Abstract
The large Neotropical family Bromeliaceae presents an outstanding example of adaptive radiation in plants, containing a wide range of terrestrial and epiphytic life-forms occupying many distinct habitats. Diversification in bromeliads has been linked to several key innovations, including water- and nutrient-impounding phytotelmata, absorptive epidermal trichomes, and the water-conserving mode of photosynthesis known as crassulacean acid metabolism (CAM). To clarify the origins of CAM and the epiphytic habit, we conducted a phylogenetic analysis of nucleotide sequences for 51 bromeliad taxa by using the plastid loci matK and the rps16 intron, combined with a survey of photosynthetic pathway determined by carbon-isotope ratios for 1,873 species representing 65% of the family. Optimization of character-states onto the strict consensus tree indicated that the last common ancestor of Bromeliaceae was a terrestrial C3 mesophyte, probably adapted to moist, exposed, nutrient-poor habitats. Both CAM photosynthesis and the epiphytic habit evolved a minimum of three times in the family, most likely in response to geological and climatic changes in the late Tertiary. The great majority of epiphytic forms are now found in two lineages: in subfamily Tillandsioideae, in which C3 photosynthesis was the ancestral state and CAM developed later in the most extreme epiphytes, and in subfamily Bromelioideae, in which CAM photosynthesis predated the appearance of epiphytism. Subsequent radiation of the bromelioid line into less xeric habitats has led to reversion to C3 photosynthesis in some taxa, showing that both gain and loss of CAM have occurred in the complex evolutionary history of this family.
The Bromeliaceae are frequently celebrated as an outstanding example of adaptive radiation in vascular plants (1, 2). They represent one of the largest families with a Neotropical distribution (3), comprising 2,885 species in 56 genera (4, 5), with an ecological range that encompasses extremes of moisture availability (from rain forests to hyperarid coastal sands), elevation (from sea level to >4,000 m), and exposure (fully exposed sites to shaded forest understories). The family contains a correspondingly rich diversity of life-forms, from soil-rooted terrestrial plants, through rosulate “tank” epiphytes with water- and nutrient-impounding phytotelmata, to extreme epiphytes completely independent of their substratum for nutrition. The evolutionary transition from terrestrial to epiphytic life-forms appears to have been closely linked to elaboration of the absorptive epidermal trichomes characteristic of the family (1, 6, 7). Indeed, about half of all bromeliads are epiphytic, and they constitute one of the most distinctive components of the Neotropical forest canopy (8, 9).
In addition to morphological specializations, another key innovation associated with the success of bromeliads in more arid habitats is the form of photosynthesis known as crassulacean acid metabolism (CAM) (10-13). In typical CAM plants, CO2 is taken up at night and temporarily stored, via a pathway involving fixation by phosphoenolpyruvate carboxylase (PEPC), in the form of malic acid in the cell vacuole. In the following light period the stomata close, malic acid is released from the vacuole and decarboxylated, and the CO2 liberated is photosynthetically reduced in the Calvin cycle (13). By restricting gas exchange with the atmosphere during the daytime, CAM plants use their available water more efficiently than C3 plants, and consequently characterize many tropical and subtropical environments with intermittent or strongly seasonal water supply. This includes epiphytic niches in the forest canopy, which can be microclimatically arid, to the extent that, amongst the 6% of flowering plants estimated to show CAM photosynthesis, there may be almost as many epiphytic as terrestrial species (13, 14).
Despite being one of the best-understood metabolic examples of an ecological adaptation in plants, relatively little is known of the evolutionary origins of the CAM pathway. Its occurrence in >30 diverse families suggests that CAM has arisen many times, but only limited work has been undertaken from a phylogenetic perspective at a finer taxonomic scale. Because of their diversity, the Bromeliaceae provide an excellent model of adaptive radiation on which to trace the origins of CAM (and the epiphytic habit) in closely related taxa. A prerequisite for such evolutionary reconstruction is a sufficiently robust phylogeny for the family based on molecular and morphological characters, given the almost complete lack of a fossil record (15). Taxonomically, Bromeliaceae have long been regarded as an isolated and natural group (3, 16), a view supported by cladistic analyses of molecular data that resolve a monophyletic Bromeliaceae within the large order Poales (17-19). The three traditionally recognized subfamilies (20, 21) all contain a mixture of C3 species and CAM species (12, 22), but the first molecular-phylogenetic analyses shed little light on CAM evolution because of limited taxon sampling and poor resolution on the trees (23-26). Moreover, CAM is common in both terrestrial and epiphytic bromeliads, so the underlying relationships between photosynthetic pathway, plant life-form, and phylogenetic lineage in the family may be complex.
To obtain further insight into the origins of CAM photosynthesis and its relationship to the evolution of epiphytism, we have derived a more detailed phylogeny for the Bromeliaceae based on nucleotide sequences of two rapidly evolving plastid loci, matK and the rps16 intron. This is combined with an extensive survey of photosynthetic pathway at the species level to determine the minimum number of times CAM may have arisen in this family. Together with evidence from present-day biogeography and ecology, this permits a reconciliation of previously conflicting hypotheses for the origins of CAM photosynthesis and the epiphytic life-form within this exceptionally diverse Neotropical family.
