Abstract
Depression is a potentially life-threatening mental disorder affecting approximately 300 million people worldwide. Despite much effort, the molecular underpinnings of clinical depression remain poorly defined, and current treatments carry limited therapeutic efficacy and potentially burdensome side effects. Recently, small noncoding RNA molecules known as microRNA (miRNA) have gained prominence as a target for therapeutic intervention, given their capacity to regulate neuronal physiology. Further, mounting evidence suggests a prominent role for miRNA in depressive molecular signaling. Recent studies have demonstrated that dysregulation of miRNA expression occurs in animal models of depression, and in the post-mortem tissue of clinically depressed patients. Investigations into depression-associated miRNA disruption reveals dramatic effects on downstream targets, many of which are thought to contribute to depressive symptoms. Furthermore, selective serotonin reuptake inhibitors, as well as other antidepressant drugs, have the capacity to reverse aberrant depressive miRNA expression and their downstream targets. Given the powerful effects that miRNA have on the central nervous system transcriptome, and the aforementioned studies, there is a compelling rationale to begin to assess the potential contribution of miRNA to depressive etiology. Here, we review the molecular biology of miRNA, our current understanding of miRNA in relation to clinical depression, and the utility of targeting miRNA for antidepressant treatment.
Keywords: depression, microRNA, miRNA, BDNF, Dicer, serotonin
Introduction
Clinical depression is a chronic mood disorder affecting nearly 300 million people worldwide, with a lifetime prevalence of approximately 19% of the population.1–3 With potentially life-threatening consequences, depression is a major health concern characterized by low mood, anhedonia, disturbance of sleep and appetite, and feelings of despair, shame, and guilt. Dysregulation of monoamine signaling has long been considered central to depressive neuropathology, but the downstream molecular etiology of clinical depression is ill defined. Consequently, the ability to quickly and effectively alleviate the multitude of clinical symptoms remains elusive. There is a pressing need for therapeutics that will quickly and selectively target the pathophysiological underpinnings of clinical depression, without the many off-target side effects associated with current antidepressants. Within this context, we describe the role that micro ribonucleic acid (miRNA) dysregulation may play in depression, and the potential therapeutic value of targeting miRNA. miRNA are a subclass of ~22 nucleotide (nt) noncoding RNA species that function principally by disrupting target messenger RNA (mRNA) expression. miRNA have garnered interest as potential therapeutic targets for an array of disorders of the central nervous system (CNS).4–6 More specifically, miRNA show remarkable potential in the treatment in mood disorders, and specifically in the treatment of clinical depression.7,8
The molecular pathophysiology of depression
Clinical depression is associated with a range of cellular and molecular deficits within the CNS. For decades, the monoamine hypothesis of depression dominated in its assertion that dysregulation of serotonergic, noradrenergic, and dopaminergic signaling was at the core of depressive etiology.9 Stemming from the clinical efficacy of first-generation monoamine oxidase inhibitors (MAOIs) and tricyclic anti-depressants (TCAs) targeting these systems, the monoamine hypothesis asserts that each depression-associated symptom stems from deficits in respective neurotransmitter signaling. This hypothesis also spawned a subsequent generation of antidepressants targeting the serotonin transporter (SERT) that are known collectively as selective serotonin reuptake inhibitors (SSRIs). Along these lines, patients with deficits in the serotonin transporter gene (5-HTT) are significantly more likely to develop depressive symptoms following stressful life events.10 Similarly, antidepressants known as norepinephrine reuptake inhibitors (NRIs) have been used to target the norepinephrine transporter (NET). This strategy can also be used in conjunction with those targeting SERT (thus, serotonin-norepinephrine reuptake inhibitors, SNRIs) and those targeting the dopamine transporter (DAT).11 Nevertheless, evidence supporting monoamine signaling as the direct precipitating factor in depressive pathogenesis is lacking.
Some progress has been made in our understanding of specific depressive pathophysiological mechanisms. The raphe nuclei, locus coeruleus, prefrontal cortex, amygdala, and hippocampus all show markedly reduced volumes and neuronal density in patients diagnosed with major depressive disorder.12–15 Dysregulation of structural plasticity, synaptic connectivity, and dendritic morphology are thought to contribute to depressive pathophysiology.16,17 Deficits in neurogenesis and neuroplasticity within the hippocampus have also been linked to dysthymia and depression-associated memory loss, and can be alleviated by increases in serotonin signaling following treatment with antidepressants.18–20 Excessive glutamatergic neurotransmission is also partially alleviated by antidepressants, and N-methyl-D-aspartate (NMDA) receptor antagonists are known to reduce depressive symptoms.21,22
Brain-derived neurotrophic factor (BDNF) signaling is of particular interest in understanding the molecular etiology of depression, and has led to the development of a neurotrophic hypothesis of depression.23 BDNF within the amygdala and the anterior cingulate cortex is protective against the depression-associated polymorphisms of the serotonin transporter.24 Concordantly, circulating levels of BDNF are markedly reduced in patients with depression, and increases in serotonin activity following SSRI treatment may contribute to a restoration of BDNF activity.25,26 Downstream second messenger signaling pathways involving mitogen-activated protein kinase/ERK (MEK)1, extracellular signal-regulated protein kinase (ERK)1/2, Raf-1, and B-Raf are altered in both animal models of depression and in the post-mortem brains of depressed suicide victims.27–32 In addition, phosphoinositide 3 (PI-3)-kinase, protein kinase A (PKA), and protein kinase C (PKC) signaling all show depression-associated down-regulation.33–36 Unsurprisingly, therefore, decreased activation of the downstream transcription factor cyclic adenosine monophosphate (cAMP) response element-binding (CREB) is a prominent feature in both post-mortem tissue and animal models of depression.37,38
In addition to neuronal deficits, glial cell dysfunction and cell loss has also been associated with depression.39,40 Further, there is increasing evidence for a neuroimmune and microglial component to depressive pathophysiology.41 In line with this idea, depressive symptoms are closely associated with the behavioral effects associated with systemic infection, and expression of cytokines such as interleukin (IL)-1, IL-6, and tumor necrosis factor (TNF)-α are increased in patients with depression.42–44 Excess glucocorticoid and cortisol activity contribute to an overactive hypothalamic–pituitary–adrenal (HPA) axis, which can both precipitate and perpetuate depressive episodes.45–47
Disturbances in circadian clock timing are also closely associated with clinical depression.48,49 Serotonergic and glutamatergic innervation within the suprachiasmatic nucleus (SCN) yields it particularly vulnerable to the dysregulation of those neurotransmitters. Projections from the SCN to the hypothalamus also exacerbate imbalance within the HPA axis. Further, depressed patients experience a decreased latency to enter rapid eye movement (REM) sleep, which lasts longer and occurs with greater frequency than in non-depressed individuals.50,51 Given that REM sleep requires reduced serotonin levels within the brainstem, it may not be surprising that such symptoms are alleviated by SSRI antidepressants.52,53 Likewise, sleep deprivation has been shown to ameliorate depressive symptoms and to induce serotonin signaling.54,55
Despite these advances in the understanding of the molecular underpinning of clinical depression, much is lacking with respect to pharmacological strategies for treatment. Hence, serotonergic and noradrenergic signaling still serve as the primary targets of modern antidepressants. These treatments yield relatively modest symptomatic relief across the population, and the therapeutic effects often do not manifest for weeks, or even months, after initiation.56 In the absence of a more nuanced understanding of the molecular mediators in clinical depression, pharmacological progress will remain stymied. Given the array of biological processes affected by, and contributing to, clinical depression, it is unlikely that treatment targeting any single aspect of depressive pathophysiology will be broadly effective. Rather, therapeutics that modulate multiple branches of depressive etiology may prove advantageous. Such ‘master’ regulators could provide systemic homeostatic control for disorders of neuronal signaling. Thus, targeting multiple nodes of depressive pathophysiology has the potential to provide relief in patients whose symptoms are unresponsive to the current standard antidepressants. As the regulatory potential of noncoding RNA becomes more fully appreciated, miRNA have begun to garner attention as potential targets for the treatment of clinical depression.
