Abstract
Vitamin B6 (pyridoxal-5'-phosphate, PLP) is linked to a variety of biological functions in prokaryotes. Here, we report that the pdxA (putative 4-hydroxy-L-threonine phosphate dehydrogenase) gene plays a pivotal role in the PLP-dependent regulation of flagellar motility, thereby altering host colonization in a leading foodborne pathogen, Campylobacter jejuni. A C. jejuni pdxA mutant failed to produce PLP and exhibited a coincident loss of flagellar motility. Mass spectrometric analyses showed a 3-fold reduction in the main flagellar glycan pseudaminic acid (Pse) associated with the disruption of pdxA. The pdxA mutant also exhibited reduced growth rates compared with the WT strain. Comparative metabolomic analyses revealed differences in respiratory/energy metabolism between WT C. jejuni and the pdxA mutant, providing a possible explanation for the differential growth fitness between the two strains. Consistent with the lack of flagellar motility, the pdxA mutant showed impaired motility-mediated responses (bacterial adhesion, ERK1/2 activation, and IL-8 production) in INT407 cells and reduced colonization of chickens compared with the WT strain. Overall, this study demonstrated that the pdxA gene affects the PLP-mediated flagellar motility function, mainly through alteration of Pse modification, and the disruption of this gene also alters the respiratory/energy metabolisms to potentially affect host colonization. Our data therefore present novel implications regarding the utility of PLP and its dependent enzymes as potent target(s) for the control of this pathogen in the poultry host.
Introduction
Campylobacter jejuni is a Gram-negative, spiral-shaped, microaerophilic bacterium that causes foodborne diarrheal illness worldwide [1], [2]. Recent epidemiological and biochemical studies have shown that Campylobacter infection is also implicated in neuropathies, including Guillain-Barré syndrome (GBS), through the production of autoantibodies induced by bacterial lipooligosaccharides [3], [4]. The chicken is the predominant natural host for this pathogen, through which Campylobacter can be transmitted to humans [5], [6], [7]. Thus, control of this pathogen in poultry habitats is associated with the global public health benefit of preventing human campylobacteriosis. However, how to control Campylobacter remains unresolved, mainly due to our lack of understanding regarding how this pathogen colonizes chickens and establishes persistent infections and how it is involved in human virulence [7]. An increased understanding of the molecular biology of Campylobacter would therefore provide valuable information for the development of therapeutic strategies and vaccines targeting this pathogen.
The differential expression of metabolic gene products in relation to pathogenesis has largely been left unexplored. However, the role of gene regulation in this phenomenon is now receiving more attention, as the metabolism of bacterial pathogens may hold important clues for understanding their life cycles and host defense mechanisms [8], [9]. Vitamin B6 (pyridoxal-5’-phosphate, PLP) is an essential metabolic cofactor with numerous functions in more than one hundred enzymatic reactions in humans [10], [11]. Among prokaryotes, the biosynthesis of PLP has been intensively studied, mainly in Escherichia coli, revealing the involvement of two pathways with seven enzymatic steps [12], [13]. For a number of years, it was tacitly assumed that such pathways are ubiquitous in all organisms. However, the biological importance of vitamin B6 for bacterial pathogenesis has only recently been thoroughly investigated in other microorganisms, including Mycobacterium tuberculosis [14], Bacillus subtilis [15], and the Campylobacter-related microorganism Helicobacter pylori [16]. In H. pylori, a study involving a pdxA mutant recently demonstrated an essential role of the pdxA gene in flagellation, likely through inactivation of the flagellin glycosylation process (decoration with pseudaminic acid) [16]. There is no evidence regarding how the pdxA gene affects the process of flagellin glycosylation in C. jejuni similarly. However, a previous biochemical analysis showed that a UDP-4-keto-6-deoxy-GlnNAc aminotransferase (Cj1294) derived from C. jejuni generates UDP-4-amino-4,6-dideoxy-alNAc with the catalytic support of PLP as a co-factor under in vitro conditions [17], which led us to the assumption that PLP biosynthesis also affects flagellation and certain types of metabolism in this pathogen, thereby altering bacterial fitness and in vivo colonization, for which flagellar motility is a prerequisite [18].
Given this background, we studied the PLP biosynthesis pathway in C. jejuni through in silico prediction and mutagenesis analyses. Biochemical and phenotypic analyses showed that the lack of the pdxA gene abolished PLP production and impaired the ability of C. jejuni to form flagella. We then focused on this mutant to characterize its biological effect(s) on host colonization through biochemical, metabolomic, and host infection approaches.
Materials and Methods
Bacterial strains and media
The bacterial strains and plasmids used in this study are listed in Table 1. C. jejuni strain 81–176 [19] was grown using routine methods in Mueller-Hinton (MH) broth or on MH agar (Becton-Dickinson, Franklin Lakes, NJ, USA) at 37°C for 24 h in a humidified CO2 AnaeroPack-Microaero gas system (Mitsubishi Gas Chemicals, Tokyo, Japan). The media were supplemented with chloramphenicol (Cm) (20 μg ml−1) or kanamycin (Km) (30 μg ml−1), as appropriate. The E. coli DH5α strain, which was used as the host for subcloning and routine DNA manipulation, was grown in LB agar or LB broth unless otherwise indicated.
Table 1. Bacterial strains and plasmids used in this study.
| Name | Description | Source/Reference |
| Bacterial strain | ||
| WT | C. jejuni wild-type (WT) strain 81–176 | [19] |
| pdxA− | C. jejuni 81–176 pdxA (CJJ81176_1253) mutant | This study |
| flaA− | C. jejuni 81–176 flaA (CJJ81176_1339) mutant | This study |
| pdxA-/+ | pdxA- strain complemented with pRY-pdxA-Km | This study |
| DH5α | E. coli strain for DNA manipulation | Sigma-Aldrich |
| Plasmid | ||
| pRY108, pRY109 | Cm- or Km-resistant C. jejuni/E. coli shuttle vector | [21] |
| pGEM-pdxA-Cm | pGEM::pdxA-cat for homologous recombination | This study |
| pGEM-flaA-Cm | pGEM::flaA-cat for homologous recombination | This study |
| pRY-pdxA-Km | pRY109::pdxAJ for complementation | This study |
Construction of C. jejuni mutants and complementation of the pdxA mutant
The 81–176 mutant, in which most of the pdxA or flaA genes were replaced with a cat cassette (encoding a Cm-resistance protein), was constructed as described previously [20]. To construct a pdxA mutant, a 500-bp fragment upstream of the 5′ end and a 500-bp fragment downstream of the 3′ end of the pdxA locus were amplified from the wild-type (WT) strain via PCR using either pdxA-s and pdxA-as-BI or pdxA-s-BI and pdxA-as primers (Table S1). After BamHI digestion, the two fragments were ligated and cloned into pGEM-T vector (Promega, Madison, WI, USA). A cat gene from the plasmid pRY109 [21] was then inserted into the BamHI site in the pGEM-T plasmid, and this allelic exchange plasmid (pGEM-pdxA-Cm, Table 1) was introduced into the genome of strain 81–176 through natural transformation [22]. Successful transformants were selected on MH agar supplemented with 5% horse blood and Cm (20 μg ml−1). Allelic replacement was confirmed via nucleotide sequencing. Disruption of the flaA gene was performed in the same manner (the oligonucleotide primers used in these procedures are listed in Table S1). The pdxAJ locus and the upstream region predicted by the Neural Network Promoter Prediction program (http://www.fruitfly.org/seq_tools/promoter.html) to contain −35 and −10 promoter binding sites were amplified via PCR using the pdxA-CF and pdxA-CR primers (Table S1). The resultant PCR fragments were cloned into the XbaI/EcoRI sites of the pRY108 plasmid [21], yielding pRY-pdxA-Km (Table 1). This plasmid was introduced into the pdxA mutant strain through natural transformation, and the transformants were recovered on MH agar containing Km (10 µg ml−1) and Cm (20 µg ml−1). The construction of this pdxA−/+ mutant strain was confirmed via PCR using the pdxA-conF and pdxA-conR primers (Table S1).
