Abstract
Background
Although the presence of genetic heterogeneity within individual patients’ tumors is established, it is unclear whether greater heterogeneity predicts worse outcome. A quantitative measure of genetic heterogeneity based on next-generation sequencing (NGS) data, mutant-allele tumor heterogeneity (MATH), was previously developed and applied to a data set on head and neck squamous cell cancer (HNSCC). Whether this measure correlates with clinical outcome was not previously assessed.
Methods
We examined the association of MATH with clinical, pathological and overall-survival data for 74 HNSCC patients for whom exome sequencing was completed.
Results
High MATH (a MATH value above the median) was significantly associated with shorter overall survival (hazard ratio 2.5; 95% CI, 1.3 to 4.8). MATH was similarly associated with adverse outcomes in clinically high risk patients with advanced stage, and in tumors classified as high risk on the basis of validated biomarkers including those negative for human papillomavirus or having disruptive TP53 mutations. In patients who received chemotherapy, the hazard ratio for high MATH was 4.1 (95%CI: 1.6 to 10.2).
Conclusions
This novel measure of tumor genetic heterogeneity is significantly associated with tumor progression and adverse treatment outcomes, supporting the hypothesis that higher genetic heterogeneity portends worse clinical outcome in HNSCC. The prognostic value of some known biomarkers may be the result of their association with high genetic heterogeneity. MATH provides a useful measure of that heterogeneity, to be prospectively validated as NGS data from homogeneously treated patient cohorts becomes available.
Keywords: head and neck cancer, intra-tumor genetic heterogeneity, tumor biomarkers, next-generation DNA sequencing, somatic mutations, overall survival
INTRODUCTION
Cancer is believed to arise from the acquisition of multiple mutations that cooperate to transform normal cells.1 Although all neoplastic cells within a cancer presumably arose from a common ancestor, the progeny of this common ancestor continue to evolve.2, 3 Hence there may be one or multiple dominant progeny subclones, and the evolutionary distance from the progenitor and the other subclones in the cancer is variable.4 The presence of multiple progeny clones within an individual tumor reflects genetic heterogeneity. While this concept is now well established,5-17 biomarkers to quantify this heterogeneity are scant.
It is likely that a greater extent of genetic heterogeneity poses a risk of worse clinical outcome, as a heterogeneous tumor might be more likely to contain subclones of cancer cells that proliferate more rapidly, are prone to metastasis, or are resistant to particular types of therapy.18-22 Until recently there had not been a simple, generally applicable measure of genetic heterogeneity to assess this risk that is suitable for use in clinical trials and in future clinical practice.
A genetically heterogeneous tumor is likely to show wide variability in mutant-allele fractions within next-generation sequencing (NGS) data, with mutations in the ancestral clone at high frequencies and subclone-specific mutations at low frequencies within mixed tumor DNA. We thus recently proposed a simple quantitative measure of genetic heterogeneity based on the variability of mutant-allele fractions,23 exploiting this consequence of multiple subclones rather than identifying and enumerating subclones directly. This heterogeneity measure, called mutant-allele tumor heterogeneity (MATH),23 is a percentage ratio of the width to the center of the distribution of mutant-allele fractions among tumor-specific mutated loci. Unlike other measures of genetic heterogeneity, MATH does not depend on pre-identifying subclonal markers or on single-cell analysis; rather, it is derived directly from the mixed-population mutant allele frequencies within a tumor. Since NGS of tumor DNA is expected to find clinical application in the near future,24 MATH could provide a clinically useful way to monitor significant, measurable genetic heterogeneity.
Based on publicly available NGS results on 74 head and neck squamous cell carcinomas (HNSCC),25 we demonstrated that poor-outcome classes of HNSCC possessed high genetic heterogeneity as measured by MATH.23 Furthermore, MATH values were unrelated to tumor mutation rates, suggesting that genetic heterogeneity involves clinically significant aspects of tumor biology beyond the accumulation of mutations. The possibility remained, however, that MATH was unrelated to clinical outcome per se, but was simply associated with certain HNSCC pathologic features.
