Abstract
The acquisition of water and nutrients by plant roots is a fundamental aspect of agriculture and strongly depends on root architecture. Root branching and expansion of the root system is achieved through the development of lateral roots and is to a large extent controlled by the plant hormone auxin. However, the pleiotropic effects of auxin or auxin-like molecules on root systems complicate the study of lateral root development. Here we describe a small-molecule screen in Arabidopsis thaliana that identified naxillin as what is to our knowledge the first non-auxin-like molecule that promotes root branching. By using naxillin as a chemical tool, we identified a new function for root cap-specific conversion of the auxin precursor indole-3-butyric acid into the active auxin indole-3-acetic acid and uncovered the involvement of the root cap in root branching. Delivery of an auxin precursor in peripheral tissues such as the root cap might represent an important mechanism shaping root architecture.
Plants have remarkable stress resistance, owing to the high flexibility in their architecture and growth pattern in response to external conditions. This flexibility is based on the continuous presence of stem cells in the meristems. The formation of new meristems persists during the entire lifespan and can be triggered by external factors, providing that the plant has continuous access to new soil resources for nutrients and water. Initially, the root system consists of a primary root originating from the embryo, but the initiation of new meristems generates lateral roots, resulting in a branched root system. Subsequent higher-order branching events give rise to the mature root architecture.
Important regulators of the process of root branching are auxins, which are a class of hormones essential for plant growth and development1,2. The most well-characterized endogenous auxin in plants is indole-3-acetic acid (IAA, 1). However, synthetic molecules such as 1-naphthalene-acetic acid (NAA, 2) or 2,4-dichlorophenoxy-acetic acid (2,4-D, 3) can elicit biochemical, molecular and physiological responses similar to those of IAA3. Endogenous IAA is biosynthesized de novo in plants from tryptophan or from tryptophan precursors. In addition, IAA can be stored as an inactive conjugate with sugar moieties, amino acids or peptides, which can be released through hydrolysis to provide free IAA1. The distribution of free auxin throughout the plant occurs via active transport, which is mediated by PIN-FORMED (PIN) efflux carriers, AUXIN-RESISTANT1 and LIKE-AUX1 influx carriers and ATP-BINDING CASSETTE transporter proteins2. Eventually, IAA and structurally related small molecules exert their effect by interacting with the TRANSPORT INHIBITOR RESPONSE1/AUXIN-BINDING F-BOX PROTEIN (TIR1/AFB) class of auxin receptors, which form part of SCF-type E3 ubiquitin ligase complexes4. Auxin serves as ‘molecular glue’ to stabilize the interaction of TIR1/AFB proteins with the transcriptional repressor auxin Aux/IAA proteins5,6. As a consequence of this stabilized interaction, Aux/IAA proteins become ubiquitinated and are targeted for proteolysis, resulting in derepression of the AUXIN RESPONSE FACTOR proteins (ARFs)7,8 and, ultimately, activation of the auxin signaling cascade.
Auxin concentrations and homeostasis are, in addition to de novo biosynthesis, conjugation and transport, also regulated by the auxin precursor indole-3-butyric acid (IBA, 4). Directional IBA transport in the root is thought to be mediated by carrier proteins such as PLEIOTROPIC DRUG RESISTANCE 8 (PDR8) and PDR9 to promote IBA efflux9,10. Because PDR8 and PDR9 have been shown to localize on the outer polar domain of epidermal and lateral root cap cells9,10, it has been suggested that these transporters move IBA from the root into the rhizosphere11. As shown by genetic evidence, IBA activity is dependent on peroxisomal import through PEROXISOMAL ABC TRANSPORTER1 (PXA1)12 and subsequent β-oxidation to IAA, which resembles the breakdown of fatty acids by two carbon units13,14. Several peroxisomal enzymes, such as the 3-ketoacyl-CoA thiolase PED, have been shown to contribute to both fatty acid and IBA β-oxidation13,15, whereas others such as the 3-hydroxyacyl-CoA dehydrogenase INDOLE-3-BUTYRIC ACID RESPONSE1 (IBR1), the acyl-CoA oxidase IBR3 and the enoyl-CoA hydratases ECH2 and IBR10 are dedicated to IBA-to-IAA conversion11,16,17.
During the process of root branching, auxin has a dominant role in every stage of lateral root development18. Lateral roots originate from a subset of xylem pole pericycle cells that undergo asymmetric cell division triggered by an auxin-dependent mechanism in the basal meristem, which leads to the induction of GATA23 expression and founder cell specification19,20. The oscillating nature of the auxin response in the basal meristem has been suggested to create the regular branching pattern observed in the Arabidopsis root system19-22. More distantly from the root tip, and also dependent on auxin signaling, a small number of adjacent pericycle founder cells undergo anticlinal divisions, giving rise to a single-layered primordium containing up to ten small cells23. Subsequent anticlinal and periclinal divisions yield a lateral root primordium, which grows through the outer cell layers and finally emerges from the primary root24. Auxin transport, the establishment of auxin maxima and auxin signaling all provide crucial regulatory input to these post-initiation events24,25.
Because auxin is involved in many developmental processes of plant growth3 and mediates several distinct processes in the root, such as cell expansion, root patterning and responses to gravity and touch, it is difficult to uncover molecular components specific to lateral root development by applying auxin or its analogs. Here, to study the development of lateral roots more selectively, we used a marker-based chemical biology approach to identify synthetic molecules that enhance auxin-regulated lateral root development. Screening of a diverse collection of chemicals led to the identification of naxillin, which is to our knowledge the first non-auxin-like synthetic molecule that induces lateral root formation. Naxillin induced an auxin response restricted to the basal meristem, the zone where pericycle cells acquire their lateral root founder cell identity. A genetic screen for naxillin resistance established that naxillin acted in the conversion of the auxin precursor IBA, a promoter of lateral root development12, into IAA. In accordance with the naxillin-induced auxin response, reporter gene expression analysis suggested that the conversion of IBA into IAA did not occur constitutively in all tissues but rather showed clear preference for specific root tissues. The use of naxillin uncovered the contribution of the root cap in root branching as an important source for IBA-derived IAA and highlighted the role of IBA-derived IAA as a specific lateral root–inducing agent.
