Abstract
In depth proteomic analyses offer a systematic way to investigate protein alterations in disease and, as such, can be a powerful tool for the identification of novel biomarkers. Here, we analyzed proteomic data from a transgenic mouse model with cardiac-specific overexpression of activated calcineurin (CnA), which results in severe cardiac hypertrophy. We applied statistically filtering and false discovery rate correction methods to identify 52 proteins that were significantly different in the CnA hearts compared to controls. Subsequent informatic analysis consisted of comparison of these 52 CnA proteins to another proteomic dataset of heart failure, three available independent microarray datasets, and correlation of their expression with the human plasma and urine proteome. Following this filtering strategy, four proteins passed these selection criteria including: myosin heavy chain 7, insulin-like growth factor-binding protein 7, annexin A2, and desmin. We assessed expression levels of these proteins in mouse plasma by immunoblotting, and observed significantly different levels of expression between healthy and failing mice for all 4 proteins. We verified antibody cross reactivity by examining human cardiac explant tissue by immunoblotting. Finally, we assessed protein levels in plasma samples obtained from 4 unaffected and 4 heart failure patients and demonstrated that all four proteins increased between 2-fold and 150-fold in heart failure. We conclude that MYH7, IGFBP7, ANXA2, and DESM are all excellent candidate plasma biomarkers of heart failure in mouse and human.
INTRODUCTION
Cardiovascular disease represents a leading cause of morbidity and mortality globally. As such, there is a pressing need for new and innovative diagnostic tests to alleviate the burden associated with this disease. The progression to overt heart failure is complex and involves the interaction of various physiological, structural, and biochemical mechanisms. The diagnosis of heart failure is most frequently made at the time of presentation of symptoms; at a stage when the heart has exhausted all ability to compensate for the injury that it has sustained. At this late stage, the disease has progressed to such an extent that the patient often requires hospital admission. The initial hospital mortality in these patients is >15% and the one-year mortality rate is >30%, with a 60% risk of being readmitted to hospital with another episode of heart failure within the year. This clinical outcome and presentation contributes to the exceptionally high costs associated with treating these patients (1, 2). The therapeutic regimen for these patients is complex and often involves combinations of many medications that are administered over a heterogeneous spectrum of disease etiologies and severity. Therefore, it is clear that a major problem in determining effective management strategies for individual patients is the lack of predictive methods for establishing optimal patient-specific therapies (3–6).
Currently there are a limited number of clinically approved biomarkers available for the management of the entire spectrum of cardiovascular diseases. These markers include: serum cholesterol total/LDL, hemoglobin A1C (for diabetes), cardiac creatine kinase, troponins (I and T), and either brain natriuretic peptide (BNP) or its precursor N-terminus proBNP (NTproBNP). Of these markers, only BNP/NTproBNP has been validated for heart failure patients (7). There are, however, a number of limitations associated with the use of BNP as a marker of heart failure, including falsely high levels in the setting of advanced age, female gender, renal disease and acute coronary syndromes, and false low levels in the setting of obesity or flash pulmonary edema (8, 9). Additionally a major caveat in the use of natriuretic peptides in clinical settings is the so-called diagnostic ‘grey zone’, which consists of BNP levels in the range of 100 to 400 pg/ml. At these levels, it is not possible to determine if the individual is suffering from heart failure, and since a significant number of high-risk patients fall within this wide range (10, 11), BNP is not useful for their diagnosis. Consequently, BNP is used predominantly for diagnosing symptomatic heart failure at an advanced stage of disease.
In the present study, we set out to determine which proteins were overexpressed in heart failure. We used a mouse model of heart failure resulting from a constitutively active form of the CnA catalytic subunit in the heart (12). Phenotypically, these transgenic mice first exhibited cardiac hypertrophy, as early as 18 days, which then progressed with fibrosis of the ventricular wall, perivascular edema in the lungs, ventricular dilation and lethal heart failure (12). We employed a systematic approach for “discovery” using ventricular samples from these mice, an approach that has been used previously by other groups in order to monitor protein expression in various diseases (13–15) and assessed subsequently the expression of candidate proteins in mouse and human plasma. Informatic analyses and validations in small patient cohorts highlighted four candidate biomarkers of cardiac heart failure.
METHODS
Ventricular Sample Preparation
The animals were analyzed at 14 weeks of age since the CnA mice exhibited significantly increased anterior and posterior wall thicknesses, end diastolic dimension, and end systolic dimension, and this was associated with a concomitant decrease in fractional shortening; and at 24 weeks of age the CnA mice are phenotypically representative of end-stage heart failure (12, 16). At 14 and 24 weeks of age, wildtype and CnA transgenic mice were euthanized via CO2 asphyxiation.
Hearts were harvested and ventricular tissue was isolated, followed by thorough rinsing with ice-cold PBS to remove any remaining blood. Tissue was pooled from six mice as described previously (13, 17). Samples were placed in an ice-cold lysis buffer (250 mM, 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2. 1 mM DTT, 1 mM PMSF). Tissue was dounce-homogenized with 15 strokes using a loose fitting pestle, after which differential centrifugation was carried out to isolate cytosolic, microsomal, and mitochondrial fractions as described previously (13). Briefly, heart lysates were centrifuged at 800xg for 15min. Next, mitochondrial and microsomal fractions were isolated by centrifuging the supernatant at 8000xg and 100,000xg, respectively. Following the final spin, we isolated the resultant supernatant, which served as the cytosolic fraction. All procedures were performed as detailed in previous publications (13).
Mass Spectrometry and Informatics
Aliquots containing 100 μg of total protein from each fraction were precipitated and subsequently reduced and alkylated gel-free shotgun liquid chromatography was performed as described previously, and where the original mass spectrometry data was obtained (13, 16).. In the present study, the tandem mass spectra were searched using the SEQUEST database search algorithm (18) to match spectra to peptide sequences in a minimally redundant manner. Following this, a STATQUEST filtering algorithm (17) was used to assign confidence levels to each protein match. Only proteins with two high-confidence supporting spectra that matched to at least two unambiguous peptides were accepted.