Here we correlate clinical, pathological, and outcome data for these 74 cases. We show that higher MATH is associated with shorter overall survival, especially in cases where treatment involved chemotherapy. The relation of MATH to outcome in this study was stronger than that of two well known poor-outcome HNSCC biomarkers, negative human papillomavirus (HPV) status,26, 27 and disruptive mutations in the TP53 tumor suppressor.28, 29 These results support the hypothesis that higher genetic heterogeneity portends worse clinical outcome in HNSCC, suggest that the prognostic value of some biomarkers may in part be due to their association with high genetic heterogeneity, and show that MATH provides a useful measure of that heterogeneity to be validated as NGS data from homogeneously treated patient cohorts becomes available.
MATERIALS AND METHODS
Clinical, pathological, and outcome data for the 74 HNSCC cases whose NGS exome-sequencing results had been reported by Stransky et al.25 were imported into R30 for analysis. Prior to surgical removal of tumor tissue, all patients had provided informed consent under protocol 99-069 approved by the University of Pittsburgh IRB. Overall survival was calculated from the date of the surgical procedure that obtained the tumor sample used for NGS. Disease staging was based on the 7th edition of the American Joint Committee on Cancer (AJCC) Manual,31 using pathological T and N classifications when available.
Numbers of tumor-specific mutations, MATH values, HPV status, total numbers of mutations, and TP53 mutation status for these tumors had previously been analyzed.23 The MATH value for each tumor was based on the distribution of mutant-allele fractions among tumor-specific mutated loci, calculated as the percentage ratio of the width (median absolute deviation, MAD, scaled by a constant factor so that the expected MAD of a sample from a normal distribution equals the standard deviation) to the center (median) of its distribution:
MATH values for these tumors ranged from 19 to 55 dimensionless units,23 with a mean value of 34, a standard deviation of 10, a median of 32, and first and third quartiles at 26 and 42 units, respectively.
Bootstrap resampling of individual NGS reads for each tumor previously indicated that each tumor’s MATH value had a typical associated standard deviation (SD) of 4 units, depending on the number of mutated loci.23 This SD arises from the sampling of individual DNA fragments among genomic loci and between mutant and reference alleles at each locus during NGS. MATH values thus are shown to two significant figures.
Relations of MATH to patient and tumor characteristics were assessed by linear models (t-tests, analysis of variance, or linear regression). Hazard ratios with respect to overall survival for MATH and for other patient and tumor characteristics were determined by Cox proportional hazards analysis (survival package in R). Significance of hazard ratios was based on the Wald test. Differences between survival curves were assessed by log-rank tests. All statistical tests were two-sided, with significance accepted at p < 0.05. The receiver operating characteristic (ROC) curve (Fig. 2b) was obtained with the nearest-neighbor method for survival data developed by Heagerty et al (survivalROC package in R, with a smoothing span of 0.1).32
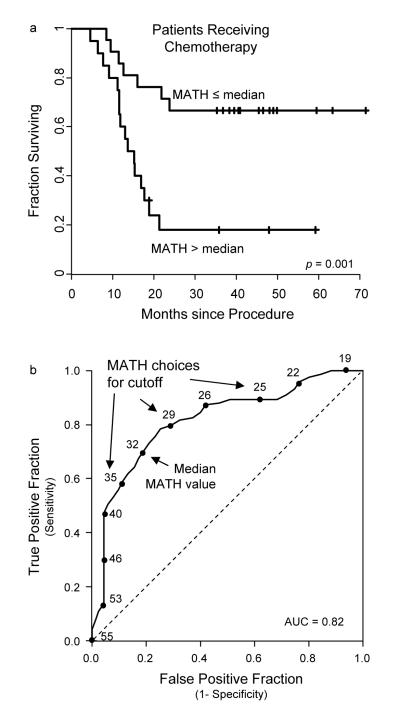
Figure 2. Relation of mutant-allele tumor heterogeneity (MATH) to outcome in HNSCC treated with chemotherapy.
(a) Survival curves as in Fig. 1, for MATH values above versus below the median in the 41 cases in which therapy involved chemotherapy (40 also involving radiation; all 41 were primary tumors). (b) Receiver operating characteristic (ROC) curve for MATH estimated from these 41 cases, showing how different choices of MATH cutoff values affect the specificity and sensitivity of outcome classification at survival times 24 months or greater. MATH value cutoffs increase from the top right to the bottom left of the solid curve, with the MATH value shown for every fourth tumor. For example, about 95% of patients whose tumors have MATH values greater than 40 die within 24 months (selectivity), but using 40 as a MATH cutoff only identifies 50% of the patients dying within 24 months (sensitivity). Using the median MATH value as the cutoff, as in panel (a), provides greater sensitivity (70%) but lower selectivity (80%). The dashed line would have been the sensitivity-specificity relation if there were no relation of MATH to outcome. The area under the curve (AUC) of 0.82 means that in 82% of random pairs of patients with one dying and one surviving, the pre-treatment MATH value for the surviving patient would be lower.