RESULTS
Identification and characterization of naxillin
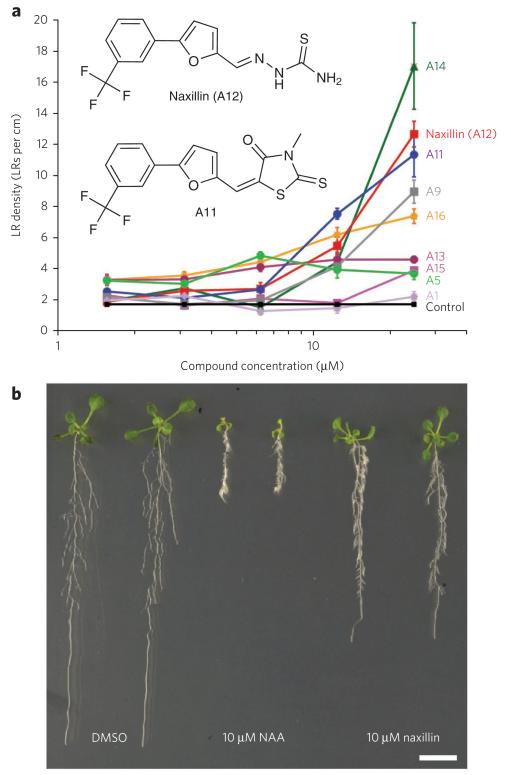
To identify new synthetic molecules that stimulate the process of lateral root development, we screened a diverse 10,000-compound library for activators of expression of CYCB1;1, a cell cycle gene that marks cellular divisions (Supplementary Results, Supplementary Table 1). In xylem pole pericycle cells, the induction of CYCB1;1 expression coincides with the formation of a new lateral root primordium and thus reports lateral root development26-28. We grew transgenic seedlings containing a construct comprising the CYCB1;1 promoter fused to β-glucuronidase (GUS) (pCYCB1;1::GUS) in a high-throughput adaptation of a previously described ‘lateral root inducible system’27. Eighty-eight molecules induced expression of CYCB1;1 in the xylem pole pericycle cells after 24 h (Supplementary Fig. 1), suggesting they were potent activators of the early stages of lateral root development. To avoid selecting auxin-like compounds that would also affect other auxin-related processes, we excluded all molecules with a chemical structure similar to that of known auxins, such as IAA, NAA, 2,4-D or sirtinol29, and retained nine hit molecules for further analysis (Fig. 1a and Supplementary Fig. 2). When grown for 5 d in the presence of these hit compounds, seedlings showed a strong increase in lateral root densities in a dose-dependent manner compared to the mock-treated control seedlings (Fig. 1a). Two of these molecules, A11 (5) and A12 (6), which we named naxillin for non-auxin-like lateral root inducer, shared a core structure (Fig. 1a and Supplementary Fig. 3). Structure-activity analysis demonstrated that the 5-[3-(trifluoromethyl)phenyl]furan substructure was required for the lateral root–inducing effect of A11 and naxillin.
Figure 1. Identification and characterization of naxillin.
(a) Dose-response analysis of lateral root (LR) density of seedlings grown on control medium until 3 d after germination and transferred to medium supplemented with the indicated hit molecule at the indicated concentration for five additional days. Insets show chemical structure of A11 and naxillin (A12). (b) Phenotype of plants grown on control medium for 3 d and then transferred to mock medium (DMSO) or medium supplemented with 10 μM NAA or 10 μM naxillin for five additional days. Scale bar, 1 cm.
A molecule that had only this core structure without additional modifications (var5) showed reduced activity compared to A11 and naxillin (Supplementary Fig. 3), indicating that modifications of the furan group increased the lateral root–inducing capacity of the compounds. Root phenotypic characterization showed that A11 and naxillin, compared to the synthetic auxin NAA, had reduced effects on primary root growth (Fig. 1b and Supplementary Fig. 4) and aerial tissue development (Supplementary Fig. 5). These data demonstrated that A11 and naxillin activate lateral roots in a more specific manner than known synthetic or natural auxins.
Naxillin activates a subset of auxin-induced transcripts
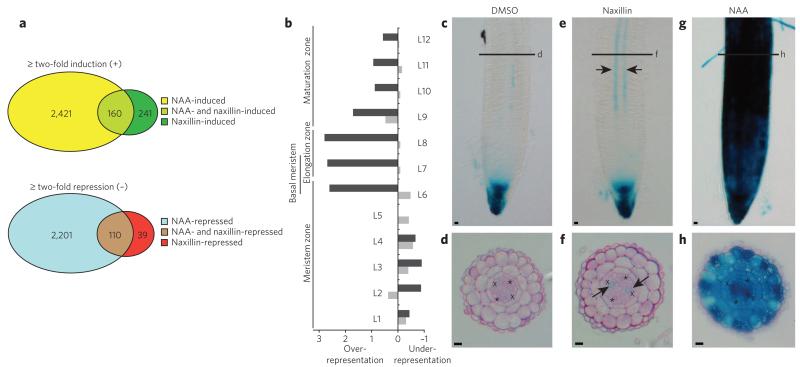
To further explore the specificity of naxillin for lateral root development, we compared the early effects of naxillin at the transcriptome level with NAA in roots of 3-d-old seedlings after 2-h and 6-h treatment. Naxillin treatment induced 401 genes, whereas NAA treatment induced 2,581 genes (Fig. 2a and Supplementary Data Set 1), suggesting a much narrower mechanism of action. A similar result was obtained for the downregulated genes, with only 149 genes downregulated by at least a factor of two by naxillin treatment, compared to 2,311 genes downregulated by NAA treatment. We found 160 and 110 genes, respectively, in the overlap between both treatments (Fig. 2a and Supplementary Data Set 1). Remarkably, many genes encoding members of the AUX/IAA, SAUR and PIN families28, were not among the naxillin-responsive genes, indicating that only a subset of auxin-regulated genes are required to selectively initiate lateral root development. In contrast, members of the GH3 family and peroxidases were strongly induced both by NAA and naxillin treatment (Supplementary Data Set 1). In addition, the GATA-type transcription factor GATA23, which functions in specifying lateral root founder cell identity20, and members of the LATERAL ORGAN BOUNDARIES (LBD) family of transcription factors (LBD33 and LBD29), which have roles during lateral root initiation as targets of ARF7 and ARF19 (ref. 30), presented naxillin induction profiles (Supplementary Data Set 1 and Supplementary Fig. 6). These examples underlined that naxillin mainly acts on the early events of lateral root development. Although no endogenous naxillin-like molecule has been reported in plants, the transcriptome analysis showed that the compound triggered a subset of the auxin-response pathways that naturally occur in plants.
