D166A, E220A, H275A, and D277A mutant ADIs exist as a monomer-dimer mixture, while C401A exists exclusively in a tetrameric form.
Strategy for ADI enzyme mutagenesis.
GAS ADI was found to share 41.3 and 36.0% sequence identity with the ADIs of M. arginini and P. aeruginosa, respectively. As the ADI active site has been well characterized in both of these species (19–22), residues for site-directed mutagenesis were selected following comparison of the GAS ADI structure to these structures. Residues D166, E220, H274, D277, and C401, all buried in the active-site pocket (Fig. 1B), were individually converted to alanine in recombinant GAS ADI, as these residues were predicted to be part of the active site or to be involved in catalysis.
Enzyme activities of wild-type and mutant ADIs.
We performed an endpoint colorimetric assay for the determination of citrulline to ascertain if the mutant ADIs had lost enzymatic activity. The conversion of arginine to citrulline was measured at 540 nm following the reaction of citrulline with 2,3-butanedione monoxime at a high temperature (95°C) in the presence of the reducing agent thiosemicarbazide, strong acids (H2SO4 and H3PO4), and catalytic quantities of Fe3+. The concentration of citrulline produced by 1 mg of wild-type ADI in 1 h was 338 mM (Fig. 1D). This concentration was significantly greater (P < 0.05) than that produced by any of the mutant ADIs, in all of which enzyme activity was abolished (<1.3 mM; Fig. 1D).
Structural characterization of mutant ADIs.
To confirm that the mutations in ADI did not alter protein structure or oligomerization, intrinsic tryptophan fluorescence, circular-dichroism (CD), and electrospray ionization (ESI)-mass spectrometry (MS) measurements were undertaken. ESI-MS was used to determine the mass-to-charge ratio of each mutant protein, which can be used to indicate a monomeric state(s). The D166A, E220A, H275A, and D277A mutant ADIs maintained the wild-type secondary structure, as indicated by intrinsic tryptophan fluorescence (Fig. 1E) and CD spectra (Fig. 1F). However, the tryptophan fluorescence and CD spectrum of the C401A mutant protein indicated an alteration in secondary structure compared to that of wild-type ADI (Fig. 1E and F). ESI-MS indicated that wild-type ADI exists as a mixture of monomers and dimers (Fig. 1G). Likewise, the D166A, E220A, H275A, and D277A mutant proteins exist as a mixture of monomeric and dimeric states (Fig. 1G). On the other hand, the C401A mutant protein existed exclusively as a tetramer (Fig. 1G). Size exclusion chromatography confirmed the finding that the C401A mutant ADI was tetrameric (data not shown). Small-angle X-ray scattering (SAXS) was used to further characterize the C401A mutant protein. The data suggest that the C401A mutant protein is a globular particle with a volume-derived molecular mass of 197.4 kDa (see Fig. S1 in the supplemental material), suggesting that it forms a stable tetrameric species in solution.
Immunogenicity of mutant ADIs.
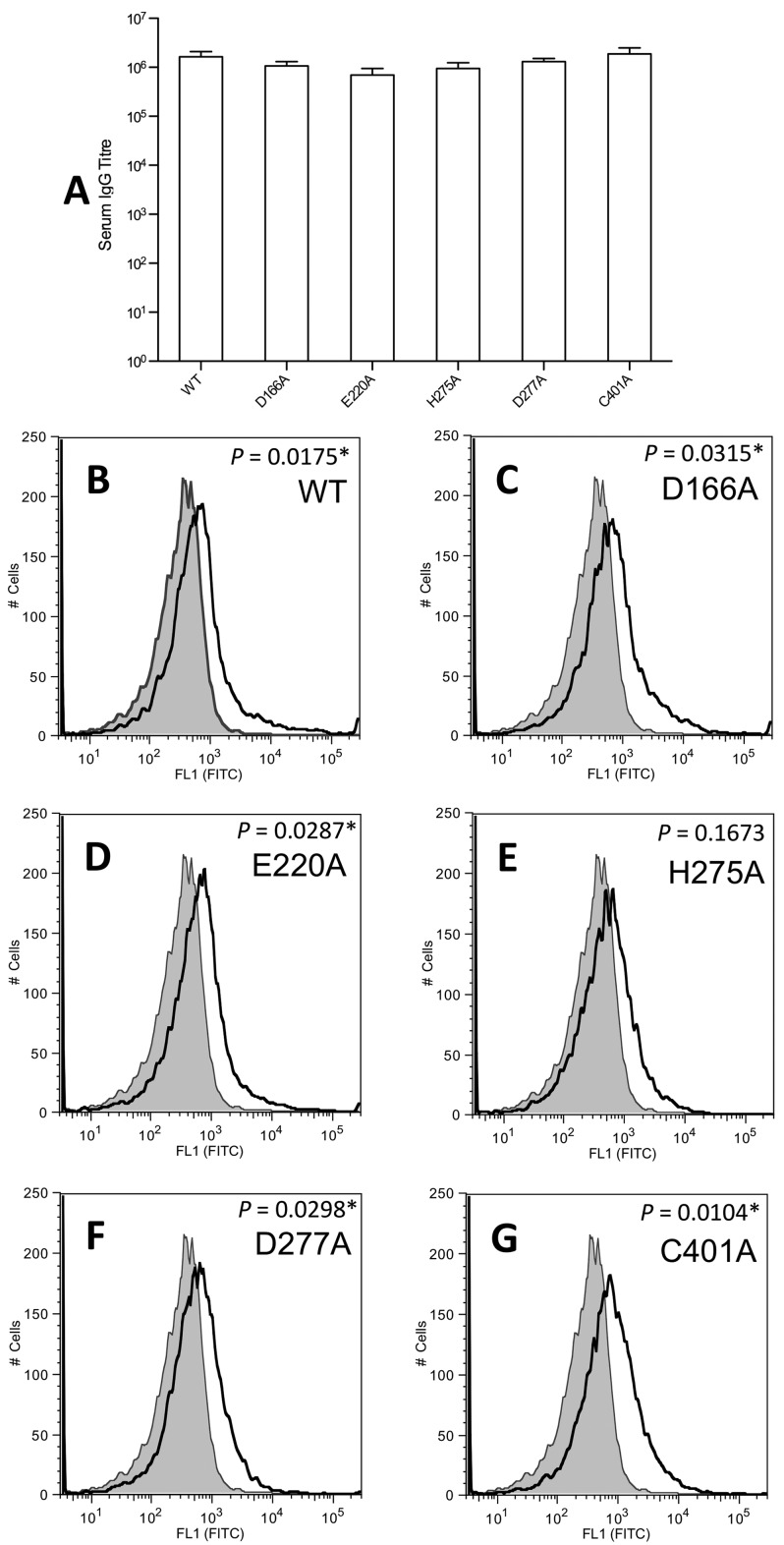
To assess whether the mutant ADIs could stimulate a humoral response capable of recognizing wild-type ADI, a murine immunization model was used. Following subcutaneous immunization with complete Freund’s adjuvant, each mutant ADI was observed to be immunogenic, stimulating a strong serum IgG response capable of recognizing wild-type ADI. When assessed by enzyme-linked immunosorbent assay (ELISA), the serum raised against each mutant ADI bound a quantity of immobilized wild-type ADI equivalent to that bound by serum generated against wild-type ADI (Fig. 2A). The average IgG titers (n = 5) for the proteins were as follows: wild-type ADI, 1,638,400; D166A mutant ADI, 1,064,960; E220A mutant ADI, 696,320; H275A mutant ADI, 942,080; D277A mutant ADI, 1,310,720; C401A mutant ADI, 1,884,160.