RESULTS
Relations of clinical characteristics to MATH and to outcomes
As shown in Table 1, the 74 patients ranged from 33 to 76 years of age (mean, 58; SD, 10; median, 57) at diagnosis. The preponderance of males and of users of tobacco and alcohol is typical of HNSCC.33 One patient was African-American; the other 73 patients were of Caucasian origin. At time of last follow-up, 39 patients were no longer alive. Median follow-up time for surviving patients was 46 months.
Table 1.
Relation of clinical variables to MATH and to Overall Survival
| Relation to MATH |
Relation to Overall Survival | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Value | Number (% of total) |
MATH ± SD |
p-value | Hazard Ratio |
95% CI | p-value |
| Gender | 0.053 | 0.87 | |||||
| Female | 20 (27%) | 30 ± 11 | |||||
| Male | 54 (73%) | 36 ± 10 | |||||
| Age at Diagnosis* | 0.64 | 0.964/yr | (0.934 - 0.995) | 0.024 | |||
| 33-50 | 17 (23%) | 36 ± 10 | |||||
| 51-56 | 18 (24%) | 32 ± 9 | |||||
| 57-65 | 19 (26%) | 34 ± 11 | |||||
| 66-76 | 20 (27%) | 35 ± 11 | |||||
| Tobacco use | 0.67 | 0.44 | |||||
| No | 9 (8%) | 33 ± 13 | |||||
| Yes | 65 (92%) | 34 ± 10 | |||||
| Alcohol use | 0.43 | 0.20 | |||||
| No | 15 (21%) | 32 ± 10 | |||||
| Yes | 58 (78%) | 34 ± 10 | |||||
| unknown | 1 (1%) | 53 | |||||
| Family cancer history | 0.086 | 0.071 | |||||
| No | 22 (30%) | 36 ± 10 | 1 | ||||
| Yes | 23 (31%) | 31 ± 9 | 0.48 | (0.21 - 1.07) | |||
| unknown | 29 (39%) | 36 ± 10 | |||||
| Tumor site | 0.29 | 0.95 | |||||
| Hypopharynx | 8 (11%) | 38 ± 10 | |||||
| Larynx | 15 (20%) | 35 ± 9 | |||||
| Oral Cavity | 38 (51%) | 35 ± 11 | |||||
| Oropharynx | 11 (15%) | 28 ± 10 | |||||
| Sinonasal | 2 (3%) | 33 ± 3 | |||||
| Recurrent tumor | 0.93 | 0.99 | |||||
| No | 67 (91%) | 34 ± 11 | |||||
| Yes | 7 (9%) | 35 ± 7 | |||||
| T classification | 0.059 | 0.28 | |||||
| 1 or 2 | 24 (32%) | 31 ± 9 | |||||
| 3 or 4 | 46 (62%) | 36 ± 10 | |||||
| unknown | 4 (5%) | 37 ± 9 | |||||
| Differentiation Grade | 0.34 | 1 | |||||
| Well | 3 (4%) | 24 ± 2 | |||||
| Moderate | 47 (64%) | 34 ± 10 | |||||
| Poor | 22 (30%) | 35 ± 12 | |||||
| unknown | 2 (3%) | 39 ± 10 | |||||
| Perineural invasion | 0.079 | 0.0092 | |||||
| No | 31 (42%) | 32 ± 9 | 1 | ||||
| Yes | 36 (49%) | 36 ± 10 | 2.60 | (1.27 - 5.35) | |||
| unknown | 7 (9%) | 35 ± 13 | |||||
| Positive tumor margins | 0.13 | 0.31 | |||||
| No | 57 (69%) | 35 ± 10 | |||||
| Yes | 11 (15%) | 30 ± 9 | |||||
| unknown | 6 (8%) | 39 ± 12 | |||||
| Tumor HPV status¶ | 0.013 | 0.21 | |||||
| Positive | 11 (15%) | 27 ± 5 | |||||
| Negative | 62 (84%) | 35 ± 10 | |||||
| unknown | 1 (1%) | 50 | |||||
| Number of mutations¶ | 0.61 | 0.82 | |||||
| 17 to 52 | 18 (24%) | 32 ± 10 | |||||
| 53 to 92 | 19 (26%) | 33 ± 9 | |||||
| 93 to 129 | 18 (24%) | 35 ± 12 | |||||
| 130 to 739 | 19 (26%) | 36 ± 10 | |||||
| TP53 mutation status¶ | 0.0065 | 0.25 | |||||
| wild type | 28 (38%) | 31 ± 8 | |||||
| non-disruptive | 16 (22%) | 32 ± 10 | |||||
| disruptive | 30 (40%) | 39 ± 11 | 0.0029 | ||||
| N classification | 0.46 | 0.00009 | |||||
| 0 or 1 | 34 (46%) | 33 ± 10 | 1 | ||||
| 2 or 3 | 36 (49%) | 35 ± 11 | 4.39§ | (2.09 - 9.21) | |||
| unknown | 4 (5%) | 37 ± 9 | |||||
| Extracapsular spread | 0.31 | 0.23 | |||||