Quantification of PLP
Bacteria were grown microaerobically in 10 ml of MH broth to mid-logarithmic phase (an OD600 of 0.6), and crude homogenates were prepared in 20 mM Tris-HCl (pH 7.4) via bead crushing. After centrifugation for 10 min at 7,000 rpm at 4°C, the PLP contents in 50 µg of protein of the lysate and serial dilutions of the lysate were measured using a vitamin B6 ELISA kit (Uscn Life Science, Houston, TX, USA) according to the manufacturer’s instructions. Fresh MH broth was also tested for the measurement of PLP.
Protein fractionation, SDS-PAGE, and immunoblotting
Membrane and cytoplasmic proteins from C. jejuni were isolated as described previously [23]. These protein samples were then separated on a 7.5% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and stained with CBB (Coomassie Brilliant Blue) to visualize the protein profiles. The proteins on the gel were simultaneously transferred to a PVDF membrane (Merck-Millipore, Billerica, MA, USA), and the FlaA protein was detected using a rabbit polyclonal antibody generated against C. jejuni flagellin [24] and an HRP-conjugated goat anti-rabbit secondary antibody (GE Healthcare, Little Chalfont, UK). The blots were developed using the ECL detection system (GE Healthcare).
Motility and growth assays
The motility of the WT, pdxA mutant, and complemented C. jejuni strains was assayed on 0.4% soft MH agar plates as previously described [25]. To measure bacterial growth, 1.4–1.9×106 cells of C. jejuni that were microaerobically grown in MH broth to an OD600 of 1.2–1.3 at 37°C, were incubated in 10 ml of fresh MH broth supplemented with or without PLP (10 mg l−1) with agitation (120 rpm) for 0, 12, 24, 36, 48, 72 h. At each time points, turbidity of the medium was measured at 600 nm.
Detection of pseudaminic acid (Pse)
(i) Derivatization of Pse
Pse was released from the 30 μg of cytosolic protein fractions from the WT and pdxA mutant strains grown in MH broth to OD600 of 0.55–0.60, using the GlycoProfileTM β-elimination kit (Sigma-Aldrich) according to the manufacturer's instructions. The released Pse was labeled with 1,2-diamino-4,5- methylenedioxybenzene (DMB) using a sialic acid fluorescence-labeling kit (TaKaRa, Shiga, Japan), and the reaction mixture was applied to a solid-phase extraction cartridge (Envi-Carb C, Supelco, Bellefonte, PA, USA). After washing with 2.5 ml of 5 mM ammonium acetate (pH 9.6), the labeled Pse was eluted using 3 ml of 45% acetonitrile/5 mM ammonium acetate (pH 9.6) and freeze-dried. Fresh MH broth was also subjected to the above sample preparation to observe the effect of background growth medium.
(ii) Liquid chromatography/mass spectrometry (LC/MS)
Chromatographic separation of DMB-labeled Pse was performed using the Paradigm MS4 HPLC system (Michrom BioResource, Auburn, CA, USA). The separated DMB-labeled Pse was applied to a C18 trap column (L-column Micro, CERI) and eluted using 0.1% formic acid/2% acetonitrile (buffer A) and 0.1% formic acid/90% acetonitrile (buffer B) with a linear gradient of 10–90% buffer B over 30 min at a flow rate of 300 μl min−1. Mass spectrometric analysis of DMB-labeled Pse was performed using a Fourier transformation ion cyclotron resonance (FT-ICR)/ion trap (IT)-type mass spectrometer (LTQ-FT) (Thermo Electron, San Jose, CA, USA) equipped with a nanoelectrospray ion source (AMR, Tokyo, Japan). The presence of DMB-Pse was determined via sequential scans consisting of selected ion monitoring (SIM, m/z 441–461) using FT-ICR-MS and data-dependent MS/MS-MS/MS/MS/MS (MSn) with IT-MS.
Detection of metabolic compounds
(i) Sample preparation
A total of 3.2–3.4×108 C. jejuni WT or pdxA mutant cells grown in MH broth to an OD600 of 0.6 were trapped on a 0.4-μm filter membrane (Merck-Millipore), washed twice with 10 ml of water, and immersed in 2 ml of methanol containing 10 μM internal standard solution 1 (Human Metabolome Technologies (HMT), Yamagata, Japan). After sonication for 30 s, 1.6 ml of each suspension was mixed with 640 µL of water and 1.6 ml of chloroform, followed by centrifugation for 5 min at 2,300×g. The 750 µl of upper aqueous layer was filtered through a 5 KDa-cutoff filter (Millipore), lyophilized, and resuspended in 25 µl of water.
(ii) Capillary Electrophoresis-Time of Flight/mass spectrometry (CE-TOF/MS)
Cationic metabolites were analyzed using a fused silica capillary tube (50 μm×80 cm) and Cation Buffer Solution (HMT) in a capillary electrophoresis system equipped with a Time-of-Flight mass spectrometer (CE-TOF/MS) and a CE-ESI-MS sprayer (Agilent Technologies, Santa Clara, CA, USA). Electrospray ionization-mass spectrometry (ESI-MS) was conducted in positive ion mode at 4,000 V. Anionic metabolites were analyzed using a fused silica capillary and Anion Buffer Solution (HMT). ESI-MS was conducted in negative ion mode at 3,500 V. In both modes, the spectrometer was scanned from m/z 50 to 1,000. The other conditions were followed the cation analysis methodology of Soga and Heiger [26].
(iii) Data analysis
Raw data were processed using the MasterHands program [27]. Signal peaks corresponding to the isotopomers of 108 compounds (including the intermediates of the glycolytic system, the intermediates of the TCA cycle, and amino acids; see Table S2 for more details) were extracted. Each obtained migration time (MT) was normalized using the values of the internal standards. The resultant relative area values were further normalized based on the sample amounts. We used duplicate sets of samples from two independent experiments.
ATP assay
To determine the intracellular ATP concentration of bacterial samples, the BacTiter-Glo Microbial Cell Viability assay kit (Promega) was used. After growing the bacteria in MH broth at 37°C under microaerobic conditions to an OD595 of 0.55–0.60, serial dilutions of all samples were prepared according to the manufacturer’s instructions. Following incubation of the samples at room temperature in a 96-well plate, luminescence was measured together with an ATP standard using GloMax Multi system (Promega), according to the manufacturer’s instructions. Simultaneously, we measured the bacterial numbers in the originally cultured MH broth by plate count.
Cell adhesion assay, IL-8 measurements, and immunoblotting
INT407 cells were seeded into 24-well culture plates (TPP) (3.0×105 cells well−1) and incubated in RPMI1640 medium (Life Technologies, Carlsbad, CA, USA) for 20 h at 37°C in a humidified CO2 incubator. The cells were then rinsed and inoculated with C. jejuni at a multiplicity of infection (m.o.i.) of 100. At 60 min post-infection, the cells were washed three times with PBS to remove non-adherent bacteria, followed by cell detachment using 0.1% saponin in PBS. Serial dilutions of the suspensions were plated onto MH agar to determine the numbers of viable, cell-associated bacteria. To measure IL-8 secretion from the INT407 cells after infection, INT407 cells were infected with C. jejuni at an m.o.i. of 100 for 0, 4, and 16 h, and the culture supernatants were used to measure IL-8 levels with a human IL-8 ELISA kit (Becton-Dickinson), according to the manufacturer’s instructions. ERK activation was examined via western blotting using tyrosine-phosphorylated and total ERK1/2 monoclonal antibodies (Cell Signaling Technology, Danvers, MA, USA).
Chicken colonization assay
Specific pathogen-free, 14-day-old white leghorn chickens (obtained from Nisseiken Co., Ltd., Japan) were orally challenged with 500 μl of MH broth containing approximately 3.0×107 WT or pdxA C. jejuni cells. The animals were euthanized at 7 and 28 days post-infection, and post-mortem cecal samples were collected after aseptic removal of the ceca. C. jejuni colonization of the cecum was examined by counting viable cells on mCCDA agar plates (Oxoid, Hampshire, UK). A control group was confirmed to be negative for Campylobacter. The above animal experiments were approved by the Committee for Animal Care and Use of the National Institute of Health Sciences, Japan.