Figure 2. Naxillin induces a subset of auxin responses in the basal meristem.
(a) Overview of the genes induced and/or repressed by NAA and naxillin treatments after 2 h or 6 h. (b) Longitudinal enrichment in NAA-responsive (light gray) and naxillin-responsive (dark gray) genes. L, longitudinal section. (c–h) Five-day-old pDR5::GUS seedlings grown on control medium (c,d) and transferred to 10 μM naxillin (e,f) or 10 μM NAA (g,h) for 8 h and stained for β-glucuronidase activity. Panels d, f and h are sections at the basal meristem zone of pDR5::GUS seedlings indicated in c, e and g respectively. Asterisks indicate phloem poles; crosses indicate xylem pole pericycle (XPP) cells; arrows in e and f indicate GUs staining in XPP cells. Scale bars, 10 μm.
Naxillin induces auxin response in the basal meristem
To reveal whether naxillin treatment affected the root transcriptome in a spatially global or more local manner, we used a bioinformatics approach to compare our transcriptome data set with the Arabidopsis high-resolution spatial map of the root transcriptome (Supplementary Methods)31. Although NAA-responsive genes were expressed homogenously in all longitudinal zones of the root, naxillin-responsive genes were over-represented by a factor of three in longitudinal regions L6 to L8, which encompass the basal meristem (Fig. 2b). Furthermore, naxillin induced the synthetic auxin-responsive marker pDR5::GUS locally in the basal meristem, specifically in xylem pole cells adjacent to the pericycle (Fig. 2c-f), whereas NAA treatment resulted in DR5 expression throughout the stele and the outer tissues of the meristematic, elongation and maturation zones (Fig. 2g,h), suggesting that naxillin affected auxin response locally in the basal meristem. Nevertheless, NAA and naxillin showed similar induction kinetics with respect to the onset of pericycle cell divisions and rate of primordia formation (Supplementary Fig. 7). These results suggested that the more restricted naxillin-induced DR5 expression pattern is sufficient to enhance lateral root development and represents the most essential auxin signaling constituent for lateral root initiation. To further validate the local effect of naxillin, we assessed the degradation of Aux/IAA proteins upon naxillin treatment in planta, which occurs rapidly in the entire root in the presence of auxin32. For that purpose, a heat shock–inducible AXR3 fused to an N-terminal GUS (pHS::AXR3-NT-GUS) transgenic line was used, in which a fast decrease of GUS activity is observed in the presence of auxin as a result of the degradation of AXR3 (also known as IAA17) (ref. 32). Absence of pHS::AXR3-NT-GUS activity was observed in the entire root upon auxin treatment. Upon naxillin treatment, however, GUS staining disappeared preferentially from the basal meristem and persisted in the very root tip and more mature tissue (Supplementary Fig. 8). Together with the transcriptome analysis, these data demonstrated that naxillin induces a specific subset of auxin responses in the basal meristem, the zone where lateral root founder cell identity is established. This mode of action might explain why, unlike auxin, naxillin shows limited interference with general root growth and development.
Naxillin does not act like a typical auxin
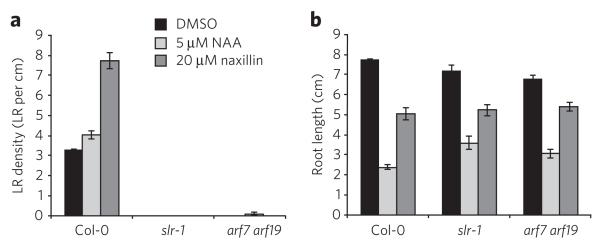
Perception of auxin is mediated by the co-receptor complex between TIR1/AFB1-3 F-box proteins and Aux/IAA proteins. This receptor complex is stabilized by auxin33. As a result, Aux/IAA proteins are ubiquitinated and targeted for proteasome-dependent degradation. Because naxillin affected the stability of AXR3-NT-GUS (Supplementary Fig. 8), we assessed the effect of naxillin on the recruitment of Aux/IAA proteins to TIR1 by pull-down assays. Unlike NAA, naxillin did not facilitate the interaction between TIR1-myc and Aux/IAA domain II peptides (Supplementary Fig. 9)32,34,35, indicating that the local auxin response induced by naxillin was not due to direct binding to the TIR1 auxin receptor. In addition, naxillin did not affect the NAA-dependent interaction of TIR1-myc and Aux/IAA domain II peptides (Supplementary Fig. 9), indicating that naxillin did not act as a competitive auxin antagonist. In contrast, naxillin, like auxin, failed to induce lateral root formation in the solitary root-1 gain-of-function Aux/IAA mutant36 or the arf7 arf19 double loss-of-function mutant37 (Fig. 3a,b), suggesting that naxillin’s function is dependent on the canonical IAA14 (also known as SOLITARY ROOT-1) and ARF7-ARF19 pathway and acts upstream of this auxin signaling pathway to induce lateral root development.
Figure 3. Naxillin and Naa have different primary targets.