| No | 19 (26%) | 33 ± 10 | |||||
| Yes | 20 (27%) | 36 ± 10 | |||||
| Nodes not taken | 9 (12%) | 31 ± 13 | |||||
| Nodes not evaluated | 26 (35%) | 34 ± 9 | |||||
| TNM Stage | 0.46 | 0.066 | |||||
| II or III | 18 (24%) | 33 ± 12 | 1 | ||||
| IV | 52 (70%) | 35 ± 10 | 2.20 | (0.95 - 5.08) | |||
| unknown | 4 (5%) | 37 ± 9 | |||||
| Additional treatment | 0.71 | 0.14 | |||||
| None | 18 (24%) | 33 ± 8 | |||||
| Radiation alone | 11 (15%) | 33 ± 12 | |||||
| Chemotherapy alone | 1 (1%) | 25 | |||||
| Chemoradiation | 40 (54%) | 35 ± 11 | |||||
| unknown | 4 (5%) | 37 ± 6 | |||||
Relations of variables to overall survival were assessed by Cox proportional hazards analysis, restricted to cases having values for the variable being considered. Hazard ratios and 95% confidence intervals (95% CI) are shown for variables having p < 0.10 by the Wald test; only those with p < 0.05 are considered significant.
Age was analyzed as a continuous variable; breakdown of MATH by age groups is provided for illustration.
HPV, human papillomavirus, as assessed via PCR by Stransky et al.25 Relations of MATH to HPV and TP53 status and to the number of tumor-specific mutations in this data set were previously reported by Mroz and Rocco23 and are presented here for reference. All tumors had been subjected to exome sequencing, so the number of mutations is proportional to tumor mutation rate, conventionally expressed as mutations per megabase of sequenced DNA.25
Evidence of non-proportional hazards; p = 0.014 in chi-square test for trend of coefficient with time. Relation of N to survival was significant in non-parametric log-rank test, p = 0.00002.
We examined the relations of tumor MATH values to clinical variables. MATH values were not significantly related to any of the variables shown in Table 1, except for the previously reported relations of high MATH to HPV-negative tumors and to tumors having disruptive mutations in the TP53 tumor-suppressor gene.23 Of note, MATH values were not significantly different between primary and recurrent tumors. Although some true relation of MATH to gender, family cancer history, T classification, or perineural invasion (PNI) cannot be ruled out in this 74-patient dataset, MATH thus does not simply represent a proxy for some other standard clinical variable.
We also examined the relations of the clinical variables listed in Table 1 to outcome. Of those variables only age at diagnosis, PNI, and N classification were significantly related to overall survival in univariate Cox proportional hazards analyses. There was no significant survival difference between patients treated for recurrent versus primary tumors. The well-established high-risk factors of negative HPV status26, 27 and disruptive TP53 mutations28, 29 were not significantly related to outcome in univariate analysis, presumably due to the relatively small number of cases or the lack of a uniform treatment regimen. Notably, the tumor mutation rate itself, as conventionally assessed by the number of mutated loci per megabase of sequenced genomic DNA,25 was also not related to outcome; as previously reported,23 genetic heterogeneity of HNSCC assessed by MATH is not significantly related to mutation rate.