Statistics and web tool
The PATRIC prediction system (http://patricbrc.vbi.vt.edu/portal/portal/patric/Home), which assesses metabolic pathways in various prokaryotes based on their genomic sequences, was used to illustrate the putative PLP and Pse biosynthesis pathways in the C. jejuni 81–176 strain. The results from the motility, growth, ATP activity, cell adhesion, IL-8 production, and chicken colonization assays were expressed as the mean ± standard deviations of at least three independent observations. The significant differences between the measurements obtained from the WT and mutant strains were determined using Student’s t-test. P values <0.05 were considered statistically significant.
Results
Disruption of the pdxA gene abolishes PLP production in C. jejuni
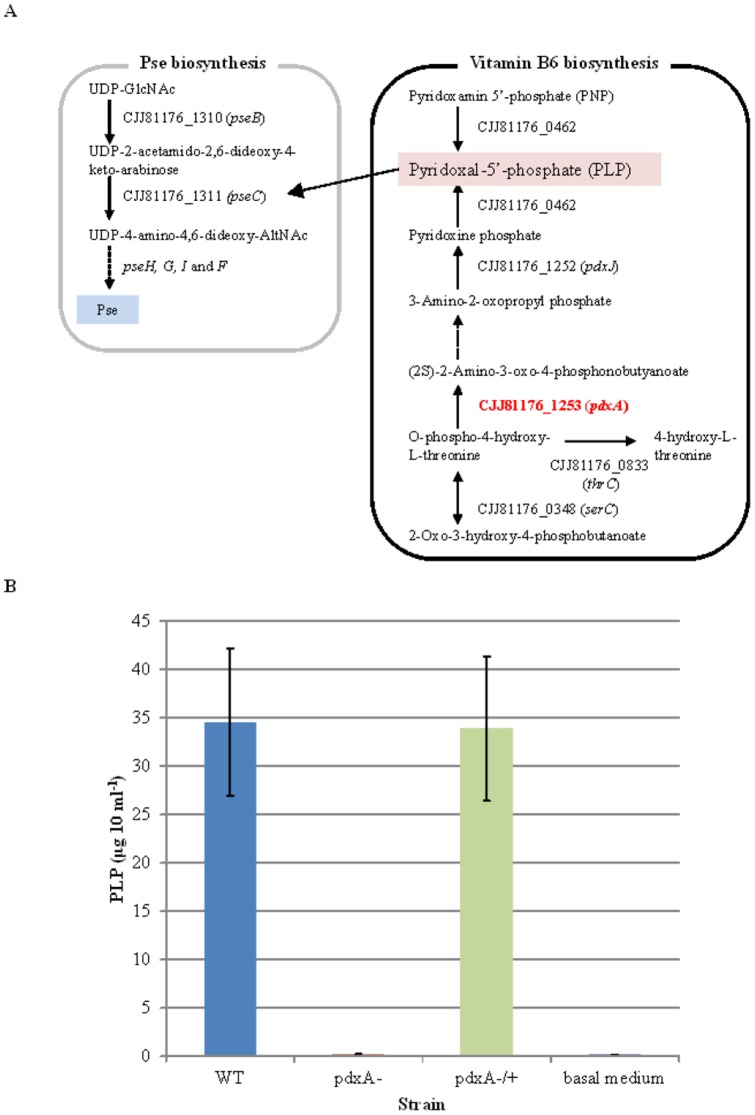
To predict the PLP biosynthesis pathway in C. jejuni 81–176, we used the in silico pathway tool PATRIC (http://www.patricbrc.org/portal/portal/patric/Home). The result of this prediction illustrated that at least five genes might constitute two independent pathways for PLP biosynthesis in this pathogen (right box, Fig. 1A). Among these genes, pdxA (CJJ81176_1253) and pdxJ (CJJ81176_1252) are known to be involved in the de novo synthesis of PLP in E. coli [12] and are, indeed, conserved in the C. jejuni genome [28]. Recently, Stahl and Stintzi [29] reported that the pdxA gene (Cj1239 in the NCTC11168 strain) may be essential for microbial growth. We therefore decided to use the pdxA gene to study the role of PLP biosynthesis in the biology of this pathogen and constructed an insertional pdxA mutant in C. jejuni strain 81–176. Biochemical assays collectively detected very less amounts of PLP (0.15±0.10 μg 10 ml−1) in the pdxA mutant than the WT strain (34.55±7.61 μg 10 ml−1), and complementation of the pdxA gene in the pdxA mutant restored PLP production (33.85±7.45 μg 10 ml−1) (Fig. 1B). Thus, we could demonstrate that the pdxA gene is truly a prerequisite for the PLP biosynthetic metabolism of this pathogen.
Figure 1. Inactivation of the pdxA gene impairs the biosynthesis of vitamin B6 (PLP) in C. jejuni.
(A) A scheme for the PLP production pathway (right box) in C. jejuni in relation to Pse biosynthesis (left box) is illustrated based on in silico pathway analysis performed using PATRIC (http://patricbrc.vbi.vt.edu/portal/portal/patric/Home). (B) The pdxA mutant produced no PLP. The C. jejuni 81–176 WT, pdxA mutant, and the complemented strains were grown in 10ml of MH broth to an OD600 of 0.60. The suspensions were then homogenized, serially diluted, and subjected to ELISA to quantify the amounts of PLP (μg 10 ml−1). The data show the mean +/− standard deviations from three independent assays.
The pdxA mutant impairs Pse production, flagellin glycosylation, and flagellation
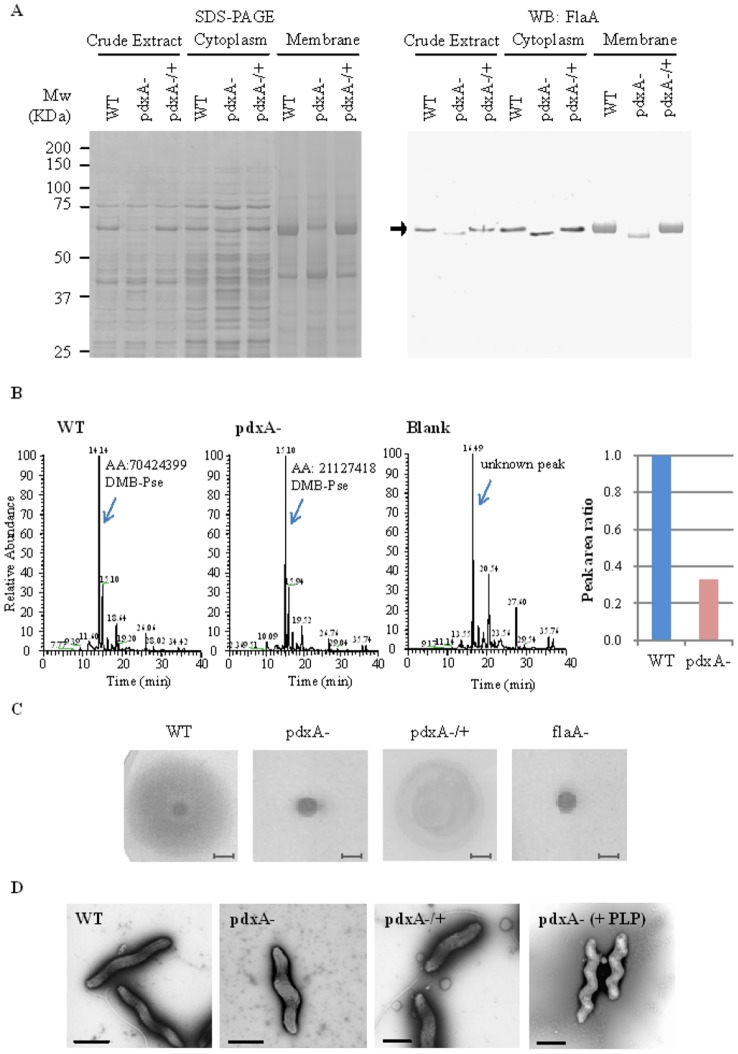
Campylobacter flagellins are decorated with O-linked glycans, which are derivatives of Pse synthesized through sequential enzymatic reactions (i.e., transamination, decarboxylation, and racemization) [30], and this type of glycomodification is a prerequisite for the biogenesis, transport, and assembly of functional flagellar filaments [31], [32]. Among components of the Pse biosynthesis process, the pseC (Cj1294/CJJ81176_1311) gene product, UDP-4-keto-deoxy-GlcNAc transaminase, is reported to require PLP to generate UDP-4-amino-4,6-dideoxy-GalNAc, a spectrometric analintermediate in the synthesis of Pse (left box, Fig. 1A) [17]. Immunoblot analyses showed the less glycosylation but expression of the flagellin A (FlaA) in cytoplasmic fraction of the pdxA mutant compared with that of WT strain (Fig. 2A), and the complementation of the pdxA gene restored the glycosylation of FlaA in the pdxA mutant (Fig. 2A). Having less detection of FlaA in the membrane fraction of the pdxA mutant than that of the WT strain (Fig. 2A), it could be considered that the less glycosylated FlaA was not transported to the surface of the pdxA mutant. In agreement, mass spectrometric analyses clearly showed that the pdxA mutant produced approximately 3-fold less Pse than the WT strain (Fig. 2B, Fig. S1, S2, S3), providing a link between pdxA, PLP, and Pse biosynthesis in this pathogen. In consistent with the fact that the glycosylation and surface expression of flagellar filaments are prerequisite for bacterial motility [31], [32], phenotypic assays clearly demonstrated that the pdxA mutant was not motile, and the complementation of the pdxA gene restored motility (Fig. 2C). Furthermore, microscopic analyses consistently showed that the pdxA mutant did not generate any flagellar filaments and that complementation of the pdxA gene restored flagellation, likely to the same level as in the isogenic WT strain (Fig. 2D). Addition of PLP did not restore the flagellar production of the pdxA mutant (Fig. 2D). Together, we were able to demonstrate that disruption of the pdxA gene impaired the glycosylation of flagellin, thereby reducing bacterial motility.