MicroRNA biogenesis and regulation
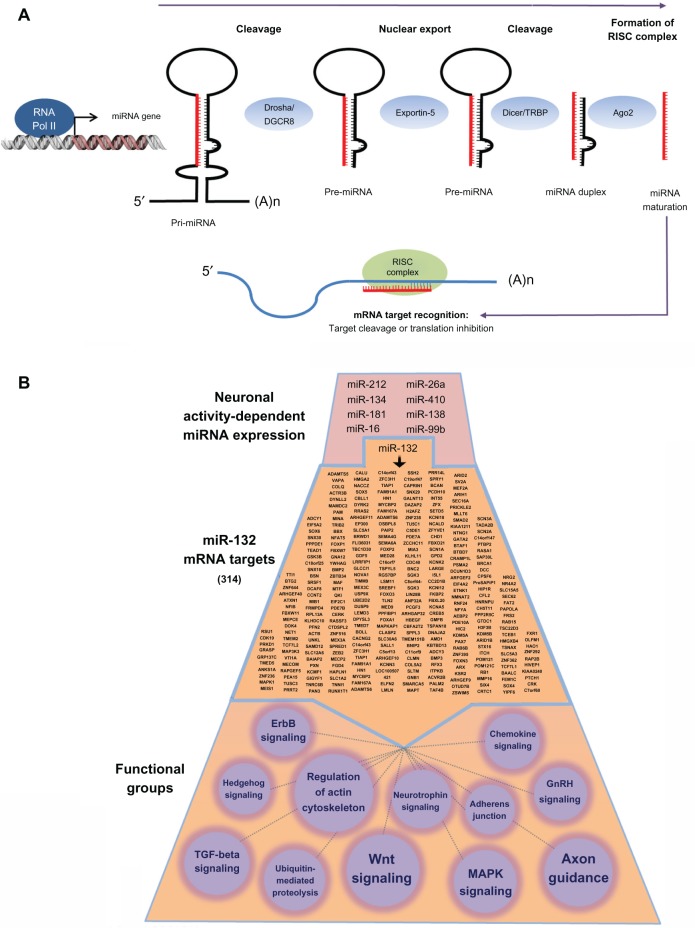
Since their discovery in Caenorhabditis elegans a decade ago, miRNA have emerged as potent regulators of cellular physiology.57–59 miRNA are members of a growing class of noncoding RNA with functional relevance in a wide array of pathological conditions. miRNA biogenesis is a multiphase process, allowing for tight control over miRNA maturation and regulatory silencing.60 In brief, miRNA are transcribed from introns or exons of noncoding RNA (or from introns of protein-coding RNA) by RNA polymerase II. These long hairpin pri-miRNA are then cleaved into ~70 nt pre-miRNA by Drosha (RNase III) before being exported from the nucleolus in an exportin-5-dependent manner. In the cytoplasm, pre-miRNA undergo final cleavage into ~22 nt, double-stranded, mature miRNA by the RNase III nuclease Dicer. One strand of the duplex is subsequently loaded into the RNA-induced silencing complex (RISC) in association with several argonaute-family proteins. Within this complex, miRNA bind to targets by complementary base pairing within the 3′ untranslated region (UTR) of mRNA. Target recognition hinges upon the 5′ seed region (nt 2–8) of each miRNA, though other factors contribute to target specify, many of which have yet to be fully described. Each miRNA has the potential to regulate hundreds of target mRNA, and thus may serve as key hubs of signaling and network regulation (Figure 1). As of publication, over 2,000 unique, mature miRNA have been identified within the human genome.61
Figure 1.
Biogenesis and miRNA functionality in neurons.
Notes: (A) miRNA are transcribed from noncoding regions of the genome by RNA polymerase II, forming a hairpin loop (pri-miRNA) that is cleaved by Drosha/DGCR8 into a ~70 nt pre-miRNA. Pre-miRNA are exported from the nucleus in an exportin-5-dependent manner before further processing by Dicer. The mature strand of the miRNA is loaded into the RISC complex, where it binds to its target mRNA to inhibit translation. (B) Depiction of the complexity of miRNA functionality. At the top of the panel, we provide a limited list of brain-enriched miRNA that are inducibly expressed by neuronal activity. To gain an appreciation of the functional effects of a single miRNA, we provide a list of miR-132 mRNA targets (314 in total: middle section). This list was generated using the TargetScan algorithm. KEGG pathways analysis (bottom section) was used to generate functional classifications of the miR132 targets. Only a subset of the classifications is provided here. Circle size denotes the relative number of genes that make up the classification (the smallest functional class is Hedgehog signaling, which comprises six genes).
Abbreviations: GnRH, gonadotropin-releasing hormone; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; RNA, ribonucleic acid; mRNA, messenger RNA; miRNA, microRNA; nt, nucleotide; RISC, RNA-induced silencing complex; TGF, transforming growth factor; RNA pol II, RNA polymerase II; RISC, RNA-induced silencing complex; TAR, trans-activation response; TRBP, TAR RNA binding protein.
The post-transcriptional repression of target mRNA by miRNA occurs in a cell- type- and tissue-specific manner, including within neurons and glia of the CNS.58,62,63 Disruption of miRNA biogenesis within neurons results in profound developmental impairment and deficits in neuronal differentiation, morphology, and signaling.64–66 miRNA have been shown to regulate a range of CNS functions, including reward feedback, circadian rhythmicity, and cognitive performance.67–69 Furthermore, the dysregulation of specific miRNA may contribute to a multitude of neuronal disorders, including schizophrenia, Alzheimer’s disease, autism, and bipolar disorder, among others.70–74 Hence, there is growing evidence for miRNA involvement in neuropathology, suggesting new avenues for therapeutic discovery.