FIG 2 .
Capacity of antisera raised against mutant ADIs to recognize recombinant and native wild-type ADIs. BALB/c mice (n = 5) were immunized via the subcutaneous route with each individual ADI protein. Negative control mice were sham immunized with TBS and adjuvant only. Mice were administered 10 µg of protein on days 0, 21, and 28. Serum was collected on day 42. (A) Serum-specific IgG antibody titers determined by ELISA by using wild-type (WT) ADI to coat the plate. TBS sham serum did not produce measurable IgG titers against ADI. The absorbance cutoff for IgG titer determination was 0.2. Error bars represent the standard error of the mean. (B to G) Quantification of native ADI on the surface of GAS M1T1 isolate 5448 by flow cytometry. The histograms obtained for sera immunized with ADI proteins are indicated with a solid line; shading indicates the histogram for TBS sham serum. Panels: B, wild-type ADI; C, D166A mutant ADI; D, E220A mutant ADI; E, H275A mutant ADI; F, D277A mutant ADI; G, C401A mutant ADI. Incubation of GAS with sera raised against wild-type ADI and the D166A, E220A, D277A, and C401A mutant ADIs resulted in a significant shift in geometric mean surface fluorescence (P < 0.05; indicated by an asterisk) in comparison with that obtained with TBS sham serum. Triplicate data from two independent replicate experiments were obtained, and representative histograms are presented.
Antisera raised against mutant ADIs bind the GAS cell surface.
To investigate the capacity of sera raised against mutant ADIs to bind native ADI on the GAS cell surface, flow cytometry was used. Antiserum raised against the surface-bound M protein was used as a positive control, while Tris-buffered saline (TBS; 0.14 M NaCl, 2.7 mM KCl, 0.05 M Tris base, pH 7.0) sham serum was used as a negative control. Preincubation of GAS with antisera generated against the M1 protein (data not shown), or the wild-type, D166A mutant, E220A mutant, D277A mutant, and C401A mutant ADIs resulted in a significant shift in geometric mean surface fluorescence in comparison to TBS sham serum (Fig. 2B to D and F to G; P < 0.05). Serum raised against H275A mutant ADI also resulted in a shift, which was not statistically significant (Fig. 2E; P > 0.05).
Mapping of linear B-cell epitopes of ADI.
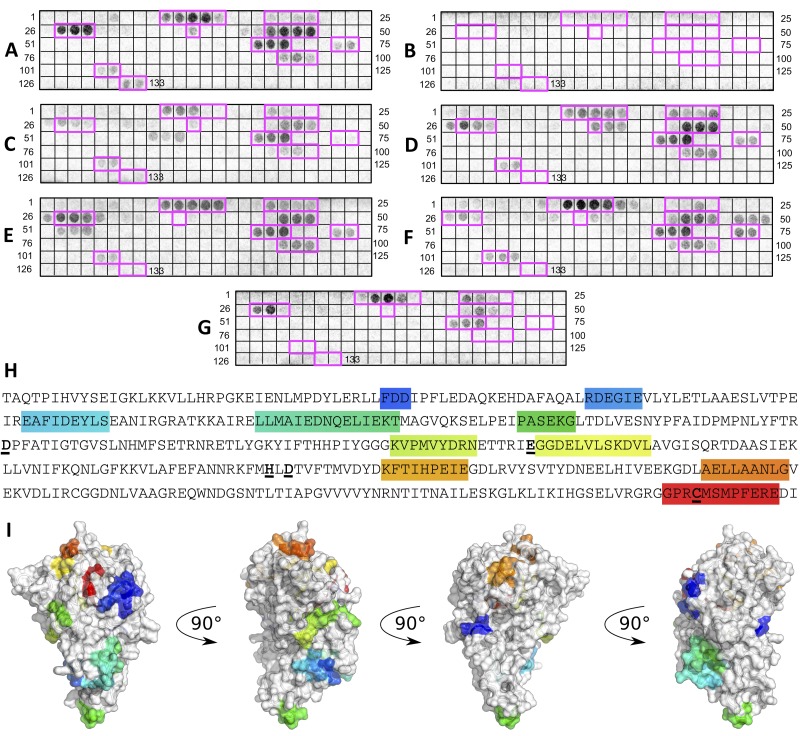
To conduct a preliminary characterization of the discrete B-cell epitopes within ADI, an overlapping peptide array membrane containing 15-mer peptides spanning the complete sequence of ADI was synthesized. Each consecutive peptide was shifted by three residues toward the C terminus of the protein. The membrane was probed with pooled antisera (n = 5) generated against wild-type ADI, stripped, and subsequently probed with a pool of TBS sham serum (n = 5). Incubation of the peptide arrays with the TBS sham serum resulted in no colorimetric reaction (Fig. 3B). In contrast, several discrete peptide spots were detected on the peptide array following probing with antiserum raised against wild-type ADI (Fig. 3A). Reactive spots are indicated within magenta boxes in Fig. 3A. Probing of the ADI peptide array with anti-wild-type ADI serum resulted in the identification of 10 linear B-cell epitopes (Fig. 3A [magenta outline] and H [highlighted]). Mapping of the immunogenic epitopes onto the structure of GAS ADI (Fig. 3I) indicated that the majority of the active-site residues (in bold in Fig. 3H) were not found in linear B-cell epitopes.