Materials and Methods
Molecular Systematics. Fifty-one species of Bromeliaceae from 27 genera were chosen to represent known diversity within and between previously recognized taxonomic groups. Species selection was biased toward subfamily Pitcairnioideae (20, 21), because this group may contain the earliest diverging lineages in the family (1, 24, 26, 27) and includes both C3 and CAM taxa (10, 12, 22). Species of seven genera of Tillandsioideae (of nine currently recognized) and eight (of 30) genera of Bromelioideae were chosen to reflect the ecological and taxonomic diversity in those groups. Three species of Rapateaceae were used as outgroups, because most molecular studies place Rapateaceae amongst the closest relatives of Bromeliaceae (18, 28).
Sequences of the matK gene (29) and rps16 intron (30) were obtained by using standard protocols for total DNA extraction, PCR amplification, and sequencing (25). These are among the most rapidly evolving plastid loci and are well suited to phylogenetic reconstruction within many angiosperm families (29-32). Details of PCR primers used are given in Table 1, and DNA sequence information, GenBank accession numbers, and voucher details are provided in Table 2, both of which are published as supporting information on the PNAS web site.
Manual alignment of sequences and all subsequent analysis was performed in paup* 4.0b6 (33). Inferred indels, where informative, were scored as additional characters following the “simple” method of Simmons and Ochoterena (34).
The matK and rps16 intron data were first analyzed separately and then simultaneously. Phylogenetic resolution can be improved by combining independent molecular data sets (32), and analysis of matK sequences for a subset of these species suggested that this locus alone would not provide sufficient resolution on the resulting trees (25, 35). Parsimony analysis used tree bisection-reconnection branch swapping and successive weights (SW) analysis (36), with iterative rounds of search followed by reweighting until tree length stabilized. To search for multiple islands of optimal trees, a random-taxon-addition-order (RTA) procedure (37) was used with 1,000 replicates, saving 30 trees per replicate. Weights were determined by using the maximum rescaled consistency index for each character on the best trees. Trees found in the final round of SW analysis were swapped to completion (using 1,000 RTA replicates) or until 80,000 trees were found. Before proceeding with the simultaneous analysis, we tested the combinability of the matK and rps16 intron data by using the incongruence length difference (ILD) test (38) to assess the statistical significance of congruence between phylogenies based on the two data partitions. The test was performed by 200 replicate parsimony analyses, each consisting of 100 RTA searches, with 100 trees saved per search. Final SW values were used.
Branch support was evaluated by 1,000 bootstrap replicates (using the SW values), saving 100 trees per replicate. This “reduced effort” procedure gives an unbiased estimate of the true bootstrap proportions (39). Support for alternative evolutionary hypotheses was evaluated by using the Kishino-Hasegawa test (40). One of the best trees consistent with each hypothesis (found by a constraint parsimony analysis with 1,000 replicate RTA searches, SW values, 30 trees saved per replicate) was compared with one of the trees found by SW analysis of the combined data set.
Photosynthetic Pathway. Photosynthetic pathway was determined from tissue carbon-isotope ratio, δ13C. This can distinguish plants that use the C3 pathway, in which the primary carboxylating enzyme is ribulose-1,5-bisphosphate carboxylase-oxygenase, from C4 or CAM plants, in which the primary carboxylating enzyme is PEPC, because of a kinetic isotope effect (41). C4 photosynthesis is not known in Bromeliaceae (1, 11), so the δ13C values could be used to distinguish CAM and C3 species. Relative natural abundance of 12C and 13C (δ13C) was determined for samples of dried shoot tissue taken from herbarium specimens as described (41, 42).
Species were classified as CAM or C3 if the δ13C value was less negative or more negative than -20.0‰, respectively. [Values more negative than -20.0‰ do not preclude the possibility of some dark CO2 fixation, but indicate that this did not make a major contribution to photosynthetic carbon gain (43).] The character states “C3” and “CAM,” and “terrestrial” and “epiphytic” were mapped onto the strict consensus tree by using macclade 3.08a (44).
Results
Molecular Systematics. For the 51 species of Bromeliaceae and 3 Rapateaceae, the aligned matK data set comprised 858 positions, 142 of which were potentially parsimony-informative, with one indel (shared by the three Puya spp.). Ingroup pairwise sequence divergence (uncorrected for multiple hits) reached a maximum of 5.3% (between Fosterella penduliflora and Guzmania monostachia). Alignment of the rps16 intron sequences required 63 gaps, which were scored as 22 indels (seven being restricted to the outgroup). This data set comprised 1135 positions, 177 of which were parsimony-informative; ingroup pairwise sequence divergence reached a maximum of 4.6% (between Ayensua uaipanensis and Pitcairnia nuda).
Estimates of phylogenetic relationships derived from the matK and rps16 intron data sets did not differ significantly (P = 0.175) based on the ILD test, so we proceeded with a simultaneous analysis. Initial (unit weight) parsimony analysis of the combined data set of 1,993 characters, including the indel characters, found optimal trees of length 753 steps (consistency index excluding uninformative characters, CI = 0.749, retention index, RI = 0.819). SW analysis converged on trees of length 495 steps (CI = 0.928, RI = 0.947) after three rounds. Swapping to completion found 756 trees in one island. The strict consensus of these trees (which is identical to that from an equal weights analysis) is shown in Fig. 1.