Relation of MATH to overall survival
In univariate analysis, higher MATH was strongly associated with shorter overall survival. We began by performing Cox proportional hazard regression of overall survival against MATH taken as a continuous variable, because MATH values spanned a range from 19 to 55 units with no obvious subgroups of MATH values.23 In this analysis, each individual tumor MATH value was related to the corresponding patient’s time to death or last follow-up, to determine how quickly the hazard of death grew as MATH values increased. Among all 74 cases, each additional unit of increase in MATH was associated with a 4.7% increased hazard of death (Table 2). This is equivalent to a hazard ratio of 5.2 between the tumors with the highest and lowest MATH values.
Table 2.
Relation of MATH to Overall Survival
| Relation to Overall Survival | |||||
|---|---|---|---|---|---|
| Analysis | deaths/ cases |
Hazard Ratio |
95% CI | p-value | |
| Univariate | 39/74 | 1.047/unit | (1.017 - 1.078) | 0.002 | |
| primary tumors | 34/67 | 1.044/unit | (1.013 - 1.075) | 0.005 | |
| Stratified by recurrence | 39/74 | 1.046/unit | (1.015 - 1.077) | 0.003 | |
| Stratified by HPV status | 39/73 | 1.051/unit | (1.018 - 1.084) | 0.002 | |
| primary tumors | 34/66 | 1.046/unit | (1.013 - 1.081) | 0.006 | |
| Univariate; HPV-negative subset | 35/62 | 1.050/unit | (1.017 - 1.083) | 0.003 | |
| primary tumors | 30/55 | 1.045/unit | (1.012 - 1.080) | 0.008 | |
| Stratified by TP53 mutation status | 39/74 | 1.048/unit | (1.016 - 1.080) | 0.003 | |
| primary tumors | 34/67 | 1.046/unit | (1.014 - 1.079) | 0.005 | |
| Univariate; disruptive TP53 subset | 15/30 | 1.088/unit | (1.031 - 1.15) | 0.002 | |
| primary tumors | 14/29 | 1.094/unit | (1.033 - 1.16) | 0.002 | |
| Stratified by PNI status | 36/67 | 1.035/unit | (1.002 - 1.068) | 0.035 | |
| primary tumors | 31/60 | 1.030/unit | (0.997 - 1.064) | 0.078 | |
| Univariate; subset with PNI | 25/36 | 1.047/unit | (1.006 - 1.089) | 0.023 | |
| primary tumors | 21/31 | 1.041/unit | (0.999 - 1.084) | 0.055 | |
| Stratified by T classification (1,2 vs. 3,4) | 36/70 | 1.043/unit | (1.012 - 1.075) | 0.006 | |
| primary tumors | 34/67 | 1.042/unit | (1.010 - 1.075) | 0.009 | |
| Univariate, subset with T > 2 | 25/46 | 1.049/unit | (1.011 - 1.088) | 0.011 | |
| primary tumors | 24/44 | 1.047/unit | (1.008 - 1.086) | 0.016 | |
| Stratified by Stage (II,III vs IV) | 36/70 | 1.047/unit | (1.015 - 1.081) | 0.004 | |
| primary tumors | 34/67 | 1.047/unit | (1.014 - 1.082) | 0.006 | |
| Univariate; subset with Stage IV | 29/52 | 1.059/unit | (1.020 - 1.10) | 0.003 | |
| primary tumors | 28/50 | 1.057/unit | (1.018 - 1.098) | 0.004 | |
| Stratified by N classification (0,1 vs. 2,3) | 36/70 | 1.048/unit | (1.016 - 1.080) | 0.003 | |
| primary tumors | 34/67 | 1.049/unit | (1.017 - 1.083) | 0.003 | |
| Univariate; subset with N > 1 (all primary) | 25/36 | 1.056/unit | (1.016 - 1.096) | 0.005 | |
| Multivariate (based on variables significantly related to outcome in univariate analyses) |
33/63 | 4 × 10−6 | |||
| MATH | 1.043/unit | (1.008 - 1.080) | 0.017 | ||
| Age | 0.946/yr | (0.910 - 0.982) | 0.003 | ||
| N >1 | 4.92§ | (2.18 - 11.1) | 0.0001 | ||
| PNI | 2.49 | (1.15 - 5.39) | 0.021 | ||
| Multivariate | primary tumors | 31/60 | 4 × 10−6 | ||
| MATH | 1.045/unit | (1.008 - 1.084) | 0.016 | ||
| Age | 0.940/yr | (0.904 - 0.978) | 0.002 | ||
| N >1 | 5.94 | (2.45 - 14.4) | 0.0001 | ||
| PNI | 2.48 | (1.11 - 5.53) | 0.027 | ||
| Univariate; cases not involving chemotherapy | 13/30 | 1.00/unit | (0.945 - 1.062) | 0.96 | |
| primary tumors | 9/24 | 0.989/unit | (0.924 - 1.059) | 0.74 | |
| Univariate; cases involving chemotherapy (all primary) | 23/41 | 1.061/unit | (1.022 - 1.10) | 0.002 | |
Results of Cox proportional hazards analysis on relations of MATH to overall survival of patients with tumor exome sequencing results reported by Stransky et al.25 Each analysis was performed on all cases having values for the variable(s) of interest, and on the subsets involving primary tumors, with the number of cases and of deaths shown. Hazard ratios are for MATH unless otherwise noted. MATH and Age were analyzed as continuous variables, so results for those variables are reported as multiplicative change in hazard per unit increase in MATH value or per year of age.
