Abstract
Bile acid synthesis not only produces physiological detergents required for intestinal nutrient absorption, but also plays a critical role in regulating hepatic and whole body metabolic homeostasis. We recently reported that over-expression of cholesterol 7α-hydroxylase (CYP7A1) in the liver resulted in improved metabolic homeostasis in Cyp7a1 transgenic (Cyp7a1-tg) mice (Li et al., Hepatology 2010; 52:678-690 & Li et al., Hepatology 2011; 53:996–1006). This study further investigated the molecular links between bile acid metabolism and lipid homeostasis. Microarray gene profiling revealed that CYP7A1 overexpression led to marked activation of the steroid response element binding protein 2 (SREBP2)-regulated cholesterol metabolic network and absence of bile acid repression of lipogenic gene expression in the liver of Cyp7a1-tg mice. Interestingly, Cyp7a1-tg mice showed significantly elevated hepatic cholesterol synthesis rates but reduced hepatic fatty acid synthesis rates, which was accompanied by increased 14C-glucose-derived acetyl-CoA incorporation into sterols for fecal excretion. Induction of SREBP2 also co-induces intronic microRNA-33a (miR-33a) in the SREBP2 gene in Cyp7a1-tg mice. Overexpression of miR-33a in the liver resulted in decreased bile acid pool, increased hepatic cholesterol content and lowered serum cholesterol in mice. This study suggests that a CYP7A1-SREBP2-miR-33a axis plays a critical role in regulation of hepatic cholesterol, bile acid and fatty acid synthesis. Antagonism of miR-33a may be a potential strategy to increase bile acid synthesis to maintain lipid homeostasis and prevent non-alcoholic fatty liver disease (NAFLD), diabetes and obesity.
Keywords: cholesterol 7α-hydroxylase, steroid response element binding protein, lipid metabolism, diabetes, non-alcoholic fatty liver disease, microRNA
Bile acids are synthesized from cholesterol exclusively in the liver (1). The rate of bile acid synthesis is mainly controlled by transcriptional regulation of cholesterol 7α-hydroxylase (CYP7A1) (1), which encodes the rate-limiting enzyme in the classic bile acid synthesis pathway. When bile acid levels increase, bile acids repress their own synthesis and stimulate biliary lipid secretion. These mechanisms allow the liver to efficiently maintain lipid homeostasis. Bile acids activate farnesoid X receptor (FXR) and the G protein-coupled receptor TGR5, and also several cell signaling pathways to regulate bile acid synthesis and lipid metabolism (1). Pharmacological activation of either FXR or TGR5 receptor has been shown to improve lipid, glucose and energy homeostasis, glucose tolerance and insulin sensitivity (2, 3). Paradoxically, loss of FXR in obese and diabetic mice reduced body weight and improved peripheral insulin sensitivity (4), and decreasing bile acid pool size with the specific FXR agonist GW4064 caused increased susceptibility to diet-induced obesity, fatty liver and hypertriglyceridemia (5). It is likely that activation of different bile acid signaling in different mouse models might have different effects on hepatic metabolism, diabetes and obesity. In Cyp7a1-tg mice, both CYP7A1 enzyme activity and bile acid pool size are doubled (6), biliary cholesterol and bile acid secretion are stimulated, and serum cholesterol is decreased, while serum triglyceride levels remain the same (7). These metabolic changes caused by increased CYP7A1 expression result in significantly improved lipid homeostasis and protection against hepatic steatosis, insulin resistance and obesity (6). Therefore, further study is necessary to understand the participation of bile acid synthesis in the regulation of metabolic homeostasis, NAFLD and diabetes.
Bile acid metabolism is closely linked to whole-body cholesterol homeostasis; bile acid synthesis and bile acid-facilitated biliary cholesterol secretion are the only significant pathways for cholesterol elimination from the body. Furthermore, the liver acquires cholesterol through dietary absorption, receptor-mediated uptake and de novo synthesis. Intracellular cholesterol/oxysterols play an important role in regulation of cholesterol synthesis through the transcriptional factor sterol response element binding protein 2 (SREBP2) (8). Upon increased intracellular cholesterol levels, SREBP2 precursor (125 kDa) forms a complex with insulin-induced gene (INSIG) and SREBP cleavage-activating protein (SCAP), which is retained in the endoplasmic reticulum (ER) membrane. When cholesterol levels decrease, SCAP escorts SREBP2 precursor to the Golgi where two steroid sensitive proteases (S1P and S2P) cleave an N-terminal fragment (68 kDa), subsequently translocating into the nuclei to activate its target genes, including LDL receptor and key genes involved in de novo cholesterol synthesis (8).
MicroRNAs (miRs) are small non-coding RNAs that, after base pairing with complementary sequences of target mRNAs, promote mRNA degradation or inhibit protein synthesis. MiR-33a, encoded by intron 16 of the SREBP2 gene, has recently been shown to regulate cellular cholesterol homeostasis (9), biliary bile acid secretion (10), and fatty acid oxidation (11). Additionally, when cellular cholesterol levels decrease, miR-33a expression is co-induced with SREBP2 mRNA. MiR-33a inhibits ABCA1 and ABCG1 to reduce cellular cholesterol efflux. Studies in mice treated with anti-miR-33a or in genetic miR-33a deficient mice showed miR-33a antagonism induced ABCA1 in macrophages and liver, increased serum HDL levels, and promoted macrophage-to-feces reverse cholesterol transport (12). Additionally, miR-33a antagonism promoted regression of atherosclerosis in mice and non-human primates (13, 14). These studies suggest that miR-33a acts in a synergistic manner with SREBP2 to regulate cellular cholesterol homeostasis.
The goal of this study is to investigate the potential impact of stimulation of bile acid synthesis on hepatic lipid metabolism using Cyp7a1-tg mice as a model. Here we report that bile acid synthesis plays an important role in integrating intracellular cholesterol sensing and homeostasis via modulating the liver SREBP2/miR-33a axis. Our study suggests antagonism of miRNA-33a to induce CYP7A1 and bile acid synthesis may be a potential therapeutic approach to treat NAFLD and diabetes.
Materials and Methods
Cyp7a1-transgenic mice
Cyp7a1 transgenic mice over-expressing rat Cyp7a1 cDNA under an ApoE3 hepatic control region have been described previously (6). “Humanized” CYP7A1 mice expressing human CYP7A1 from a BAC clone on a mouse cyp7a1 knockout background were generated as described previously (15). Mice were maintained under a 12-hr light (6 am - 6 pm) and 12-hr dark (6 pm - 6 am) cycle. Male wild type and Cyp7a1-tg mice were fed chow or Western diet (Harlan-Teklad 88137; 42% fat calories, 0.2 % cholesterol) for 4 months. The Institutional Animal Care and Use Committee approved all animal protocols.