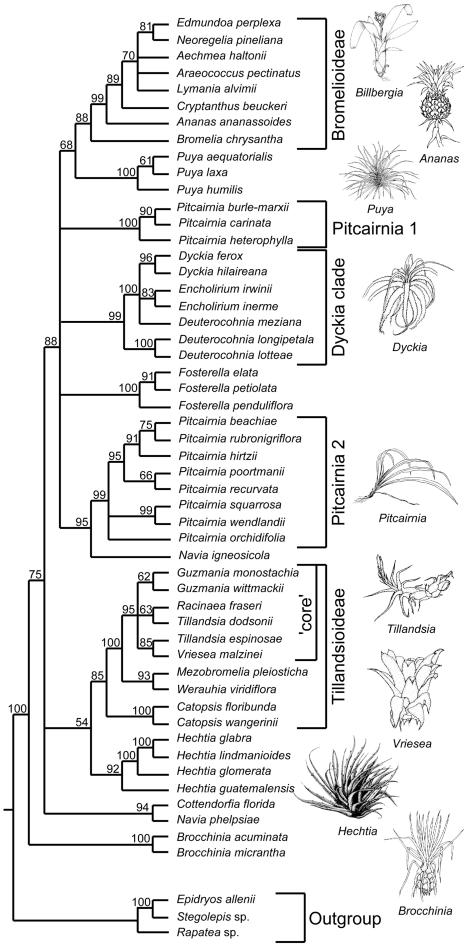
Fig. 1.
Strict consensus of 756 trees found during the final round of SW analysis of the combined matK plus rps16 intron data set for 51 species of Bromeliaceae. The tree was rooted on the branch separating Rapateaceae and Bromeliaceae. Bootstrap values are indicated above the relevant branches; clades referred to in the text are bracketed alongside representative life-forms. [Illustrations reproduced with permission from ref. 1 (Copyright 2000, Cambridge Univ. Press) and ref. 45 (Copyright 1997, Missouri Botanical Garden).]
This analysis strongly supports the monophyly of Bromeliaceae (bootstrap value of 100%). Other well supported groups (bootstrap values >80%) include the two subfamilies Bromelioideae and Tillandsioideae, and the genera Brocchinia, Catopsis, Dyckia, Encholirium, Fosterella, Hechtia, and Puya. Deuterocohnia meziana does not group with other Deuterocohnia species but is part of a robust clade including Dyckia and Encholirium, confirming that this genus is not monophyletic (24-26). Pitcairnia species are placed in two well supported groups, Pitcairnia 1 (3 spp.) and Pitcairnia 2 (8 spp.). Navia igneosicola is sister to Pitcairnia 2, whereas N. phelpsiae groups with Cottendorfia. In addition, there is support for Brocchinia as sister to the rest of the family, Puya as sister to Bromelioideae, and a derived clade comprising Bromelioideae and all of the Pitcairnioideae sampled except for Brocchinia, Hechtia, Cottendorfia, and Navia phelpsiae (the “DFPPB” clade). Within Tillandsioideae, Catopsis and then Mezobromelia plus Werauhia are strongly supported as successive sister groups to a core clade comprising members of Guzmania, Racinaea, Tillandsia, and Vriesea.
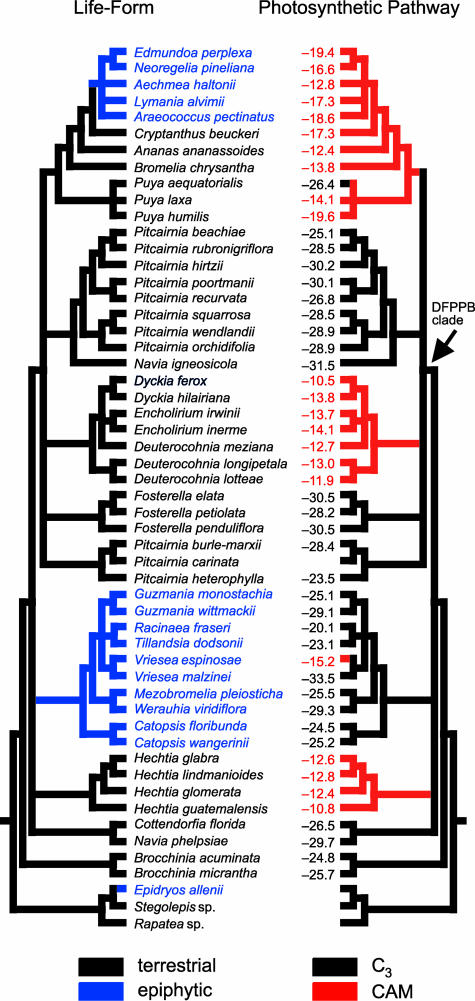
Photosynthetic Pathway. Carbon-isotope ratios for the 51 species used to construct the molecular phylogeny are optimized by using parsimony onto the strict consensus tree in Fig. 2 together with plant life-form. A Kishino-Hasegawa test rejects the hypothesis of a single origin for CAM (P < 0.001). A sister-group relationship between Hechtia and Tillandsioideae receives only weak bootstrap support, and indeed the present data do not rule out a placement of Hechtia as sister to the DFPPB clade (P = 0.05), but Hechtia as sister to Tillandsioideae is corroborated by maximum likelihood and neighbor-joining analyses of trnL intron sequence (26). A sister-group relationship between Fosterella and the Dyckia clade also receives weak bootstrap support but is not present in the strict consensus tree. Thus, CAM photosynthesis occurs in four clades (Fig. 2): Hechtia, the “core” Tillandsioideae, the Dyckia clade, and Puya + Bromelioideae. In contrast, epiphytes are distributed in two main clades, Tillandsioideae (virtually all of which are epiphytic or lithophytic) and Bromelioideae [≈50% of which are epiphytic (1, 20, 45)]. Two of ≈20 species of Brocchinia can grow epiphytically, although this is only one of several life-forms found in this ecologically diverse genus (1, 2, 46).