Evidence of non-proportional hazards for N; p = 0.048 in chi-square test for trend of coefficient of N with time. Relations of the other 3 variables to overall survival were similar in analysis stratified by N to allow for this non-proportionality; in that stratified analysis, global chi-square test had p = 0.96.
To determine whether MATH could be used to classify patients into high- and low-risk groups, we then compared patients whose tumors had MATH values above versus below the median value of 32 units. Among all 74 cases, the hazard ratio associated with MATH above the median was 2.46 (95%CI: 1.26 - 4.79; p = 0.008, Wald test); survival curves are shown in Fig. 1a.
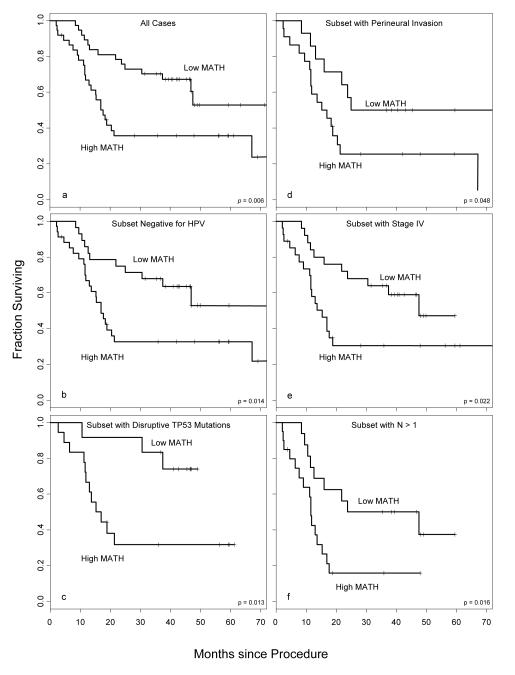
Figure 1. Relation of mutant-allele tumor heterogeneity (MATH) to outcome in clinically defined subsets of HNSCC.
Each panel shows overall survival curves for patients whose tumors had MATH above (“High MATH”) versus below (“Low MATH”) the overall median value of 32 units. Survival is calculated from the time of the surgical procedure that obtained the tumor sample subjected to NGS by Stransky et al.25 Crosses represent the last follow-up times for surviving patients. p-values are for log-rank tests. Panels represent different subsets of cases. The numbers of cases and numbers of deaths in each subset analysis are shown in Table 2. (a) Comparison of High-MATH and Low-MATH groups for all 74 cases. (b) Subset with HPV-negative tumors. (c) Subset in which tumors had disruptive mutations in the TP53 gene; all these tumors were also HPV-negative. (d) Subset with documented perineural invasion. (e) Subset with Stage IV disease. (f) Subset with N classification of 2 or 3; all these cases were Stage IV.
The relation of MATH both to HPV status and to TP53 mutation status (Table 1) raised the possibility that MATH might not be related to outcomes within groups defined by those variables. Critically, as shown in Table 2, MATH as a continuous variable was still related to outcome when cases were stratified by HPV status or by TP53 status. MATH was also significantly related to outcome when classes were stratified by perineural invasion status or by N classification (Table 2), each of which was also significantly related to outcome (Table 1), or by T classification or TNM stage (Table 2).