(a,b) Lateral root density (a) and primary root length (b) of wild-type (Col-0), slr-1 and arf7 arf19 seedlings germinated on control medium and transferred to DMSO, 5 μM NAA or 20 μM naxillin for five additional days. Error bars represent mean ± s.e.m.
Naxillin activity required IBA-derived IAA
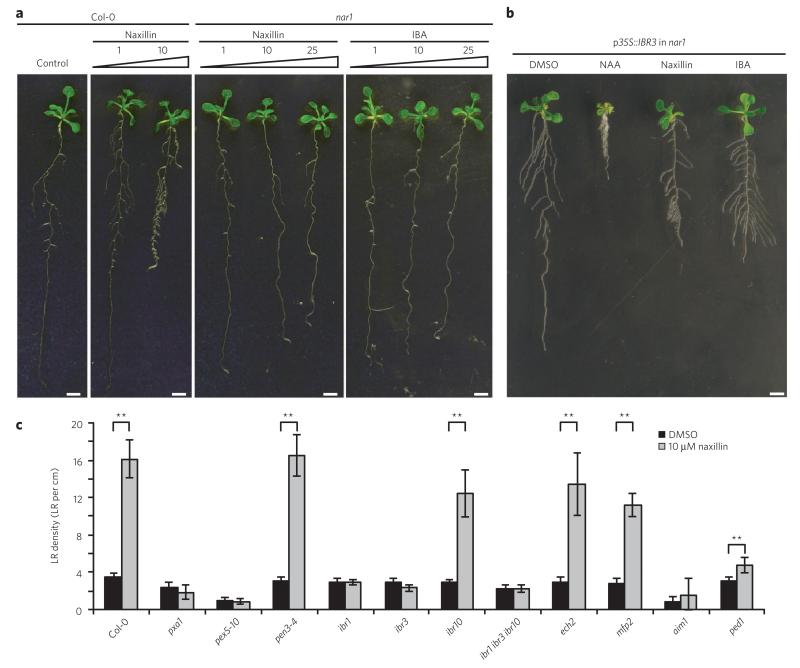
To gain further insight into naxillin’s mode of action, we screened an ethyl methane sulfonate (EMS)-mutagenized population for mutants resistant to the lateral root–inducing effect of naxillin. One mutant showed complete resistance to naxillin-induced lateral root development (Fig. 4a), which we have termed naxillin resistant 1 (nar1). Positional cloning located the mutation in a 300-kb interval on chromosome 3 between markers F24P17 and F17A9, a region containing 88 genes (Supplementary Fig. 10). Sequence analysis revealed a missense mutation in the IBR3 gene, which encodes a protein involved in the conversion of IBA to IAA17. The naxillin-resistant phenotype of the nar1 mutant could be restored by ectopic overexpression of IBR3 (ref. 17) (Fig. 4b and Supplementary Fig. 11), confirming that a defect in IBR3 function caused the naxillin-resistant phenotype in the nar1 mutant. Consistent with a defect in IBR3, nar1 roots were resistant to IBA (Fig. 4a) and did not show the typical DR5 expression pattern in the basal meristem, which was present upon naxillin and IBA treatment in wild-type seedlings but not in the ibr3 mutant background (Supplementary Fig. 12). These data indicate that the transcriptional auxin response in the basal meristem induced by naxillin and IBA is crucial to enhance lateral root development and suggest that this response requires IBA-to-IAA conversion.
Figure 4. Naxillin acts in the IBA-to-IAA conversion pathway.
(a) Phenotype of 3-d-old nar1 mutant seedlings transferred to naxillin or IBA at indicated concentrations (in μM) for five more days. (b) Complementation of the nar1 mutant with p35S::IBR3. Three-day-old plants were transferred to medium containing 10 μM NAA, 10 μM naxillin or 10 μM IBA for five more days. (c) Lateral root density of wild-type and mutant plants germinated on control medium for 3 d and transferred to medium supplemented with naxillin for five more days. Error bars represent means ± s.d. Asterisks indicate significant differences as calculated using a standard Student’s t-test (**P < 0.001). Scale bars, 1 cm.
Previously, ibr3 was identified in a screen for IBA-resistant root elongation and was implicated in the peroxisomal β-oxidation pathway in which IBA is converted to IAA16,17. IBR3 was suggested to act as an acyl-CoA oxidase in the IBA-to-IAA conversion pathway17. In addition to IBR3, other genes involved in IBA-to-IAA conversion (IBR1, IBR10, ECH2 and PED1) have been identified11,16,38. Moreover, genes involved in cellular (PEN3, also known as PDR8) and peroxisomal (PXA1) IBA transport and in import of peroxisomal matrix proteins in the peroxisomes (PEX5) can affect IBA response10,12,13. PXA1 is required for IBA uptake into the peroxisomes before IBA is converted to IAA12. Mutations in PXA1 conferred resistance to naxillin (Fig. 4c), suggesting that naxillin activity required IBA uptake into the peroxisomes. Disruption of peroxisomal protein import in the pex5-10 mutant (ref. 39) or reduction of IBA-to-IAA conversion in the ibr1-2 mutant17 also led to naxillin resistance (Fig. 4c), supporting the requirement of peroxisomal IBA-to-IAA conversion for naxillin activity. Conversely, the pen3-4 mutant showed sensitivity to naxillin at 10 μM (Fig. 4c) and hypersensitivity to both IBA10 and naxillin at 3 μM (Supplementary Fig. 13), which is in accordance with the earlier reported IBA hyperaccumulation in the pen3-4 mutant10. Remarkably, although mutations in IBA conversion genes IBR1, IBR3 and PED1 caused resistance to both IBA and naxillin, mutations in IBR10 and ECH2 led to IBA but not naxillin resistance (Fig. 4c and Supplementary Fig. 14). In accordance, the local DR5 expression pattern in the basal meristem was induced by naxillin in the ibr10 mutant background, whereas it was not induced by IBA (Supplementary Fig. 12). The naxillin sensitivity of the ibr10 and ech2 mutants suggests that naxillin might act at the level of IBR10 or ECH2, which have been suggested to be enoyl-CoA hydratases11,16 (Supplementary Fig. 14). In Arabidopsis, two additional peroxisomal proteins, ABNORMAL INFLORESCENCE MERISTEM 1 (AIM1) and MULTIFUNCTIONAL PROTEIN 2 (MFP2), act in the process of β-oxidation of fatty acids and have enoyl-CoA hydratase activity40,41. Analysis of aim1-1 (ref. 41) and mfp2-2 (ref. 42) mutants demonstrated that naxillin promoted lateral root development in the mfp2-2 mutant but not in the aim1-1 mutant (Fig. 4c), suggesting that naxillin is dependent on AIM1 as an enoyl-CoA hydratase, rather than on IBR10 and ECH2, to convert IBA to IAA (Supplementary Fig. 14). Taken together, mutant analyses suggested that naxillin requires the endogenous IBA conversion pathway and thus acts through IBA-derived IAA to promote the development of lateral roots. Indeed, this conclusion is in accordance with the highly similar lateral root phenotypes of IBA- and naxillin-treated plants (Fig. 4b and Supplementary Fig. 12).