Figure 2. The pdxA mutant shows impaired pseudaminic acid (Pse) production and flagellar motility.
(A) The pdxA mutant shows less glycosylation of FlaA. SDS-PAGE and western blotting were conducted to detect the C. jejuni FlaA protein. Crude extracts and subcellular (cytoplasmic and membrane) fractions were extracted from C. jejuni and visualized using CBB staining in an SDS-polyacrylamide gel (left panel). Western blot analyses were simultaneously performed to detect the FlaA protein (arrow, right panel). (B) The pdxA mutant shows reduced Pse production. The left panel shows an extracted ion chromatogram at m/z 441.0–461.0 obtained through SIM of DMB-labeled Pse from the WT and pdxA mutant strains (arrowed). The extracted ion chromatogram of blank sample (fresh MH broth) was simultaneously subjected to confirm the absence of Pse. AA, peak area in arbitrary units. Each ion signal is expressed as a relative percentage of the WT-derived sample (set to 100%) from two independent tests (right panel). MSn data were shown in Fig. S1, S2, S3. (C) The disruption of the pdxA gene impairs motility of C. jejuni. The WT, pdxA mutant, pdxA-complemented (pdxA−/+), and flaA mutant (flaA-) strains were spotted and incubated onto 0.4% soft agar. Scale bars represent 3 mm. The motility of pdxA mutant was also assayed in the supplementation of 10 mg l−1 of PLP (pdxA− + PLP). (D) The pdxA mutant is aflagellated. Electron micrographs of the C. jejuni WT, pdxA mutant with or without supplementation of PLP (10 mg l−1), pdxA- complemented strains. The scale bars represent 1 μm.
The pdxA mutant exhibits altered Pse biosynthetic metabolism
Considering that PLP mediates a variety of enzymatic processes [33], a comparative metabolomic analysis was performed to characterize/confirm the types of metabolism that might be related to PLP activity. CE-TOF/MS (capillary electrophoresis time-of-flight/mass spectrometry) analysis detected 99 metabolic compounds extracted from the WT and pdxA mutant strains, among which the levels of 10 and 6 compounds were either increased or reduced >2-fold, respectively, in the pdxA mutant compared with the WT strain (Table 2 and more detailed information in Table S2). In support of the link between the presence of the pdxA gene and Pse production, the pdxA mutant exhibited greater amounts of UDP-glucose (a Pse precursor, Fig. 1A left panel) and a PLP precursor, pyridoxamine-5’-phosphate (PNP, Fig. 1A right panel), which showed concentrations that were at 3.6-fold and 2.5-fold higher than were exhibited by the WT strain, respectively (Table 2). Thus, these data clearly indicated an essential role of the pdxA gene in Pse biosynthesis in C. jejuni.
Table 2. Representative metabolites that are altered between the C. jejuni WT and pdxA mutant strains.
| Compound name | m/z *1 | MT *2 | Relative Area | Ratio *3 (pdxA−/WT) | |
| WT | pdxA− | ||||
| Increased | |||||
| UDP-glucose/galactose | 565.05 | 8.22 | 2.25E-04 | 7.99E-04 | 3.55 |
| Azelaic acid | 187.10 | 11.56 | 2.84E-04 | 9.31E-04 | 3.28 |
| 2-Amino-2-(hydroxymethyl)-1,3-propanediol | 122.08 | 7.83 | 7.14E-04 | 2.01E-03 | 2.82 |
| cis-Aconitic acid | 173.01 | 26.16 | 3.36E-03 | 9.17E-03 | 2.73 |
| ATP | 505.99 | 11.16 | 4.37E-04 | 1.16E-03 | 2.66 |
| GDP | 442.02 | 10.11 | 1.91E-04 | 5.09E-04 | 2.66 |
| Pyridoxamine-5'-phosphate (PNP) | 249.06 | 9.78 | 2.25E-04 | 5.55E-04 | 2.46 |
| β-Alanine | 90.06 | 6.96 | 6.85E-04 | 1.65E-03 | 2.40 |
| Glycine | 76.04 | 7.87 | 4.58E-03 | 1.04E-02 | 2.27 |
| Isocitrate | 191.02 | 26.91 | 3.00E-03 | 6.39E-03 | 2.13 |
| Decreased | |||||
| NADP+ | 742.07 | 8.92 | 5.3E-03 | 2.6E-03 | 0.49 |
| ppGpp_divalent | 300.47 | 13.69 | 4.3E-03 | 2.0E-03 | 0.48 |
| Asparagine | 133.06 | 9.80 | 1.3E-03 | 5.0E-04 | 0.40 |
| Agmatine | 131.13 | 4.94 | 5.5E-03 | 1.1E-03 | 0.19 |
The detected metabolites exhibiting >2.0-fold differences between the WT and pdxA- strains are shown. Each mean represents average from two independent tests. Candidate compounds are identified based on the detection peak (m/z)*1 and migration time (MT)*2 through HTM database. *3 Relative mean of the pdxA-/WT ratio. Full lists are shown in Table S2.