The role of microRNA in depression
Could disruption of normal miRNA regulation result in a heightened susceptibility to clinical depression? Mutations within the target mRNA 3′ UTR, as well as within the miRNA itself, can result in impaired regulatory function. Likewise, even small changes in levels of miRNA expression can lead to both deviations from a homeostatic norm and profound molecular disruption. Aberrant biogenesis, shuttling, or regulatory binding of miRNA by Dicer, Drosha, RISC, or other processing proteins also has the potential to disrupt miRNA repression. Thus, there are multiple points at which disrupted miRNA signaling could initiate, or exacerbate, depressive pathophysiology.
Several miRNA–mRNA interactions have been found to be altered in animal models and in patients with clinical depression. A polymorphism within miR-30e is positively correlated with depression and its symptomatic onset.75 miR-30e is a known tumor suppressor (via inhibition of cell growth) and is also associated with the development of schizophrenia.76,77 In addition, the mood stabilizers lithium and sodium valproate each modified the expression of a number of miRNA in Wistar rats.78 Seven miRNAs were down-regulated in both treatment groups (let-7b, let-7c, miR-128a, miR-24a, miR-30c, miR-34a, miR-221), with only miR-144 found to be up-regulated in both lithium- and valproate-treated animals. Collectively, these miRNA-target mRNAs are involved in PI 3-kinase, PKC, mitogen-activated protein kinase (MAPK), and immune response signaling pathways. Of note, electroconvulsive shock (ECS) is used as a therapy for the most severe cases of depression, and ECS-mediated neuronal activity induced 119 different miRNA within the hippocampus in animals.79 Some global expression profiling of neuronal miRNA expression has been performed in the context of bipolar disorder, revealing several key alterations in miRNA expression.80,81 However, similar studies in major depression patients are only in their earliest stages.82,83 Hence, more such studies would provide needed insight into the differential expression of miRNA in patients with clinical depression.
MicroRNA in serotonergic signaling
miRNA may play a role in modulating depressive neurophysiology via the regulation of serotonergic signaling.84 For example, miR-16 has been shown to target the serotonin transporter, SERT.85,86 As described above, SERT is the predominant mediator of SSRI antidepressants, and, as such, its regulation by miR-16 is of particular interest. Indeed, Wnt pathway-dependent increases in miR-16 within the raphe nuclei after treatment with fluoxetine (Prozac®; provided by Dr M Bouhassira, Eli Lilly, Indianapolis, IN, USA) suppressed SERT translation, leading to increased serotonergic activity and an antidepressive behavioral outcome.86 Interestingly, evidence suggests that miR-16 is involved in fate determination of serotonergic and noradrenergic cells, the latter of which show increased miR-16 expression that suppresses translation of SERT mRNA within the locus coeruleus.85 After fluoxetine treatment, cells within the locus coeruleus showed decreased miR-16 (in contrast to the effect within the raphe nuclei), illustrating the region-specific nature of SSRI treatment. In addition to serotonin transporters, several serotonin receptors are targets of miRNA. A polymorphism within the 3′ UTR of the serotonergic 5-HT1B receptor has been shown to prevent its down-regulation by miR-96, thus increasing receptor expression and membrane integration.87 In addition, miR-195 regulates the 5-HT2 A serotonin receptor and potentially the 5-HT4 receptor.70,88 Taken together, these studies suggest there are multiple means by which disruption of miRNA function could lead to an imbalance of serotonergic signaling, which could contribute to the multifaceted pathophysiology of clinical depression.
MicroRNA and the neurotrophic hypothesis of depression
While the above discussion of miRNA regulation of serotonergic signaling in models of depression reinforces the monoamine hypothesis, several other miRNA targets may also influence depressive pathophysiology. BDNF is under the regulatory control of a number of miRNA, including miR-30a-5p whose expression increases in cells treated with the SSRI antidepressant drug paroxetine.89–91 In addition to targeting the serotonin receptors mentioned above, miR-195 also targets BDNF and glutamate receptors.88 Increases in BDNF are thought to indirectly mediate the antidepressant effects of SSRIs, so the over-expression of miRNA-targeting BDNF after drug treatment potentially suggests homeostatic feedback to maintain BDNF within a favorable range. Drugs that induce BDNF expression, without concordant increases in inhibitory miRNA, may prove to be more specific and efficient for antidepressant treatment than current antidepressants.
Expression of the neuronal plasticity-associated transcription factor CREB has also been reported to be reduced in the temporal cortex of depressed patients, while increases in CREB within the hippocampus yield an antidepressant effect.37,38,92 Indeed, SSRIs are known to increase CREB expression in an array of brain regions and to reverse the impairment in CREB function observed in animal models of depression.93,94 Several miRNA with increased expression in models of depression are predicted to target CREB, including miR-22, miR-200b, miR-211, and miR-300.95 Of note, miR-124 is known to regulate serotonin-dependent synaptic plasticity by targeting CREB expression.96 Given the role of CREB in neuronal plasticity and the neurotrophic hypothesis that prominently features downstream targets of BDNF, miRNAs associated with CREB-regulated transcription may serve as avenues for therapeutic investigation.
BDNF is known to contribute to a number of regulatory feedback loops within neurons, including one involving miR-132.97–99 miR-132 is induced by BDNF and is expressed in a CREB-dependent manner that couples synaptic activity to dendritic morphogenesis.89,100–103 miR-132 is also induced by light within the SCN, regulating the expression of the per1 (Period circadian protein homolog 1) clock gene and modulating the capacity of light to entrain circadian rhythmicity.67,104 As described above, disruption of circadian timing is known to affect mood and depressive physiology. Thus, miR-132 regulates activity-dependent neuroplasticity as well as modulating sleep/wake cycles, both of which have the potential to affect depressive neurophysiology.
Circadian and stress-triggered dysregulation of miRNA regulation
In addition to miR-132, several other miRNA are involved in regulation of time. Indeed, interruption of miRNA biogenesis disrupts circadian signaling, and numerous miRNA have been shown to oscillate in a diurnal manner.105,106 miR-219 expression is also regulated in a circadian-dependent manner through direct binding of Circadian Locomotor Output Cycles Kaput (CLOCK)/BMAL (Brain and muscle ARNT like 1) to its promoter region.67 Other miRNA directly target components of the circadian clock-signaling pathway, such as the miR-192/194 cluster, which inhibits the PER gene family.107 Of particular note, mutations within miR-182 increase vulnerability to depression, potentially as a subsequent symptom of sleep dysregulation.108 miR-182 is predicted to target the circadian regulatory gene, Clock, and exhibits diurnal oscillation within the retina.106,108 Thus, miRNA may modulate the depressive effect of circadian disruption and could serve as therapeutic targets to restore normal sleep patterns and stabilize mood.