IBA and naxillin act in root cap cells
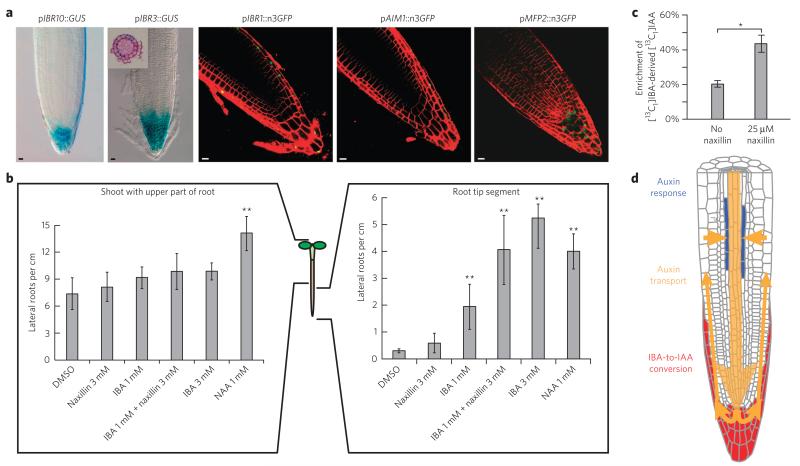
In addition to IBA-to-IAA conversion, naxillin activity was also dependent on the IAA14 (also known as SOLITARY ROOT-1) and ARF7-ARF19 auxin signaling pathway (Fig. 3a,b). NAA and IAA showed an overall effect on plant growth and development, whereas IBA and naxillin had a much more subtle effect on lateral root development (Fig. 4b and Supplementary Fig. 5). The latter phenotype implies a localized auxin effect of both IBA and naxillin, which could be attributed to tissue-specific IBA-to-IAA conversion, resulting in the production of IBA-derived IAA in root tissues predominantly involved in lateral root development. To assess whether IBA-to-IAA conversion might be more represented in specific zones or tissues of the root than elsewhere, we analyzed expression patterns of conversion genes. IBR1, IBR3, IBR10, AIM1 and MFP2 promoter expression domains all overlapped in the root tip of the primary root, more specifically in root cap cells (Fig. 5a and Supplementary Fig. 15), suggesting peroxisomal IBA-to-IAA conversion might show preference for the root cap.
Figure 5. IBA and naxillin act locally in the root cap.
(a) pIBR10::GUS; pIBR3::GUS; pIBR1::n3GFP; pAIM1::n3GFP and pMFP2::n3GFP expression patterns, showing overlapping expression in root cap cells of the primary root. Inset, sections at the root cap of pIBR3::GUS seedlings. Scale bars, 10 μm. (b) Lateral root density of wild-type plants germinated on control medium for 5 d and transferred to medium supplemented with IBA and/or naxillin at indicated concentrations (in μM) for five more days (NAA was used as a control). before transfer, seedlings were left intact, the apical part of the root was removed (shoot with upper part of root; left) or the shoot and basal part of the root were removed (root tip segment; right) as indicated. Error bars represent means ± s.d. Statistical significance was calculated using a standard two-sided t-test (**P < 0.001) (c) Short-term treatment of Arabidopsis seedlings with naxillin increases the flux of IBA to free IAA in root tips. Five-day-old seedlings were incubated with medium containing either 10 μM [13C1]IBA or 10 μM [13C1]IBA and 25 μM naxillin for 2 h. The percentage enrichment of [13C1]IAA derived from [13C1]IBA in the free IAA pool was determined. Error bars represent s.d. observed in four biological replicates, and the asterisk indicates the statistically significant (standard two-sided t-test, P = 0.028) difference between the untreated and naxillin-treated samples. (d) Hypothetical model for the selective action of IBA and naxillin toward lateral root development.
The predominant expression of conversion genes in the root cap and the dependence of both naxillin and IBA on the conversion highlighted the root tip as essential for the lateral root–inducing activity of both compounds. To further test the hypothesis that IBA and naxillin have a localized effect at the root tip, we applied both compounds to seedlings whose root apical meristem was removed. IBA and naxillin did not enhance lateral root development in seedlings lacking a root apex (Fig. 5b). In contrast, in seedlings with removed shoot and upper root parts, IBA strongly promoted lateral root development, whereas naxillin had only minor effects (Fig. 5b). However, when naxillin-treated samples were also supplied with IBA at a concentration insufficient to increase lateral root number, naxillin significantly (P < 0.001) enhanced lateral root development. These experiments confirmed that IBA and naxillin act at the root tip and also suggest that naxillin requires IBA as an endogenous factor to enhance lateral root development.