Microarray and pathway analysis
MouseRef-8 v2.0 Expression BeadChip kit (BD-202-0202, Illumina, San Diego, CA) was used for microarray analysis. The raw microarray data were log2 transformed and processed with background correction and quintile normalization. Quality control analyses were applied to detect outlier samples. The expression signals with an Illumina detection threshold < 0.05 across all samples were used. Linear models and empirical Bayes method in Limma (16) were used to access the differential expression between the control and transgenic groups. Those genes that satisfied the false discovery rate (FDR) adjusted p-value < 0.05 or raw p-value of <0.001, whichever was more stringent (Benjamini-Hochberg method), and fold change threshold of 1.5 was identified for inclusion in the functional pathway and network analysis. Functional profiling of differentially affected biological processes and pathways between transgenic and control mice were evaluated using publicly available tools (e.g., NCBI, Ensemble, FatiGO, FatiScan, MGI, and KEGG) and commercial pathway analysis databases such as Metacore (Metacore™, www.genego.com) and Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com). The consensus findings from these tools were used to interpret and understand the biological mechanisms behind the data. Microarray data have been deposited to NCBI’s GEO repository (accession #GSE38872).
Measurement of cholesterol and fatty acid synthesis rate
Mice were fasted for 4 hr and were ip. injected with 10 µCi [1-14C]-sodium acetate (Perkin Elmer, Waltham, MA). Mice were sacrificed 30 min after injection and ~250 mg liver was rinsed in ice-cold 1xPBS. Liver tissue was saponified in 2.2 ml mixture of 50% KOH: 95% ethanol (1/10, v/v) at 70°C overnight. 3H-cholesterol (1 µCi) was added to the same tube as a recovery control. Sterols were extracted in 3 ml hexane, dried, and re-dissolved in 300 µl mixture of acetone: ethanol (1/1, v/v). Sterols were then precipitated with 1 ml digitonin (0.5% in 95% ethanol) overnight at room temperature. Saponified fatty acids were acidified and extracted with petroleum ether.
Measurement of fecal sterol excretion
Wild type and Cyp7a1-tg mice were ip. injected with a single dose of glucose (8 g/kg) containing 5 µCi [1-14C] glucose (Perkin Elmer, Waltham, MA) as an isotope tracer. Mice were allowed free access to standard chow and water, and feces were collected for 3 consecutive days. Fecal samples were then homogenized in 5 ml mixture of 50% KOH: 95% ethanol (1/10, v/v) at 70 °C overnight. 3H-cholesterol (1 µCi) was added to the same tube as a recovery control. Neutral sterols were separated from bile acids by extraction in 6 ml hexane, dried and re-dissolved in 1 ml mixture of acetone: ethanol (1/1, v/v), and were then precipitated with 3 ml of digitonin (0.5% in 95% ethanol) overnight at room temperature.
Recombinant adenovirus
Adenovirus-expressing miR-33a was purchased from Applied Biological Materials, Inc. (Richmond, BC, Canada). Adeno-null that does not express a gene product was purchased from Vector Biolabs (Philadelphia, PA). Adenovirus was administered at 2×109 pfu/mouse via tail vein. Experiments were carried out 7 days post-injection.
Statistical analysis
Results were expressed as mean ± S.E. unless noted. Statistical analysis was performed by Student’s t-test; p < 0.05 indicates statistical significance.
Results
Microarray gene expression profiling in the liver of Cyp7a1-tg mice revealed a tight link between bile acid synthesis and cholesterol metabolism
To obtain molecular insight into the role of bile acid synthesis in maintaining hepatic lipid homeostasis, we used microarray gene profiling to identify differentially expressed genes in the liver of chow-fed and Western high fat diet-fed Cyp7a1-tg and wild type mice. Under chow-fed conditions, 77 genes were identified as differentially expressed with a 2-fold induction or a 50% inhibition, while under Western diet-fed conditions 144 genes were differentially expressed (Supplemental Fig 1A). There were 52 differentially expressed genes that were identified by comparison under both chow-fed condition and Western diet-fed condition. Since the expressions of these 52 genes are likely genotype-dependent but independent of dietary condition, we speculate that some of these genes may be responsible for the improved metabolic homeostasis in Cyp7a1-tg mice. Remarkably, of all 35 up-regulated genes in Cyp7a1-tg mice, most genes are clustered in cholesterol metabolism, with 12 of the top 13 up-regulated genes directly involved in cholesterol biosynthesis, esterification, transport and regulation (Table 1). Ingenuity Pathway Analysis (IPA) identified sterol biosynthesis as the top differentially regulated pathway in Cyp7a1-tg mice, followed by tryptophan metabolism, LPS/IL-1-mediated inhibition of RXR function, bile acid synthesis, and metabolism of xenobiotics by cytochrome P450 (Supplemental Table 1). Some of the results were confirmed by quantitative real-time PCR analysis. Table 2 shows real-time PCR analysis of expression of key regulatory genes in cholesterol metabolism, bile acid synthesis and detoxification, and fatty acid metabolism in chow-fed and Western diet-fed wild type and Cyp7a1-tg mouse liver. HMG-CoA reductase and HMG-CoA synthase gene expression was induced more than 10-fold in chow-fed and high fat diet-fed Cyp7a1-tg mice compared to wild type mice. Both microarray and real-time PCR detected higher SREBP2 mRNA in Cyp7a1-tg mice (Table 1 & 2), and mature SREBP2 protein was markedly increased in the liver of Cyp7a1-tg mice (Supplemental Fig. 2). Other SREBP2 induced genes such as LDL receptor, CYP51, and PCSK9 were also induced. Taken together, these data support activation of a SREBP2-regulated cholesterol metabolic network in Cyp7a1-tg mice. It is well known that SREBP2 maturation is repressed by cholesterol. Consistently, all SREBP2 target genes were down regulated upon feeding wild type mice a cholesterol-rich Western diet (Table 1 and 2). Interestingly, Western diet feeding did not repress induction of cholesterologenic genes in Cyp7a1-tg mice (Table 1 and 2), suggesting that increasing bile acid synthesis has a dominant positive effect on hepatic cholesterol synthesis.
Table 1.
Microarray analysis of genes related to cholesterol and fatty acid metabolism
| Rank | gene | WT + C | Tg + C | WT + WD | Tg + WD |
|---|---|---|---|---|---|
| 1 | SQLE | 1 | 10.60* | 0.49* | 14.80* |
| 2 | FDPS | 1 | 8.60* | 0.51* | 13.40* |
| 3 | CYP51 | 1 | 7.50* | 0.45* | 10.00* |
| 4 | SC4MOL | 1 | 6.60* | 0.39* | 8.70* |
| 5 | PCSK9 | 1 | 6.50* | 0.55* | 10.30* |
| 6 | LSS | 1 | 4.90* | 0.66* | 7.60* |
| 7 | IDI1 | 1 | 4.90* | 0.68* | 6.80* |
| 8 | MVD | 1 | 4.40* | 0.84 | 8.80* |
| 9 | NSDHL | 1 | 4.40* | 0.74* | 5.40* |
| 10 | LOC100040592 | 1 | 4.00* | 0.78 | 5.90* |
| 11 | HSD3B5 | 1 | 3.70* | 0.88* | 3.40* |
| 13 | HSD17B7 | 1 | 3.40* | 0.47* | 4.10* |
| 15 | ApoA4 | 1 | 3.00* | 2.70* | 2.30* |
| 16 | PMVK | 1 | 2.90* | 0.73* | 4.20* |
| 19 | StarD4 | 1 | 2.60* | 0.56* | 4.20* |
| 20 | DHCR7 | 1 | 2.50* | 0.72* | 4.0* |
| 25 | INSIG1 | 1 | 2.30* | 0.75* | 2.7* |
| 35 | ACAT2 | 1 | 2.00* | 1.1 | 3.1* |
| LDLR | 1 | 1.20 | 1.0 | 1.4 | |
| SREBP2 | 1 | 1.50 | 0.85 | 1.8* | |
| CES1 | 1 | 0.37* | 1.5* | 0.6* | |
| PPARγ | 1 | 0.40* | 1.2 | 0.3* |
Note WT: wild type; Tg: Cyp7a1-tg; C: chow; WD: Western diet. n=4. A
indicates statistical significance, p < 0.05; vs. WT+C group.