In addition to the miRNA response to sleep dysregulation, miRNA are involved in the cellular response to stress, which is a known trigger of clinical depression.109,110 Both acute and chronic stress alters miRNA expression within the amygdala and the hippocampus in a region-specific manner (eg, Let-7, miR-9, miR-26, miR-30, and miR-124, miR-132/212, miR-134, miR-183).111–114 Among these, miR-134 and miR-183 are both increased following acute stress, but are decreased and unchanged, respectively, after exposure to chronic stressors.111 Both miRNA target splicing factor SC35, which promotes the processing of stress-induced acetylcholinesterase (AChE)-R rather than its more common variant (AChE-S).115,116 Thus, disruption of miR-134 and miR-183 expression would bias AChE RNA splicing toward a heightened stress response. Furthermore, excessive AChE-R is accompanied by over-expression of miR-132, which in turn suppresses translation of its target AChE-S, yielding cognitive impairment and disrupted nocturnal activity.117
Some measure of transcriptional regulation is expected as an adaptive response to stress, but excessive stress leads to dysfunction on a cellular and behavioral level. Prolonged or extreme stress can induce maladaptive glucocorticoid levels that contribute to oxidative stress and impaired neural function, all of which are commonly associated with depressive pathophysiology.118 miR-18 and miR-124a both down-regulate the glucocorticoid receptor, which also has the potential to be targeted by a range of other miRNA.119,120 Of note, glucocorticoid impairs miR-132 expression in a BDNF-dependent manner, suggesting a potential mechanism of stress-induced and miRNA-mediated depression.121 Together, these data indicate that antidepressant treatment has the capacity to reverse stress-induced changes to miRNA expression, which may serve to couple stress response signaling with changes in neuronal plasticity.122 A list of depression-relevant miRNAs is included in Table 1.
Table 1.
Depression-associated microRNAs
| miRNA | Species | Relevant target(s) | Depression association/functional role | References |
|---|---|---|---|---|
| miR-30e | Human | Unknown | Polymorphism in depressed patients Tumor suppressor Inhibition of cell growth | 74–76 |
| miR-30a-5p | Human | BDNF | Increased expression after paroxetine treatment | 87–89 |
| let-7b, let-7c, miR-128a, miR-24a, miR-30c, miR-34a, miR-221, miR-144 | Rat | Diverse | Altered expression after lithium treatment PI-3-kinase, PKC, MAPK, and immune response signaling pathways | 77 |
| miR-16 | Mouse | SERT | Serotonergic signaling Altered expression after fluoxetine treatment |
83,84 |
| miR-96 | Human | 5-HT1B receptor | Serotonergic signaling | 85 |
| miR-195 | Human | 5-HT2A receptor | Serotonergic signaling | 86 |
| 5-HT4 receptor BDNF | ||||
| miR-22, miR-200b, miR-211, and miR-300 | Rat | CREB | Transcriptional regulation of neuronal plasticity and morphology | 93 |
| miR-124 | Aplysia P19 cells | CREB GCR |
Modulates serotonin-dependent synaptic plasticity | 94,117 |
| miR-132 | Mouse | p300 AChE |
BDNF feedback loop, CREB-regulated, down-regulated by glucocorticoid, regulates neuronal morphology, circadian rhythmicity | 66,95,98,99, 102,115,119 |
| miR-219 | Mouse | Unknown | CLOCK/BMAL-dependent circadian expression | 66 |
| miR-192/194 | Cell culture | Period family | Modulation of circadian timing | 105 |
| miR-182 | Mouse Human |
Clock | Increases vulnerability to depression, circadian rhythmicity | 104,106 |
| miR-134, miR-183 | Rat | SC35 | Altered expression after stress, promote stress-induced AChE-R | 109 |
| miR-18 | P19 cells | GCR | Stress and depression | 117 |
Abbreviations: AChE, acetylcholinesterase; ARNT, aryl hydrocarbon receptor nuclear translocator; AChE, acetylcholinesterase; BDNF, brain-derived neurotrophic factor; BMAL, brain and muscle ARNT like 1; CREB, cAMP response element-binding; CLOCK, Circadian Locomotor Output Cycles Kaput; GCR, glucocorticoid receptor; MAPK, mitogen-activated protein kinase; RNA, ribonucleic acid ; miRNA, microRNA; PI-3, phosphoinositide 3; PKC, protein kinase C; SERT, serotonin transporter.
MicroRNA as a target of depression therapy
The limitations of current monoaminergic-associated anti-depressants suggest the need for alternative and/or more comprehensive approaches to the treatment of clinical depression. Given the multifaceted etiology of depression, novel therapeutics must take into account the many aspects of the molecular imbalances observed. The involvement of miRNA across many facets of depressive pathophysiology makes them intriguing targets of antidepressant treatment. miRNA may serve as fine-tuned homeostatic regulators that target multiple nodes of depressive dysfunction (Table 1). Given that some current antidepressants already modulate miRNA expression in a manner that is consistent with a beneficial effect, a more direct targeting of these miRNA may prove efficacious. Notably, the first phase II clinical trial involving miRNA is currently underway, with others not far behind.4,123–125 Indeed, the number of pharmaceutical companies with miRNA-based workflows is also on the rise (eg, MiRNA Therapeutics, Regulus, RXi Pharmaceuticals, Santaris).
As described in The role of microRNA in depression section, both increases and decreases in miRNA functionality are associated with depressive pathophysiology. miRNA over-expression leads to the inappropriate silencing of hundreds of downstream targets. The use of antisense or ‘sponge’ technology to bind specific nt sequences would selectively suppress the action of pathologically abundant miRNA.126 As such, antagomirs (cholesterol-conjugated 2-O-methyl RNA antisense oligonucleotides) are currently being used to target aberrant miRNA expression in a range of disease models.123,127,128 Conversely, impairment of miRNA expression leads to excessive translation of target mRNA, detrimentally affecting neuronal physiology. While the lack of global miRNA dysregulation in models of clinical depression suggests that miRNA biogenesis and repression mechanisms remain largely intact, some mutations in genes required for miRNA processing have been found.129 Further, mutations within specific miRNA promoters, seed sequences, or the 3′ UTR of target mRNA may result in deficiency of regulation by specific miRNA. In the case of dysfunction with the miRNA itself, artificial miRNA mimetics may prove useful in restoring normal transcriptional regulation. The capacity of miRNA to maintain subtle regulatory balance is attractive in disorders where both increases and decreases of signaling activity can be causitive.68 In the case of excessive protein expression, custom small interfering RNA (siRNA) constructs can preferentially target specific protein-coding mRNA, even in the absence of endogenous miRNA regulation.130 Indeed, the monoaminergic transporters DAT and SERT have both been effectively targeted using siRNA treatment, yielding an antidepressive effect.131–133
Though the therapeutic potential of miRNA-based strategies is promising, significant challenges remain if the development of miRNA-mediated antidepressant therapy is to prove a meaningful endeavor. The capacity of miRNA to regulate multiple targets allows for therapeutic potential across several aspects of chemical imbalance, but caution must be taken to avoid unintended effects of altered miRNA expression. Small changes in miRNA activity can have an amplified effect on both direct and indirect downstream targets, many of which may be unrelated to the pathophysiology at hand. The involvement of miRNA in several signaling feedback loops, as well as their role in fine-tuning regulatory pathways, indicates the importance of careful dosing of any miRNA-associated therapeutics.67,97 miRNA may have a buffered effect on downstream targets such that minor changes in miRNA expression therapeutically hone signaling pathways, but yield massive dysregulation if expressed beyond a threshold of tolerance.134 Hence, each new treatment will require extensive toxicity screening due to the potential for multiple unintended miRNA effects.