Fig. 2.
Most-parsimonious reconstruction of the evolution of life-form and photosynthetic pathway in Bromeliaceae, based on relationships supported by bootstrap analysis of the combined matK plus rps16 intron data set. Carbon-isotope ratios (δ13C values in ‰) are shown for the taxa analyzed. The derived character-states “epiphytic” and “CAM” are highlighted in blue and red, respectively. Two species of Brocchinia can grow epiphytically (not shown), probably representing a further independent origin of epiphytism in Bromeliaceae.
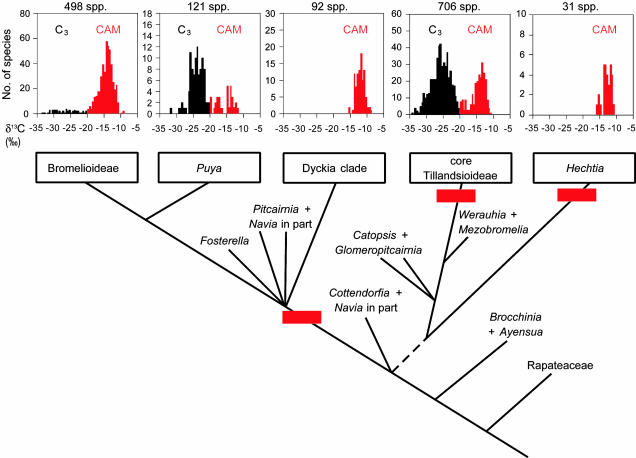
Because taxon sampling for sequencing of the plastid loci was necessarily restricted, a more complete survey of δ13C values in the family was undertaken that included 55 genera (i.e., all except the monotypic bromelioid genus Pseudaechmea L.B.Sm. & Read) and 1,873 species (65% of the estimated total in the family). Of these species, 826 (44%) were found to be CAM plants, all of which were in genera that can be ascribed to the four CAM lineages in Fig. 2. Taking into account the detailed ecological information available for the family and the range of species sampled, it is very unlikely that other lineages containing CAM plants exist in the Bromeliaceae.
The relative abundance of CAM species in the major phylogenetic lineages deduced from this study are summarized in Fig. 3. Both Hechtia and the Dyckia clade [Deuterocohnia (including Abromeitiella), Dyckia, and Encholirium] are comprised solely of CAM species. Amongst the Tillandsioideae, 28% (223 of 788 species sampled) were identified as CAM plants: these are effectively all restricted to the genus Tillandsia, because the four species of Vriesea (of 135 sampled) identified as CAM plants are taxa that should probably be realigned into Tillandsia (47). CAM is also well represented in Puya and Bromelioideae, which were resolved as sister groups (Fig. 1), as found in the ndhF (24) and trnL (26) phylogenies. In Puya, a minority of species sampled (24%) are CAM plants, whereas in Bromelioideae the great majority are CAM (91%).
Fig. 3.
Summary of numbers of C3 and CAM species in the five lineages within Bromeliaceae found to contain CAM taxa. Phylogenetic inferences are based on Fig. 1, together with information on Ayensua (26) and Glomeropitcairnia (26, 53). Horizontal red bars indicate the minimum number of independent origins of CAM. The closest relative of Hechtia was not resolved by the present analysis; the dashed line indicates its affinities based on analysis of trnL intron sequence data (26).
The position of Brocchinia (sister to the rest of the family) supports previous suggestions that C3 photosynthesis is plesiomorphic in Bromeliaceae (10, 12). The monotypic Ayensua uaipanensis formed a robust clade with Brocchinia (94% bootstrap support) based on rps16 intron sequence data (data not shown), a relationship supported by trnL intron sequence analysis (26); however, matK sequence could not be obtained. With Brocchinia + Ayensua at the base, the phylogeny indicates a minimum of three independent origins of CAM within the family (Fig. 3): one ancestral to Hechtia, one in the core Tillandsioideae, and a third in the DFPPB clade. On the basis of the matK + rps16 intron data set alone, it is not possible to reject the hypothesis that CAM had only a single origin in the DFPPB clade, but an ndhF phylogeny also resolved Dyckia in a pitcairnioid clade distinct from Puya + Bromelioideae (24) (Fig. 2), suggesting that CAM evolved independently in these two lineages.