FIG 3 .
Determination of linear B-cell epitopes and immunogenic peptide mapping of wild-type ADI. Spots 1 to 133 are 15-residue-long peptides encompassing the ADI sequence immobilized on cellulose membrane. Each peptide is shifted by 3 amino acids toward the C terminus. (A to G) Membranes probed with pooled serum from BALB/c mice (n = 5) subcutaneously immunized as follows: A, wild-type ADI; B, TBS (sham); C, D166A mutant ADI; D, E220A mutant ADI; E, H275A mutant ADI; F, D277A mutant ADI; G, C401A mutant ADI. (H) Linear B-cell epitopes are highlighted in rainbow colors, starting with blue at the N terminus and ending with red at the C terminus. Individual residues targeted by site-directed mutagenesis are in bold with underlining. (I) Immunogenic epitopes in the structure of GAS ADI are highlighted in rainbow colors.
Effect of site-directed mutagenesis on the immunogenic B-cell epitopes of ADI.
Following stripping, the membrane was further probed with pools of antisera (n = 5) raised against each individual mutant ADI. For sera raised against the D166A (Fig. 3C), E220A (Fig. 3D), and H275A (Fig. 3E) mutant proteins, the same pattern of linear epitopes was recognized as when anti-wild-type ADI serum was used to probe the array (Fig. 3A). Serum generated against the D277A mutant protein recognized the same immunogenic epitopes as wild-type ADI serum, with an additional string of three peptides (Fig. 3F; peptides 48 to 50). For the sera raised against the D166A, H275A, D277A, and C401A mutant proteins, the peptide spots containing the respective mutations (Fig. 3) did not result in colorimetric development. The serum raised against the E220A mutant protein, however, did react with the peptide spots containing the E220 residue (Fig. 3D). Colorimetric development following probing of the peptide array with anti-C401A mutant ADI serum (Fig. 3G) resulted in substantially fewer developed spots than probing with anti-wild-type ADI serum (Fig. 3A). The anti-C401A mutant ADI serum reacted with only five of the immunogenic epitopes recognized by anti-wild-type ADI serum.
Comparison of GAS ADI to human PAD4.
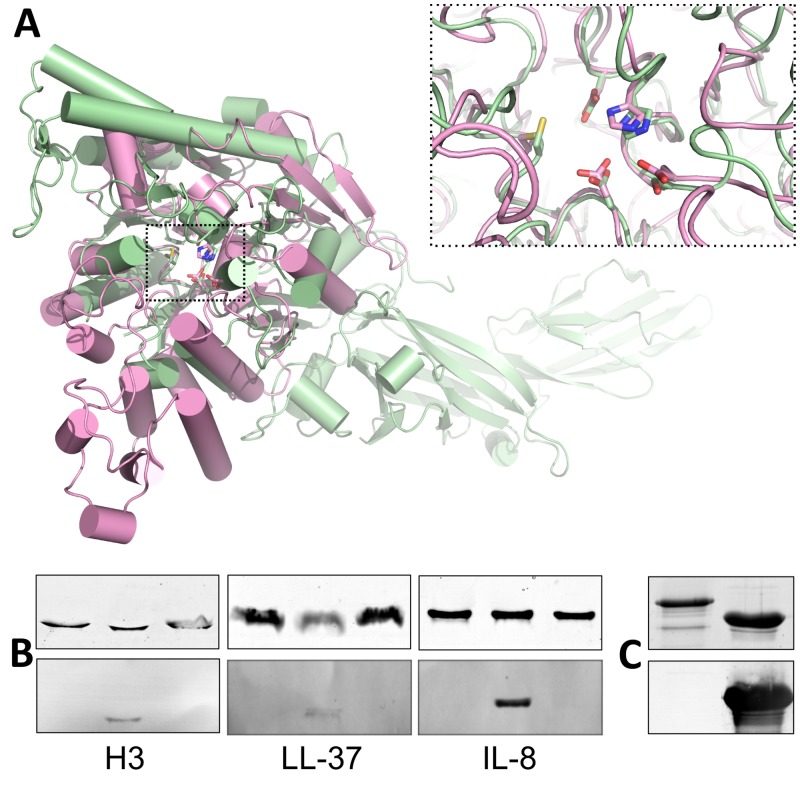
Peptidyl ADI 4 (PAD4) is known to convert arginine residues to citrulline in histones (23) and chemokines (24) within the host. To determine if ADI could be molecularly mimicking the function of human PAD4, we first compared the structure of GAS ADI to that of PAD4 (Protein Data Bank accession no. 2DW5) (25). These superpose with an RMSD of 2.8 Å over 216 residues, with the majority of the deviation coming from the larger α-helical elements present in PAD4. In contrast, the active-site residues of the two structures overlap with an RMSD of <1.0 Å (Fig. 4A). Western blotting indicated that recombinant human PAD4 successfully citrullinated host targets in vitro (Fig. 4B). However, recombinant wild-type ADI did not citrullinate host targets of PAD4 in vitro, including histone H3, LL-37, or interleukin-8 (IL-8) (Fig. 4B). Antiserum raised against wild-type ADI did not recognize human PAD4 in Western blot assays (Fig. 4C). Additionally, incubation of anti-wild-type ADI serum with immobilized PAD4 in an ELISA did not result in a measurable response (data not shown).
FIG 4 .
Comparison of GAS ADI to human PAD4. (A) Structural alignment of GAS ADI (pink) with human PAD4 (green). The superposition is centered on the α/β-propeller domains, omitting the orthogonal bundle from ADI and the two N-terminal all-β domains from PAD4. ADI active-site residues D166, E220, H275, D277, and C401 align closely with the corresponding residues in PAD4. (B) Unlike human PAD4, GAS ADI does not citrullinate host targets, histone H3, LL-37, or IL-8 in vitro. Host targets were incubated with 5 µM ADI (left lane), 500 nM PAD4 (middle lane), or neither (right lane) for 1 h at 37°C. The reaction was immediately stopped by incubation with SDS-PAGE loading buffer for 15 min at 95°C. Samples containing 1.5 µg of target protein were separated by 15% PAGE and either stained with Coomassie blue (top) or transferred to nitrocellulose membrane for Western blotting with antibodies for the detection of citrulline (bottom). (C) Anti-ADI serum does not recognize recombinant human PAD4 in a Western blot assay. Samples containing 2 µg of ADI (right lane) or PAD4 (left lane) were separated by 15% PAGE and either stained with Coomassie blue (top) or transferred to nitrocellulose membrane for Western blotting (bottom).