Table 2.
Gene expression analysis by quantitative real-time PCR
| Gene | WT + C | Tg + C | WT + WD | Tg + WD |
|---|---|---|---|---|
|
Cholesterol metabolism | ||||
| SREBP2 | 1±0.18 | 3.10±0.25* | 0.55±0.04* | 3.23±0.4* |
| LDLR | 1±0.27 | 2.70±0.27* | 0.55±0.04* | 2.40±0.10* |
| CYP51 | 1±0.26 | 14.7±1.25* | 0.27±0.03* | 19.3±1.27* |
| HMGCoAR | 1±0.3 | 10.8±2.20* | 0.36±0.03* | 13.7±0.50* |
| HMGCS1 | 1±0.20 | 7.99±2.18* | 0.77±0.06 | 19.55±1.21* |
| ABCG8 | 1±0.3 | 1.80±0.20* | 2.60±0.03* | 2.70±0.30* |
| PCSK9 | 1±0.3 | 15.7±1.6* | 0.30±0.01* | 25.7±2.40* |
| Insig1 | 1±0.3 | 6.90±1.10* | 0.60±0.02* | 8.60±10.0* |
| StarD4 | 1±0.3 | 4.90±0.48* | 0.30±0.02* | 5.10±0.57* |
| CES1 | 1±0.1 | 0.23±0.04* | 1.17±0.10 | 0.40±0.04* |
|
Bile acid synthesis/detoxification | ||||
| BSEP | 1±0.14 | 1.10±0.05 | 2.40±0.30* | 3.57±0.45* |
| CYP3A11 | 1±0.10 | 0.84±0.10 | 1.00±0.10 | 0.90±0.20 |
| SULT2a2 | 1±0.20 | 1.78±0.27* | 1.38±0.40 | 0.70±0.40 |
| MRP3 | 1±0.07 | 0.70±0.100 | 0.90±0.08 | 0.60±0.01* |
| CYP7A1 | 1±0.02 | 0.04±0.001* | 0.67±0.10* | 0.02±0.001* |
| CYP8B1 | 1±0.12 | 0.05±0.001* | 0.28±0.01* | 0.05±0.001* |
| SHP | 1±0.20 | 0.90±0.160 | 2.35±0.73* | 3.8±10* |
|
Fatty acid metabolism | ||||
| SREBP1 | 1±0.03 | 1.66±0.03* | 4.53±0.90* | 1.60±0.30* |
| FAS | 1±0.06 | 3.80±0.30* | 1.70±0.13* | 5.60±0.30* |
| ACC | 1±0.28 | 1.90±0.30* | 0.80±0.14 | 1.90±0.20* |
| ChREBP | 1±0.20 | 1.56±0.16* | 1.96±0.13* | 0.98±0.19 |
| L-PK | 1±0.10 | 1.86±0.15* | 2.10±0.20* | 0.90±0.16 |
| CD36 | 1±0.05 | 0.18±0.06* | 1.02±0.10 | 0.24±0.03* |
| PPARγ | 1±0.15 | 0.17±0.03* | 1.50±0.08* | 0.20±0.03* |
| PPARα | 1±0.15 | 1.14±0.25 | 1.73±0.15* | 0.92±0.30 |
Note WT: wild type; Tg: Cyp7a1-tg; C: chow; WD: Western diet. n=4. A
indicates statistical significance, p < 0.05; vs. WT+C group.
Dissociation of hepatic bile acid metabolism and lipogenic gene expression from fatty acid synthesis in Cyp7a1-tg mice
In Cyp7a1-tg mice, endogenous mouse CYP7A1 and sterol 12α-hydroxylase (CYP8B1) mRNA levels were decreased as the result of increased bile acid feedback (Table 2). However, FXR target genes small heterodimer partner (SHP), involved in regulation of bile acid synthesis, and canalicular bile salt export pump (BSEP), involved in bile acid efflux, were not identified by microarray analysis and their mRNA levels were not induced (Table 2). Solute transporter 2a2 (SULT2a1), involved in efflux of sulfoconjugated xenobiotics and bile acids was increased in Cyp7a1-tg mice indicating increased excretion of conjugated bile acids and xenobiotics. Multidrug resistant protein 3 (MRP3, ABCC3), the basolateral efflux transporter of conjugated bile acid expressed under cholestatic condition was reduced in hepatocytes of western diet-fed Cyp7a1-tg mice (Table 2), consistent with no cholestatic injury in these mice. The mRNA levels of SREBP1c, fatty acid synthase (FAS) and acetyl CoA carboxylase (ACC) were not changed in chow-fed Cyp7a1-tg mice over wild type controls. However, real-time PCR detected a ~66% induction of SREBP1c, ~4-fold induction of FAS, and ~2-fold induction of ACC in Cyp7a1-tg mice (Table 2). These data also did not indicate differential expression of any fatty acid synthesis genes and IPA did not identify fatty acid metabolism as a top regulated pathway. Interestingly, mRNA levels of CD36, a major hepatic fatty acid transporter, were reduced in Cyp7a1-tg. Peroxisome proliferator activated receptor γ (PPARγ), involved in induction of hepatic fatty acid synthesis, was markedly reduced in both chow and high fat diet-fed Cyp7a1-tg mice. Liver pyruvate kinase (L-PK) and carbohydrate response element binding protein (CnREBP), involved in lipogenesis, were increased in chow-fed but decreased in Western diet-fed Cyp7a1-tg mice compared to respective wild type mice. These data suggest that reduced free fatty transport to hepatocytes and fatty acid synthesis in hepatocytes may prevent hepatic steatosis in Cyp7a1-tg mice.