The C. jejuni pdxA mutant exhibits altered respiratory/energy metabolism
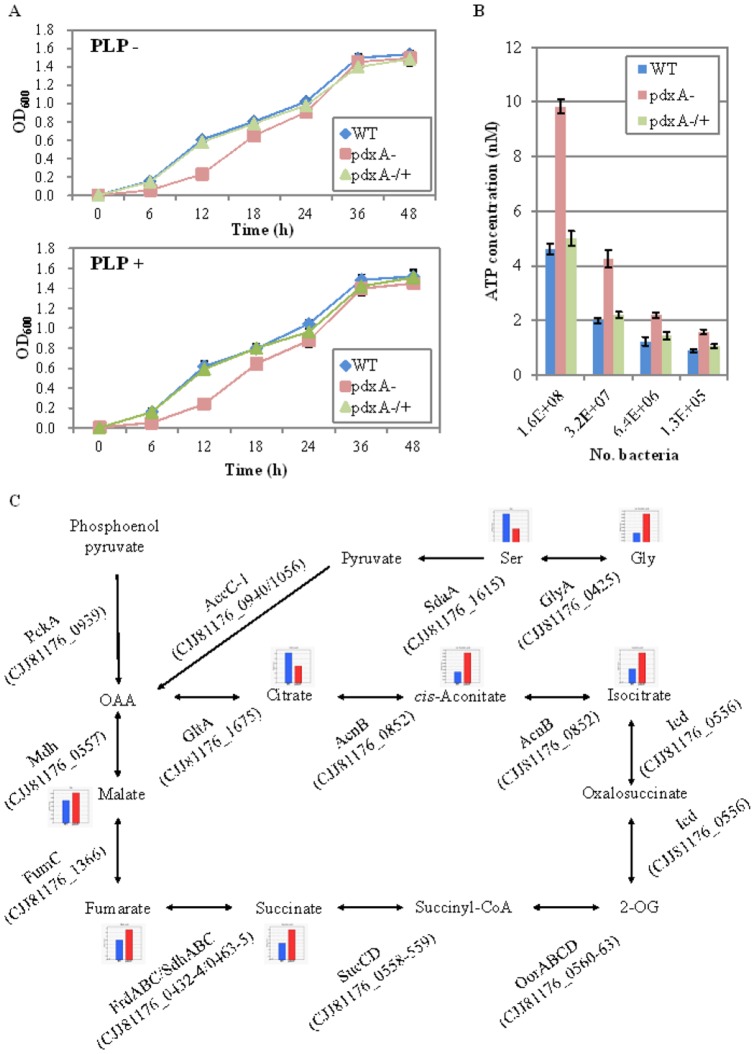
As an additional characteristic, we notified that the C. jejuni pdxA mutant exhibited growth defect compared with the WT strain, but this mutant showed successive growth in the absence of PLP, indicating that the pdxA gene was not essential for the growth of this pathogen (Fig. 3A, left panel). Different from H. pylori pdxA mutant [16], the addition of PLP did not restore growth of the C. jejuni pdxA mutant (Fig. 3A). To investigate the metabolisms associated with the altered growth kinetics of the C. jejuni pdxA mutant, we thus focused indicators of significant alterations in energy/respiratory metabolisms because of the pivotal role of these metabolisms in the growth in this pathogen [34]. The metabolomic data showed that the pdxA mutant indeed produced greater amounts of ATP and GDP (by 2.7-fold each) and, hence, decreased amounts of NADP+ (0.5-fold) compared with the levels in the WT strain (Table 2). In agreement with these findings, the pdxA mutant exhibited an approximately 2.14-fold greater amount of ATP compared with the WT strain (when 1.6×108 cells were assayed; Fig. 3B). Energy metabolism is well known to depend on the respiratory cycle. Corroborating this fact, the pdxA mutant showed alterations in the concentrations of TCA cycle intermediates including cis-aconitic acid (2.7-fold), isocitrate (2.1-fold), succinate (2.1-fold), malate (1.5-fold), citrate (0.6-fold), and serine (0.5-fold, a major carbon source for the respiratory cycle in this pathogen [35]) compared with the WT strain (Fig. 3C, Table 2, Table S2). Thus, we were able to show that the C. jejuni pdxA mutant exhibited altered growth and respiratory/energy metabolism.
Figure 3. The C. jejuni pdxA mutant shows altered growth kinetics and respiratory/energy metabolism.
(A) Growth curves of C. jejuni 81–176 WT, pdxA−, and the complemented mutant strains in MH broth not supplemented (left panel) or supplemented (right panel) with PLP (10 mg l−1). (B) Intracellular ATP levels of C. jejuni 81–176 WT, pdxA−, and the complemented mutant strains. ATP contents of four serial dilutions of the bacteria (shown as CFU 100 μl−1) under investigation were measured. The results are shown as means ± SD of data from triplicate wells of a representative experiment. (C) Focused dynamics of the C. jejuni TCA-cycle pathway. The pathway, the relative mean concentrations of the related metabolites in the WT (blue bars) and the pdxA mutant (red bars) strains, and the genes associated with the enzymatic conversion of each metabolite were illustrated with the PATRIC pathway analysis program.
The pdxA mutant exhibits impaired in vitro cell adhesion and chicken colonization
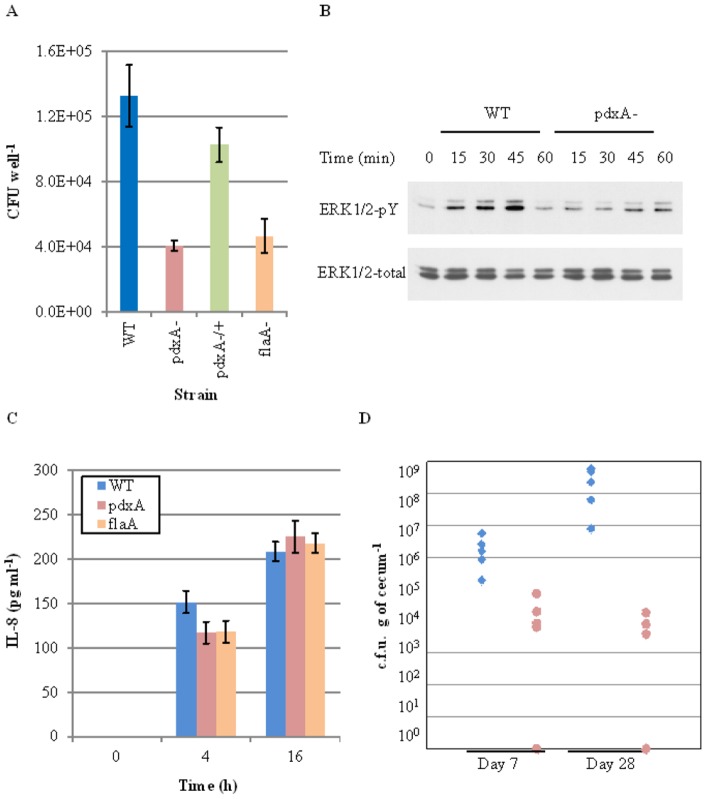
C. jejuni requires flagellum-mediated motility for establishing the early phase of infection both in vitro [36] and in vivo [37], [38]. Accordingly, when INT407 cells were infected with the pdxA mutant, WT, and flaA mutant strains, the cell adhesion score of the pdxA mutant was found to be almost identical to that of the flaA mutant, exhibiting a 3-fold reduction compared with the WT strain at 1 h post infection (p.i.) (Fig. 4A). Following adherence, C. jejuni activates ERK1/2 MAPK signaling, which stimulates the production of interleukin (IL)-8 in INT407 cells [39]. In agreement with the above cell adhesion scores, the pdxA mutant caused delayed phosphorylation of ERK1/2 MAPK compared with the WT strain (Fig. 4B), which was similar to the flaA mutant [39]. Additionally, similar to the flaA mutant [39], the pdxA mutant induced significantly less IL-8 production in the INT407 cells compared with the WT strain at 4 h p.i. (p<0.05, Fig. 4C). A prolonged period after infection (18 h p.i.) resulted in normalization of the IL-8 secretion observed in response to all of the bacterial strains tested (Fig. 4C). These data indicated that the pdxA gene is a prerequisite for cell adhesion, with the mutant exhibiting delayed activation of ERK1/2 signaling and impaired IL-8 production in intestinal epithelial cells. We also tested the colonization ability of the pdxA mutant in chickens because this host represents the most important reservoir of the pathogen for human infection [5] as well as the fact that the flagellin mutants exhibited less colonization in chicken [38]. At 7 and 28 days after infection, the pdxA mutant exhibited approximately 100-fold (2.14±2.92×104 CFU/g at day 7) and 4.6×104-fold (6.00±7.48×103 CFU/g at day 28) decreases in the colonization of chicken cecum tissues compared with the parental strain (2.14±2.12×106 CFU/g at day 7 and 2.74±2.55×108 CFU/g at day 28, both of which were significantly different (p<0.05) from the pdxA mutant-infected animals) (Fig. 4D). Together, we were able to demonstrate that disruption of the pdxA gene impaired the colonization of chicken intestine by C. jejuni.
