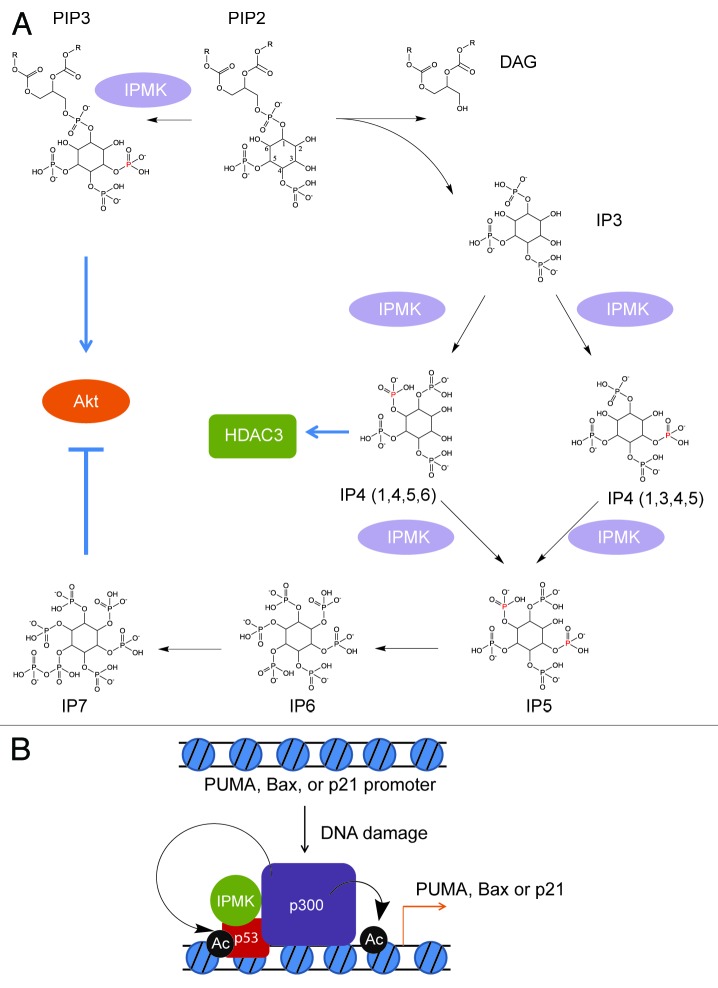
The tumor suppressor p53 is a major transcription factor that induces genes regulating cell cycle arrest and death and which is inactivated in about half of all human cancers. Despite considerable research on p53, mechanisms regulating its activation have not been fully elucidated. We have recently established that the enzyme inositol polyphosphate multikinase (IPMK) is a transcriptional co-activator of p53.1 Independent of its catalytic activity, IPMK binds p53 and stimulates its binding to the acetyltransferase p300, increasing its acetylation activity, which augments transcriptional activity and p53-associated cell death.
IPMK is a remarkably pleiotropic enzyme. Its first characterized enzyme activity involves phosphorylation of inositol phosphates, acting as the rate-limiting enzyme in generation of inositol pentakisphosphate and thus of higher inositol phosphates, especially the energetic inositol pyrophosphates (Fig. 1A).2 Recently, Watson et al. described a potential role for IP4 (1,4,5,6) in the activation of HDAC3.3 IPMK is also a major PI3 kinase, which acts together with the wortmannin-sensitive p110/p85 PI3 kinase to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3) that activates Akt and protein synthesis.4 Interestingly, IPMK also displays physiologic roles independent of its catalytic activity. Both wild-type and catalytically inactive IPMK stabilize the mTOR-1 complex to facilitate protein translation.5
Figure 1. Inositol polyphosphate multikinase (IPMK) is a multifunctional enzyme with PI3-kinase, IP3-kinase and catalytically independent activities. (A) IPMK is a PI3-kinase which phosphorylates PIP2 into PIP3, serving as a physiologic activator of Akt/PKB. It also possesses IP3-kinase activity, converting IP3 into IP4 (1,3,4,5) or IP4 (1,4,5,6). Whether the phosphorylation of IP3 at the 3-position or the 6-position is physiologically regulated remains to be fully elucidated. One of the IP4 isoforms, IP4 (1,4,5,6), has been co-crystallized at the interface of HDAC3 and the DAD domain of NCor/SMRT, enhancing HDAC3 activity in vitro.3 IPMK subsequently phosphorylates IP4 into IP5, and is the rate-limiting enzyme for this metabolite. Thus, IPMK serves as the gate-keeping enzyme for the synthesis of all higher inositol polyphosphate species, including inositol pyrophosphates, which have been implicated in diverse physiologic processes. (B) Independent of catalytic activity, IPMK binds to p53 and enhances its association with p300. The enhancement of p300s histone acetyltransferase activity by IPMK leads to increased acetylation of p53 and histone H3, as well as p53 association to target promoters. This augmentation of p53-dependent gene transcription enhances cell death.