In our study here, we first summed high confidence spectral counts for each protein across all subcellular fractions to determine protein abundance. Each experimental run had the potential of containing bias and noise; thus, to address this, we normalized the data. Here, data were separated into 100 bins on the basis of spectral count value distribution. Then we used LOWESS, locally weighted linear regression, to normalize each bin. This procedure divides the data into a number of overlapping intervals and fits using polynomial function of the form:
Next, to determine significance levels of each protein identified, an ANOVA test was carried out. Here, we generated two linear models; the first assessed both control and disease states as well as the two time-points (14 and 24 weeks) and subcellular localization (cytosol, microsome, mitochondria I, and mitochondria II), the second modeled only time and localization as established previously (13). The output from the two models was compared using the ANOVA test, with our null hypothesis being that there is no difference between the two models. A low p-value suggests that there is a difference between the two models, indicating that the disease state is involved in the differential protein expression seen.
Following assignment of p-values, we subjected a subset of proteins with nominal p-values < 0.05, to a false discovery rate (FDR) correction using the Benjamini-Hochberg calculation. This analysis led to the discovery of a subset of 52 proteins meeting these criteria. This modeling allowed us to make direct comparisons against our earlier proteomic studies, which used the similar calculations and statistically filtering (13).
This subset of 52 proteins was subsequently mapped against databases including: another model of mouse heart failure originating from a phospholamban point mutation (13); the human urinary Proteome (19), and the ‘high confidence’ plasma proteome (20), to provide insight into the clinical utility of these proteins. We also cross referenced our data against available Gene Expression Omnibus (GEO) microarrays including: isoproterenol-induced cardiomyopathy in the mouse (GSE18801) (21); analysis of mouse heart left ventricles with pressure overload LV hypertrophy caused by transverse aortic constriction (GSE2459) (22); and microarray analysis of alpha-tropomyosin mutant models of familial cardiac hypertrophy (GSE4678) (23). We chose these datasets since they represent three consistent models of mouse hypertrophy. Samples were analyzed on the Affymetrix U74A version 2 microarray, Affymetrix Mouse Genome 430 version 2 microarray, or Affymetrix Mouse Expression 430A microarray for GSE2459, GSE18801, GSE4678, respectively. Probe IDs were converted to common gene names for comparison between different microarray chips and probe intensities were normalized. There were three biological replicates of the hypertrophy model in GSE18801, a gene was considered for further comparison if it had a detected p-value of less than 0.05 in at least two out of the three replicates, and the same approach was applied to GSE4678, in order to confidently ensure presence over background. In GSE2459, five out of six replicates with a detected p-value of less than 0.05 were considered for comparison to the control. The ratio of diseased to control were used for comparison to avoid problems associated with probe intensity and chip to chip variations.
Assessment of Protein Abundance in Mouse Plasma
Wild-type and CnA mice were CO2 asphyxiated at 14 weeks, and ~1ml of blood was taken from the hearts and immediately placed on ice. Plasma was collected in microtainers with K2EDTA (BD Falcon), and centrifuged at 900g for 30 minutes. The supernatant consisted of plasma, and was stored at −80°C. Plasma was later subjected to albumin removal using albumin-depletion columns (Calbiochem). Here, 75μl of plasma was added to 750μl of binding buffer. Columns were prewashed with 750μl of fresh binding buffer. Diluted samples were then applied to the columns, and the 1.5ml flow through were collected and diluted with one final 750μl washing step. The resulting 2.25ml albumin depleted samples were collected and stored at −20°C. These samples were later concentrated using Amicon Ultra 4 centrifugal filter devices (Millipore). Briefly, albumin depleted samples were centrifuged at 2500g for 30mins at 4°C. The resultant samples were quantified and 25μg of protein was subjected to immunoblot analysis. CnA heart lysate and albumin enriched samples were used as controls in immunoblot experiments. Also, GAPDH levels were assessed in the following samples: albumin depleted samples, albumin enriched samples, and immunoglobin enriched samples (results not shown).
Assessment of Cross-Reactivity of Antibodies in Human Cardiac Explants
Tissue from human cardiac explants was obtained from patients who had undergone cardiac transplantation for advanced heart failure secondary to dilated cardiomyopathy, and their collection and use were approved by the Research Ethics Board of the University Health Network, Toronto, Ontario. Left ventricular samples were subjected to differential centrifugation using a benchtop centrifuge according to our cellular fractionation procedure (13) for 30min. In this study, we used the cytosolic, soluble ventricular fraction to assess the cross-reactivity of our antibodies.
Assessment of Protein Abundance in Human Plasma
Patients with symptomatic NYHA class III/IV heart failure and elevated levels of BNP were identified through the Canadian Heart Failure Network at the Toronto General Hospital. After obtaining informed consent, blood samples were obtained from patients with heart failure, as well as healthy individuals over the age of 35. Clinical data was available for patients, whereas echocardiography was not available for the healthy cohort. Maxi Albumin/IgG removal columns were used to deplete these plasma samples (Calbiochem). Columns were first washed with 1X Binding Buffer. Next, 100μl of plasma was diluted with 10X Binding Buffer. Diluted samples were added to the pre-washed column. Depleted samples were then diluted by allowing 2ml of 1X Binding Buffer to flow through. The resultant samples were concentrated as previously described and stored at −20°C until further use. Quantification of proteins of interest was performed and 25μg of protein was subjected to immunoblot analysis. As controls, CnA heart lysates were assessed as well (results not shown). We also attempted to assess protein levels in the samples that were not subjected to albumin/IgG depletion since proteins may bind to albumin and become depleted during the albumin removal. However, in these cases, we were not able to observe any specific bands aside from quite extensive IgG/albumin contamination throughout the entire membrane. As such, we focused on the albumin/IgG depleted samples.
Human Protein Atlas (proteinatlas.org)
To examine the subset of proteins we identified, we mined the Protein Atlas confocal database for protein expression in human cardiac sections for DESM, MYH7, IGFBP7 and ANXA2. Protein Atlas version 10.0 (released 2012.09.12) was used (24–28). For this analysis, we were able to find cardiac sections from three patients (Patient IDs # 2224, 2523 and 3732 with staining against the four markers. Antibodies used in the Protein Atlas were: MYH7 (HPA001239) rabbit polyclonal from Sigma-Aldrich; DESM (CAB000034) mouse monoclonal from DAKO; IGFBP7 (HPA002196) rabbit polyclonal from Sigma Aldrich; IGFBP7 (CAB020668) rabbit polyclonal from SDIX (Newark, Delaware); and ANXA2 (CAB004311) mouse monoclonal from Santa Cruz Biotech. The cancer expression correlations were made using the available cancer pathology database of the Atlas (25, 26).