DISCUSSION
Structural biology is being increasingly used for the rational design and optimization of vaccine antigens. Knowledge of the structure of a given antigen may allow researchers to design subunits, peptides, or other vaccine constituents based on a particular region. Structural information can also facilitate inactivation or detoxification of antigens possessing enzyme activity or toxic effects. Other recent studies using structural biology for the optimization of GAS vaccine candidates include SLO (14) and JJo (26), a chimeric protein based on the J14 epitope of the M1 protein. Researchers have also turned to a structural approach for the optimization of vaccine antigens effective against other streptococcal species, including group B Streptococcus (27) and meningococcus (28). The present study paired a structural biology approach with linear epitope mapping to facilitate the optimization of the GAS vaccine antigen ADI.
Previous studies undertaking site-directed mutagenesis of recombinant ADIs from M. hominis (29) and P. aeruginosa (22) identified a number of mutations that led to a complete loss of enzyme activity. In this study, we identified residues in GAS ADI (D166, E220, H275, D277, and C401) whose mutation to alanine resulted in a complete abrogation of enzymatic activity. On the basis of these results and the X-ray crystal structure of ADI, we propose that residues Glu220, His275, and Cys401 form the catalytic triad of GAS ADI.
The crystal structure of ADI indicated that the residues of the active site are shielded within a pocket. This observation suggests that the active site may not be readily accessible to antisera recognizing conformational epitopes. Nonetheless, antibodies recognizing linear epitopes may bind to active-site residues. To ensure that mutation of active-site residues did not alter the recognition and binding profile of anti-ADI sera, we immunized mice with the wild-type and mutant ADIs. This generated high-titer antisera capable of recognizing native ADI on the surface of GAS. Subsequently, we probed an ADI peptide array with serum raised against wild-type ADI to reveal the linear B cell epitopes. Ten linear epitopes were identified. Mutation of the D166, H275, and D277 residues did not affect the pattern of epitopes recognized by anti-ADI sera. The E220 and C401 residues fell directly adjacent to or within linear epitopes of ADI. Therefore, mutation of E220 or C401 in an optimized vaccine preparation is less desirable, as this may alter the epitopes targeted and subsequently recognized by host-derived antibodies. In addition, the serum generated against the H275A mutant ADI had reduced binding to the surface of whole GAS (when assessed by flow cytometry). For this reason, mutation of the H275 residue is less advisable for the preparation of an optimized vaccine preparation.
Analysis of the structure of the mutant ADIs indicated that the D166A, E220A, H275A, and D277A mutant ADIs had secondary-structure and oligomerization properties equivalent to those of wild-type ADI. On the other hand, the mutation of cysteine 401 to alanine had an unexpected effect. The C401A mutant protein lost its enzyme activity but also showed some change in secondary structure compared to that of wild-type ADI and adopted a solely tetrameric state, with no monomer or dimer detected. Wild-type ADI did not form any tetramers but rather existed only as a monomer-dimer mixture. When antiserum raised against the C401A mutant protein was used to probe the peptide array, only 5 of the 10 immunogenic epitopes detected by anti-wild-type ADI serum were detected. Presumably, the conformation disruption, in addition to the change in oligomeric state, altered the epitopes recognized by the antiserum. We conclude that the C401A mutation would be a less attractive choice for optimization of ADI as a GAS vaccine candidate.
ADI does not possess a human ortholog. Our previous study reported that ADI does not react with human heart tissue or human sera obtained from individuals living in regions with endemic disease (10). Prior to further development of ADI as a GAS vaccine candidate, we wanted to confirm that anti-ADI serum does not cross-react with host proteins possessing structural similarity. The active site of ADI is highly conserved among GAS ADI, M. arginini ADI, P. aeruginosa ADI, and human PAD4. Given the level of conservation in the active sites of GAS ADI and human PAD4, we assessed whether recombinant wild-type ADI could citrullinate known host-derived targets of PAD4, including histone H3, the chemokine IL-8, and the antimicrobial peptide LL-37. While recombinant PAD4 citrullinated each of these targets in vitro, recombinant ADI did not. Thus, it appears that GAS ADI does not mimic the functionality of PAD4 during infection of the host. Even though GAS ADI and human PAD4 show some structural similarities in the active-site region, they do not share significant amino acid sequence identity or perform the same biological function. Western blotting and ELISAs indicated that anti-ADI sera did not recognize human PAD4 in vitro. Therefore, the administration of optimized ADI in a GAS vaccine preparation is not expected to generate antibodies that bind to or interfere with the function of PAD4 in the host.
ADI has been reported to protect GAS from the low internal pH of host cells, specifically, to shield streptococcal cells on the respiratory surfaces or within phagolysosomes (11, 12, 30). Similarly, in Listeria monocytogenes, ADI has been found to promote extracellular acid tolerance (31). ADI has previously been reported to influence GAS virulence factor expression, including the expression of the hyaluronic acid capsule and pyrogenic exotoxin SpeB (32). Through the modulation of the expression of such virulence factors, ADI is also thought to influence the internalization of GAS into epithelial cells (32). GAS ADI is also thought to play a role in the inhibition of human peripheral blood mononuclear cell (PBMC) proliferation (11, 33). By doing so, ADI may endow GAS with the ability to downregulate or modulate the host immune response to infection. The inclusion of an optimized mutant ADI in a future GAS vaccine formulation may not only lead to opsonophagocytosis of GAS, as reported previously (10), but may also disrupt ADI modulation of host PBMC proliferation and GAS virulence factor regulation at the site of infection.
We suggest that two of our designed mutant ADIs, the D166A and D277A mutant proteins, are ideal for inclusion in a GAS vaccine. The secondary structure and oligomerization state of these mutant proteins remained equivalent to those of the wild-type protein. These residues do not fall within linear B cell epitopes. Antisera raised against the D166A and D277A mutant proteins were of high titer, recognize the same immunogenic epitopes as wild-type ADI, and detect native ADI associated with the surface of GAS.
MATERIALS AND METHODS
Bacterial strains and plasmids used in this study.
GAS M1T1 strain 5448 (emm1) has been described elsewhere (34). Escherichia coli strain BL21 Star (DE3) (Invitrogen) was used for protein expression.
Construction of ADI expression vector and site-directed mutagenesis.
