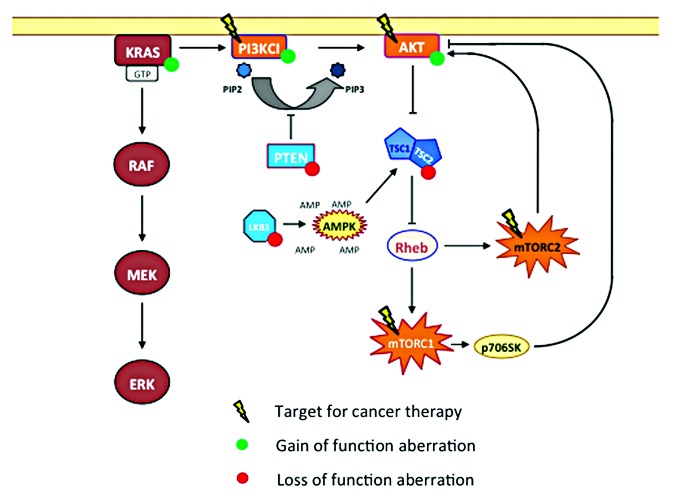
Increased signaling through the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway occurs in diverse malignancies.1 In cancer, the PI3K/AKT/mTOR pathway can be activated by mutations in several oncogenes such as PIK3CA, PIK3R1, AKT, TSC1/2, LKB1 and PTEN (Fig. 1). Most activating mutations occur in the helical or kinase domain of the PIK3CA gene.
Figure 1. PI3K/AKT/mTOR pathway, targets for anticancer therapy and most common locations for gain-of-function aberrations (green) or loss-of-function aberrations (red).
Preclinical models demonstrated that mutations in PIK3CA have oncogenic potential and can also be associated with sensitivity to PI3K/AKT/mTOR inhibitors.2-4 In a human non-small cell lung cancer (NSCLC) xenograft model, PIK3CA mutation H1047R was associated with response to the dual PI3K and mTOR kinase inhibitor BEZ235.3 Similarly, breast cells with the same mutation demonstrated reduced proliferation compared with breast cells with wild-type (wt) PIK3CA.4 In addition, early clinical data from histology-independent protocols suggested that PIK3CA mutations could be associated with sensitivity to therapies targeting PI3K/AKT/mTOR signaling in subsets of patients with advanced cancers. The reported response rate of these patients in early-phase clinical trials was approximately 30%, which is less than that with some other molecularly matched therapies, but significantly more than the traditional response rates of 4% to 6% observed in this patient population.5
Preclinical models also demonstrated that KRAS mutations can be associated with resistance to PI3K/AKT/mTOR targeted therapies. A human NSCLC xenograft model with a KRAS G12D mutation demonstrated resistance to the dual PI3K and mTOR kinase inhibitor BEZ235, but had a good response to the MEK inhibitor AZD6244 or combination of BEZ235 and AZD6244.3 Similarly, several cell lines with simultaneous PIK3CA and KRAS mutations demonstrated relative resistance to the pan-PI3K inhibitor PX-866, whereas cell lines with a PIK3CA mutation only were sensitive to it.2 Finally, colorectal cancer cell lines with simultaneous PIK3CA and KRAS mutations demonstrated resistance to the mTOR inhibitor everolimus, which was eliminated by restoration of wt KRAS status, and those observations were confirmed in a human colon cancer xenograft model.4 These data are particularly interesting because patients with PIK3CA mutations and advanced cancers are twice as likely to have simultaneous KRAS mutations (34% vs. 21%, p = 0.047).1 Of note, in early-phase clinical trials enrolling patients with advanced cancers with PIK3CA and KRAS mutations in codon 12 or 13, treatment with PI3K/AKT/mTOR inhibitors led to lower response rates compared with patients without simultaneous KRAS mutations (response rate of 0% vs. 23%, p = 0.046).6
It is also plausible that not all PIK3CA mutations equally predict response to PI3K/AKT/mTOR inhibitors. Interestingly, observations from early clinical studies demonstrated that patients with advanced cancer and a H1047R mutation have higher response rates to PI3K/AKT/mTOR inhibitors than patients with other PIK3CA mutations (38% vs. 10%, p = 0.018).1,6
Early clinical experience suggests that single-agent PI3K/AKT/mTOR inhibitors are seldom effective compared with combinations (response rate of 0% vs. 29%, p = 0.002; progression-free survival of 3.1 vs. 1.8 mo; p = 0.004).6 There are several possible explanations for this. First, tumor heterogeneity might play role. It has been demonstrated that DNA isolated from three different areas of a small breast cancer sample had three different results for PIK3CA status (H1047R, wild-type, E542K, respectively).7 Second, preclinical experiments in cell lines with PIK3CA mutations demonstrated that sensitivity to single-agent inhibition can be dependent on BIM (a pro-apoptotic Bcl-2 family protein) levels, because low levels of BIM prevent cancer cells from undergoing apoptosis in response to targeted therapy but not to chemotherapy.8 Third, activation of collateral pathways through KRAS or other proteins (MET, MYC, etc.) is not effectively abrogated by inhibition of the single pathway.
PIK3CA mutations do not seem to have a common taxonomy across diverse tumor types except for an association with KRAS mutations, at least in some tumor types.1 However, therapeutic targeting with PI3K/AKT/mTOR pathway inhibitors in cancers with an activated PI3K/AKT/mTOR pathway demonstrated efficacy in preclinical and early clinical experiments; this has implications for cancer treatment, because many drugs targeting the PI3K/AKT/mTOR signaling pathway are currently in clinical development.
Support
Research support by Novartis, Roche, Trovagene, Transgenomic, Biocartis; Consultant advisory board: Trovagene.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25118
References
- 1.Janku F, Wheler JJ, Naing A, Stepanek VM, Falchook GS, Fu S, et al. PIK3CA mutations in advanced cancers: characteristics and outcomes. Oncotarget. 2012;3:1566–75. doi: 10.18632/oncotarget.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ihle NT, Lemos R, Jr., Wipf P, Yacoub A, Mitchell C, Siwak D, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69:143–50. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–66. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10:558–65. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73:276–84. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont Jensen J, Laenkholm AV, Knoop A, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in Breast Cancer. Clin Cancer Res. 2011;17:667–77. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 8.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–65. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]