The inability of nt constructs to cross the blood–brain barrier also remains problematic for therapeutic delivery. Indeed, many miRNA therapies currently under investigation in animal models employ highly invasive delivery methods that would be appropriate only in the most extreme cases of human disease. However, progress on this front is also being made with the use of adeno-associated viruses (AAVs) that have been shown to be a safe, and increasingly effective, means of gene delivery.135–137 Moreover, integration of viral-vector technology with the use of cell-type specific promoters could prove useful given the brain-region specific nature of molecular dysregulation in clinical depression (and indeed of miRNA themselves).85
Despite these challenges, miRNA should remain a significant focus in the development of novel antidepressant treatments. Of great interest would be the additional use of high-throughput approaches to look at the global dysregulation of miRNA in animal models of depression, as well as in the post-mortem brains of depressed patients.83 Some high-throughput screens of miRNA derived from the blood work of depressed patients has already taken place.82,138 Continuing to prioritize such studies would help identify potential miRNA biomarkers of clinical depression, and allow for screening of patient susceptibility. Similarly, profiling serum miRNA changes in patients with varying degrees of responsiveness to current antidepressants could help predict efficacy of treatment options for individuals. Such screens would allow for more informed, patient-driven treatments and reduce negative off-target effects. Such screens have found success in an array of disorders and thus hold promise for more accurately predicting patient vulnerability and resilience in clinical depression.139–142 Clearly, these applications may be more immediately attainable than the use of miRNA as direct therapeutics for clinical depression. The therapeutic application of the research findings mentioned here will require a more detailed understanding of the signaling networks that regulate, and are regulated by, miRNA in depressed patients. Nevertheless, miRNAs hold great potential in the treatment of clinical depression.
Acknowledgments
The authors thank Ryan Hansen for his perspective and input on the manuscript. National Institutes of Health Grant numbers: F31-MH096460-01, NS066345.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Vos T, Flaxman AD, Naghavi M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade L, Caraveo-Anduaga JJ, Berglund P, et al. The epidemiology of major depressive episodes: results from the International Consortium of Psychiatric Epidemiology (ICPE) Surveys. Int J Methods Psychiatr Res. 2003;12(1):3–21. doi: 10.1002/mpr.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessler RC, McGonagle KA, Zhao S, et al. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Arch Gen Psychiatry. 1994;51(1):8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- 4.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803(11):1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Hunsberger JG, Austin DR, Chen G, Manji HK. MicroRNAs in mental health: from biological underpinnings to potential therapies. Neuromolecular Med. 2009;11(3):173–182. doi: 10.1007/s12017-009-8070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol Psychiatry. 2012;17(4):359–376. doi: 10.1038/mp.2011.162. [DOI] [PubMed] [Google Scholar]
- 7.Dwivedi Y. Evidence demonstrating role of microRNAs in the etiopathology of major depression. J Chem Neuroanat. 2011;42(2):142–156. doi: 10.1016/j.jchemneu.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mouillet-Richard S, Baudry A, Launay JM, Kellermann O. MicroRNAs and depression. Neurobiol Dis. 2012;46(2):272–278. doi: 10.1016/j.nbd.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Nutt DJ. Relationship of neurotransmitters to the symptoms of major depressive disorder. J Clin Psychiatry. 2008;69(Suppl E1):4–7. [PubMed] [Google Scholar]
- 10.Caspi A, Sugden K, Moffitt TE, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 11.Tian JW, Jiang WL, Zhong Y, et al. Preclinical pharmacology of TP1, a novel potent triple reuptake inhibitor with antidepressant properties. Neuroscience. 2011;196:124–130. doi: 10.1016/j.neuroscience.2011.08.064. [DOI] [PubMed] [Google Scholar]
- 12.Hercher C, Turecki G, Mechawar N. Through the looking glass: examining neuroanatomical evidence for cellular alterations in major depression. J Psychiatr Res. 2009;43(11):947–961. doi: 10.1016/j.jpsychires.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Kempton MJ, Salvador Z, Munafò MR, et al. Structural neuroimaging studies in major depressive disorder. Meta-analysis and comparison with bipolar disorder. Arch Gen Psychiatry. 2011;68(7):675–690. doi: 10.1001/archgenpsychiatry.2011.60. [DOI] [PubMed] [Google Scholar]
- 14.Arnone D, McIntosh AM, Ebmeier KP, Munafò MR, Anderson IM. Magnetic resonance imaging studies in unipolar depression: systematic review and meta-regression analyses. Eur Neuropsychopharmacol. 2012;22(1):1–16. doi: 10.1016/j.euroneuro.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Sheline YI, Gado MH, Kraemer HC. Untreated depression and hippocampal volume loss. Am J Psychiatry. 2003;160(8):1516–1518. doi: 10.1176/appi.ajp.160.8.1516. [DOI] [PubMed] [Google Scholar]
- 16.Hajszan T, MacLusky NJ, Leranth C. Short-term treatment with the antidepressant fluoxetine triggers pyramidal dendritic spine synapse formation in rat hippocampus. Eur J Neurosci. 2005;21(5):1299–1303. doi: 10.1111/j.1460-9568.2005.03968.x. [DOI] [PubMed] [Google Scholar]
- 17.Honer WG, Falkai P, Chen C, Arango V, Mann JJ, Dwork AJ. Synaptic and plasticity-associated proteins in anterior frontal cortex in severe mental illness. Neuroscience. 1999;91(4):1247–1255. doi: 10.1016/s0306-4522(98)00679-4. [DOI] [PubMed] [Google Scholar]
- 18.Malberg JE, Eisch AJ, Nestler EJ, Duman RS. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci. 2000;20(24):9104–9110. doi: 10.1523/JNEUROSCI.20-24-09104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs BL, van Praag H, Gage FH. Adult brain neurogenesis and psychiatry: a novel theory of depression. Mol Psychiatry. 2000;5(3):262–269. doi: 10.1038/sj.mp.4000712. [DOI] [PubMed] [Google Scholar]
- 20.Horan WP, Pogge DL, Borgaro SR, Stokes JM, Harvey PD. Learning and memory in adolescent psychiatric inpatients with major depression: a normative study of the California Verbal Learning Test. Arch Clin Neuropsychol. 1997;12(6):575–584. [PubMed] [Google Scholar]
- 21.Hashimoto K. The role of glutamate on the action of antidepressants. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(7):1558–1568. doi: 10.1016/j.pnpbp.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Li N, Liu RJ, Dwyer JM, et al. Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biol Psychiatry. 2011;69(8):754–761. doi: 10.1016/j.biopsych.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duman RS. Synaptic plasticity and mood disorders. Mol Psychiatry. 2002;7(Suppl 1):S29–S34. doi: 10.1038/sj.mp.4001016. [DOI] [PubMed] [Google Scholar]