The full-length sagP gene encoding ADI was amplified from GAS strain 5448 previously (10). The sagP gene was cloned into the pET151 expression vector (Invitrogen) in accordance with the manufacturer’s instructions. Individual residues were targeted for site-directed mutagenesis on the basis of their positions in the GAS ADI structure and following comparison to ADI structures from M. arginini (19) and P. aeruginosa (20) in which the active site had been defined. Residues of interest (D166, E220, H275, D277, and C401) were mutated to alanine by QuikChange XL site-directed mutagenesis (Stratagene) (for the primers used, see Table S2 in the supplemental material) as previously described (35).
Recombinant protein expression and purification.
Expression plasmids were transformed into E. coli strain BL21 Star (DE3), and recombinant protein expression was performed as previously described (35). Cells were frozen at −80°C and lysed with 20 ml of lysis buffer per liter of original culture (20 mM Tris [pH 7.6], 300 mM NaCl, 3 mM 2-mercaptoethanol [2-ME], 10% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM imidazole, 1 mg/ml lysozyme, 1 Complete protease inhibitor cocktail [Roche] tablet). Lysate was sonicated for 10 20-s intervals with alternating rest periods of 20 s and placed on a suspension mixer (Ratek Instruments) for 30 min at 4°C. Lysate was centrifuged at 20,000 × g for 1 h at 4°C. Ni-nitrilotriacetic acid (NTA) resin (Merck Millipore) was added to the cleared lysate, a 2-ml bed volume per 20 ml of lysate, and incubated on a suspension mixer (Ratek Instruments) for 1 h at 4°C. The mixture was applied to an Econo-Pac chromatography column (Bio-Rad), and the lysate passed through the resin by gravity flow. The resin was washed with 20-ml aliquots of wash buffer (20 mM Tris [pH 7.6], 150 mM NaCl, 3 mM 2-ME, 5% [vol/vol] glycerol, 1 mM PMSF) each with an increasing concentration of imidazole, 20, 50, and 100 mM, respectively. Recombinant protein was eluted by using 1-ml fractions of elution buffer (20 mM Tris [pH 7.6], 150 mM NaCl, 3 mM 2-ME, 5% [vol/vol] glycerol, 1 mM PMSF, 250 mM imidazole). Fractions containing protein were pooled and dialyzed at 4°C overnight against 20 mM Tris (pH 7.6) and 150 mM NaCl. Protein concentration was determined with the NanoDrop 2000 (Thermo Scientific) and the bicinchoninic acid protein assay kit (Sigma-Aldrich). The extinction coefficient of ADI, 26,485 M−1 cm−1, was obtained from the ProtParam tool on the Expasy website (http://web.expasy.org/protparam/).
Recombinant protein purification and His tag cleavage for crystallography.
Expression of wild-type ADI, cell lysis, and Ni-NTA purification were performed as described above. Following purification, fractions containing protein were pooled and tobacco etch virus protease was added (0.1 mg per liter of starting culture) and the mixture was dialyzed against a combination of 20 mM Tris (pH 7.6), 150 mM NaCl, 3 mM 2-ME, and 10% glycerol at 4°C overnight. A second Ni-NTA purification was performed to separate cleaved ADI from uncleaved ADI. Ni-NTA resin (Merck Millipore), 1-ml bed volume, was added to the dialyzed sample prior to 1 h of incubation on the suspension mixer (Ratek Instruments) at 4°C. The mixture was applied to an Econo-Pac chromatography column (Bio-Rad), and the sample passed through by gravity flow. The resin was washed with 10-ml aliquots of wash buffer (20 mM Tris [pH 7.6], 150 mM NaCl, 3 mM 2-ME, 10% [vol/vol] glycerol) each with an increasing concentration of imidazole, 5, 10 20, 50, and 250 mM, respectively. Fractions containing cleaved protein only were concentrated with an Amicon Ultra-15 centrifugal filter unit with a 10-kDa molecular mass cutoff (Merck Millipore). Concentrated protein was passed through a HiLoad 26/60 Superdex 200 gel filtration column (GE Life Sciences) with s200 buffer (20 mM Tris [pH 7.6], 150 mM NaCl, 5 mM dithiothreitol [DTT]). Fractions corresponding to the elution peak were pooled, concentrated to 6 mg/ml with the aforementioned Amicon filter device, and snap-frozen in liquid N2.
Crystallization and data collection.
Initial searches for crystallization conditions were performed as 100 nl of well solution plus 100 nl of protein hanging-drop vapor diffusion experiments with a 75-µl reservoir at 18°C, with sparse-matrix crystallization screens, set up with a TTP Labtech mosquito crystal liquid-handling robot. Conditions were further refined as hanging-drop vapor diffusion experiments with 1 µl of well solution added to 1 µl of protein equilibrated against a 500-µl reservoir at 18°C. Diffracting ADI crystals formed in 0.1 M bis-Tris (pH 5.5)–20% PEG 3350–0.3 M lithium sulfate. Crystals were transferred to a 100% Paratone drop before vitrification in liquid nitrogen. Diffraction experiments were conducted at the Australian Synchrotron MX1 beam line at 100 K.
Structure determination.
Data were integrated and scaled by XDS and XSCALE, respectively (36). Manual inspection of spot predictions during integration indicated diffraction from two distinct lattices. These were separately integrated and scaled together with a concomitant improvement of merging statistics. Molecular replacement was performed in PHASER (37) with ADI from M. arginini as the search model (19). Initial automated rebuilding was performed with Phase_and_build from the PHENIX software suite (38), followed by manual rounds of rebuilding in COOT (39) and refinement in phenix.refine (38) or Buster (40). The final model was validated with Phenix.model_vs_data (41) and Molprobity (42).
SAXS.
Purified C401A mutant ADI was dialyzed for 18 h into a mixture of 20 mM Tris (pH 7.6), 150 mM NaCl, and 1 mM DTT. A dilution series was prepared from this for concentrations between 1.48 and 0.31 mg/ml. SAXS data were collected on a Pilatus 1M detector at the SAXS/WAXS beam line of the Australian Synchrotron (Melbourne, Australia). The sample-to-detector distance was 1.6 m, and the wavelength was 1.12713 Å, yielding a range of momentum transfer of 0.011 < q < 0.500 Å − 1, where q = 4π × sin(θ)/λ and 2θ is the scattering angle. Sample volumes of 50 µl were exposed at 298K while flowing through a 1.5-mm-diameter quartz capillary at a rate of 1 µl/s. Images were taken every 1 s, and data reduction was performed in scatterBrain (http://www.synchrotron.org.au/index.php/aussyncbeamlines/saxswaxs/software-saxswaxs). Images were summed, normalized to the transmitted intensity, averaged, buffer subtracted, and converted to an absolute scale against the scattering cross-section of water.