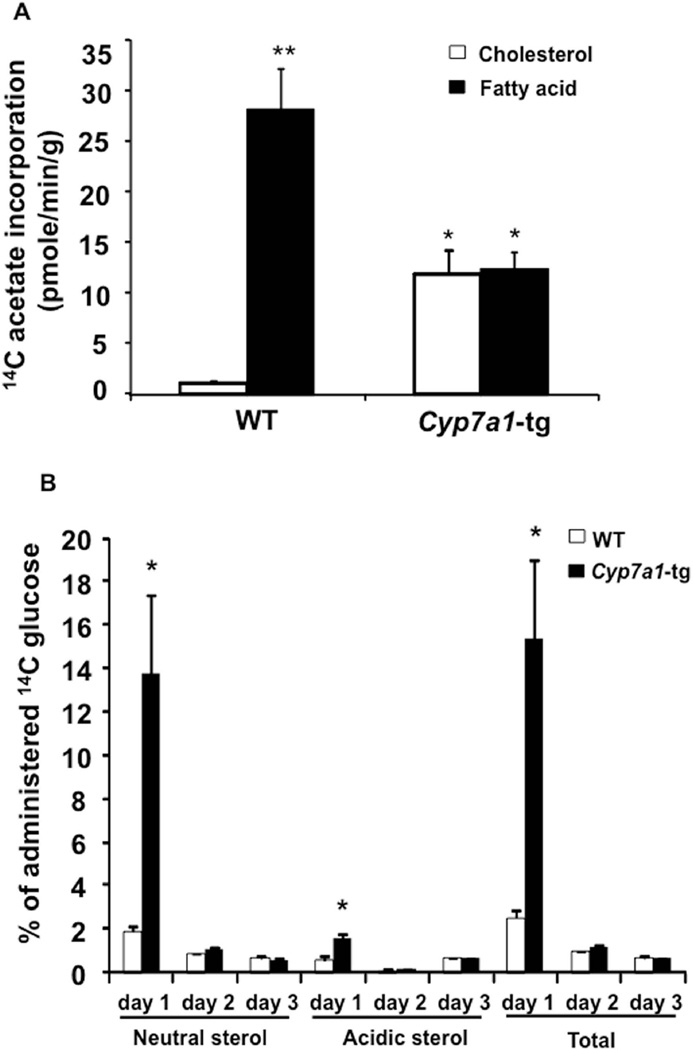
Given that induction of hepatic bile acid synthesis in Cyp7a1-tg mice is associated with increased expression of cholesterologenic and lipogenic genes, we injected 14C-labeled sodium acetate to chow-fed WT and Cyp7a1-tg mice to study the hepatic fatty acid and cholesterol synthesis rate. As estimated by pmole of 14C-acetate incorporated into fatty acids and sterols, Fig 1A shows that acetyl-CoA was mainly used for fatty acid synthesis in WT liver. Interestingly, cholesterol synthesis rate was increased ~12-fold, while fatty acid synthesis rate was decreased ~60% in Cyp7a1-tg mice, resulting in approximately equal incorporation of 14C-acetate into cholesterol and fatty acids.
Figure 1. Cyp7a1-tg mice have increased cholesterol synthesis and decreased fatty acid synthesis in the liver.
A. Hepatic cholesterol and fatty acid synthesis rates were measured as described in Materials and Methods. The activities of 3H and 14C were determined in a scintillation counter. The cholesterol and fatty acid synthesis rates were expressed as 14C radioactivity derived from 14C-acetate (adjusted by internal recovery standard 3H-cholesterol radioactivity) and incorporated into cholesterol or fatty acids per minute per gram liver tissue. B. Wild type and Cyp7a1-tg mice were ip. injected with a single dose of glucose (8 g/kg) containing 5 µCi [1-14C] glucose, and fecal 14C sterol amount was estimated as described in Materials and Methods. The fecal acidic sterols (bile acids) and neutral sterols (cholesterol) derived from 14C-glucose were estimated by measuring the radioactivity of 14C incorporated, adjusted by internal standard 3H-cholesterol radioactivity, and expressed as percentage of total administrated 14C activity. Results are expressed as mean ± SE; “*” indicates p < 0.05 vs. WT. “**” indicates p < 0.05 vs. 14C incorporation into cholesterol in WT mice, n = 4-5.
During the postprandial state, acetyl-CoA derived from glycolysis is used for both lipogenesis and cholesterologenesis. Induction of cholesterol synthesis provides cholesterol substrate to stimulate CYP7A1 activity and bile acid synthesis, and subsequently stimulates fecal excretion of cholesterol and bile acids. To test the potential contribution of this route to hepatic lipid metabolism, we administered 14C-glucose to mice and measured 14C radioactivity in fecal neutral and acidic sterols. Fig 1B shows that fecal 14C radioactivity in neutral, acidic and total sterols was markedly and rapidly increased in day 1 in Cyp7a1-tg mice compared to wild type mice. Fecal samples from Cyp7a1-tg mice contained significantly higher 14C radioactivity, accounting for ~15% of 14C-glucose administered compared to wild type mice feces, which contained only ~2% of 14C-glucose administered. In addition, the majority of fecal 14C radioactivity was recovered as neutral sterols. Fecal acidic sterols (bile acids) were increased 2-fold in Cyp7a1-tg mice. In summary, these data suggest that in Cyp7a1-tg mice, stimulation of bile acid synthesis may divert acetyl-CoA from fatty acid synthesis to cholesterol synthesis and secretion to decrease lipogenesis.
MiR-33a is induced in Cyp7a1-tg mice and inhibits CYP7A1 mRNA and bile acid synthesis in wild type mice
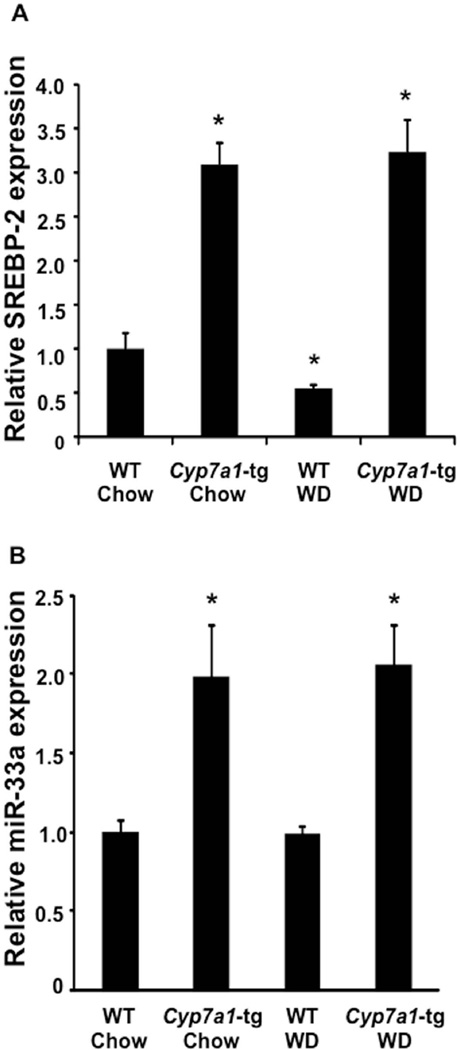
Since SREBP2 and miR-33a are co-expressed by the SREBP2 gene, we hypothesized that miR-33a might be also induced in Cyp7a1-tg mice to participate in the regulation of cholesterol metabolism. Indeed, hepatic miR-33a expression was co-induced with SREBP2 in Cyp7a1-tg mice under both chow and Western diet-feeding conditions (Fig 2A & B) (9). These results suggest that increasing bile acid synthesis in Cyp7a1-tg mice may induce miR-33a expression by inducing cholesterol-regulated SREBP2 expression.
Figure 2. Induction of CYP7A1 induces SREBP2 and miR-33a expression in Cyp7a1-tg mice.
Wild type and Cyp7a1-tg mice were fed a standard chow diet or Western diet for 4 months. The mRNA expression of (A) SREBP2 and (B) miR-33a was measured by real-time PCR. Results are expressed as mean ± S.E. “*” indicates p < 0.05 vs. WT of same diet, n=4.