Discussion
Plants showing CAM photosynthesis are widely believed to have evolved from C3 ancestors, but the exact circumstances under which the major CAM lineages arose are not well understood. The highly dispersed taxonomic distribution of CAM photosynthesis, which occurs in 33 families and an estimated 16,000 species of vascular plants (13), suggests it has arisen on multiple occasions. In families such as the Agavaceae, Cactaceae, and Didiereaceae, almost all species have the capacity for CAM and thus exhibit the presumed apomorphic character-state (13, 48). Other families such as the Aizoaceae, Bromeliaceae, Crassulaceae, and Orchidaceae contain large numbers of both C3 and CAM species, so these may be more informative for reconstructing the origins of the CAM pathway, providing such an analysis can be supported by an appropriately resolved phylogeny.
Although the Bromeliaceae have been much studied with respect to their ecological diversity and life-forms, taxonomic relationships within the family have remained controversial (1, 3, 20, 24, 45). Previous molecular-phylogenetic studies have suffered from relatively poor resolution because of low sequence divergence within the family, but the matK and rps16 loci used in the present study gave a well-resolved phylogeny when analyzed as a combined data set. Our results support a basal separation of Brocchinia + Ayensua (24, 26), distinctive C3 taxa that are geographically restricted to the escarpment of the Guayana Shield (1, 2, 46, 49). Further, the family Rapateaceae, which appears the most likely sister group and was used to root the bromeliad tree, also has a distribution centered on the Guayana Shield and consists largely of terrestrial C3 herbs of wet, infertile soils, frequently cooccurring with Brocchinia (2, 28, 42). Given the absence of CAM and rarity of epiphytism in the other 17 families making up the order Poales (9, 13, 19), this strongly suggests that both C3 photosynthesis and a terrestrial growth habit are plesiomorphic in Bromeliaceae.
Within Bromeliaceae, the epiphytic life-form and CAM photosynthesis have clearly arisen multiple times independently, so their origins must be sought in the evolutionary history of separate lineages. In the family as a whole, there is a strong correlation between habitat aridity and the occurrence of CAM (10-12, 50), but CAM is widespread in both terrestrial and epiphytic species, and in all three major subfamilies. Our survey of carbon-isotope ratios suggests that most genera of the largely terrestrial Pitcairnioideae are exclusively either CAM (Hechtia, Dyckia, Encholirium, Deuterocohnia) or C3 (Brocchinia, Navia, Steyerbromelia, Brewcaria, Cottendorfia, Lindmania, Connellia, Pitcairnia, Fosterella), and that only Puya contains both C3 and CAM species. The matK plus rps16 phylogeny confirms earlier suggestions (23-26) that Pitcairnioideae as traditionally circumscribed are paraphyletic, although formal taxonomic revision should await clarification of the phylogenetic relationships of four other rare genera of Guayana Shield endemics, Steyerbromelia, Brewcaria, Lindmania, and Connellia. Among the CAM taxa, Hechtia had been considered closely related to the other xeromorphic pitcairnioids with succulent, spiny leaves (21, 27), but the present analysis suggests that CAM and the associated vegetative characters are independently derived in Hechtia and the DFPPB clade (Fig. 2). Hechtia has a notably disjunct distribution, its 51 species being restricted to northern Central America, Mexico, and southern Texas (20). CAM very likely arose in this taxon in the same arid-zone habitats that fostered evolution of the Agavaceae and Cactaceae, two of the most distinctive Neotropical families of terrestrial CAM plants (48, 51). In contrast, the CAM taxa in the Dyckia clade (Figs. 2 and 3), comprising Deuterocohnia, Dyckia, and Encholirium, are all centered on xeric habitats in the southern Andes, Argentina, and south and eastern Brazil (20).
The matK plus rps16 phylogeny confirms the monophyly of the two other bromeliad subfamilies, Tillandsioideae and Bromelioideae, consistent with phylogenies derived from ndhF (24) and trnL intron (26) sequences with somewhat different taxon sampling. Tillandsioideae are almost wholly epiphytic or lithophytic, but C3 photosynthesis is clearly plesiomorphic in the subfamily (Figs. 2 and 3). CAM photosynthesis is restricted to Tillandsia s.l., a very large (≈540 spp.) and diverse genus containing both C3 and CAM species (12, 20, 22, 52, 53). The epiphytic habit also reaches its most extreme form in this genus, approximately half of which (the so-called “atmospheric” species) lack water-impounding phytotelmata, have root systems reduced to holdfasts, and are entirely dependent for water and nutrient uptake on absorptive trichomes that cover the shoot (1). All of the atmospheric species of Tillandsia are CAM plants, suggesting that CAM may have been a key innovation enabling the adaptive radiation of this genus into more xeric habitats.
The other monophyletic subfamily, Bromelioideae, was resolved as sister group to the genus Puya. This relationship was also found in the ndhF (24) and trnL (26) phylogenies, and is supported by putative synapomorphies such as leaf morphology and trichome structure (1). A sister-group relationship of Puya and Bromelioideae has important implications for the origins of CAM photosynthesis. Puya is a large genus (195 spp.) of terrestrial, often xeromorphic plants commonly found on open slopes of the Andean cordillera, but only a minority (24%) appear to be CAM plants. Whether the last common ancestor of the Puya + Bromelioideae lineage already possessed CAM photosynthesis, or whether CAM arose more than once in this clade, should be testable by a phylogenetic analysis with greater sampling density in these taxa. Within Bromelioideae, epiphytism is clearly the derived condition, but >90% of the subfamily are CAM species (and the basally diverging genera Bromelia and Ananas entirely so). Nevertheless, there has been considerable ecological diversification within the subfamily. Several CAM species of Aechmea are found in relatively shaded, humid habitats (11, 54, 55); epiphytic genera such as Nidularium, Ronnbergia, and Wittrockia contain both C3 and CAM species; and four small genera that are phylogenetically more derived, Fascicularia, Greigia, Fernseea, and Ochagavia (26), contain exclusively C3 species. Thus, although CAM is ancestral in the subfamily, there is evidence for reversion from CAM to C3 photosynthesis as certain lineages radiated into more mesic habitats.