The ATSAS 2.5 software package was used for subsequent analyses (43–45). Guinier analysis was performed for q × Rg = <1.3 with AUTORG, and data sets were examined for concentration dependence and linearity. P(r) distributions were obtained by indirect transformation in AUTOGNOM, and molecular weights were estimated from these distributions with SAXS MoW (46).
The 0.71-mg/ml data set was selected for comparison and modeling over a range of 0.019 < q < 0.300 Å − 1. Predicted scattering from the crystallographic tetramer was calculated with CRYSOL (47) by using default parameters. Rigid-body models were generated with SASREF7 in the ATSAS Online suite (48) by using four copies of a single monomer from the crystal structure, P1 symmetry, and no additional restraints.
Colorimetric assay for determination of citrulline.
The determination of citrulline produced from ADI acting on arginine was performed in accordance with the established protocols (49). The amount of citrulline produced by wild-type ADI was compared to that produced by each mutant ADI by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test (P < 0.05).
CD measurements.
CD measurements were performed as previously described (35).
Nano-ESI-MS.
MS was performed on a SYNAPT HDMS mass spectrometer (Waters) with a nano-ESI source. Instrument conditions were as follows: capillary voltage, 1.5 kV; sample cone, 150 V; extraction cone, 4 V; transfer collision energy, 8 V. Wild-type and mutant ADIs were buffer exchanged into 200 mM ammonium acetate (pH 6.8) with a Superdex 200 10/300 G analytical size exclusion column (GE Healthcare) prior to MS. In all experiments, 2 µl of each protein was electrosprayed from gold-coated borosilicate glass capillaries prepared in house. All spectra were analyzed with MassLynx V4.1 software with minimal smoothing applied to the raw spectral data.
Intrinsic tryptophan fluorescence.
Tryptophan fluorescence spectra were acquired with a Cary Eclipse fluorescence spectrophotometer (Varian). The excitation wavelength was set at 295 nm, and the emission spectra were recorded from 300 to 450 nm. The slit width for both excitation and emission spectra was set at 5 nm. Wild-type and mutant ADI were prepared to a final concentration of 10 µM in 25 mM Tris–150 mM NaCl (pH 7.4). Proteins were incubated at room temperature for 15 min prior to analysis.
Subcutaneous immunization of BALB/c mice and ELISA.
Immunization of BALB/c mice and subsequent ELISA were performed as previously described (10). Negative control mice were sham immunized with TBS and adjuvant only. The experimental protocols described in this report were performed with approval from the University of Queensland Animal Ethics Committee (approval number SCMB/114/10) and complied with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (National Health and Medical Research Council, Australia).
Flow cytometry.
Flow cytometry was performed as previously described (10).
Analysis of peptide arrays.
An overlapping peptide spot membrane of ADI was constructed from an acid-stable AC-S01-type amino-pegylated cellulose membrane (AIMS Scientific Products GmbH, Braunschweig, Germany). A series of 15-mer synthetic peptides, each offset by 3 residues, were assembled at defined intervals on the membrane. The membrane was rehydrated with 100% ethanol, washed with TBS for 5 min, and blocked overnight at 4°C with membrane blocking solution (MBS; 2% skim milk and 0.2% Tween 20 in TBS). The membrane was washed for 5 min with TBS containing 0.05% Tween 20 (TBST). The membrane was subsequently probed with a pool of mouse antiserum raised against each ADI protein (or TBS sham serum) diluted 1:100 in MBS for 3 h with gentle rocking at room temperature. Following three TBST washes, the membrane was incubated with a 1:2,000 dilution of goat anti-mouse IgG-alkaline phosphatase conjugate (Sigma, St. Louis, MO) for 1.5 h at room temperature. Two TBST washes were performed, followed by two 5-min washes with citrate-buffered saline (NaCl at 8 g/liter, KCl at 0.2 g/liter, citric acid at 10.51 g/liter, pH 7.0). The membranes were developed with color-developing solution [50 µl of 1 M magnesium chloride per 10 ml, 40 µl of 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt per 10 ml, and 60 µl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide per 10 ml] for 10 min, and the reaction was terminated by washing with phosphate-buffered saline. The developed membrane array was scanned with a GS-800 calibrated densitometer (Bio-Rad Laboratories, Hercules, CA). Following reaction of the first antiserum, the membrane was stripped. In short, the membrane was washed twice with water (5 min) and incubated in an excess of dimethylformamide (DMF) until the blue color of spot signals dissolved. All washing steps were performed for 5 min. The DMF step was repeated, followed by three water washes. Membranes were then washed three times with stripping mixture A (8 M urea, 1% SDS, 0.5% 2-ME, pH 7.0) in a sonication bath at 40°C, followed by washing three times with stripping mixture B (10% acetic acid, 50% ethanol). Finally, the membrane was subjected to three washes with 100% ethanol, followed by one wash with TBS. Only the more intensely labeled peptide spots were considered reactive.
Reaction of ADI and PAD4 with host targets.
The citrullination assay for ADI and PAD4 was performed as previously described (50). PAD4 (Cayman Chemicals), recombinant histone H3 (Cayman Chemicals), IL-8 (Merck Millipore), and LL-37 (AnaSpec) were all purchased commercially.
Western blot analysis.
Western blotting was performed as previously described (51). Citrulline residues in target proteins were detected with the Anti-Citrulline (Modified) Detection Kit (Merck Millipore).
Protein structure accession number.
The coordinates and structural factors described in this report have been deposited in the Protein Data Bank and assigned accession number 4BOF.
SUPPLEMENTAL MATERIAL
SAXS data for C401A mutant ADI are consistent with a stable tetrameric complex different from that observed in the wild-type crystal structure. (A) Experimental scattering from the C401A mutant protein as I(q) versus q on an absolute logarithmic scale and with 1σ error bars shown in gray. The calculated scattering from the crystallographic tetramer is shown in red, fitting a χ2 value of 6.61. That of a refined tetrameric rigid-body model is shown in orange, fitting a χ2 value of 0.99. (B) Arrangement of subunits in the best-fitting tetrameric rigid-body model. Component monomers are shown as surfaces over a cartoon representation and are colored alternating yellow and orange. (C) Pairwise interatomic distance distribution, P(r), derived from the scattering via indirect transformation. The distance vector (r) has a maximum of 134 Å, and the particle’s P(r)-derived radius of gyration (Rg) is 38.5 Å. (D) Guinier transformation of the data as ln[I(q)] versus q2 over a range of q × Rg = <1.3, demonstrating linearity across all concentrations. Download
Crystallographic data collection and refinement statistics.