To test whether naxillin increased the rate of IBA-to-IAA conversion, we monitored the rate at which [13C1]IBA was converted to [13C1]IAA in the absence and presence of naxillin in 0.5-cm root tip segments. After 2 h in the presence of naxillin, there was a two-fold increase in the amount of [13C1]IAA formed (Fig. 5c), indicating that naxillin indeed stimulated the formation of IAA from IBA. These measurements provided biochemical evidence for naxillin’s stimulatory effect on the IBA-to-IAA conversion in the root tip and highlights the importance of root tip–derived IAA for lateral root initiation.
DISCUSSION
Colonization of the soil by plants occurs through root branching, a developmental process that is predominantly controlled by the plant hormone auxin. Auxin and auxin-like molecules have been extensively used to study root branching at the molecular level. Auxin treatments, however, affect the entire plant in various ways depending on the organ, tissue or cell type. One of the most obvious auxin effects is the abrupt arrest of root elongation, which compromises the study of lateral root formation because of its dependence on intact primary root growth. As shown by the absence of a strong deceleration of primary root growth in combination with the massive induction of lateral roots resulting from naxillin treatment, naxillin was clearly more selective toward lateral root development than auxins. This observation was further corroborated by naxillin’s smaller effect on the transcriptome compared to that of auxin, its restricted site of action and its specificity to induce the GATA23 transcription factor, which was previously described to be a key component of the lateral root founder cell specification process20. Notably, the overall transcriptional changes caused by naxillin treatment were less pronounced compared to those of auxin treatment. As all tissue types of the root were sampled and naxillin has tissue-specific action, the weaker effect of naxillin is most likely due to dilution with the nonresponsive tissues, which is not the case for auxin. To induce lateral root formation, naxillin clearly worked upstream of the canonical auxin signaling pathway and was dependent on a functional IBA-to-IAA conversion pathway. All of the naxillin-resistant mutants seemed also to be resistant to the lateral root–inducing effect of IBA. Previous studies have suggested a role for this endogenous auxin precursor in lateral root initiation because a block in the conversion of IBA to IAA resulted in plants with fewer lateral roots17. Remarkably, mutations in either IBR10 or ECH2, which may catalyze the hydration step of enoyl-CoA during IBA-to-IAA conversion11, led to IBA resistance but retained sensitivity to naxillin, suggesting that naxillin is able to complement a defective IBR10- or ECH2-dependent hydratase activity. One explanation for this result could be that naxillin lowered the substrate specificity of other enzymes with hydratase activity, thereby ‘recruiting’ other hydratases to promote the IBA-to-IAA conversion. In addition to IBR10 and ECH2, two multifunctional peroxisomal proteins with both enoyl-CoA hydratase and hydroxyacyl-CoA dehydrogenase activity, AIM1 and MFP2 (ref. 40), have been identified in Arabidopsis. A full IBA response requires IBR10, ECH2 and AIM1 (refs. 11,13), whereas naxillin depends on activity of AIM1 but not ECH2 and IBR10, suggesting that naxillin acts at the enoyl-CoA hydratase step and might indeed recruit enoyl-CoA hydratases such as AIM1, but not ECH2 and IBR10, to enhance the conversion of endogenous IBA to IAA. Notably, chromeceptin, a small molecule that shares the trifluoronated methylphenyl substructure with naxillin, interacts with a mammalian multifunctional protein (MFP2) implicated in peroxisomal β-oxidation43. Cells with silenced MFP2 expression showed reduced sensitivity to chromeceptin43, similar to aim1-1 mutants being unresponsive to naxillin. It is therefore tempting to speculate that naxillin might interact with multifunctional peroxisomal proteins such as AIM1 in Arabidopsis. Taken together, these observations suggest that naxillin acts as a regulator of IBA-to-IAA conversion at the level of enoyl-CoA hydration and leads to more efficient usage of the endogenous IBA pool to promote lateral root development via IBA-derived IAA.
The fact that IBA application did not enhance the development of lateral roots in IBA-to-IAA conversion mutants indicates that IBA does not act directly on lateral root development but requires conversion to IAA to exert its effect. Thus, the action of both IBA and naxillin eventually channels through the canonical auxin signaling pathway. This observation raises the question of how application of IBA or naxillin can be more selective toward lateral root development. Expression analysis of several genes involved in the peroxisomal β-oxidation of IBA to IAA demonstrated that these expression patterns overlap in the root cap, suggesting that this organ is an important site of IBA-to-IAA conversion. As naxillin acts in the IBA-to-IAA conversion pathway, its root-inducing effect might therefore rely on the supply of endogenous IBA to the root cap.
Recently, PEN3 has been suggested to be an IBA efflux protein10 that is localized on outward membranes of the epidermis and root cap cells. Remarkably, the pen3 mutant is more sensitive to naxillin, suggesting that reduced efflux, and thus increased endogenous IBA, allows more naxillin-dependent IBA-to-IAA conversion to occur. In this model, once IBA is converted to IAA, the IBA-derived IAA can enter the endogenous flow of auxin and be directed toward the basal meristem to elicit a local auxin response leading to root branching (Fig. 5d). It is tempting to speculate that such a mechanism, by which plants can make use of an extra source of auxin in the form of a precursor that is locally released as IAA in the root cap, could contribute to the adaptability of plants to respond to ever-changing soil conditions. This mechanism would allow the plant to modify plant root architecture by affecting root branching, with a minimal effect on other processes such as primary root elongation.
By promoting the root cap conversion of the endogenous auxin precursor IBA into free IAA, naxillin is to our knowledge the first synthetic molecule that acts on rooting by specifically affecting auxin homeostasis. Given the reduced side effects typical of auxin treatment, naxillin represents a valuable tool to further decipher the molecular networks involved in lateral root branching.