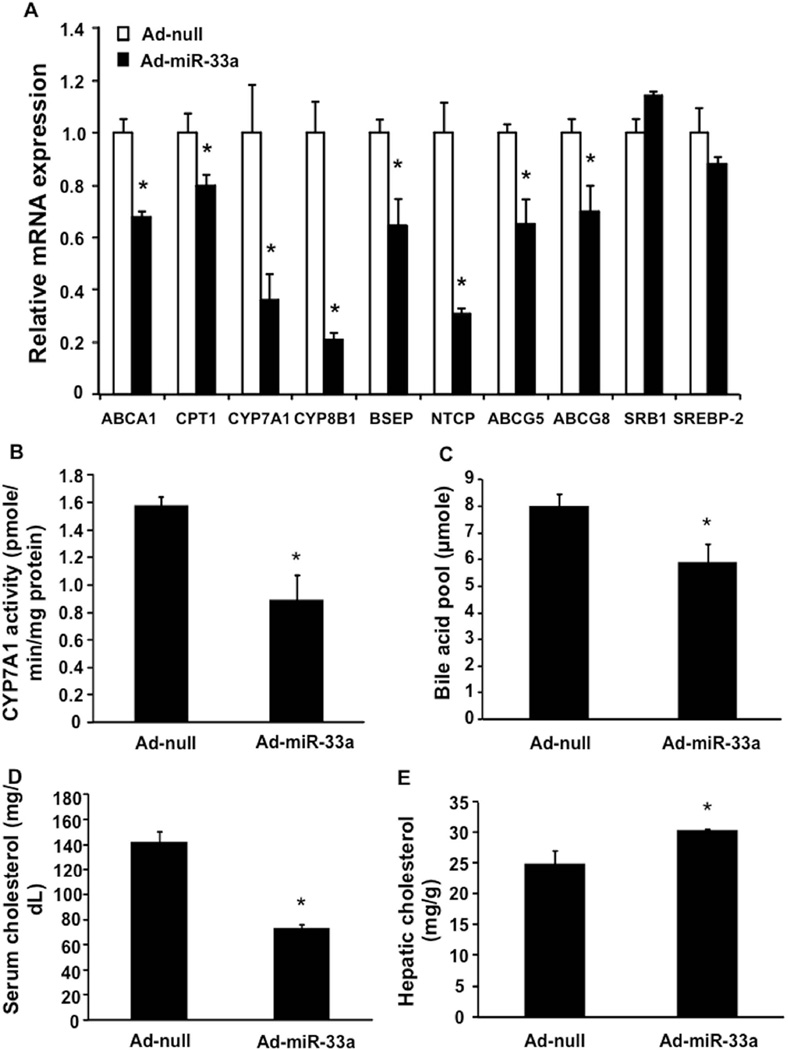
To further test if miR-33a regulates bile acid metabolism, we used adenovirus-mediated gene delivery to over-express miR-33a specifically in the wild type mouse liver (Supplemental Fig 3). The mRNA analysis by real-time PCR showed that over-expression of miR-33a reduced the mRNA expression of CYP7A1 and CYP8B1 and Na+-dependent taurocholate co-transport peptide (NTCP), the basolateral bile acid uptake transporter (Fig 3A). The mRNA levels of BSEP, ABCG5 and ABCG8 were also reduced by miR-33a (Fig 3A). As a positive control, miR-33a inhibited ABCA1 and carnitine: palmitoyl-CoA transferase 1 (CPT1) mRNA (Fig 3A) (9, 11). Consistent with down-regulation of CYP7A1 mRNA, miR-33a over-expression reduced microsomal CYP7A1 enzyme activity by ~40% (Fig 3B) and total bile acid pool size by ~25% (Fig 3C). In addition, miR-33a reduced total serum cholesterol levels by ~50% (Fig 3D), but increased hepatic cholesterol content by ~20% (Fig 3E). Such changes in serum and hepatic cholesterol levels are likely resultant from inhibition of both ABCA1 and CYP7A1.
Figure 3. Effects of hepatic miR-33a over-expression on bile acid and cholesterol metabolism in mice.
Wild type C57BL6J mice were administered adenovirus expressing miR-33a (Ad-miR-33a) or control adenovirus (Ad-null) via tail vein injection and were sacrificed after 7 days. A. Hepatic mRNA expression was measured by real-time PCR. B. Hepatic CYP7A1 enzyme activity. C. Total bile acid pool size. D. Serum cholesterol. E. Hepatic cholesterol. Results are expressed as mean ± S.E. “*” indicates p < 0.05 vs. Ad-null controls, n=4.
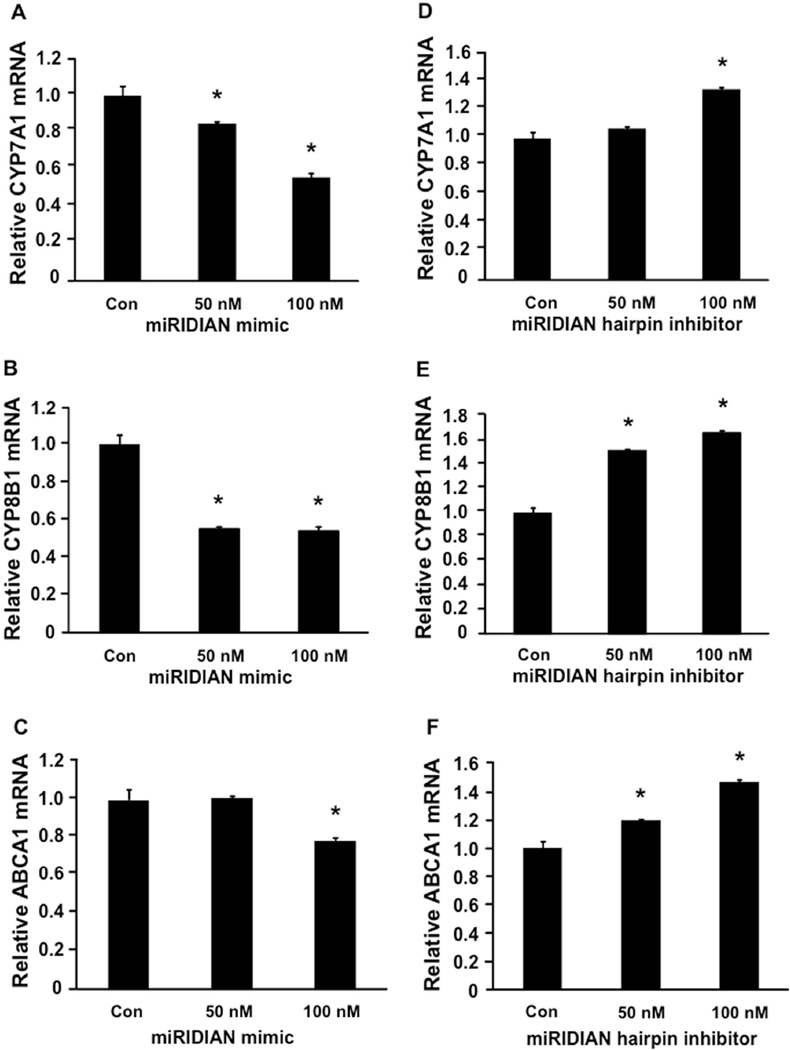
To investigate if miR-33a regulation of CYP7A1 is conserved in the human CYP7A1 gene, we used adenovirus-mediated gene delivery to over-express miR-33a in “humanized” mice, which express the human CYP7A1 gene. As shown in Supplemental Fig 4A and 4C, miR-33a over-expression resulted in ~40% reduction of human CYP7A1 mRNA and ~25% reduction in total bile acid pool size. As a positive control, miR-33a repressed ABCA1 mRNA in the “humanized” CYP7A1 mouse liver (Supplemental Fig 4B). We next transfected miR-33a mimic or miR-33a hairpin inhibitor into HepG2 cells to confirm our in vivo observations. As show in Fig 4 A, B and C, miR-33a mimic dose-dependently decreased mRNA levels of CYP7A1 and CYP8B1, and ABCA1 (positive control). In addition, antagonism of miR-33a by a miR-33a hairpin inhibitor dose-dependently increased the mRNA levels of CYP7A1, CYP8B1 and ABCA1 (Fig 4D, E and F). In summary, these data suggest that miR-33a regulates CYP7A1 and bile acid synthesis, and may coordinately regulate hepatic cholesterol and bile acid homeostasis.