Hints that IPMK might have a transcriptional function stemmed from the earliest identification of an IPMK homolog in yeast, which is part of a transcriptional complex regulating arginine-linked genes, and so was designated Arg82.6 Such a nuclear function in mammals was presaged by observations that a major portion of cellular IPMK is concentrated in the nucleus.7
Evidence that IPMK might influence p53 function came from our observation that the two proteins physiologically bind, especially following apoptotic stimuli.1 Overexpression of IPMK increases transcriptional activity of p53 as reflected in mRNA levels for canonical p53 targets such as PUMA, Bax and p21. These actions are physiologically relevant, as genetic depletion of IPMK decreases levels of PUMA, Bax and p21 transcripts. IPMK is a component of the p53 transcriptional complex, since chromatin immunoprecipitation (ChIP) analysis reveals association of IPMK with p53 as well as PUMA, Bax and p21 promoters (Fig. 1B).
How might IPMK promote p53 actions? The histone acetyltransferase p300 is a major mediator of p53 actions, acetylating p53 and acting as a transcriptional co-activator. We discovered that IPMK enhances the binding of p53 to p300 and augments p53 acetylation, which is associated with activation of p53 and enhanced transcriptional activity. More importantly, depleting IPMK leads to a profound reduction of p53-p300 binding as well as p53 acetylation. These interactions are direct, as purified IPMK doubles the acetylation by p300 of purified p53. IPMK also regulates acetylation of histones by p300 at p53 target genes. Thus, the binding of p300 to the promoter of p21 is markedly diminished in primary mouse embryonic fibroblasts from IPMK-knockout mice. Together, these observations indicate that IPMK stimulates p53’s transcriptional activity by increasing p300-mediated acetylation of p53 and of histones at the promoters of p53 targets.
These molecular actions of IPMK impact the apoptotic influences of p53. We showed that IPMK overexpression markedly stimulates p53-dependent but not p53-independent cell death. p53-dependent cell death requires binding of IPMK to p53, as a dominant-negative construct, which prevents such binding, markedly reduces p53 signaling and associated apoptosis.
Remarkably, all these influences of IPMK upon p53 signaling are independent of IPMK’s catalytic activity, as mutants of the enzyme that abolish catalytic activity retain the ability to regulate p53. Interestingly, the arginine transcriptional response in yeast by IPMK is also independent of the enzyme’s catalytic activity.8
Our observations may have therapeutic implications. The ability of a dominant-negative construct to block IPMK’s regulation of p53 suggests that low-molecular weight drugs acting similarly might prevent p53 activation. In disorders such as Huntington’s disease and stroke, cell death has been linked to p53, so that drugs selectively preventing p53-IPMK binding may prove beneficial.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25119
References
- 1.Xu R, Sen N, Paul BD, Snowman AM, Rao F, Vandiver MS, et al. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013;6:ra22. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–56. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–40. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maag D, Maxwell MJ, Hardesty DA, Boucher KL, Choudhari N, Hanno AG, et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci U S A. 2011;108:1391–6. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–21. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bechet J, Greenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–9. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 7.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–9. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 8.Bosch D, Saiardi A. Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem. 2012;287:38347–55. doi: 10.1074/jbc.M112.384255. [DOI] [PMC free article] [PubMed] [Google Scholar]