Many earlier authors have speculated on the evolutionary origins of epiphytism in Bromeliaceae, and the matK plus rps16 phylogeny, together with other molecular studies (24, 26), helps to reconcile some previously conflicting views. Schimper (6) originally proposed that tropical epiphytes evolved from terrestrial ancestors in the relatively moist, shaded forest understory, with some forms migrating up the forest profile and eventually colonizing the canopy. Although this model is applicable to other families of rainforest epiphytes, Tietze (56) and Pittendrigh (45) proposed a radically different explanation for the origin of epiphytic bromeliads, suggesting they may have entered the forest as relatively light-demanding forms derived from terrestrial ancestors adapted to open habitats, subsequently diversifying into a variety of microhabitats including the shaded understory. The present results support the notion that the ancestral bromeliad was a terrestrial C3 plant of exposed but relatively moist environments (3, 10, 45), perhaps similar to those occupied by present-day Brocchinia (1, 2). These habitats may also have been nutrient-poor, possibly providing a strong selective pressure for the evolution of absorptive trichomes (1, 10, 57). Apart from occasional examples of epiphytism in Brocchinia (2), the epiphytic life-form has become widespread in two main lineages. The last common ancestor of Tillandsioideae, although a C3 plant, had already acquired an epiphytic growth habit, and this subfamily has remained wholly epiphytic (or lithophytic), with the appearance of CAM being limited to more xeric forms in the genus Tillandsia. In contrast, the last common ancestor of Bromelioideae already possessed CAM, and epiphytism, as well as reversion to C3 photosynthesis, has been a later development in certain taxa. The phylogenetic reconstruction does not reveal any evidence for shade-tolerant terrestrial forms amongst the immediate ancestors of these two subfamilies. This lends support to the Tietze-Pittendrigh model, suggesting that the progenitors of epiphytic bromeliads were plants adapted to relatively exposed habitats, and that the species now found in the forest understory are secondarily shade-adapted.
The low degree of nucleotide sequence divergence found for four loci are consistent with the Bromeliaceae being relatively young. But in the almost complete absence of a fossil record, with the exception of a single report of Tillandsia-type pollen from the Upper Eocene (15), it is not yet possible to assign a precise chronology to the family's evolutionary history. The Neotropical distribution of bromeliads suggests an origin some time after the break-up of West Gondwana and the reduction of biological exchange between Africa and South America ≈85 million years ago (Ma) (58). An emergence of Bromeliaceae by the early Tertiary is also suggested by the appearance of other Poales in the fossil record by 70-55 Ma (59). Some major events in the family's evolution may have occurred much more recently. For example, the mainly Andean distribution of Puya (20), and the abundance of Tillandsioideae in northern Peru, Ecuador, and Colombia, suggests that diversification may have been associated with the emergence of new habitats during periods of Andean orogeny in the Miocene and Pliocene (60, 61), as proposed for epiphytic Lycopodiaceae (62). Progressive aridification and declining CO2 concentrations during the Tertiary (60, 63, 64) would have gradually favored the emergence of CAM photosynthesis in Bromeliaceae, perhaps in a manner similar to the Miocene expansion of grasses showing C4 photosynthesis (65, 66). Firmer conclusions about the chronology of events will only be possible once the molecular phylogeny can be calibrated against other evidence. However, this study suggests that combined information from a variety of sources may also be valuable in tracing the origins of CAM photosynthesis in other groups of vascular plants.
Supplementary Material
Acknowledgments
We thank E. Leme (Rio de Janeiro, Brazil) and the directors of the Missouri Botanical Garden (St. Louis), Marie Selby Botanical Gardens (Sarasota, FL), New York Botanical Garden (Bronx, NY), Palmengarten (Frankfurt, Germany), United States National Herbarium, Smithsonian Institution (Washington, DC), and Venezuelan Botanic Garden (Caracas, Venezuela) for access to their collections, and R. Duno, B. Holst, H. Luther, J. Solomon, S. Wookey, L. Giles, R. Terry, O. Huber, E. Medina, E. Olivares, and L. Pond for assistance and advice. This work was supported by an award from the Andrew W. Mellon Foundation and by the Smithsonian Tropical Research Institute.
Abbreviations: CAM, crassulacean acid metabolism; RTA, random-taxon-addition-order; SW, successive weights.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. can be found in Table 2, which is published as supporting information on the PNAS web site).
References
- 1.Benzing, D. H. (2000) Bromeliaceae: Profile of an Adaptive Radiation (Cambridge Univ. Press, Cambridge, U.K.).