Figure 4. Deletion of the pdxA gene impairs in vitro cellular responses and in vivo colonization.
(A) INT407 cells were infected for 1 h with the C. jejuni WT, pdxA−, pdxA−/+, and flaA− strains. The number of cell-adherent bacteria was measured by counting the plates after washing three times with PBS. (B) ERK1/2 activation upon infection. Western blotting was performed to detect the levels of phosphorylated and total ERK1/2 in the lysates from infected cells. (C) IL-8 production in INT407 cells was measured at 4 h and 16 h p.i. via ELISA. The data are presented in sections A and C as the mean values ± standard deviations from samples run in duplicate in at least three experiments. (D) Disruption of the pdxA gene reduces the colonization of the chicken cecum by C. jejuni. Groups of 14-day-old chickens (n = 10 per group) were orally inoculated with approximately 3×107 CFU of WT or pdxA mutant C. jejuni. At 1 week and 4 weeks p.i., the ceca were aseptically removed from the infected animals (n = 5 for each time point) and homogenized. Serial dilutions of the suspensions were plated on mCCDA agar to count CFU numbers. The closed diamonds and open circles represent the numbers of WT and pdxA mutant CFUs recovered from the animals, respectively.
Discussion
Here, we examined the role of the PLP synthetic pathway in the biology of C. jejuni. Disruption of the pdxA gene clearly impaired PLP production. Mass spectrometric and biochemical analyses revealed a reduced production and glycosylation of flagellins in the pdxA mutant, which is likely to impair bacterial motility. Having the altered growth by disruption of the pdxA gene in this pathogen, we then performed comparative metabolomic approaches, further revealing the association of pdxA gene to energy/respiratory metabolisms. We finally showed that The pdxA mutant exhibited decreased cell adhesion-dependent responses in vitro and in vivo host colonization.
Based on the in silico pathway prediction for the PLP biogenesis, we selected the pdxA as a putative essential gene for the PLP production in this pathogen. A mutation of the pdxA gene impaired production of PLP in C. jejuni 81–176, supporting our prediction. The reduced Pse production in the pdxA mutant was also supportable to a previous study demonstrating the essentiality of PLP in the Pse production in C. jejuni [17]. The reduced production, but not complete loss of Pse in the pdxA mutant might be explained by the fact that small amounts of PLP (0.14±0.07 μg 10ml−1) were also detected from basal MH broth (Fig. 1B). Perhaps, the residual Pse peak in the pdxA mutant might be stem from residual PLP in the medium.
Since flagellin glycosylation is prerequisite for the biogenesis, transport, and assembly of flagellar filaments in this pathogen [31], [32] and which thereby alters the motility and host colonization of this pathogen [37], [38], it was plausible that the decreased Pse levels in the pdxA mutant, affected flagellar glycosylation, thereby altering transport of flagellin to the bacterial surface. Phenotypic and infection studies indeed showed impaired motility and host colonization of C. jejuni by disruption of the pdxA gene, supporting the idea that the reduced motility of the pdxA mutant was mainly due to the altered network between PLP and Pse.
We identified a link between PLP and the Pse modification system in C. jejuni 81–176, in agreement with the previously reported essential role of the pdxA gene in flagellar glycosylation in a closely related pathogen, H. pylori [16]. Moreover, the less glycosylation of FlaA protein in the cytoplasm of pdxA mutant was in agreement with the previous report demonstrating that the C. jejuni pseC mutant expressed unglycosylated FlaA in the cytoplasm [40]. Unlike H. pylori, however, the C. jejuni pdxA mutant could grow without supplementation of PLP, and the addition of PLP did not restore the motility and growth of C. jejuni pdxA mutant. These suggest the distinct metabolic impacts of PLP to the growth and/or viability between H. pylori and C. jejuni. A protein-protein network prediction tool, STRING database (http://string.embl.de/) indeed shows differential networks of the pdxA gene between the two pathogens (Fig. S4).
Campylobacter exhibits unique nutritional requirements, and it has been thought to only utilize amino acids and TCA cycle intermediates as carbon sources for energy production [41]. The TCA cycle is a sequential process involving enzymatic reactions in which a two-carbon acetyl unit is oxidized to CO2 and H2O to provide energy in the form of high-energy phosphate bonds. The different types of energy metabolism observed in the WT and pdxA mutant strains therefore suggested a possible link of PLP with these types of metabolism. Representative metabolites that were significantly altered by inactivation of the pdxA gene were thus discussed below.
(i) Serine/Glycine
This pathogen exhibits a complete TCA cycle [42], and serine is particularly useful as a nutritional substrate that can be catabolized for growth and colonization in the chicken gut [37], [43], [44]. The decreased serine level detected would appear to be connected to glycine metabolism because E. coli serine hydroxymethyltransferase (GlyA) catalyzes the reversible interconversion of the amino acids serine and glycine using one-carbon tetrahydrofolate and PLP [45]. Thus, it could be considered that the imbalance between serine and glycine in the pdxA mutant might associate with the altered functionality of GlyA due to the lack of PLP.
(ii) Citrate/cis-aconitate/isocitrate
These TCA intermediates are interconverted by aconitases [46], among which AcnB functions as the major TCA cycle enzyme in E. coli [47], [48]. Considering that C. jejuni 81–176 also harbors an acnB gene (CJJ81176_0852), the imbalance in these three TCA intermediates in the pdxA mutant might be due to reduced AcnB activity. AcnB forms an iron-sulfur cluster, thereby affecting its enzymatic activity [49]. Iron depletion has been shown to inhibit AcnB activity in E. coli [49], suggesting that the pdxA mutant might exhibit an altered iron metabolism and/or iron-sulfide cluster formation and, thus, reduced AcnB activity.
(iii) Agmatin
Agmatin is a decarboxylation product of arginine that is involved in the urea cycle, the synthesis of creatine, and the generation of nitric oxide in eukaryotes [50]. The unaltered levels of arginine between the WT and pdxA mutant strains suggested that arginine decarboxylase (SpeA) might also require PLP for its activation. In support of this concept, E. coli SpeA shows a PLP-binding affinity [51], and a recent structural analysis showed that C. jejuni SpeA contains potent PLP-binding residues, similar to those of E. coli [52].
(iv) β-alanine/asparagine
In contrast to the above three examples, the pdxA mutant exhibited an increased level of β-alanine, a precursor of coenzyme A (CoA), compared with the WT strain. β-alanine is mainly synthesized via the decarboxylation of L-aspartate in E. coli [53]. In this regard, the decreased levels of asparagine observed in the pdxA mutant suggested that asparaginase (AnsB), which is capable of deaminating periplasmic asparagine to aspartate [54], might be inactivated in this mutant, thereby causing the accumulation of asparagine, a precursor of β-alanine.
(v) Glycolate
The pdxA mutant displayed decreased production of glycolate (hydroxylacetic acid), one of the smallest alpha-hydroxy acids (AHA). This metabolite is synthesized from 3-hydroxypyruvate (3HP) through reaction with glycoaldehyde, followed by decarboxylation, which requires PLP in E. coli [55], providing a possible reason for the decreased glycolate detected in the pdxA mutant.
Further studies will be necessary to elucidate the molecular impacts of PLP activity on the infection process in this pathogen through in-depth functional and/or structural analyses of each enzymatic reaction. Nevertheless, the data obtained in the present study provide the first evidence that biologically links PLP to the respiratory/energy metabolism as well as the flagellar glycosylation system, affecting the host colonization of C. jejuni.
It is likely that a number of factors could contribute to the colonization of chickens by C. jejuni (i.e., flagellum-mediated motility, chemotaxis, amino acid metabolism, energy metabolism, and iron utilization) [18]. The in vivo growth of C. jejuni has been argued to depend mainly on the availability of free amino and keto acids scavenged from the host or the intestinal microbiota [56]. The data reported herein therefore suggest that in addition to the decreased motility of the pdxA mutant, the altered levels of respiratory/energy metabolism might also participate in the impaired colonization of the chicken gut by this mutant. In vivo metabolic profiling of this pathogen would improve our understanding of the molecular basis underlying its adaptation to and interaction with the host and microbiota during infection.