Furthermore, MATH was related to outcome within the known or expected worse-outcome subsets of cases defined by each of these variables (HPV-negative, disruptive TP53 mutation, presence of PNI, stage IV, N 2 or 3, T 3 or 4). This was true both for MATH as a continuous variable (Table 2) and for classes based on MATH above versus below the median (Figure 1, b-f). These significant relations of MATH to outcome in stratified or subset analyses were maintained, except for groups defined by PNI, when analysis was restricted to the 67 cases with primary tumors (Table 2). MATH was also significantly related to outcome in a multivariate analysis that incorporated all 4 variables found significant in univariate outcome analysis (Table 2).
Genetic heterogeneity might be expected to bear different relations to outcome depending on the type of therapy used. Thus we evaluated the relation of MATH to outcome within subsets of patients defined by therapy. MATH was not significantly related to outcome in the cases with either no adjuvant therapy or with radiation alone as adjuvant (Table 2), although the small number of such cases means that some relation of MATH to outcome in those treatment settings cannot be ruled out.
In contrast, the relation of MATH to outcome was clearly seen in the cases in which systemic chemotherapy, usually combined with radiation, was received (Table 2, last row). In these 41 cases, all involving primary tumors, the hazard of death associated with MATH as a continuous variable increased 6.1% per unit, equivalent to a hazard ratio of 8.4 between the tumors with the highest and the lowest MATH values. In terms of classification, the hazard ratio for MATH above the median was 4.1 (95%CI: 1.6 to 10.2; Figure 2a). The ROC curve of Figure 2b shows the tradeoff between sensitivity and specificity at different choices of the MATH classification cutoff for these 41 chemotherapy cases. Thus the relation of higher MATH to worse outcome was most pronounced for patients who received chemotherapy.
DISCUSSION
These results provide direct evidence, based on novel genomic analysis, that high genetic heterogeneity is related to shorter overall survival. This result is consistent with the long-standing hypothesis that high genetic heterogeneity is a risk factor for worse outcome in cancer.18-22 Although the mechanisms linking high genetic heterogeneity to shorter overall survival cannot be determined from these data, our results are consistent with the hypothesis that genetically heterogeneous tumors are more likely to contain subclones of cancer cells resistant to chemoradiation therapy .
A primary role of intra-tumor genetic heterogeneity in determining clinical outcome may shed some light on the relations of disruptive TP53 mutations and HPV-negative status to worse outcomes in HNSCC.28, 29 Insofar as TP53 mutations impair both DNA repair processes and the removal of cells that develop additional mutations,1 early clonal expansion of TP53-mutated cells would be predicted to lead to increased genetic heterogeneity as measured by MATH. The relation of high MATH specifically to disruptive but not to non-disruptive TP53 mutations suggests that disruptive mutations, as defined by the nature and site of the mutation in the p53 protein,28 are most likely to impair both DNA repair and the removal of cells with newly mutated genomes and thus to promote genetic heterogeneity. Furthermore, high MATH was still associated with shorter overall survival within the subset of cases involving disruptive TP53 mutations or when cases were stratified by TP53 status (Table 2). These results suggest that a major influence of disruptive TP53 mutations on outcome may be their tendency to increase genetic heterogeneity. Similarly, HPV-negative tumors have greater genetic heterogeneity than do HPV-positive tumors, consistent with lower genetic heterogeneity as a reason for better outcomes in HPV-positive HNSCC, which are typically treated with concurrent chemoradiation.
These results raise questions about the processes that lead to high genetic heterogeneity within a tumor. The lack of relation of mutation rate to MATH and to outcome indicates that mutations alone are not enough. Rather, additional processes must allow the development and survival of genetically distinct subclones. Disruptive TP53 mutations are evidently involved in some cases, yet processes other than mutations in TP53 can lead to high genetic heterogeneity. Nearly one-third (12 of 37) of the tumors within the top half of MATH values had no TP53 mutation, disruptive or otherwise. Additional heterogeneity-inducing mechanisms thus need to be identified. Insofar as high genetic heterogeneity is a cause of shorter survival, therapies that target these mechanisms or the resulting heterogeneity itself may represent novel therapeutic approaches.