- 2.Givnish, T. J., Sytsma, K. J., Smith, J. F., Hahn, W. J., Benzing, D. H. & Burkhardt, E. M. (1997) in Molecular Evolution and Adaptive Radiation, eds. Givnish, T. J. & Sytsma, K. J. (Cambridge Univ. Press, Cambridge, U.K.), pp. 259-311.
- 3.Smith, L. B. (1934) Bot. Jahrb. 66, 446-468. [Google Scholar]
- 4.Luther, H. E. (2000) An Alphabetical List of Bromeliad Binomials (The Bromeliad Society International, Sarasota, FL).
- 5.Taylor, D. C. & Robinson, H. (1999) Harvard Pap. Bot. 4, 203-217. [Google Scholar]
- 6.Schimper, A. F. W. (1888) Die epiphytische Vegetation Amerikas (Gustav Fischer, Jena, Germany).
- 7.Mez, C. (1904) Jahrb. Wiss. Bot. 40, 157-229. [Google Scholar]
- 8.Ozanne, C. M. P., Anhuf, D., Boulter, S. L., Keller, M., Kitching, R. L., Körner, C., Meinzer, F. C., Mitchell, A. W., Nakashizuka, T., Dias, P. L. S., et al. (2003) Science 301, 183-186. [DOI] [PubMed] [Google Scholar]
- 9.Kress, W. J. (1989) in Vascular Plants as Epiphytes: Evolution and Ecophysiology, ed. Lüttge, U. (Springer, Berlin), pp. 234-261.
- 10.Medina, E. (1974) Evolution (Lawrence, Kans.) 28, 677-686. [Google Scholar]
- 11.Griffiths, H. & Smith, J. A. C. (1983) Oecologia 60, 176-184. [DOI] [PubMed] [Google Scholar]
- 12.Smith, J. A. C. (1989) in Vascular Plants as Epiphytes: Evolution and Ecophysiology, ed. Lüttge, U. (Springer, Berlin), pp. 109-138.
- 13.Winter, K. & Smith, J. A. C., eds. (1996) Crassulacean Acid Metabolism: Biochemistry, Ecophysiology, and Evolution (Springer, Berlin).
- 14.Winter, K., Wallace, B. J., Stocker, G. C. & Roksandic, Z. (1983) Oecologia 57, 129-141. [DOI] [PubMed] [Google Scholar]
- 15.Benton, M. J., ed. (1993) The Fossil Record 2 (Chapman & Hall, London).
- 16.Dahlgren, R., Clifford, H. T. & Yeo, P. F. (1985) The Families of the Monocotyledons: Structure, Evolution, and Taxonomy (Springer, Berlin).
- 17.Gilmartin, A. J. & Brown, G. K. (1987) Syst. Bot. 12, 493-500. [Google Scholar]
- 18.Chase, M. W., Soltis, D. E., Soltis, P. S., Rudall, P. J., Fay, M. F., Hahn, W. H., Sullivan, S., Joseph, J., Molvray, M., Kores, P. J., et al. (2000) in Monocots: Systematics and Evolution, eds. Wilson, K. L. & Morrison, D. A. (CSIRO, Melbourne), pp. 3-16.
- 19.The Angiosperm Phylogeny Group (2003) Bot. J. Linn. Soc. 141, 399-436. [Google Scholar]
- 20.Smith, L. B. & Downs, R. J. (1979 1974) Flora Neotropica (Hafner, New York), Vol. 14, Pts. 1-3.
- 21.Smith, L. B. & Till, W. (1998) in The Families and Genera of Vascular Plants, ed. Kubitzki, K. (Springer, Berlin), Vol. 4, pp. 74-99. [Google Scholar]
- 22.Martin, C. E. (1994) Bot. Rev. 60, 1-82. [Google Scholar]
- 23.Ranker, T. A., Soltis, D. E., Soltis, P. S. & Gilmartin, A. J. (1990) Syst. Bot. 15, 425-434. [Google Scholar]
- 24.Terry, R. G., Brown, G. K. & Olmstead, R. G. (1997) Am. J. Bot. 84, 664-670. [PubMed] [Google Scholar]
- 25.Crayn, D. M., Terry, R. G., Smith, J. A. C. & Winter, K. (2000) in Monocots: Systematics and Evolution, eds. Wilson, K. L. & Morrison, D. A. (CSIRO, Melbourne), pp. 569-579.
- 26.Horres, R., Zizka, G., Kahl, G. & Weising, K. (2000) Plant Biol. 2, 306-315. [Google Scholar]
- 27.Varadarajan, G. S. & Gilmartin, A. J. (1988) Syst. Bot. 13, 283-293. [Google Scholar]
- 28.Givnish, T. J., Evans, T. M., Zjhra, M. L., Patterson, T. B., Berry, P. E. & Sytsma, K. J. (2000) Evolution (Lawrence, Kans.) 54, 1915-1937. [DOI] [PubMed] [Google Scholar]
- 29.Hilu, K. W. & Liang, H. P. (1997) Am. J. Bot. 84, 830-839. [PubMed] [Google Scholar]
- 30.Oxelman, B., Lidén, M. & Berglund, D. (1997) Plant Syst. Evol. 206, 393-410. [Google Scholar]
- 31.Kelchner, S. A. (2002) Am. J. Bot. 89, 1651-1669. [DOI] [PubMed] [Google Scholar]
- 32.Soltis, D. E. & Soltis, P. S. (1998) in Molecular Systematics of Plants II: DNA Sequencing, eds. Soltis, D. E., Soltis, P. S. & Doyle, J. J. (Kluwer, Norwell, MA), pp. 1-42.