In summary, this is the first report to demonstrate a functional role of the pdxA gene in altering the motility of and colonization of chickens by a leading foodborne pathogen, Campylobacter jejuni, including the demonstration of a novel link between PLP and flagellar glycosylation. PLP-dependent enzymes are likely to represent approximately 4% of the enzymes present in mammals [57], which attracted our interest in the investigation of PLP functions in terms of potential drug targets. Indeed, certain PLP-dependent enzymes are increasingly being identified as potential drug targets for the treatment of protozoan diseases [58], [59]. As poultry animals are the predominant reservoirs of this pathogen for human infection, our data reveal new prospects for potent targeting of PLP and its dependent enzymes to modulate the dynamics of and control this pathogen in livestock animals.
Supporting Information
Mass spectrum of DMB-labeled pseudaminic acid (Pse) acquired from the arrowed peaks in an extracted ion chromatogram at m / z 441-2-461.2 obtained through SIM of DMB-labeled Pse from the C. jejuni 81–176 wild type (WT), pdxA mutant (pdxA-), and fresh MH broth (blank) samples shown in Fig. 2B .
(TIF)
MSn spectra of DMB-labeled pseudaminic acid (Pse) from the 81–176 wild type (WT). (A) the MS/MS spectrum acquired from the molecular ion [M + H]+ (m/z 451.2) of peak (arrowed) in Fig. S1; (B) the MS/MS/MS spectrum acquired from the product ion (m/z 433.1) in the MS/MS; (C) the MS/MS/MS/MS spectrum acquired from the product ion (m/z 415.1) in the MS/MS/MS; (D) Fragmentation of DMB-labeled Pse. In addition to the DMB-labeled Pse, some ms/ms peaks were also detected. To indicate the molecular mass of these peaks, green ticks were used (to distinguish from the mass peaks).
(TIF)
MSn spectra of DMB-labeled pseudaminic acid (Pse) from the 81–176 pdxA mutant. (A) the MS/MS spectrum acquired from the molecular ion [M + H]+ (m/z 451.2) of peak (arrowed) in Fig. S1; (B) the MS/MS/MS spectrum acquired from the product ion (m/z 433.1) in the MS/MS; (C) the MS/MS/MS/MS spectrum acquired from the product ion (m/z 415.0) in the MS/MS/MS; (D) Fragmentation of DMB-labeled Pse. In addition to the DMB-labeled Pse, some ms/ms peaks were also detected. To indicate the molecular mass of these peaks, green ticks were used (to distinguish from the mass peaks).
(TIF)
STRING network analysis. Protein-protein network analysis was carried out using the STRING database (http://string.embl.de/). Protein entries from C. jejuni strain 81–176 or H. pylori strain G27 were used for the identification of putative protein-protein associations of PdxA to other bacterial proteins according to the guideline of the database.
(TIF)
Oligonucleotide primers used in this study.
(XLSX)
Metabolic compounds in C. jejuni identified by CE-MS analysis.
(XLS)
Acknowledgments
We thank Tomoya Ekawa for the technical assistance of animal experiments.
Funding Statement
This work was supported in part through funding from a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (22780275), and a grant from Ministry of Health, Labour, and Welfare, Japan (H24-shokuhin-ippan-009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Friedman CR, Neimann J, Wegener HC, Tauxe RV (2000) Epidemioogy of Camylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd edition: ASM Press. 121–138.
- 2. Rautelin H, Haenninen ML (2000) Campylobacters: the most common bacterial enteropathogens in the Nordic countries. Ann Med 32: 440–445. [DOI] [PubMed] [Google Scholar]
- 3.Yuki N (2010) Human gangliosides and bacterial lipo-oligosaccharides in the development of autoimmune neuropathies. Methods Mol Biol 600: 51–65. Review. [DOI] [PubMed]
- 4. Vucic S, Kiernan MC, Cornblath DR (2009) Guillain-Barré syndrome: an update. J Clin Neurosci 16: 733–741. [DOI] [PubMed] [Google Scholar]
- 5. Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, et al. (2012) Poultry as a host for the zoonotic pathogen Campylobacter jejuni . Vector Borne Zoonotic Dis 12: 89–98. [DOI] [PubMed] [Google Scholar]
- 6. Moore JE, Corcoran D, Dooley JS, Fanning S, Lucey B, et al. (2005) Campylobacter . Vet Res 36: 351–382. [DOI] [PubMed] [Google Scholar]
- 7. Young KT, Davis LM, Dirita VJ (2007) Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5: 665–679. [DOI] [PubMed] [Google Scholar]
- 8. Jamshidi N, Palsson BØ (2007) Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muñoz-Elías EJ, McKinney JD (2006) Carbon metabolism of intracellular bacteria. Cell Microbiol 8: 10–22. [DOI] [PubMed] [Google Scholar]
- 10.Drewke C, Leistner E (2001) Biosynthesis of vitamin B6 and structurally related derivatives. In Vitamins and Hormones, vol. 63 (Litwack, G., ed.), 121–155, Academic Press, San Diego. [DOI] [PubMed]
- 11. Eliot AC, Kirsch J (2004) Pyridoxal phosphate enzymes: mechanistic, structural and evolutionary considerations. Annu Rev Biochem 73: 383–415. [DOI] [PubMed] [Google Scholar]
- 12. Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, et al. (2007) Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem J 407: 1–13. [DOI] [PubMed] [Google Scholar]
- 13. Safo MK, Musayev FN, Schirch V (2005) Structure of Escherichia coli pyridoxine 5'-phosphate oxidase in a tetragonal crystal form: insights into the mechanistic pathway of the enzyme. Acta Crystallogr D Biol Crystallogr 61: 599–604. [DOI] [PubMed] [Google Scholar]
- 14. Dick T, Manjunatha U, Kappes B, Gengenbacher M (2010) Vitamin B6 biosynthesis is essential for survival and virulence of Mycobacterium tuberculosis . Mol Microbiol 78: 980–988. [DOI] [PubMed] [Google Scholar]
- 15. Raschle T, Amrhein N, Fitzpatrick TB (2005) On the two components of pyridoxal 5'-phosphate synthase from Bacillus subtilis . J Biol Chem 280: 32291–32300. [DOI] [PubMed] [Google Scholar]
- 16. Grubmann A, Phillips A, Thibonnier M, Kaparakis-Liaskos M, Johnson C, et al. (2010) Vitamin B6 is required for full motility and virulence in Helicobacter pylori . mBIO 1: e00112–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Obhi RK, Creuzenet C (2005) Biochemical characterization of the Campylobacter jejuni Cj1294, a novel UDP-4-keto-6-deoxy-GlcNAc aminotransferase that generates UDP-4-amino-4,6-dideoxy-GalNAc. J Biol Chem 280: 20902–20908. [DOI] [PubMed] [Google Scholar]
- 18. Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U (2010) Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol 300: 205–211. [DOI] [PubMed] [Google Scholar]
- 19. Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ (1988) Experimental Campylobacter jejuni infection in humans. J Infect Dis 157: 472–479. [DOI] [PubMed] [Google Scholar]
- 20. Asakura H, Yamasaki M, Yamamoto S, Igimi S (2007) Deletion of peb4 gene impairs cell adhesion and biofilm formation in Campylobacter jejuni . FEMS Microbiol Lett 275: 278–285. [DOI] [PubMed] [Google Scholar]
- 21. Yao R, Alm RA, Trust TJ, Guerry P (1993) Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130: 127–130. [DOI] [PubMed] [Google Scholar]
- 22. Guerry P, Yao R, Alm RA, Burr DH, Trust TJ (1994) Systems of experimental genetics for Campylobacter species. Methods Enzymol 235: 474–481. [DOI] [PubMed] [Google Scholar]
- 23. Guerry P, Alm RA, Power ME, Logan SM, Trust TJ (1991) Role of two flagellin genes in Campylobacter motility. J Bacteriol 173: 4757–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hobb RI, Fields JA, Burns CM, Thompson SA (2009) Evaluation of procedures for outer membrane isolation from Campylobacter jejuni . Microbiology 155: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Asakura H, Churin Y, Bauer B, Boettcher JP, Bartfeld S, et al. (2010) Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol Microbiol 78: 1130–1144. [DOI] [PubMed] [Google Scholar]
- 26. Soga T, Heiger DN (2000) Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 72: 1236–1241. [DOI] [PubMed] [Google Scholar]