- 24.Pezawas L, Meyer-Lindenberg A, Goldman AL, et al. Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Mol Psychiatry. 2008;13(7):709–716. doi: 10.1038/mp.2008.32. [DOI] [PubMed] [Google Scholar]
- 25.Sen S, Duman R, Sanacora G. Serum brain-derived neurotrophic factor, depression, and antidepressant medications: meta-analyses and implications. Biol Psychiatry. 2008;64(6):527–532. doi: 10.1016/j.biopsych.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry. 2001;50(4):260–265. doi: 10.1016/s0006-3223(01)01083-6. [DOI] [PubMed] [Google Scholar]
- 27.Qi X, Lin W, Wang D, Pan Y, Wang W, Sun M. A role for the extracellular signal-regulated kinase signal pathway in depressive-like behavior. Behav Brain Res. 2009;199(2):203–209. doi: 10.1016/j.bbr.2008.11.051. [DOI] [PubMed] [Google Scholar]
- 28.Todorovic C, Sherrin T, Pitts M, Hippel C, Rayner M, Spiess J. Suppression of the MEK/ERK signaling pathway reverses depressionlike behaviors of CRF2-deficient mice. Neuropsychopharmacology. 2009;34(6):1416–1426. doi: 10.1038/npp.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Aberrant extracellular signal-regulated kinase (ERK)1/2 signalling in suicide brain: role of ERK kinase 1 (MEK1) Int J Neuropsychopharmacol. 2009;12(10):1337–1354. doi: 10.1017/S1461145709990575. [DOI] [PubMed] [Google Scholar]
- 30.Gourley SL, Wu FJ, Kiraly DD, et al. Regionally specific regulation of ERK MAP kinase in a model of antidepressant-sensitive chronic depression. Biol Psychiatry. 2008;63(4):353–359. doi: 10.1016/j.biopsych.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dwivedi Y, Rizavi HS, Conley RR, Pandey GN. ERK MAP kinase signaling in post-mortem brain of suicide subjects: differential regulation of upstream Raf kinases Raf-1 and B-Raf. Mol Psychiatry. 2006;11(1):86–98. doi: 10.1038/sj.mp.4001744. [DOI] [PubMed] [Google Scholar]
- 32.Dwivedi Y, Rizavi HS, Roberts RC, Conley RC, Tamminga CA, Pandey GN. Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. J Neurochem. 2001;77(3):916–928. doi: 10.1046/j.1471-4159.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- 33.Dwivedi Y, Rizavi HS, Teppen T, et al. Lower phosphoinositide 3-kinase (PI 3-kinase) activity and differential expression levels of selective catalytic and regulatory PI 3-kinase subunit isoforms in prefrontal cortex and hippocampus of suicide subjects. Neuropsychopharmacology. 2008;33(10):2324–2340. doi: 10.1038/sj.npp.1301641. [DOI] [PubMed] [Google Scholar]
- 34.Shelton RC, Sanders-Bush E, Manier DH, Lewis DA. Elevated 5-HT 2A receptors in postmortem prefrontal cortex in major depression is associated with reduced activity of protein kinase A. Neuroscience. 2009;158(4):1406–1415. doi: 10.1016/j.neuroscience.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelton RC, Hal Manier D, Lewis DA. Protein kinases A and C in postmortem prefrontal cortex from persons with major depression and normal controls. Int J Neuropsychopharmacol. 2009;12(9):1223–1232. doi: 10.1017/S1461145709000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey GN, Dwivedi Y, Pandey SC, Conley RR, Roberts RC, Tamminga CA. Protein kinase C in the postmortem brain of teenage suicide victims. Neurosci Lett. 1997;228(2):111–114. doi: 10.1016/s0304-3940(97)00378-9. [DOI] [PubMed] [Google Scholar]
- 37.Dwivedi Y, Rao JS, Rizavi HS, et al. Abnormal expression and functional characteristics of cyclic adenosine monophosphate response element binding protein in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60(3):273–282. doi: 10.1001/archpsyc.60.3.273. [DOI] [PubMed] [Google Scholar]
- 38.Chen AC, Shirayama Y, Shin KH, Neve RL, Duman RS. Expression of the cAMP response element binding protein (CREB) in hippocampus produces an antidepressant effect. Biol Psychiatry. 2001;49(9):753–762. doi: 10.1016/s0006-3223(00)01114-8. [DOI] [PubMed] [Google Scholar]
- 39.Cotter DR, Pariante CM, Everall IP. Glial cell abnormalities in major psychiatric disorders: the evidence and implications. Brain Res Bull. 2001;55(5):585–595. doi: 10.1016/s0361-9230(01)00527-5. [DOI] [PubMed] [Google Scholar]
- 40.Rajkowska G, Miguel-Hidalgo JJ. Gliogenesis and glial pathology in depression. CNS Neurol Disord Drug Targets. 2007;6(3):219–233. doi: 10.2174/187152707780619326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnadas R, Cavanagh J. Depression: an inflammatory illness? J Neurol Neurosurg Psychiatry. 2012;83(5):495–502. doi: 10.1136/jnnp-2011-301779. [DOI] [PubMed] [Google Scholar]
- 42.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9(1):46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howren MB, Lamkin DM, Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: a meta-analysis. Psychosom Med. 2009;71(2):171–186. doi: 10.1097/PSY.0b013e3181907c1b. [DOI] [PubMed] [Google Scholar]
- 44.Dowlati Y, Herrmann N, Swardfager W, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67(5):446–457. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Pace TW, Miller AH. Cytokines and glucocorticoid receptor signaling. Relevance to major depression. Ann N Y Acad Sci. 2009;1179:86–105. doi: 10.1111/j.1749-6632.2009.04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Syvälahti E, Eskola J, Ruuskanen O, Laine T. Nonsuppression of cortisol in depression and immune function. Prog Neuropsychopharmacol Biol Psychiatry. 1985;9(4):413–422. doi: 10.1016/0278-5846(85)90195-2. [DOI] [PubMed] [Google Scholar]
- 47.Meltzer HY, Lowy MT, Koenig JI. The hypothalamic-pituitary-adrenal axis in depression. Adv Biochem Psychopharmacol. 1987;43:165–182. [PubMed] [Google Scholar]
- 48.Monteleone P, Martiadis V, Maj M. Circadian rhythms and treatment implications in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(7):1569–1574. doi: 10.1016/j.pnpbp.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 49.Mendlewicz J. Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS Drugs. 2009;23(Suppl 2):15–26. doi: 10.2165/11318630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Kupfer DJ, Foster FG, Reich L, Thompson SK, Weiss B. EEG sleep changes as predictors in depression. Am J Psychiatry. 1976;133(6):622–626. doi: 10.1176/ajp.133.6.622. [DOI] [PubMed] [Google Scholar]
- 51.Kupfer DJ. REM latency: a psychobiologic marker for primary depressive disease. Biol Psychiatry. 1976;11(2):159–174. [PubMed] [Google Scholar]
- 52.Kaf-De St Hilaire S, Merica H, Gaillard JM. The effects of indalpine – a selective inhibitor of 5-HT uptake – on rat paradoxical sleep. Eur J Pharmacol. 1984;98(3–4):413–418. doi: 10.1016/0014-2999(84)90290-5. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson AN, Pascoe PA. 5-Hydroxytryptamine and noradrenaline uptake inhibition: studies on sleep in man. Neuropharmacology. 1986;25(10):1079–1083. doi: 10.1016/0028-3908(86)90153-x. [DOI] [PubMed] [Google Scholar]
- 54.Bliss EL. Sleep in schizophrenia and depression: studies of sleep loss in man and animals. Res Publ Assoc Res Nerv Ment Dis. 1967;45:195–210. [PubMed] [Google Scholar]
- 55.Vogel GW, Vogel F, McAbee RS, Thurmond AJ. Improvement of depression by REM sleep deprivation. New findings and a theory. Arch Gen Psychiatry. 1980;37(3):247–253. doi: 10.1001/archpsyc.1980.01780160017001. [DOI] [PubMed] [Google Scholar]