Figure 4. Regulation of CYP7A1 mRNA by miR-33a in HepG2 cells.
HepG2 cells were transfected with miRIDIAN miR-33a mimic (A-C) or miRIDIAN miR-33a hairpin inhibitor (D-F) at 50nM and 100nM for 48 hr. Respective controls were used to equally adjust the total amount transfected among samples. mRNA expression was measured by real-time PCR, 48 hr after transfection. Assays were performed in triplicate and expressed as mean ± S.D. “*” indicates p < 0.05 vs. controls.
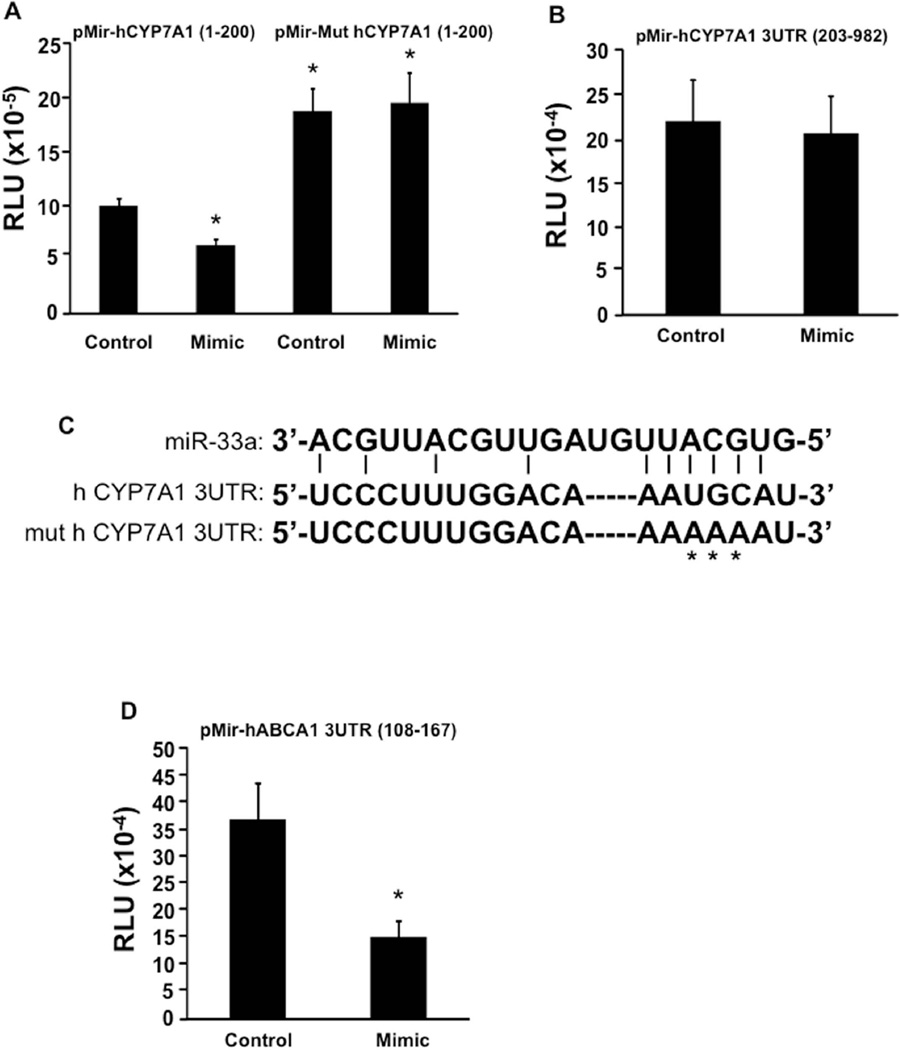
Identification of miR-33a target sequence in the 3’-untranslated region (3’-UTR) of human CYP7A1 mRNA
MicroRNA-mediated target gene repression commonly occurs via binding to the 3’-UTR in target mRNAs, which usually results in degradation of the mRNA and inhibition of protein translation. To identify the potential miR-33a target site in the human CYP7A1 3’-UTR, we cloned two human CYP7A1 3’-UTR fragments (nt 1-200, and nt 203-982) downstream of a luciferase gene in the pMir REPORT luciferase vector and tested the effect of miR-33a mimic on reporter activity. Co-transfection of miR-33a mimic in transient transfection assay of pMir-hCYP7A1 (1–200) reporter in HepG2 cells resulted in ~40% inhibition of the reporter activity (Fig 5A), but showed no effect on the pMir-hCYP7A1 (203–982) reporter activity (Fig 5B). These results suggest that the nt 1-200 region of the human CYP7A1 3’-UTR may contain a potential miR-33a target site. Analysis of the sequence in this region identified a putative seed-match sequence for miR-33a binding (Fig 5C). Mutations of this putative seed-match sequence resulted in elevated basal reporter activity and abolished the inhibitory effect of miR-33a mimic on the mutant reporter (Fig 5A). As a positive control, miR-33a mimic repressed the ABCA1 3’-UTR reporter activity as expected (Fig 5D). These results suggest that a putative miR-33a binding site located in the 3’-UTR of human CYP7A1 mRNA is functional in mediating the inhibitory effect of miR-33a. In vitro and in vivo studies in both wild type and humanized CYP7A1-tg mice showed that this miR-33a-mediated regulatory mechanism is functionally conserved in humans and mice. However, we have not identified a functional miR-33a target site in the mouse cyp7a1 mRNA 3’UTR.
Figure 5. Identification of a putative miR-33a target site in human CYP7A1 3’-UTR region.
A. Effect of miR-33a mimic on wild type (pMir-hCYP7A1 (1–200)) and mutant (pMir-hCYP7A1 (1–200)) reporter activity in HepG2 cells. B. Effect of miR-33a mimic on human pMir-hCYP7A1 (203–982) reporter activity in HepG2 cells. C. Putative miR-33a target site in the human CYP7A1 3’-UTR. D. Effect of miR-33a mimic on human ABCA1 3’-UTR reporter activity in HepG2 cells. Assays were performed in triplicate and expressed as mean ± S.D. “*” indicates p > 0.05 vs. controls.