Primers used for ADI site-directed mutagenesis.
ACKNOWLEDGMENTS
A.H. is a recipient of a DAAD Research Grant for Doctoral Candidates, Young Academics and Scientists. This work was supported by National Health and Medical Research Council of Australia (NHMRC) development grant APP1000512, Preclinical Studies of Group A Streptococcal Vaccine Candidates (M.J.W., B.K., and M.R.B.). B.K. and M.J.W. are NHMRC research fellows.
A.H., M.R.B., and M.J.W. have an intellectual property interest in the antigens described in this report.
Footnotes
Citation Henningham A, Ericsson DJ, Langer K, Casey LW, Jovcevski B, Chhatwal GS, Aquilina JA, Batzloff MR, Kobe B, Walker MJ. 2013. Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus. mBio 4(4):e00509-13. doi:10.1128/mBio.00509-13.
REFERENCES
- 1. Henningham A, Barnett TC, Maamary PG, Walker MJ. 2012. Pathogenesis of group A streptococcal infections. Discov. Med 13:329–342 [PubMed] [Google Scholar]
- 2. Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9:724–736 [DOI] [PubMed] [Google Scholar]
- 3. Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henningham A, Gillen CM, Walker MJ. 2013. Group A streptococcal vaccine candidates: potential for the development of a human vaccine. Curr. Top. Microbiol. Immunol. 368:207–242 [DOI] [PubMed] [Google Scholar]
- 5. Kotloff KL, Corretti M, Palmer K, Campbell JD, Reddish MA, Hu MC, Wasserman SS, Dale JB. 2004. Safety and immunogenicity of a recombinant multivalent group A streptococcal vaccine in healthy adults: phase I trial. JAMA 292:709–715 [DOI] [PubMed] [Google Scholar]
- 6. McNeil SA, Halperin SA, Langley JM, Smith B, Warren A, Sharratt GP, Baxendale DM, Reddish MA, Hu MC, Stroop SD, Linden J, Fries LF, Vink PE, Dale JB. 2005. Safety and immunogenicity of 26-valent group A Streptococcus vaccine in healthy adult volunteers. Clin. Infect. Dis. 41:1114–1122 [DOI] [PubMed] [Google Scholar]
- 7. McMillan DJ, Drèze PA, Vu T, Bessen DE, Guglielmini J, Steer AC, Carapetis JR, Van Melderen L, Sriprakash KS, Smeesters PR. 2013. Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin. Microbiol. Infect. 19:E222–E229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smeesters PR, McMillan DJ, Sriprakash KS, Georgousakis MM. 2009. Differences among group A Streptococcus epidemiological landscapes: consequences for M protein-based vaccines? Expert Rev. Vaccines 8:1705–1720 [DOI] [PubMed] [Google Scholar]
- 9. Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. 2009. Global emm type distribution of group A Streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 9:611–616 [DOI] [PubMed] [Google Scholar]
- 10. Henningham A, Chiarot E, Gillen CM, Cole JN, Rohde M, Fulde M, Ramachandran V, Cork AJ, Hartas J, Magor G, Djordjevic SP, Cordwell SJ, Kobe B, Sriprakash KS, Nizet V, Chhatwal GS, Margarit IY, Batzloff MR, Walker MJ. 2012. Conserved anchorless surface proteins as group A streptococcal vaccine candidates. J. Mol. Med. 90:1197–1207 [DOI] [PubMed] [Google Scholar]
- 11. Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curran TM, Lieou J, Marquis RE. 1995. Arginine deiminase system and acid adaptation of oral streptococci. Appl. Environ. Microbiol. 61:4494–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ji Y, Carlson B, Kondagunta A, Cleary PP. 1997. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65:2080–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiarot E, Faralla C, Chiappini N, Tuscano G, Falugi F, Gambellini G, Taddei A, Capo S, Cartocci E, Veggi D, Corrado A, Mangiavacchi S, Tavarini S, Scarselli M, Janulczyk R, Grandi G, Margarit I, Bensi G. 2013. Targeted amino acid substitutions impair streptolysin O toxicity and group A Streptococcus virulence. mBio 4:e00387-12. 10.1128/mBio.00387-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bensi G, Mora M, Tuscano G, Biagini M, Chiarot E, Bombaci M, Capo S, Falugi F, Manetti AG, Donato P, Swennen E, Gallotta M, Garibaldi M, Pinto V, Chiappini N, Musser JM, Janulczyk R, Mariani M, Scarselli M, Telford JL, Grifantini R, Norais N, Margarit I, Grandi G. 2012. Multi high-throughput approach for highly selective identification of vaccine candidates: the group A Streptococcus case. Mol. Cell. Proteomics 11:M111.015693. 10.1074/mcp.M111.015693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turner CE, Kurupati P, Wiles S, Edwards RJ, Sriskandan S. 2009. Impact of immunization against SpyCEP during invasive disease with two streptococcal species: Streptococcus pyogenes and Streptococcus equi. Vaccine 27:4923–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gillen CM, Courtney HS, Schulze K, Rohde M, Wilson MR, Timmer AM, Guzman CA, Nizet V, Chhatwal GS, Walker MJ. 2008. Opacity factor activity and epithelial cell binding by the serum opacity factor protein of Streptococcus pyogenes are functionally discrete. J. Biol. Chem. 283:6359–6366 [DOI] [PubMed] [Google Scholar]
- 18. Ulrich RG. 2008. Vaccine based on a ubiquitous cysteinyl protease and streptococcal pyrogenic exotoxin A protects against Streptococcus pyogenes sepsis and toxic shock. J. Immune Based Ther. Vaccines 6:8. 10.1186/1476-8518-6-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das K, Butler GH, Kwiatkowski V, Clark AD, Yadav P, Arnold E. 2004. Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism. Structure 12:657–667 [DOI] [PubMed] [Google Scholar]
- 20. Galkin A, Kulakova L, Sarikaya E, Lim K, Howard A, Herzberg O. 2004. Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target. J. Biol. Chem. 279:14001–14008 [DOI] [PubMed] [Google Scholar]
- 21. Galkin A, Lu X, Dunaway-Mariano D, Herzberg O. 2005. Crystal structures representing the Michaelis complex and the thiouronium reaction intermediate of Pseudomonas aeruginosa arginine deiminase. J. Biol. Chem. 280:34080–34087 [DOI] [PubMed] [Google Scholar]