METHODS
Compound screening and growth conditions
A commercial 10,000-compound library (DIVERSet, ChemBridge Corporation) was screened for induction of pCYCB1;1::GUS expression in xylem pole pericycle cells. About three seeds of this marker line in Arabidopsis (L.) Heynh. Col-0 background were sown in 96-well filter plates (Multiscreen HTS MSBVS1210; Millipore) in liquid medium derived from standard Murashige and Skoog (Duchefa) medium and supplemented with 10 μM of the auxin transport inhibitor naphthylphthalamic acid (NPA, Duchefa), resulting in a primary root devoid of lateral roots and allowing synchronization of lateral root development. Subsequently, seeds were incubated in a growth chamber under continuous light (110 μE m−2 s−1 photosynthetically active radiation, supplied by cool-white fluorescent tungsten tubes; Osram) at 22 °C. Three days after germination, the liquid NPA medium was removed and replaced with fresh liquid medium. Compounds were added to the 96-well plates to a final concentration of 50 μM for 24 h. Plants incubated in 2% (v/v) DMSO (Sigma) or 10 μM NAA (Sigma) were used as negative and positive control, respectively. Next, all plants were incubated in GUS buffer as described28 and analyzed for GUS staining in xylem pole pericycle cells. Only compounds that showed similar staining profiles in all seedlings were considered. For all subsequent phenotypic analyses, plants were grown on square plates (Greiner Labortechnik) with solid medium derived from standard MS medium under the same conditions supplemented with compounds dissolved in DMSO as described previously44.
Naxillin, derivatives and other compounds
Confirmed activator hit molecules were purchased from ChemBridge Corporation (ID for A1: 5627285, A5: 5263355, A9: 5467678, A11: 5853934, naxillin (A12): 5856819, A13: 5919797, A14: 6389571, A15: 6142645, A16: 6519229, var2: 5889244), Asinex (var1: BAS00126433), Sigma (var3: 526673, var4: 12810) or Vitas-M Lab (var5: STK096837). Large quantities of naxillin or (E)-2-((5-(3-(trifluoromethyl)-phenyl)furan-2-yl)methylene)hydrazine-carbothioamide were synthesized according to the procedure shown in ref. 45. We dissolved 5-[3-(trifluoromethyl)-phenyl]furfural (4.0022 g, 16.65 mmol, 1 equiv.) and thiosemicarbazide (1.5207 g, 16.5 mmol, 1 equiv.) in anhydrous methanol under argon and refluxed the solution for 16 h. The solvent was removed in vacuo, and the crude product was recrystallized from hexanes in ethyl acetate to yield the product as a yellow solid (4.2733 g, 13.65 mmol, 83% purity): melting point 191.2–192.0 °C; 1H NMR (d6-DMSO): 11.56 (s, 1H), 8.32 (s, 1H), 8.19–8.14 (m, 2H), 8.00 (s, 1H), 7.87 (s, 1H), 7.67–7.63 (m, 2H), 7.34 (d, J = 2.5, 1H), 7.10 (d, J = 2.5, 1H);13C NMR (d6-DMSO): 177.8, 152.8, 149.8, 131.7, 130.5, 130.1, 129.9 (q, JC-F = 31.8), 127.7, 124.3 (q, JC-F = 3.7), 124.0 (q, JC-F = 72.5), 120.2 (q, JC-F = 3.7), 115.2, 110.0; IR (KBr) 3,413 cm−1, 3,233 cm−1, 3,152 cm−1, 1,600 cm−1. High-resolution mass spectra (ESI) calculated for C13H10F3N3NaOS [M + Na]+, 336.0389; found, 336.0372.
Dose-response analysis
Five-day-old plants germinated on solid medium were transferred to solid medium plates supplemented with several concentrations of the indicated compounds and grown for five additional days, and emerged lateral roots were counted.
Pull-down experiments
Aux/IAA domain II peptides were purchased from Thermo Electron, and pull-down experiments were done as described previously35.Peptide data: mass calculated for the peptide by MS: 2,209.6, experimental 2,210.4; HPLC: >91%.
IBA-to-IAA flux experiment
Col-0 Arabidopsis seeds were surface sterilized, vernalized at 4 °C for 2 d and sown on 40-μm nylon mesh. After allowing the seedlings to grow with the plates in a vertical position for 5 d, the nylon mesh containing seedlings was transferred to growth medium containing either 10 μM [13C1]IBA or 10 μM [13 C1]IBA and 25 μM naxillin and incubated for 2 h. The root tips (5- to 8-mm segment) from 80–100 seedlings were harvested, flash frozen in liquid nitrogen and stored at −80 °C until free IAA was extracted and analyzed. The synthesis of [13C1]IBA and the solid phase extraction and gas chromatography–selected reaction monitoring-MS analysis of free IAA was performed as described previously46. The quinolinium ions with m/z 130 produced from the molecular ions m/z 189 were monitored for unlabeled IAA, and the quinolinium ions with m/z 131 produced from the molecular ions with m/z 190 were monitored for [13C1]IAA. The enrichment of [13C1]IAA derived from [13C1]IBA was determined as the increase in the ratio of m/z 131 relative to m/z 130. The natural abundance of [13C1]IAA was determined using the unlabeled IAA standard (Sigma) and is corrected for in the data presented.
EMS screening and positional cloning
About 30,000 seedlings from 21 EMS-mutagenized pools were germinated on standard MS medium. To exclude effects on germination, these plants were subsequently transferred to 25 μM naxillin 3 d after germination. Plants resistant to the lateral root–inducing effect of naxillin were selected after 5 more days. Before positional cloning, mutants were back-crossed to Col-0 plants and selected again for the resistant phenotype. For PCR-based positional cloning using simple sequence length polymorphism markers, the mutant was crossed with Ler plants and subsequently selfed. Forty-five resistant F2 seedlings were used to map the mutation to chromosome 3 between BAC NGA172 (0.7 Mb) and F17A9 (2.4 Mb). Ninety more F2 seedlings were used to fine map the mutation to a 300-kb interval between F24P17 (1.94 Mb) and F17A9 (2.24 Mb) containing 88 genes. A candidate gene approach and sequencing were used to identify a new point mutation at the start of exon 16 of the IBR3 gene, resulting in an arginine-to-tryptophan substitution. Primers used for mapping and sequencing are listed in Supplementary Table 2.