Discussion
In this study, we used Cyp7a1-tg mice as an experimental model to demonstrate that stimulating bile acid synthesis significantly impacts hepatic lipid metabolism and homeostasis, and to elucidate the underlying molecular mechanism for bile acid signaling in preventing diet-induced hepatic steatosis, insulin resistance and obesity. We demonstrate that stimulating de novo bile acid synthesis results in decreased lipogenesis via mechanisms independent of hepatic FXR signaling. This study unveiled complex links between bile acid, cholesterol and fatty acid metabolism. We also uncovered a novel role for miR-33a in coordinated regulation of hepatic bile acid and cholesterol metabolism. We found that in response to increased conversion of cholesterol to bile acids, SREBP2 is induced to stimulate cholesterol synthesis to provide a substrate to CYP7A1, and miR-33a is co-induced to reduce CYP7A1 mRNA translation. This feed-forward activation of CYP7A1 enzyme activity by cholesterol and feedback inhibition of CYP7A1 translation by miR-33a provide a rapid post-transcriptional mechanism for regulation of bile acid synthesis to maintain hepatic lipid homeostasis.
We first showed that a 2-3 fold stimulation of hepatic CYP7A1 enzyme activity resulted in marked induction of cholesterol synthetic genes and de novo cholesterol synthesis rate in Cyp7a1-tg mice (6). Stimulation of cholesterol catabolism to bile acids resulted in activation of SREBP2 and all SREBP2-regulated genes in cholesterol metabolism (17). The ER is a cholesterol-poor organelle (18), and intracellular cholesterol/oxysterol levels are critical in regulation of the SREBP2-mediated cholesterol metabolism network. Thus, co-localization of CYP7A1 and SREBP2 in the ER provides advantages for efficient sensing of intracellular cholesterol metabolism, which further suggests that CYP7A1 enzyme activity may play a key regulatory role in cholesterol metabolism. It is well known that cholesterol synthesis increases during the postprandial state to meet increasing demands for cholesterol (19). We recently reported that feeding rapidly and markedly induced CYP7A1 mRNA expression and increased CYP7A1 enzyme activity by ~2-fold in mice (15). Furthermore, CYP7A1 mRNA expression peaked 3 hr after refeeding, while HMG-CoA reductase mRNA was minimally affected at 3 hr, but increased by ~12-fold 6 hr after refeeding (15). We hypothesize that rapid nutritional induction of CYP7A1 may play a role in stimulating postprandial cholesterol synthesis and lipid homeostasis. Upon food intake, bile acids released into the intestine induce fibroblast growth factor 15 (FGF15), which may be transported to hepatocytes to inhibit bile acid synthesis (20). This mechanism may reduce CYP7A1 to basal levels after the postprandial period. Postprandial increase in bile acid synthesis is also supported by a recent report that serum bile acid concentrations increased after oral glucose challenge in patients with normal glucose tolerance, but this response was blunted in patients with impaired glucose tolerance (21). Interestingly, Roux-en Y gastric bypass rapidly improved insulin resistance and glucose tolerance and is associated with higher serum bile acid levels (22, 23). Reduced bile acid circulation back to the liver in bypass patients may stimulate bile acid synthesis and signaling, which stimulates energy metabolism and glucagon-like peptide 1 (GLP-1) to improve insulin sensitivity and reduce weight.
This study unexpectedly revealed that marked induction of CYP7A1 enzyme activity in mouse liver resulted in dissociation of SREBP1c-dependent lipogenic gene expression and hepatic fatty acid synthesis rate. Increased SREBP1c and its targets FAS and ACC in Cyp7a1-tg mice, (despite a 2-3 fold enlarged bile acid pool) suggests that CYP7A1 enzyme activity, presumably by modulating cholesterol catabolism, may have a predominant role in SREBP1c maturation over the repressive effect of bile acids on SREBP1c-regulated lipogenesis. Further, these results suggest that reduced hepatic fatty acid synthesis rate in Cyp7a1-tg mice is unlikely a direct result of transcriptional repression of hepatic lipogenic genes by the bile acid/FXR/SHP pathway as previously reported (24). Circulating bile acids modulate peripheral energy expenditure, which could indirectly affect hepatic lipogenesis in Cyp7a1-tg mice (6, 25). Our results here support such a mechanism that stimulation of bile acid and cholesterol synthesis could have a negative impact on de novo lipogenesis by limiting cellular acetyl-CoA availability for fatty acid synthesis. Activation of bile acid and cholesterol synthesis, and decreasing lipogenesis, may result in a metabolic shift that directs acetyl-CoA towards sterol synthesis for biliary excretion and fecal elimination. It should be noted that our study using Cyp7a1-tg mice as a model does not necessarily contradict results from other bile acid-treated experimental models because we have shown that increasing de novo bile acid synthesis did not result in bile acid accumulation in the liver, likely due to efficient bile acid secretion.