- 22. Lu X, Li L, Wu R, Feng X, Li Z, Yang H, Wang C, Guo H, Galkin A, Herzberg O, Mariano PS, Martin BM, Dunaway-Mariano D. 2006. Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides insight into structural determinants of function. Biochemistry 45:1162–1172 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306:279–283 [DOI] [PubMed] [Google Scholar]
- 24. Proost P, Loos T, Mortier A, Schutyser E, Gouwy M, Noppen S, Dillen C, Ronsse I, Conings R, Struyf S, Opdenakker G, Maudgal PC, Van Damme J. 2008. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J. Exp. Med. 205:2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. 2006. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry 45:11727–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Georgousakis MM, Hofmann A, Batzloff MR, McMillan DJ, Sriprakash KS. 2009. Structural optimisation of a conformational epitope improves antigenicity when expressed as a recombinant fusion protein. Vaccine 27:6799–6806 [DOI] [PubMed] [Google Scholar]
- 27. Nuccitelli A, Cozzi R, Gourlay LJ, Donnarumma D, Necchi F, Norais N, Telford JL, Rappuoli R, Bolognesi M, Maione D, Grandi G, Rinaudo CD. 2011. Structure-based approach to rationally design a chimeric protein for an effective vaccine against group B Streptococcus infections. Proc. Natl. Acad. Sci. U. S. A. 108:10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scarselli M, Arico B, Brunelli B, Savino S, Di Marcello F, Palumbo E, Veggi D, Ciucchi L, Cartocci E, Bottomley MJ, Malito E, Lo Surdo P, Comanducci M, Giuliani MM, Cantini F, Dragonetti S, Colaprico A, Doro F, Giannetti P, Pallaoro M, Brogioni B, Tontini M, Hilleringmann M, Nardi-Dei V, Banci L, Pizza M, Rappuoli R. 2011. Rational design of a meningococcal antigen inducing broad protective immunity. Sci. Transl. Med. 3:91ra62. 10.1126/scitranslmed.3002234 [DOI] [PubMed] [Google Scholar]
- 29. Wei Y, Zhou H, Sun Y, He Y, Luo Y. 2007. Insight into the catalytic mechanism of arginine deiminase: functional studies on the crucial sites. Proteins 66:740–750 [DOI] [PubMed] [Google Scholar]
- 30. Curran TM, Ma Y, Rutherford GC, Marquis RE. 1998. Turning on and turning off the arginine deiminase system in oral streptococci. Can. J. Microbiol. 44:1078–1085 [DOI] [PubMed] [Google Scholar]
- 31. Ryan S, Begley M, Gahan CG, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11:432–445 [DOI] [PubMed] [Google Scholar]
- 32. Marouni MJ, Ziomek E, Sela S. 2003. Influence of group A streptococcal acid glycoprotein on expression of major virulence factors and internalisation by epithelial cells. Microb. Pathog. 35:63–72 [DOI] [PubMed] [Google Scholar]
- 33. Degnan BA, Palmer JM, Robson T, Jones CE, Fischer M, Glanville M, Mellor GD, Diamond AG, Kehoe MA, Goodacre JA. 1998. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 66:3050–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51:123–134 [DOI] [PubMed] [Google Scholar]
- 35. Cork AJ, Jergic S, Hammerschmidt S, Kobe B, Pancholi V, Benesch JL, Robinson CV, Dixon NE, Aquilina JA, Walker MJ. 2009. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J. Biol. Chem. 284:17129–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. 2012. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68:368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Afonine PV, Grosse-Kunstleve RW, Chen VB, Headd JJ, Moriarty NW, Richardson JS, Richardson DC, Urzhumtsev A, Zwart PH, Adams PD. 2010. Phenix.model_vs_data: a high-level tool for the calculation of crystallographic model and data statistics. J. Appl. Crystallogr. 43:669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. 2006. ATSAS, p 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39:277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36:1277–1282 [Google Scholar]
- 45. Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. 2012. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45:324–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fischer H, De Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. 2009. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43:101–109 [Google Scholar]
- 47. Svergun D, Barberato C, Koch MHJ. 1995. CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28:768–773 [Google Scholar]
- 48. Petoukhov MV, Svergun DI. 2005. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knipp M, Vasák M. 2000. A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal. Biochem. 286:257–264 [DOI] [PubMed] [Google Scholar]
- 50. Horikoshi N, Tachiwana H, Saito K, Osakabe A, Sato M, Yamada M, Akashi S, Nishimura Y, Kagawa W, Kurumizaka H. 2011. Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene. Acta Crystallogr. D Biol. Crystallogr. 67:112–118 [DOI] [PubMed] [Google Scholar]
- 51. Cole JN, Aquilina JA, Hains PG, Henningham A, Sriprakash KS, Caparon MG, Nizet V, Kotb M, Cordwell SJ, Djordjevic SP, Walker MJ. 2007. Role of group A Streptococcus HtrA in the maturation of SpeB protease. Proteomics 7:4488–4498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SAXS data for C401A mutant ADI are consistent with a stable tetrameric complex different from that observed in the wild-type crystal structure. (A) Experimental scattering from the C401A mutant protein as I(q) versus q on an absolute logarithmic scale and with 1σ error bars shown in gray. The calculated scattering from the crystallographic tetramer is shown in red, fitting a χ2 value of 6.61. That of a refined tetrameric rigid-body model is shown in orange, fitting a χ2 value of 0.99. (B) Arrangement of subunits in the best-fitting tetrameric rigid-body model. Component monomers are shown as surfaces over a cartoon representation and are colored alternating yellow and orange. (C) Pairwise interatomic distance distribution, P(r), derived from the scattering via indirect transformation. The distance vector (r) has a maximum of 134 Å, and the particle’s P(r)-derived radius of gyration (Rg) is 38.5 Å. (D) Guinier transformation of the data as ln[I(q)] versus q2 over a range of q × Rg = <1.3, demonstrating linearity across all concentrations. Download
Crystallographic data collection and refinement statistics.
Primers used for ADI site-directed mutagenesis.