RESULTS
Establishment of the High-Confidence Candidate Dataset
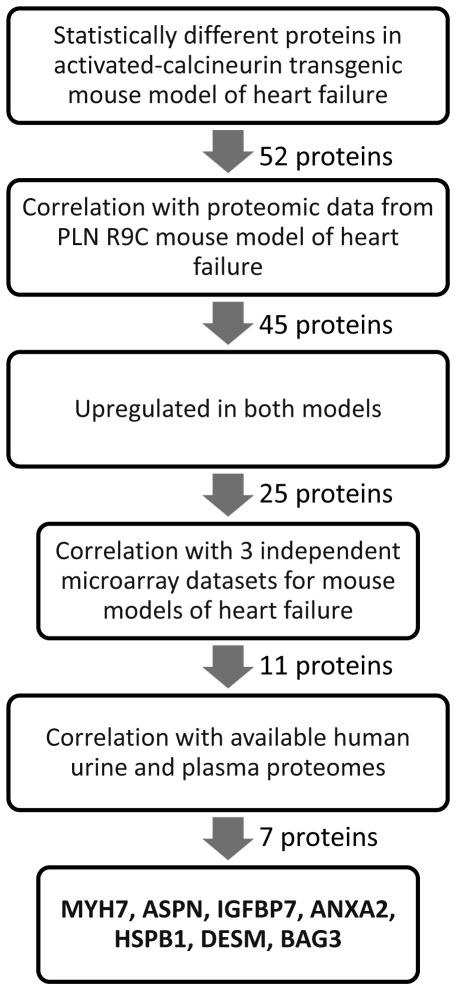
Our large-scale proteomic study investigating protein changes in cardiac hypertrophy uncovered 5397 proteins in WT and CnA ventricular samples (16). These proteins were reanalyzed using a linear statistical model to determine levels of statistical confidence of each match as described previously (13). Two models were constructed with the only difference being the control and disease state. This analysis revealed 474 nominally significant proteins (p<0.05), with 286 up-regulated, and 188 down-regulated proteins in CnA mice. Following this, a false discovery rate was incorporated to minimize the likelihood of errors in our analysis. The FDR calculation led to a final list of 52 proteins that had a corrected p-value of <0.05, with 29 proteins up-regulated with 23 down-regulated in the CnA mouse heart (Fig 1).
Figure 1. Correlation of Proteomic Dataset with Other Models of Cardiomyopathy.
Proteomics data for the top 52 protein candidates in the CnA model was compared to the R9C model of dilated cardiomyopathy. Twenty five proteins showed concordance with up-regulation in both CNA and R9C, which were further compared to GEO microarrays: isoproterenol-induced cardiomyopathy and exercise-induced cardiac hypertrophy (GSE18801), left ventricular hypertrophy model (GSE2459), and microarray analysis of gene expression during mild and severe cardiac hypertrophy (GSE4678).
Cross-Referencing with Available Databases
To aid in the selection of possible candidates, we next examined the list of 52 proteins and correlated them with our existing heart failure proteome dataset (13) and with the available microarray datasets. The 52 proteins could be mapped against 45 proteins in the R9C database; seven of the CnA proteins were not detected in the R9C data. Twenty-five of the convergent proteins showed consistent trends of upregulated expression (Fig 1). These 25 linked proteins were then analyzed against three microarray datasets at the gene name level as well as compared against published human urinary proteome (19), and the ‘high-confidence’ human plasma proteome (20). Of the 25, we were not able to detect expression of 14 of these candidates in either the arrays or the proteomes. However, for the remaining 11, we were able to identify upregulated mRNA levels for all of these candidates, and seven of these proteins were found in the urinary and/or the plasma proteome. These seven proteins that passed all these correlation criteria were: IGFBP7, ANXA2, MYH7, ASPN, HSPB1, DESM, and BAG3, ie. these proteins were found to be present in both heart failure proteomes, upregulated in the independent microarrays and expressed in the urine proteome, the plasma proteome, or both. A schematic of the entire workflow is presented in Fig 2.
Figure 2. Schematic Diagram Describing Protein Target Selection.
Protein expression from the CnA dataset was mapped against the PLN R9C dataset, a model of dilated cardiomyopathy. Forty-five proteins were present in both datasets. Of these 45 proteins, 25 were upregulated in both conditions, and these proteins were compared subsequently to 3 independent microarray datasets of heart failure. Eleven proteins showed up-regulation in at least one of the microarrays, and these were further mapped to the urinary proteome and high-confidence plasma proteome. Seven proteins were present in either the urinary or plasma proteome. Intensity of the bands is indicated by the color bar, N/D indicates the gene/protein was not detected in the dataset.
Expression in Mouse Plasma
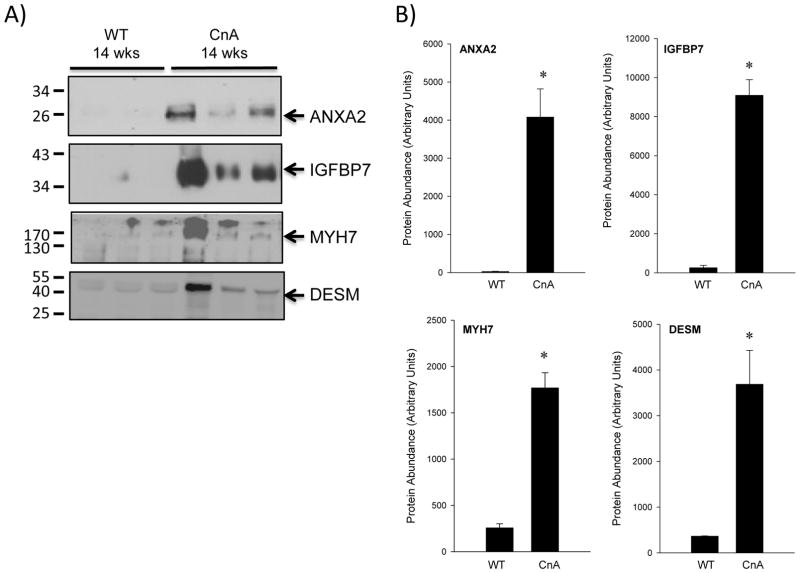
Since we had determined expression levels of many of these proteins in cardiac tissues previously (16), our first set of experiments here was to determine whether these highly ranked candidates were expressed in the plasma of the mouse. Although BAG3 and HSPB1 filtered through our criteria, we did not pursue them given that they are abundant heat shock response proteins and have been shown to be affected in numerous other non-cardiac conditions where they are commonly up-regulated to prevent the aggregation of newly translated polypeptides. We did not investigate ASPN further as we were not able to obtain a specific antibody against ASPN. Accordingly, we investigated the expression levels of ANXA2, IGFBP7, MYH7, and DESM. For these studies, we used plasma obtained from wild-type and CnA transgenic mouse blood. We included a positive control of the CnA mouse heart lysate since that was where we originally identified the protein (results not shown). In these studies, ANXA2 and IGFBP7 showed very low levels/absent expression in wild-type samples (26±8 A.U. and 262±116 A.U., respectively), while MYH7 and DESM exhibited a higher level of expression in the wild-type, 258±43 and 366±8, respectively. However, all 4 proteins were shown to increase significantly in the CnA plasma samples (Figure 3B). ANXA2 levels increased to 4079±739 (151-fold over WT) and IGFBP7 to 9095±799 (35-fold over WT). More modest increases were observed for MYH7 (1771±163; ~7-fold over WT) and DESM (3690±739; ~10-fold over WT). Statistical evaluation of expression patterns revealed significant (P<0.05) overexpression of all candidates (ANXA2, IGFBP7, DESM, and MYH7).