These results also raise questions about how therapy might affect intra-tumor heterogeneity. In particular, if a particular mode of therapy selects for one or a few subclones from a tumor, genetic heterogeneity would be decreased initially but might increase later as new subclones arise. Although the average MATH value of the 7 recurrent tumors in this study did not differ significantly from that of the 67 primary tumors (Table 1), the small number of cases, the variety of prior treatments (1 surgery alone, 4 plus radiation, 2 plus chemoradiation), and the lack of corresponding pre-treatment specimens mean that further studies are required to determine both how therapy affects heterogeneity in HNSCC and the clinical implications of heterogeneity in the setting of recurrent disease.
The relation of genetic heterogeneity to outcome was surprisingly strong for this relatively small number of cases, which included both primary and recurrent tumors and which were not part of a controlled-treatment study design. This group of patients was evidently too small or too heterogeneous to show a significant relation of HPV status or of disruptive TP53 mutations to outcome, despite the well established relation of these classifications to outcome in studies of HNSCC that were larger or involved homogeneous treatment regimens.26-29 In contrast, MATH was significantly related to outcome not only on its own but also within the already high-risk groups defined by those and by other variables (Table 2, Figure 1).
MATH values were not significantly related to N classification, the best single prognostic variable in this data set, or to TNM stage. MATH was related to outcome both when cases were stratified by N classification or stage and when analysis was restricted to the subsets of high-N and high-stage cases. These results thus support the use of MATH as an independent prognostic marker.
As NGS becomes widely used in clinical oncology, calculating MATH from the tumor-specific mutant-allele fractions in NGS results will provide a clinically relevant measure of genetic heterogeneity. MATH is not specific to HNSCC; it can be calculated from NGS results on any type of tumor that has an adequate number of tumor-specific mutations. This method of analyzing genetic heterogeneity thus also gives a concrete and straightforward way to test hypotheses about genetic heterogeneity and outcomes in other types of cancer. The present study indicates that the type of genetic heterogeneity captured by MATH values is related to HNSCC outcomes. Future controlled studies will determine the clinical usefulness of MATH as a prognostic biomarker in HNSCC and in other types of cancer.
Acknowledgments
Funding sources: The National Institute of Dental and Craniofacial Research (R01 DE022087; RC2DE020958), the National Cancer Institute (R21 CA119591), the Cancer Prevention Research Institute of Texas (RP100233). and the Bacardi MEEI Biobank Fund
Footnotes
Human subjects: Informed consent was obtained from all human subjects and/or guardians, under protocol 99-069 approved by the University of Pittsburgh IRB.
Financial disclosures: Mass. General Hospital has filed a patent application based on subject matter discussed in this manuscript, with Edmund A. Mroz and James W. Rocco listed as inventors. There are no other disclosures to report by any of the authors.
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasa Y, Michor F. Evolutionary dynamics of intratumor heterogeneity. PLoS One. 2011;6:e17866. doi: 10.1371/journal.pone.0017866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jovanovic L, Delahunt B, McIver B, Eberhardt NL, Grebe SK. Most multifocal papillary thyroid carcinomas acquire genetic and morphotype diversity through subclonal evolution following the intra-glandular spread of the initial neoplastic clone. J Pathol. 2008;215:145–154. doi: 10.1002/path.2342. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Wang K, Jensen TD, Li S, Bolund L, Wiuf C. Tumor heterogeneity in neoplasms of breast, colon, and skin. BMC Res Notes. 2010;3:321. doi: 10.1186/1756-0500-3-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 11.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russnes HG, Navin N, Hicks J, Borresen-Dale AL. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest. 2011;121:3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SP, Morin RD, Khattra J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 15.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Hou Y, Yin X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakansson L, Trope C. On the presence within tumours of clones that differ in sensitivity to cytostatic drugs. Acta Pathol Microbiol Scand A. 1974;82:35–40. doi: 10.1111/j.1699-0463.1974.tb03825.x. [DOI] [PubMed] [Google Scholar]
- 19.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 20.Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 21.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 23.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R. Next-generation sequencing in the clinic: are we ready? Nat Rev Genet. 2012;13:818–824. doi: 10.1038/nrg3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 27.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner HD, Sandulache VC, Ow TJ, et al. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res. 2012;18:290–300. doi: 10.1158/1078-0432.CCR-11-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Development Core Team . R: A language and environment for statistical computing Vienna. R Foundation for Statistical Computing; Austria: 2011. [Google Scholar]
- 31.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7 th ed Springer; New York: 2009. [Google Scholar]
- 32.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 33.Marur S, Forastiere AA. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2008;83:489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]