- 27. Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M (2010) Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6: 78–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ehrenshaft M, Bliski P, Li MY, Chingnell CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci U S A 96: 9374–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stahl M, Stintzi A (2011) Identification of essential genes in C. jejuni genome highlights hyper-variable plasticity regions. Funct Integr Genomics 11: 241–257. [DOI] [PubMed] [Google Scholar]
- 30. Logan SM (2006) Flagellar glycosylation – a new component of the motility repertoire? Microbiology 152: 1249–1262. [DOI] [PubMed] [Google Scholar]
- 31. Ewing CP, Andreishcheva E, Guerry P (2009) Functional characterization of flagellin glycosylation in Campylobacter jejuni 81–176. J Bacteriol 191: 7086–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, et al. (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol 60: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Percudani R, Peracchi A (2003) A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep 4: 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weerakoon DR, Borden NJ, Goodson CM, Grimes J, Olson JW (2009) The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb Pathog 47: 8–15. [DOI] [PubMed] [Google Scholar]
- 35. Guccione E, Hitchcock A, Hall SJ, Mulholland F, Shearer N, et al. (2010) Reduction of fumarate, mesaconate and crotonate by Mfr, a novel oxygen-regulated periplasmic reductase in Campylobacter jejuni . Environ Microbiol 12: 576–591. [DOI] [PubMed] [Google Scholar]
- 36. Grant CC, Konkel ME, Cieplak W Jr, Tompkins LS (1993) Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun 61: 1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hendrixson DR, DiRita VJ (2004) Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol 52: 471–484. [DOI] [PubMed] [Google Scholar]
- 38. Nachamkin I, Yang XH, Stern NJ (1993) Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol 59: 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watson RO, Galán JE (2005) Signal transduction in Campylobacter jejuni-induced cytokine production. Cell Microbiol 7: 655–665. [DOI] [PubMed] [Google Scholar]
- 40. McNally DJ, Hui JP, Aubry AJ, Mui KK, Guerry P, et al. (2006) Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81–176 using a focused metabolomics approach. J Biol Chem. 281: 18489–18498. [DOI] [PubMed] [Google Scholar]
- 41. Line JE, Hiett KL, Guard-Bouldin J, Seal BS (2010) Differential carbon source utilization by Campylobacter jejuni 11168 in response to growth temperature variation. J Microbiol Methods 80: 198–202. [DOI] [PubMed] [Google Scholar]
- 42. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, et al. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403: 665–668. [DOI] [PubMed] [Google Scholar]
- 43.Kelly DJ, Hughes NJ, Poole RK (2001) Microaerobic Physiology: Aerobic Respiration, Anaerobic Respiration, and Carbon Dioxide Metabolism. In: Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter pylori: Physiology and Genetics. Washington DC: ASM Press; 2001. Chapter 10. [PubMed]
- 44. Velayudhan J, Jones MA, Barrow PA, Kelly DJ (2004) L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni . Infect Immun 72: 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malerba F, Bellelli A, Giorgi A, Bossa F, Contestabile R (2007) The mechanism of addition of pyridoxal 5’-phosphate to Escherichia coli apo-serine hydroxymethyltransferase. Biochem J 404: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beinert H, Kennedy MC (1993) Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J 7: 1442–1449. [DOI] [PubMed] [Google Scholar]
- 47. Bradbury AJ, Gruer MJ, Rudd KE, Guest JR (1996) The second aconitase (AcnB) of Escherichia coli . Microbiology 142: 389–400. [DOI] [PubMed] [Google Scholar]
- 48. Cunningham L, Gruer MJ, Guest JR (1997) Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli . Microbiology 143: 3795–3805. [DOI] [PubMed] [Google Scholar]
- 49. Varghese S, Tang Y, Imlay JA (2003) Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol 185: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reyes A, Karl I, Klahr S (1994) The role of arginine in health and disease. Am J Physiol 267: 331–346. [DOI] [PubMed] [Google Scholar]
- 51. Wu WH, Morris DR (1973) Biosynthetic arginine decarboxylase from Escherichia coli. Subunit interactions and the role of magnesium ion. J Biol Chem 248: 1696–1699. [PubMed] [Google Scholar]
- 52. Forouhar F, Lew S, Seetharaman J, Xiao R, Acton TB, et al. (2010) Structures of bacterial biosynthetic arginine decarboxylases. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1562–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cronan JE Jr (1980) Beta-alanine synthesis in Escherichia coli . J Bacteriol 141: 1291–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hofreuter D, Novik V, Galán JE (2010) Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4: 425–433. [DOI] [PubMed] [Google Scholar]
- 55. Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD (2010) Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5’-phosphate synthesis. Mol Syst Biol 6: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keeney KM, Finlay BB (2011) Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr Opin Microbiol 14: 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mozzarelli A, Bettati S (2006) Exploring the pyridoxal 5'-phosphate-dependent enzymes. Chem Rec 6: 275–287. [DOI] [PubMed] [Google Scholar]
- 58. Kappes B, Tews I, Binter A, Macheroux P (2011) PLP-dependent enzymes as potential drug targets for protozoan diseases. Biochim Biophys Acta 1814: 1567–1576. [DOI] [PubMed] [Google Scholar]
- 59. Müller IB, Wu F, Bergmann B, Knöckel J, Walter RD, et al. (2010) Poisoning pyridoxal 5-phosphate-dependent enzymes: a new strategy to target the malaria parasite Plasmodium falciparum . PLoS One 4: e4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectrum of DMB-labeled pseudaminic acid (Pse) acquired from the arrowed peaks in an extracted ion chromatogram at m / z 441-2-461.2 obtained through SIM of DMB-labeled Pse from the C. jejuni 81–176 wild type (WT), pdxA mutant (pdxA-), and fresh MH broth (blank) samples shown in Fig. 2B .
(TIF)
MSn spectra of DMB-labeled pseudaminic acid (Pse) from the 81–176 wild type (WT). (A) the MS/MS spectrum acquired from the molecular ion [M + H]+ (m/z 451.2) of peak (arrowed) in Fig. S1; (B) the MS/MS/MS spectrum acquired from the product ion (m/z 433.1) in the MS/MS; (C) the MS/MS/MS/MS spectrum acquired from the product ion (m/z 415.1) in the MS/MS/MS; (D) Fragmentation of DMB-labeled Pse. In addition to the DMB-labeled Pse, some ms/ms peaks were also detected. To indicate the molecular mass of these peaks, green ticks were used (to distinguish from the mass peaks).
(TIF)
MSn spectra of DMB-labeled pseudaminic acid (Pse) from the 81–176 pdxA mutant. (A) the MS/MS spectrum acquired from the molecular ion [M + H]+ (m/z 451.2) of peak (arrowed) in Fig. S1; (B) the MS/MS/MS spectrum acquired from the product ion (m/z 433.1) in the MS/MS; (C) the MS/MS/MS/MS spectrum acquired from the product ion (m/z 415.0) in the MS/MS/MS; (D) Fragmentation of DMB-labeled Pse. In addition to the DMB-labeled Pse, some ms/ms peaks were also detected. To indicate the molecular mass of these peaks, green ticks were used (to distinguish from the mass peaks).
(TIF)
STRING network analysis. Protein-protein network analysis was carried out using the STRING database (http://string.embl.de/). Protein entries from C. jejuni strain 81–176 or H. pylori strain G27 were used for the identification of putative protein-protein associations of PdxA to other bacterial proteins according to the guideline of the database.
(TIF)
Oligonucleotide primers used in this study.
(XLSX)
Metabolic compounds in C. jejuni identified by CE-MS analysis.
(XLS)