Figure 3. Assessment of Protein Expression in Mouse Plasma.
Expression of ANXA2, IGFBP7, MYH7, and DESM was evaluated in mouse plasma. (A) Three control samples and three CnA samples were assessed by immunoblot. As a positive control, CnA heart lysate was included (not shown). Molecular weights are indicated on the left. (B) Following immunoblot analysis, quantification of signals at the appropriate molecular weights was. Shown are densities obtained in the WT and CnA samples and are represented in arbitrary units. Mean and standard error of the mean (SEM) are reported; *, p<0.05, t-test.
Cross-Reactivity in Human Cardiac Tissue
To test whether the antibodies we used would cross-react against the corresponding human protein, we isolated human cardiac tissue along with mouse WT and diseased cardiac tissue, and performed immunoblotting to determine if we could detect these 4 proteins. In these immunoblot studies, we were able to visualize the correct molecular weight proteins with the antibodies in both mouse and human cardiac tissues (Fig 4), and thus confirmed cross reactivity, along with confirming the upregulation of protein levels in disease mouse hearts (Fig 4).
Figure 4. Antibody Cross-Reactivity in Human Tissue.
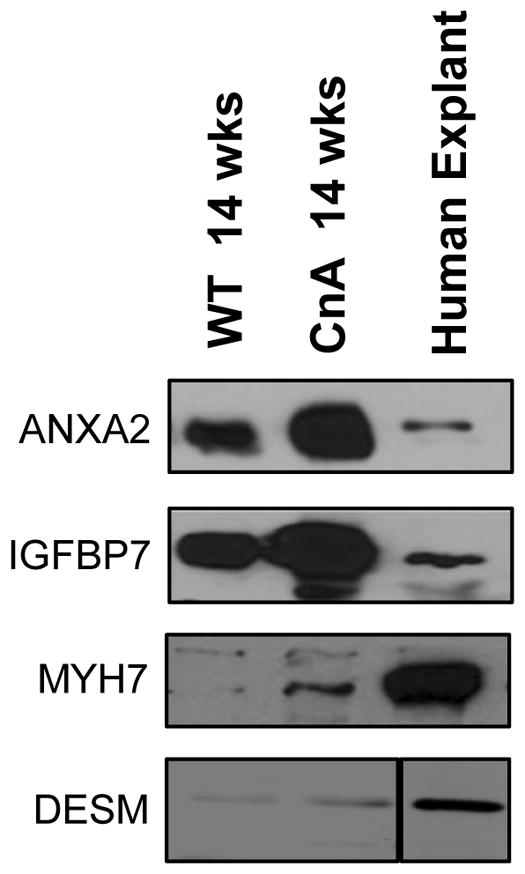
The cross-reactivity of antibodies used in mouse samples was tested in human explants obtained from dilated cardiomyopathic patients. Shown are representative immunoblots of these human cardiac tissues along with ventricular tissues obtained from wild-type and CnA mice.
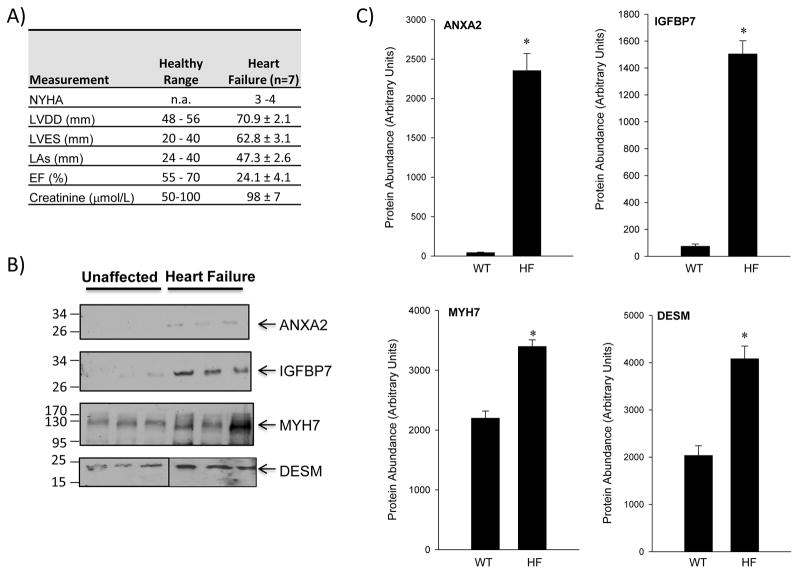
Expression in Human Plasma
The expression levels of ANXA2, IGFBP7, and MYH7 and DESM were studied in plasma samples taken from patients who had heart failure (class III/IV), along with high levels of BNP, (Fig 5A). Similar to the mouse plasma analyses, ANXA2 and IGFBP7 showed very low levels of expression in unaffected samples (44±6 and 75±17 A.U., respectively), whereas MYH7 and DESM showed higher expression with 2200±118 and 2037±207 A.U., respectively (Figure 5B and C). In the heart failure patient plasma samples, ANXA2 levels were 2354±218 (54-fold over unaffected) and IGFBP7 levels were 1505±98 (21-fold over WT). MYH7 levels were 3398±111 (~1.5-fold over unaffected) and DESM levels were increased to 4083±269 (~2-fold over unaffected). Statistical evaluation of expression patterns revealed significant (P<0.05) changes in expression for all candidates (ANXA2, IGFBP7, DESM, and MYH7).