- 33.Swofford, D. L. (1998) paup*: Phylogenetic Analysis using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 34.Simmons, M. P. & Ochoterena, H. (2000) Syst. Biol. 49, 369-381. [PubMed] [Google Scholar]
- 35.Reinert, F., Russo, C. A. M. & Salles, L. O. (2003) Biol. J. Linn. Soc. 80, 261-268. [Google Scholar]
- 36.Farris, J. S. (1969) Syst. Zool. 18, 374-385. [Google Scholar]
- 37.Maddison, D. R. (1991) Syst. Zool. 40, 315-328. [Google Scholar]
- 38.Farris, J. S., Källersjö, M., Kluge, A. G. & Bult, C. (1994) Cladistics 10, 315-319. [DOI] [PubMed] [Google Scholar]
- 39.Mort, M. E., Soltis, P. S., Soltis, D. E. & Mabry, M. L. (2000) Syst. Biol. 49, 160-171. [DOI] [PubMed] [Google Scholar]
- 40.Kishino, H. & Hasegawa, M. (1989) J. Mol. Evol. 29, 170-179. [DOI] [PubMed] [Google Scholar]
- 41.Osmond, C. B., Allaway, W. G., Sutton, B. G., Troughton, J. H., Queiroz, O., Lüttge, U. & Winter, K. (1973) Nature 246, 41-42. [Google Scholar]
- 42.Crayn, D. M., Smith, J. A. C. & Winter, K. (2001) Plant Biol. 3, 569-576. [Google Scholar]
- 43.Winter, K. & Holtum, J. A. M. (2002) Plant Physiol. 129, 1843-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maddison, W. P. & Maddison, D. R. (1999) macclade (Sinauer, Sunderland, MA), Version 3.08a.
- 45.Pittendrigh, C. S. (1948) Evolution (Lawrence, Kans.) 2, 58-89. [DOI] [PubMed] [Google Scholar]
- 46.Holst, B. K. (1997) in Flora of the Venezuelan Guayana, eds. Berry, P. E., Holst, B. K. & Yatskievych, K. (Missouri Botanical Garden, St. Louis), Vol. 3, pp. 548-676. [Google Scholar]
- 47.Grant, J. R. (1993) Phytologia 75, 170-175. [Google Scholar]
- 48.Gibson, A. C. & Nobel, P. S. (1986) The Cactus Primer (Harvard Univ. Press, Cambridge, MA).
- 49.Varadarajan, G. S. & Gilmartin, A. J. (1988) Syst. Bot. 13, 294-299. [Google Scholar]
- 50.Medina, E., Delgado, M., Troughton, J. H. & Medina, J. D. (1977) Flora 166, 137-152. [Google Scholar]
- 51.Hershkovitz, M. A. & Zimmer, E. A. (1997) Taxon 46, 217-232. [Google Scholar]
- 52.Benzing, D. H. & Renfrow, A. (1971) Bull. Torrey Bot. Club 98, 322-327. [Google Scholar]
- 53.Terry, R. G., Brown, G. K. & Olmstead, R. G. (1997) Syst. Bot. 22, 333-345. [Google Scholar]
- 54.Skillman, J. B., Garcia, M. & Winter, K. (1999) Ecology 80, 1584-1593. [Google Scholar]
- 55.Pierce, S., Winter, K. & Griffiths, H. (2002) Plant Cell Environ. 25, 1181-1189. [Google Scholar]
- 56.Tietze, M. (1906) Z. Naturwiss. 78, 1-50. [Google Scholar]
- 57.Pierce, S., Maxwell, K., Griffiths, H. & Winter, K. (2001) Am. J. Bot. 88, 1371-1389. [PubMed] [Google Scholar]
- 58.Goldblatt, P., ed. (1993) Biological Relationships between Africa and South America (Yale Univ. Press, New Haven, CT).
- 59.Grass Phylogeny Working Group (2001) Ann. Mo. Bot. Gard. 88, 373-457. [Google Scholar]
- 60.Gentry, A. H. (1982) Ann. Mo. Bot. Gard. 69, 557-593. [Google Scholar]
- 61.Burnham, R. J. & Graham, A. (1999) Ann. Mo. Bot. Gard. 86, 546-589. [Google Scholar]
- 62.Wikström, N., Kenrick, P. & Chase, M. (1999) Plant Syst. Evol. 218, 221-243. [Google Scholar]
- 63.Pearson, P. N. & Palmer, M. R. (2000) Nature 406, 695-699. [DOI] [PubMed] [Google Scholar]
- 64.Willis, K. J. & McElwain, J. C. (2002) The Evolution of Plants (Oxford Univ. Press, Oxford).
- 65.Cerling, T. E., Harris, J. M., MacFadden, B. J., Leakey, M. G., Quade, J., Eisenmann, V. & Ehleringer, J. R. (1997) Nature 389, 153-158. [Google Scholar]
- 66.Pagani, M., Freeman, K. H. & Arthur, M. A. (1999) Science 285, 876-879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.