- 56.Georgotas A, McCue RE, Cooper TB, Nagachandran N, Friedhoff A. Factors affecting the delay of antidepressant effect in responders to nortriptyline and phenelzine. Psychiatry Res. 1989;28(1):1–9. doi: 10.1016/0165-1781(89)90192-3. [DOI] [PubMed] [Google Scholar]
- 57.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 58.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 59.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 60.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 61.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J, Krichevsky A, Grad Y, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7(12):911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 64.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 65.Cuellar TL, Davis TH, Nelson PT, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105(14):5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis TH, Cuellar TL, Koch SM, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28(17):4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng HY, Papp JW, Varlamova O, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. miRNA-132: a dynamic regulator of cognitive capacity. Brain Struct Funct. 2013;218(3):817–831. doi: 10.1007/s00429-012-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hollander JA, Im HI, Amelio AL, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466(7303):197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46(2):263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 71.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 72.Ghahramani Seno MM, Hu P, Gwadry FG, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380:85–97. doi: 10.1016/j.brainres.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 73.Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 2011;3(2):10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69(2):188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu Y, Liu H, Li F, et al. A polymorphism in the microRNA-30e precursor associated with major depressive disorder risk and P300 waveform. J Affect Disord. 2010;127(1–3):332–336. doi: 10.1016/j.jad.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 76.Perkins DO, Jeffries CD, Jarskog LF, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schepeler T, Holm A, Halvey P, et al. Attenuation of the beta-catenin/TCF4 complex in colorectal cancer cells induces several growth-suppressive microRNAs that target cancer promoting genes. Oncogene. 2012;31(22):2750–2760. doi: 10.1038/onc.2011.453. [DOI] [PubMed] [Google Scholar]
- 78.Zhou R, Yuan P, Wang Y, et al. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34(6):1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eacker SM, Keuss MJ, Berezikov E, Dawson VL, Dawson TM. Neuronal activity regulates hippocampal miRNA expression. PLoS One. 2011;6(10):e25068. doi: 10.1371/journal.pone.0025068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim AH, Reimers M, Maher B, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124(1–3):183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rong H, Liu TB, Yang KJ, et al. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45(1):92–95. doi: 10.1016/j.jpsychires.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 82.Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol. 2013;23(7):602–611. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 83.Oved K, Morag A, Pasmanik-Chor M, et al. Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics. 2012;13(10):1129–1139. doi: 10.2217/pgs.12.93. [DOI] [PubMed] [Google Scholar]
- 84.Millan MJ. MicroRNA in the regulation and expression of serotonergic transmission in the brain and other tissues. Curr Opin Pharmacol. 2011;11(1):11–22. doi: 10.1016/j.coph.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 85.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329(5998):1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 86.Launay JM, Mouillet-Richard S, Baudry A, Pietri M, Kellermann O. Raphe-mediated signals control the hippocampal response to SRI antidepressants via miR-16. Transl Psychiatry. 2011;1:e56. doi: 10.1038/tp.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry. 2009;14(4):381–389. doi: 10.1038/mp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo AY, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst Biol. 2010;4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Numakawa T, Richards M, Adachi N, Kishi S, Kunugi H, Hashido K. MicroRNA function and neurotrophin BDNF. Neurochem Int. 2011;59(5):551–558. doi: 10.1016/j.neuint.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 90.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17(19):3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Angelucci F, Croce N, Spalletta G, et al. Paroxetine rapidly modulates the expression of brain-derived neurotrophic factor mRNA and protein in a human glioblastoma-astrocytoma cell line. Pharmacology. 2011;87(1–2):5–10. doi: 10.1159/000322528. [DOI] [PubMed] [Google Scholar]
- 92.Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased temporal cortex CREB concentrations and antidepressant treatment in major depression. Lancet. 1998;352(9142):1754–1755. doi: 10.1016/S0140-6736(05)79827-5. [DOI] [PubMed] [Google Scholar]
- 93.Thome J, Sakai N, Shin K, et al. cAMP response element-mediated gene transcription is upregulated by chronic antidepressant treatment. J Neurosci. 2000;20(11):4030–4036. doi: 10.1523/JNEUROSCI.20-11-04030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Böer U, Alejel T, Beimesche S, et al. CRE/CREB-driven up-regulation of gene expression by chronic social stress in CRE-luciferase transgenic mice: reversal by antidepressant treatment. PLoS One. 2007;2(5):e431. doi: 10.1371/journal.pone.0000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smalheiser NR, Lugli G, Rizavi HS, et al. MicroRNA expression in rat brain exposed to repeated inescapable shock: differential alterations in learned helplessness vs non-learned helplessness. Int J Neuropsychopharmacol. 2011;14(10):1315–1325. doi: 10.1017/S1461145710001628. [DOI] [PubMed] [Google Scholar]
- 96.Rajasethupathy P, Fiumara F, Sheridan R, et al. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63(6):803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10(12):1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 98.Cheng A, Wang S, Cai J, Rao MS, Mattson MP. Nitric oxide acts in a positive feedback loop with BDNF to regulate neural progenitor cell proliferation and differentiation in the mammalian brain. Dev Biol. 2003;258(2):319–333. doi: 10.1016/s0012-1606(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 99.Obrietan K, Gao XB, Van Den Pol AN. Excitatory actions of GABA increase BDNF expression via a MAPK-CREB-dependent mechanism: a positive feedback circuit in developing neurons. J Neurophysiol. 2002;88(2):1005–1015. doi: 10.1152/jn.2002.88.2.1005. [DOI] [PubMed] [Google Scholar]