Figure 5. Protein Expression in Human Plasma Samples.
(A) Immunoblots of ANXA2, IGFBP7, MYH7 and DESM levels were assessed in unaffected and heart failure patient samples. (B) Quantification of proteins targeted in human plasma was performed to evaluate expression levels. Shown are band densities represented in arbitrary units. Mean and SEM are reported; *, p<0.05, t-test.
Expression in Human Cardiac Sections
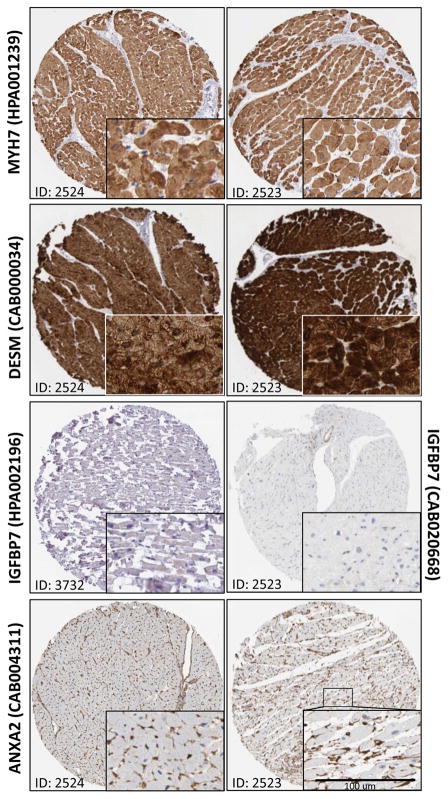
We examined protein expression patterns in human cardiac sections for DESM, MYH7, IGFBP7 and ANXA2 using the Protein Atlas confocal database (Fig 6). A high level of cardiac staining was observed for both MYH7 and DESM in two separate patient biopsies (Fig 6, upper panels). Higher magnification images show clear expression of both proteins that appears to be restricted to the cardiomyocytes. By contrast, staining of IGFBP7 using two separate antibodies failed to demonstrate a sufficient degree of stain in either section. ANXA2 was present in both patient cardiac biopsies, although it appears to be restricted to the endothelial cell lining of the vascular beds (Fig 6, lower panel).
Figure 6. Protein Expression in Human Cardiac Ventricular Sections.
Immunohistochemistry validation using the Human Protein Atlas database was performed and representative images were obtained for MYH7 (antibody HPA001239), DESM (antibody CAB000034), IGFBP7 (antibodies HPA002196 and CAB020668), and ANXA2 (antibody CAB004311). Patient IDs of #2424, #2423 and #3732 are shown. Inset, higher magnification images for sections. Scale bar is 100 μm.
Expression in Human Cancer Expression Profiles
Finally, we utilized the Protein Atlas to assess for potential specificity of these markers to a cardiac pathology. Notwithstanding the fact that caution needs to be used when interpreting large scale datasets such as the Atlas, pathology slides examining candidate protein expression in 20 different types of human cancer, including breast, colorectal, lung and prostate cancer, from 216 different patients, can be assessed by a qualitative antibody expression grading scale. This scale represents antibody labeling determined qualitatively to be either: Strong, Moderate, Weak, or Negative (25, 26). The overall cancer tissue staining statistics can be assessed and highlight the fraction of patient samples with the various grading scale. Specifically, DESM was found to be 100% Negative in all cancer patient sections analyzed, ie. it was never expressed in any of the cancer slides in the Protein Atlas (Table 1). In a consistent pattern, the results for MYH7 showed 1% of cancers showed ‘Strong’ staining: 1% ‘Moderate’, 6% ‘Weak’, and 92 % ‘Negative’. IGFBP7 was found expressed in: 5% Strong, 13% Moderate, 17% Weak, and 65% Negative. ANXA2 showed the greatest levels in cancer sections whereby: 8% of slides showed ‘Strong’ antibody staining, 62% had ‘Moderate’, and 22% showed ‘Weak’, with the remaining 8% classified as ‘Negative’.
Table 1. Cancer expression correlations using the cancer pathology database of the Protein Atlas.
Human pathology slides examining candidate protein expression in 20 different types of human cancer, including breast, colorectal, lung and prostate cancer, can be assessed by a qualitative antibody expression grading scale. The overall cancer tissue staining statistics over all sections was assessed and highlights the fraction of patient samples with the various grading scale. The overall cancer tissue staining statistics show the fraction of patient samples with either Strong, Moderate, Weak or Negative staining, using the available antibodies to the proteins (DESM, MYH7, IGFBP7 and ANXA2)
| Strong | Moderate | Weak | Negative | |
|---|---|---|---|---|
| DESM | - | - | - | 100% |
| MYH7 | 1% | 1% | 6% | 92% |
| IGFBP7 | 5% | 13% | 17% | 65% |
| ANXA2 | 8% | 62% | 22% | 8% |
DISCUSSION
Establishment of Biomarkers of Heart Failure
Taking into consideration the overall public health burden of advanced heart failure, there is an urgent need for new methods of diagnosing heart failure in the earlier stages, prior to requiring urgent care (29). In the present study, proteomic profiling during progression of heart failure resulting from cardiac hypertrophy offered insight into proteins associated with this disease. The complex interplay of pathological mechanisms during the progression of heart failure can be exploited to determine not only the presence of myocardial damage, but may also act as a prognostic indicator of heart failure (30, 31). For example, inflammatory pathways, myocardial injury and stress, matrix and cellular remodeling, oxidative stress pathways, neurohormonal activation, and the cardio-renal syndrome are all adverse processes that can provide insight onto the degree of damage to the myocardium in patients with heart failure (30). These proteins can potentially be used as pre-clinical diagnostic biomarkers for patients who may be susceptible to heart failure. Our informatics analysis led to the prioritization of 52 hypertrophy-associated protein candidates down to 4 proteins, including ANXA2, IGFBP7, MYH7, and DESM.