Cloning and construction of expression vectors
Donor vectors were created by cloning 2-kb promoter sequences for IBR1 and IBR10 into pDONR221. Expression clones were constructed by cloning the required fragments into pK7GWFS7 (ref. 47,48). We cloned 2-kb promoter sequences for IBR3, AIM1 and MFP2 using ligation-independent cloning as described into the pGIIB-LIC-n3GFP-NOSt vector49. Primers used are listed in Supplementary Table 2.
Histochemical and histological analysis and microscopy
The GUS assays were performed as described previously20. For microscopic analysis, samples were cleared by mounting in 90% (v/v) lactic acid (Acros Organics). All samples were analyzed by differential interference contrast microscopy (BX51, Olympus). For anatomical sections, GUS-stained samples were fixed overnight and embedded as described previously20. Fluorescence imaging of roots was performed with an Axiovert 100M confocal laser scanning microscope with software package LSM 510 version 3.2 (Zeiss). For excitation of GFP, the 488-nm line of an argon laser was used.
Supplementary Material
Acknowledgments
We thank J. Cohen and J. Normanly for helpful discussions, and E. Feraru and M. Feraru for help with mapping the nar1 mutant. This work was supported by the Interuniversity Attraction Poles Programme (IUAP VI/33) initiated by the Belgian State Science Policy Office, the Special Research Fund of Ghent University, a long-term Federation of European Biochemical Societies fellowship (pre- and post-doctoral fellowships to B.D.R.), the Robert A. Welch Foundation (C-1309 to B.B.), the US National Institutes of Health (R00-GM089987-03 to L.C.S.), the US National Science Foundation (DBI-1039655 to P.J.O., R.H. and R.B.; DBI-0923960 to P.J.O. and R.H.) and the Research Foundation Flanders (FWO, research project 3G002911). S.V. is a postdoctoral fellow of the Research Foundation-Flanders. D.A. is a postdoctoral fellow of the FWO. D.A. and L.N. are part of the VIB Compound Screening Facility.
Footnotes
Additional information. Details on microarray analysis, statistical data processing and calculating over-representation are listed in Supplementary Methods.
Author contributions
B.D.R., D.A., L.N. and L.J. performed chemical genetics screening; X.L. and P.O. performed the IBA-to-IAA conversion experiment; P.O., R.H. and R.B. synthesized naxillin; B.P. performed statistical analysis on data sets; M.F.N. helped with sectioning; A.G. and I.A.G. performed enzyme activity assays; S.K. performed in vitro pull-down experiment; B.D.R., D.A. and W.X. performed all other experiments; B.D.R., D.A., P.O., S.V., L.C.S., B.B., D.I. and T.B. conceived experiments and wrote the paper.
competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information and chemical compound information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 3.De Rybel B, Audenaert D, Beeckman T, Kepinski S. The past, present, and future of chemical biology in auxin research. ACS Chem. Biol. 2009;4:987–998. doi: 10.1021/cb9001624. [DOI] [PubMed] [Google Scholar]
- 4.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Gray WM, Muskett PR, Chuang HW, Parker JE. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilfoyle TJ, Hagen G. Auxin response factors. Curr. Opin. Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruzicka K, et al. Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc. Natl. Acad. Sci. USA. 2010;107:10749–10753. doi: 10.1073/pnas.1005878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strader LC, Bartel B. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell. 2009;21:1992–2007. doi: 10.1105/tpc.109.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strader LC, et al. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell. 2011;23:984–999. doi: 10.1105/tpc.111.083071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- 13.Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strader LC, Culler AH, Cohen JD, Bartel B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010;153:1577–1586. doi: 10.1104/pp.110.157461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-Dichloro-phenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics. 2008;180:237–251. doi: 10.1534/genetics.108.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zolman BK, Nyberg M, Bartel B. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 2007;64:59–72. doi: 10.1007/s11103-007-9134-2. [DOI] [PubMed] [Google Scholar]
- 18.Péret B, et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 19.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 20.De Rybel B, et al. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski M, et al. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6:e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Risueno MA, et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 24.Swarup K, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- 25.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 26.Himanen K, et al. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Himanen K, et al. Transcript profiling of early lateral root initiation. Proc. Natl. Acad. Sci. USA. 2004;101:5146–5151. doi: 10.1073/pnas.0308702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanneste S, et al. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell. 2005;17:3035–3050. doi: 10.1105/tpc.105.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science. 2003;301:1107–1110. doi: 10.1126/science.1084161. [DOI] [PubMed] [Google Scholar]
- 30.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 32.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 33.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 34.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 35.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 36.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 37.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:4561–4566. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arent S, Christensen CE, Pye VE, Norgaard A, Henriksen A. The multifunctional protein in peroxisomal β-oxidation: structure and substrate specificity of the Arabidopsis thaliana protein MFP2. J. Biol. Chem. 2010;285:24066–24077. doi: 10.1074/jbc.M110.106005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richmond TA, Bleecker AB. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell. 1999;11:1911–1924. doi: 10.1105/tpc.11.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rylott EL, et al. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment. Plant J. 2006;45:930–941. doi: 10.1111/j.1365-313X.2005.02650.x. [DOI] [PubMed] [Google Scholar]
- 43.Choi Y, et al. Chemical genetic identification of the IGF-linked pathway that is mediated by STAT6 and MFP2. Chem. Biol. 2006;13:241–249. doi: 10.1016/j.chembiol.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 44.De Rybel B, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du X, et al. Synthesis and structure-activity relationship study of potent trypanocidal thio semicarbazone inhibitors of the trypanosomal cysteine protease cruzain. J. Med. Chem. 2002;45:2695–2707. doi: 10.1021/jm010459j. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Cohen JD, Gardner G. Low-fluence red light increases the transport and biosynthesis of auxin. Plant Physiol. 2011;157:891–904. doi: 10.1104/pp.111.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karimi M, Depicker A, Hilson P. Recombinational cloning with plant gateway vectors. Plant Physiol. 2007;145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilson P, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Rybel B, et al. A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 2011;156:1292–1299. doi: 10.1104/pp.111.177337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.