- 100.Remenyi J, Hunter CJ, Cole C, et al. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J. 2010;428(2):281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- 101.Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102(45):16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Impey S, Davare M, Lesiak A, et al. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43(1):146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wayman GA, Davare M, Ando H, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105(26):9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alvarez-Saavedra M, Antoun G, Yanagiya A, et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20(4):731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kadener S, Menet JS, Sugino K, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23(18):2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organspecific miRNA cluster. J Biol Chem. 2007;282(34):25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 107.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276(19):5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 108.Saus E, Soria V, Escaramís G, et al. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19(20):4017–4025. doi: 10.1093/hmg/ddq316. [DOI] [PubMed] [Google Scholar]
- 109.Claes S. Glucocorticoid receptor polymorphisms in major depression. Ann N Y Acad Sci. 2009;1179:216–228. doi: 10.1111/j.1749-6632.2009.05012.x. [DOI] [PubMed] [Google Scholar]
- 110.Calabrese F, Molteni R, Racagni G, Riva MA. Neuronal plasticity: a link between stress and mood disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S208–S216. doi: 10.1016/j.psyneuen.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 111.Meerson A, Cacheaux L, Goosens KA, Sapolsky RM, Soreq H, Kaufer D. Changes in brain MicroRNAs contribute to cholinergic stress reactions. J Mol Neurosci. 2010;40(1–2):47–55. doi: 10.1007/s12031-009-9252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rinaldi A, Vincenti S, De Vito F, et al. Stress induces region specific alterations in microRNAs expression in mice. Behav Brain Res. 2010;208(1):265–269. doi: 10.1016/j.bbr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 113.Uchida S, Nishida A, Hara K, et al. Characterization of the vulnerability to repeated stress in Fischer 344 rats: possible involvement of microRNA-mediated down-regulation of the glucocorticoid receptor. Eur J Neurosci. 2008;27(9):2250–2261. doi: 10.1111/j.1460-9568.2008.06218.x. [DOI] [PubMed] [Google Scholar]
- 114.Uchida S, Hara K, Kobayashi A, et al. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J Neurosci. 2010;30(45):15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zimmerman G, Soreq H. Readthrough acetylcholinesterase: a multifaceted inducer of stress reactions. J Mol Neurosci. 2006;30(1–2):197–200. doi: 10.1385/JMN:30:1:197. [DOI] [PubMed] [Google Scholar]
- 116.Meshorer E, Bryk B, Toiber D, et al. SC35 promotes sustainable stress-induced alternative splicing of neuronal acetylcholinesterase mRNA. Mol Psychiatry. 2005;10(11):985–997. doi: 10.1038/sj.mp.4001735. [DOI] [PubMed] [Google Scholar]
- 117.Shaltiel G, Hanan M, Wolf Y, et al. Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct Funct. 2013;218(1):59–72. doi: 10.1007/s00429-011-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Madrigal JL, García-Bueno B, Caso JR, Pérez-Nievas BG, Leza JC. Stress-induced oxidative changes in brain. CNS Neurol Disord Drug Targets. 2006;5(5):561–568. doi: 10.2174/187152706778559327. [DOI] [PubMed] [Google Scholar]
- 119.Vreugdenhil E, Verissimo CS, Mariman R, et al. MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology. 2009;150(5):2220–2228. doi: 10.1210/en.2008-1335. [DOI] [PubMed] [Google Scholar]
- 120.Turner JD, Alt SR, Cao L, et al. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem Pharmacol. 2010;80(12):1860–1868. doi: 10.1016/j.bcp.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 121.Kawashima H, Numakawa T, Kumamaru E, et al. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165(4):1301–1311. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 122.O’Connor RM, O’Leary OF, Dinan A, Gokul A, Dinan TG, Cryan JF. Stress-induced alterations in hippocampal and amygdalar microR-NAs: effects of lithium. European Neuropsychopharmacology. 2010;20(August):S166–S167. [Google Scholar]
- 123.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA. 2009;106(44):18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 129.He Y, Zhou Y, Xi Q, et al. Genetic variations in microRNA processing genes are associated with susceptibility in depression. DNA Cell Biol. 2012;31(9):1499–1506. doi: 10.1089/dna.2012.1660. [DOI] [PubMed] [Google Scholar]
- 130.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 131.Hoyer D, Thakker DR, Natt F, et al. Global down-regulation of gene expression in the brain using RNA interference, with emphasis on monoamine transporters and GPCRs: implications for target characterization in psychiatric and neurological disorders. J Recept Signal Transduct Res. 2006;26(5–6):527–547. doi: 10.1080/10799890600929663. [DOI] [PubMed] [Google Scholar]
- 132.Thakker DR, Natt F, Hüsken D, et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci USA. 2004;101(49):17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thakker DR, Natt F. Hüsken D, et al. siRNA-mediated knockdown of the serotonin transporter in the adult mouse brain. Mol Psychiatry. 2005;10(8):782–789. 714. doi: 10.1038/sj.mp.4001687. [DOI] [PubMed] [Google Scholar]
- 134.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43(9):854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chtarto A, Bockstael O, Tshibangu T, Dewitte O, Levivier M, Tenenbaum L. A next step in adeno-associated virus (AAV)-mediated gene therapy for neurological diseases: regulation and targeting. Br J Clin Pharmacol. 2013 Jan 18; doi: 10.1111/bcp.12065. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gray SJ, Nagabhushan Kalburgi S, McCown TJ, Jude Samulski R. Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther. 2013;20(4):450–459. doi: 10.1038/gt.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Samaranch L, Salegio EA, San Sebastian W, et al. Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther. 2012;23(4):382–389. doi: 10.1089/hum.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Belzeaux R, Bergon A, Jeanjean V, et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry. 2012;2:e185. doi: 10.1038/tp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mo MH, Chen L, Fu Y, Wang W, Fu SW. Cell-free circulating miRNA biomarkers in cancer. J Cancer. 2012;3:432–448. doi: 10.7150/jca.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Karolina DS, Tavintharan S, Armugam A, et al. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab. 2012;97(12):E2271–E2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 141.Zhao C, Dong J, Jiang T, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6(8):e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leuenberger N, Jan N, Pradervand S, Robinson N, Saugy M. Circulating microRNAs as long-term biomarkers for the detection of erythropoiesis-stimulating agent abuse. Drug Test Anal. 2011;3(11–12):771–776. doi: 10.1002/dta.370. [DOI] [PubMed] [Google Scholar]