MYH7 is a slow molecular ATPase involved in muscle contraction (32). Our proteomic data indicated that MYH7 protein expression was upregulated ~19 fold in cardiac hypertrophy (Fig 1) (16). It is of interest that MYH7 shares considerable properties in common with another marker of myocardial injury, namely troponin I and T. Cardiac troponin I has been used clinically for many years as a marker of myocardial injury (33, 34) and shows an exceptionally high level of specificity. However, TnT and TnI are useful only in the myocardial infarction setting, and not in the diagnosis or management of heart failure. Cardiac troponins I/T are components of the cardiac contractile apparatus, similar to MYH7, and cardiac troponin I has been shown to be released from the myocyte via a necrotic pathway (35). Additionally, a recent study by Wen and colleagues has identified myosin light chain 2 (MLC2) and myosin heavy chain 11 (MYH11) to be circulating markers in Chagas Disease; a tropical parasitic disease where many patients develop cardiac dysfunction (36) Therefore, given the many similarities between these proteins, it is not entirely surprising that MYH7 was identified and found to be up-regulated in the cardiac patient plasma.
DESM is a cytoskeletal protein that was shown to be up-regulated in our model of heart failure likely as result of the large-scale myocyte architecture changes that occur in this disease. Desmin has been previously implicated as having a role in hypertrophic cardiomyopathy (HCM). For instance, DESM has been found to increase in cardiac tissues from transgenic mouse models of HCM (37) and in patients with end-stage idiopathic dilated heart failure (38) Our data showing an up-regulation of DESM in both cardiac tissue and in the plasma lends support to the model that this protein likely communicates mechanical stress signals from the extracellular matrix to the nucleus. Well described during DCM is an alteration in cell size, nucleus shape, and variations in contractile apparatus (39, 40). There could potentially be an increase in transduction of signals from the extracellular space to stress bearing filaments like DESM, within the cell, leading to an eventual increase of this protein in the circulation.
ANXA2 was another top candidate with increased levels in the cardiac tissue from the CnA mouse model of heart failure, with a very substantial up-regulation in the plasma of both mouse and human. The ANXA family of proteins are activated through Ca2+, enabling them to participate in organization of membrane domains and membrane recruitment platforms (41). As such, abnormal increases in Ca2+ within the cell may lead to an increase in the level of activity of ANXA2. Interestingly, several proteins in the ANXA family have been shown to be involved in cardiac disease. In their large-scale microarray analysis, Hwang et al. demonstrated in an increase in the expression of lipocortin, an annexin protein, in a model of dilated cardiomyopathy (42). This family of calcium handling proteins was also studied in patients with end stage heart failure. Indeed, protein expression of not only ANXA2, but also ANXA5 and ANXA6 was elevated in failing hearts compared to controls, indicating a correlation of these proteins in heart failure (43). Our data highlighting the ubiquitous expression of ANXA2 throughout most tissues, its presence in the vascular beds, and the fact that it is found upregulated in most cancer profiles suggests this protein has a clear role in the vascularization processes associated with cardiovascular remodeling and cancer progression. As such, it is quite likely to be present in the plasma of patients presenting with any diseases associated with vascular remodeling.
IGFBP7 was the final protein found to be upregulated in the plasma of mouse and human heart failure samples. Generally, the family of insulin binding proteins bind IGFs in the plasma and modulate their action by increasing their half-life or aiding in the delivery of these small proteins to the appropriate receptor (44). Insulin-like growth factors function by regulating a number of pathological states, including the promotion of abnormal cell proliferation or conversely the inhibition of cell death (45). Our proteomic findings demonstrated a ~6 fold increase in expression of this high confidence protein during cardiac hypertrophy, with nearly a 150-fold increase in its detection in human plasma from heart failure patients. It is of interest that a recent report indicated that IGFBP7 plasma expression was associated with human heart failure (46). In their study, Dinh et al. assessed 281 patients with normal ejection fraction compared to 78 patients with heart failure with a preserved ejection fraction, and showed that IGBP7 plasma levels were able to distinguish the patient groups. What is particularly important is that the levels of NT-ProBNP levels were not statistically different between these groups, highlighting the early predictive power of IGFBP7 over NT-ProBNP. (46)
To improve the clinical diagnosis of heart failure, it is essential to develop single biomarkers or multiple marker panels that will either improve upon BNP/NTproBNP, or compensate for its deficiencies. Ideally, new generations of biomarkers will identify patients who are at risk for developing heart failure, enabling the implementation of preventive strategies prior to the development of symptoms. Candidate patient groups that would most benefit from early diagnosis includes i) patients with ischemic heart disease following acute coronary syndromes, as up to 50% of this patient population is at risk of developing heart failure, however there is no indication as to which 50% will progress (47); ii) patients with a family history of heart disease and those suffering from hypertension or diabetes; and iii) patients that suffer from a new type of heart failure known as diastolic heart failure or heart failure with preserved ejection fraction. This disease is distinguished from other types of heart failure in that the heart retains a normal or smaller than normal chamber size and maintains a normal ejection fraction. Diastolic heart failure is rapidly emerging as one of the predominant forms of the disease, and has an equally poor clinical outcome (48, 49).
In conclusion, our study ultimately uncovered 4 proteins, ANXA2, DESM, MYH7, and IGFBP7, which were significantly increased in human patient plasma samples. Ultimately, additional follow-up studies examining: additional independent studies, the expression of these proteins in unique patient populations, and validation of their meeting a clinical utility in predicting patient outcomes or directing patient therapy are required.
LIMITATIONS
The inclusion of greater patient populations, examining at-risk patients without overt cardiac disease, and examining plasma-based expression of additional proteins might show improved specificity and sensitivity in detecting cardiac disease phenotypes at early stages of progression and highlight the degree of patient to patient variability. Additionally, we concentrated on the four proteins with available antibodies. However, current targeted mass spectrometry methods now allow for analysis of proteins without this requirement.
Acknowledgments
This project was funded by the Heart and Stroke Foundation of Ontario (T-6281 and NS-6636) to AOG; CIHR to AOG (MOP-106538); the Heart and Stroke Richard Lewar Centre of Cardiovascular Excellence; and the Ontario Research Fund - Global Leadership Round in Genomics and Life Sciences to AOG and PPL (GL2-01012). AOG was a New Investigator of the Heart and Stroke Foundation of Canada and is a Canada Research Chair in Cardiovascular Proteomics and Molecular Therapeutic.