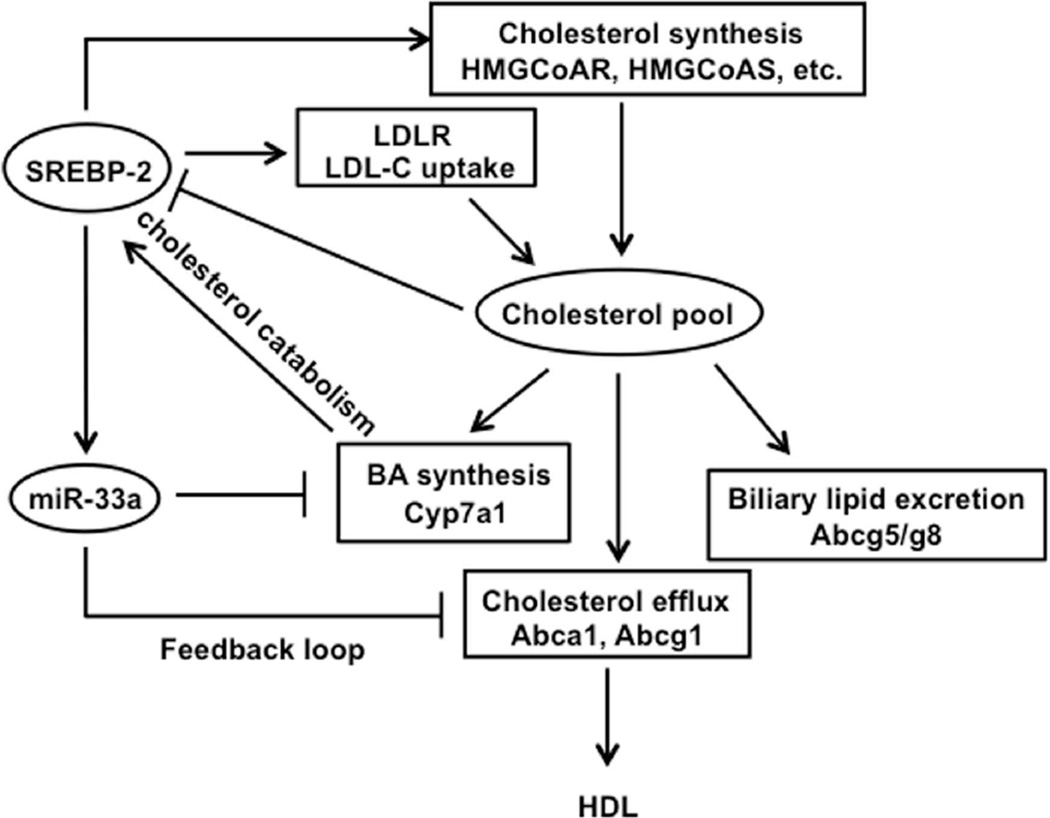
Finally, this study identified a novel miR-33a-mediated repression of CYP7A1, as a result of SREBP2 induction, could be part of the feedback loop to reduce bile acid synthesis. The recent discovery of co-expression of SREBP2 and miR-33a, as well as down-regulation of ABCA1 by miR-33a, provided the first evidence that miR-33a down-regulates cellular cholesterol efflux to HDL in response to decreased cellular cholesterol levels to maintain hepatic lipid homeostasis (9). Our study provides further evidence that miR-33a inhibition of CYP7A1 and bile acid synthesis may also contribute to maintaining cholesterol homeostasis. Cholesterol/oxysterols might also repress miR-33a levels to increase CYP7A1 expression as well as cholesterol efflux transporters (9). Fig 6 shows a proposed mechanism for the regulation of cholesterol homeostasis by a CYP7A1/SREBP2/miR-33a axis based on this study and the well recognized mechanism for maintaining cholesterol homeostasis and pool by intracellular cholesterol or oxysterol levels (8). Increased CYP7A1 enzyme activity results in increased cholesterol catabolism and decreased intracellular cholesterol, which leads to proteolytic activation of SREBP2 and subsequent stimulation of de novo cholesterol synthesis and LDLR-mediated cholesterol uptake to reduce serum cholesterol. Simultaneously, SREBP2 activation of its own gene transcription co-induces miR-33a, which down-regulates cholesterol efflux transporters and bile acid synthesis. These changes result in increased intrahepatic cholesterol, which subsequently represses SREBP2 and miR-33a expression. This mechanism integrates bile acids and cholesterol metabolism to control lipid homeostasis at both transcriptional and posttranscriptional levels. Thus, CYP7A1 may play a central role in sensing intracellular cholesterol levels by converting excess hepatic cholesterol to bile acids, thus activating SREBP2 and miR-33a, which inhibits CYP7A1 translation as a rapid feedback mechanism.
Fig 6.
Regulation of cholesterol and bile acid metabolism by the CYP7A1/SREBP2/miR-33a axis. This figure illustrates the coordinated regulation of cholesterol and bile acid metabolism based on this study. Under conditions when cellular cholesterol decreases or cholesterol catabolism increases, SREBP2 is activated. To prevent cellular cholesterol from decreasing, SREBP2 induces LDLR-mediated LDL-C uptake into hepatocytes. In addition, SREBP2 induces a number of key genes to stimulate de novo cholesterol synthesis. On the other hand, SREBP2 binds to its own gene promoter to induce SREBP2 gene transcription, which also results in co-induction of miR-33a. Increased miR-33a down-regulates ABCA1 and ABCG1 to reduce cellular cholesterol efflux to HDL. Meanwhile, miR-33a inhibits CYP7A1 and bile acid synthesis to inhibit cholesterol catabolism. Under conditions when cellular cholesterol increases, cholesterol represses these SREBP2-miR-33a regulated pathways, while cholesterol feed-forward induces CYP7A1 (in mouse) to convert cholesterol into bile acids and to stimulate biliary cholesterol secretion via ABCG5/G8 heterodimers. This feed-forward activation of CYP7A enzyme activity by cholesterol and feedback inhibition of CYP7A1 translation by miR-33a provide a rapid post-transcriptional mechanism for regulation of bile acid synthesis to maintain hepatic lipid homeostasis.
Inducing CYP7A1 activity by targeting miR-33a may be a potential therapeutic approach to improve metabolic homeostasis. This study suggests the cardio-protective effects of miR-33a antagonism can be attributed not only to stimulating HDL biogenesis but bile acid synthesis, the final step in macrophage-to-feces reverse cholesterol transport. In this study, we also showed the mRNA of CYP8B1, NTCP, and BSEP were repressed upon miR-33a over-expression in mice, indicating that miR-33a antagonism also stimulates enterohepatic bile acid circulation. Interestingly, a recent study reported that BSEP and ATP8B1 (a canalicular phospholipids flippase) are targets of miR-33a (10), and antagonism of miR-33a increased biliary bile acid and phospholipid secretion in mice. Therefore, miR-33a antagonism may be a potential strategy for increasing bile acid synthesis to treat NAFLD, diabetes and obesity.
Supplementary Material
Acknowledgements
This work was supported by grants R37DK058379 and R01DK044442 to JYLC from NIDDK, NIH. JF is an awardee of the National Research Service Award (F32 DK096784). We acknowledge microarray analysis by Banu Gopalan and Jie Na (Cleveland Clinic Foundation Genomic Core).
Abbreviations used
- ABC
ATP binding cassette
- ACC
acetyl CoA carboxylase
- BSEP
bile salt export pump
- CYP7A1
cholesterol 7α-hydroxylase
- CYP8B1
sterol 12α-hydroxylase
- ER
endoplasmic reticulum
- FAS
fatty acid synthase
- FXR
farnesoid X receptor
- INSIG
insulin-induced gene
- miR
microRNA
- NAFLD
non-alcoholic fatty liver disease
- NTCP
sodium taurocholate cotransporting polypeptide
- SCAP
SREBP cleavage-activating protein
- SREBP
steroid response element binding protein
- 3’ UTR
3’ untranslated region.
References
- 1.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile Acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 3.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR, Dumont J, et al. Farnesoid X Receptor Deficiency Improves Glucose Homeostasis in Mouse Models of Obesity. Diabetes. 2011 doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, Mataki C, et al. Lowering bile acid pool size with a synthetic FXR agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem. 2011 doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JY. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, et al. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, et al. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, et al. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem. 2012;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 17.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soccio RE, Breslow JL. Intracellular cholesterol transport. Arterioscler Thromb Vasc Biol. 2004;24:1150–1160. doi: 10.1161/01.ATV.0000131264.66417.d5. [DOI] [PubMed] [Google Scholar]
- 19.Tomkins GM, Chaikoff IL. Cholesterol synthesis by liver. I. Influence of fasting and of diet. J Biol Chem. 1952;196:569–573. [PubMed] [Google Scholar]
- 20.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, Carr SA, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patti ME, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, Badman MK, et al. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 2009;17:1671–1677. doi: 10.1038/oby.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonen M, Dali-Youcef N, Kaminska D, Venesmaa S, Kakela P, Paakkonen M, Hallikainen M, et al. Conjugated bile acids associate with altered rates of glucose and lipid oxidation after Roux-en-Y gastric bypass. Obes Surg. 2012;22:1473–1480. doi: 10.1007/s11695-012-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.