Footnotes
Conflict of interest. Drs Liu and Gramolini hold a patent entitled “Use of IGFBP-7 in the assessment of heart failture”
References
- 1.Jong P, Gong Y, Liu PP, Austin PC, Lee DS, Tu JV. Care and outcomes of patients newly hospitalized for heart failure in the community treated by cardiologists compared with other specialists. Circulation. 2003;108:184–191. doi: 10.1161/01.CIR.0000080290.39027.48. [DOI] [PubMed] [Google Scholar]
- 2.Jong P, Vowinckel E, Liu PP, Gong Y, Tu JV. Prognosis and determinants of survival in patients newly hospitalized for heart failure: a population-based study. Arch Intern Med. 2002;162:1689–1694. doi: 10.1001/archinte.162.15.1689. [DOI] [PubMed] [Google Scholar]
- 3.Arnold JM, Liu P, Demers C, Dorian P, Giannetti N, Haddad H, Heckman GA, Howlett JG, Ignaszewski A, Johnstone DE, et al. Canadian Cardiovascular Society consensus conference recommendations on heart failure 2006: diagnosis and management. Can J Cardiol. 2006;22:23–45. doi: 10.1016/s0828-282x(06)70237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee DS, Mamdani MM, Austin PC, Gong Y, Liu PP, Rouleau JL, Tu JV. Trends in heart failure outcomes and pharmacotherapy: 1992 to 2000. Am J Med. 2004;116:581–589. doi: 10.1016/j.amjmed.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Ross H, Howlett J, Arnold JM, Liu P, O’Neill BJ, Brophy JM, Simpson CS, Sholdice MM, Knudtson M, Ross DB, et al. Treating the right patient at the right time: access to heart failure care. Can J Cardiol. 2006;22:749–754. doi: 10.1016/s0828-282x(06)70290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan AT, Yan RT, Liu PP. Narrative review: pharmacotherapy for chronic heart failure: evidence from recent clinical trials. Ann Intern Med. 2005;142:132–145. doi: 10.7326/0003-4819-142-2-200501180-00013. [DOI] [PubMed] [Google Scholar]
- 7.Doust JA, Glasziou PP, Pietrzak E, Dobson AJ. A systematic review of the diagnostic accuracy of natriuretic peptides for heart failure. Arch Intern Med. 2004;164:1978–1984. doi: 10.1001/archinte.164.18.1978. [DOI] [PubMed] [Google Scholar]
- 8.Doust J, Lehman R, Glasziou P. The role of BNP testing in heart failure. Am Fam Physician. 2006;74:1893–1898. [PubMed] [Google Scholar]
- 9.Cameron SJ, Green GB. Cardiac biomarkers in renal disease: the fog is slowly lifting. Clin Chem. 2004;50:2233–2235. doi: 10.1373/clinchem.2004.041566. [DOI] [PubMed] [Google Scholar]
- 10.Arnold JM, Howlett JG, Dorian P, Ducharme A, Giannetti N, Haddad H, Heckman GA, Ignaszewski A, Isaac D, Jong P, et al. Canadian Cardiovascular Society Consensus Conference recommendations on heart failure update 2007: Prevention, management during intercurrent illness or acute decompensation, and use of biomarkers. Can J Cardiol. 2007;23:21–45. doi: 10.1016/s0828-282x(07)70211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maisel A, Mueller C, Adams K, Jr, Anker SD, Aspromonte N, Cleland JG, Cohen-Solal A, Dahlstrom U, DeMaria A, Di Somma S, et al. State of the art: using natriuretic peptide levels in clinical practice. Eur J Heart Fail. 2008;10:824–839. doi: 10.1016/j.ejheart.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gramolini AO, Kislinger T, Alikhani-Koopaei R, Fong V, Thompson NJ, Isserlin R, Sharma P, Oudit GY, Trivieri MG, Fagan A, et al. Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Mol Cell Proteomics. 2008;7:519–533. doi: 10.1074/mcp.M700245-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Donahue MP, Rose K, Hochstrasser D, Vonderscher J, Grass P, Chibout SD, Nelson CL, Sinnaeve P, Goldschmidt-Clermont PJ, Granger CB. Discovery of proteins related to coronary artery disease using industrial-scale proteomics analysis of pooled plasma. Am Heart J. 2006;152:478–485. doi: 10.1016/j.ahj.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, Yin X, Fernandez-Caggiano M, Willeit P, Puntmann VO, Aldama-Lopez G, Shah AM, et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- 16.Bousette N, Chugh S, Fong V, Isserlin R, Kim KH, Volchuk A, Backx PH, Liu P, Kislinger T, MacLennan DH, et al. Constitutively active calcineurin induces cardiac endoplasmic reticulum stress and protects against apoptosis that is mediated by α-crystallin-B. Proc Natl Acad Sci U S A. 2010;107:18481–18486. doi: 10.1073/pnas.1013555107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 18.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 19.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 21.Galindo CL, Skinner MA, Errami M, Olson LD, Watson DA, Li J, McCormick JF, McIver LJ, Kumar NM, Pham TQ, et al. Transcriptional profile of isoproterenol-induced cardiomyopathy and comparison to exercise-induced cardiac hypertrophy and human cardiac failure. BMC physiology. 2009;9:23. doi: 10.1186/1472-6793-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirotsou M, Dzau VJ, Pratt RE, Weinberg EO. Physiological genomics of cardiac disease: quantitative relationships between gene expression and left ventricular hypertrophy. Physiological Genomics. 2006;27:86–94. doi: 10.1152/physiolgenomics.00028.2006. [DOI] [PubMed] [Google Scholar]
- 23.Rajan S, Williams SS, Jagatheesan G, Ahmed RP, Fuller-Bicer G, Schwartz A, Aronow BJ, Wieczorek DF. Microarray analysis of gene expression during early stages of mild and severe cardiac hypertrophy. Physiological Genomics. 2006;27:309–317. doi: 10.1152/physiolgenomics.00072.2006. [DOI] [PubMed] [Google Scholar]
- 24.Kampf C, Bergman J, Oksvold P, Asplund A, Navani S, Wiking M, Lundberg E, Uhlen M, Ponten F. A tool to facilitate clinical biomarker studies--a tissue dictionary based on the Human Protein Atlas. BMC medicine. 2012;10:103. doi: 10.1186/1741-7015-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 26.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nature Biotechnology. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 28.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005;4:384–393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Thom T, Haase N, Rosamond W, Howard VJ, Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O’Donnell C, Kittner S, et al. Heart disease and stroke statistics--2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;113:e85–151. doi: 10.1161/CIRCULATIONAHA.105.171600. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad T, Fiuzat M, Felker GM, O’Connor C. Novel biomarkers in chronic heart failure. Nat Rev Cardiol. 2012;9:347–359. doi: 10.1038/nrcardio.2012.37. [DOI] [PubMed] [Google Scholar]
- 31.Azuaje FJ, Dewey FE, Brutsaert DL, Devaux Y, Ashley EA, Wagner DR. Systems-based approaches to cardiovascular biomarker discovery. Circ Cardiovasc Genet. 2012;5:360–367. doi: 10.1161/CIRCGENETICS.112.962977. [DOI] [PubMed] [Google Scholar]
- 32.Kesidis N, Metaxas TI, Vrabas IS, Stefanidis P, Vamvakoudis E, Christoulas K, Mandroukas A, Balasas D, Mandroukas K. Myosin heavy chain isoform distribution in single fibres of bodybuilders. Eur J Appl Physiol. 2008;103:579–583. doi: 10.1007/s00421-008-0751-5. [DOI] [PubMed] [Google Scholar]
- 33.Wang TJ. Significance of circulating troponins in heart failure: if these walls could talk. Circulation. 2007;116:1217–1220. doi: 10.1161/CIRCULATIONAHA.107.721845. [DOI] [PubMed] [Google Scholar]
- 34.Horwich TB, Patel J, MacLellan WR, Fonarow GC. Cardiac troponin I is associated with impaired hemodynamics, progressive left ventricular dysfunction, and increased mortality rates in advanced heart failure. Circulation. 2003;108:833–838. doi: 10.1161/01.CIR.0000084543.79097.34. [DOI] [PubMed] [Google Scholar]
- 35.Wu AH, Ford L. Release of cardiac troponin in acute coronary syndromes: ischemia or necrosis? Clin Chim Acta. 1999;284:161–174. doi: 10.1016/s0009-8981(99)00078-9. [DOI] [PubMed] [Google Scholar]
- 36.Wen JJ, Zago MP, Nunez S, Gupta S, Burgos FN, Garg NJ. Serum proteomic signature of human chagasic patients for the identification of novel potential protein biomarkers of disease. Mol Cell Proteomics. 2012;11:435–452. doi: 10.1074/mcp.M112.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam L, Tsoutsman T, Arthur J, Semsarian C. Differential protein expression profiling of myocardial tissue in a mouse model of hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2009 doi: 10.1016/j.yjmcc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Pawlak A, Gil RJ, Kulawik T, Pronicki M, Karkucinska-Wieckowska A, Szymanska-Debinska T, Gil K, Lagwinski N, Czarnowska E. Type of desmin expression in cardiomyocytes - a good marker of heart failure development in idiopathic dilated cardiomyopathy. J Int Med. 2012;272:287–297. doi: 10.1111/j.1365-2796.2012.02524.x. [DOI] [PubMed] [Google Scholar]
- 39.Scholz D, Diener W, Schaper J. Altered nucleus/cytoplasm relationship and degenerative structural changes in human dilated cardiomyopathy. Cardioscience. 1994;5:127–138. [PubMed] [Google Scholar]
- 40.Mirza M, Marston S, Willott R, Ashley C, Mogensen J, McKenna W, Robinson P, Redwood C, Watkins H. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J Biol Chem. 2005;280:28498–28506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 41.Hayes MJ, Longbottom RE, Evans MA, Moss SE. Annexinopathies. Subcell Biochem. 2007;45:1–28. doi: 10.1007/978-1-4020-6191-2_1. [DOI] [PubMed] [Google Scholar]
- 42.Hwang JJ, Allen PD, Tseng GC, Lam CW, Fananapazir L, Dzau VJ, Liew CC. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics. 2002;10:31–44. doi: 10.1152/physiolgenomics.00122.2001. [DOI] [PubMed] [Google Scholar]
- 43.Benevolensky D, Belikova Y, Mohammadzadeh R, Trouve P, Marotte F, Russo-Marie F, Samuel JL, Charlemagne D. Expression and localization of the annexins II, V, and VI in myocardium from patients with end-stage heart failure. Lab Invest. 2000;80:123–133. doi: 10.1038/labinvest.3780016. [DOI] [PubMed] [Google Scholar]
- 44.Kim HS, Nagalla SR, Oh Y, Wilson E, Roberts CT, Jr, Rosenfeld RG. Identification of a family of low-affinity insulin-like growth factor binding proteins (IGFBPs): characterization of connective tissue growth factor as a member of the IGFBP superfamily. Proc Natl Acad Sci U S A. 1997;94:12981–12986. doi: 10.1073/pnas.94.24.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuvians TP, Gashaw I, Hasenfus A, Hacherhacker A, Winterhager E, Grobholz R. Differential expression of IGF components and insulin receptor isoforms in human seminoma versus normal testicular tissue. Neoplasia. 2005;7:446–456. doi: 10.1593/neo.04643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinh W, Barroso MC, Futh R, Lankisch M, Klein RM, Gulker J, Bufe A, Scheffold T, Nickl W, Seyfarth M. Serum insulin like growth factor-I and its binding protein-7: novel promising biomarker in heart failure with preserved ejection fraction. Clin Res Cardiol. 2012;101:P1288. [Google Scholar]
- 47.Flaherty JD, Bax JJ, De Luca L, Rossi JS, Davidson CJ, Filippatos G, Liu PP, Konstam MA, Greenberg B, Mehra MR, et al. Acute heart failure syndromes in patients with coronary artery disease early assessment and treatment. J Am Coll Cardiol. 2009;53:254–263. doi: 10.1016/j.jacc.2008.08.072. [DOI] [PubMed] [Google Scholar]
- 48.Bhatia RS, Tu JV, Lee DS, Austin PC, Fang J, Haouzi A, Gong Y, Liu PP. Outcome of heart failure with preserved ejection fraction in a population-based study. N Engl J Med. 2006;355:260–269. doi: 10.1056/NEJMoa051530. [DOI] [PubMed] [Google Scholar]
- 49.Owan TE, Hodge DO, Herges RM, Jacobsen SJ, Roger VL, Redfield MM. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N Engl J Med. 2006;355:251–259. doi: 10.1056/NEJMoa052256. [DOI] [PubMed] [Google Scholar]