Abstract
FLASH/Casp8AP2 is a huge multifunctional protein involved in multiple cellular processes, reaching from death receptor signaling to regulation of histone gene transcription and histone mRNA processing. Previous work has shown that FLASH localizes to Cajal bodies and promyelocytic leukemia (PML) bodies. However, the function of its nuclear body association remains unclear. Here we demonstrate that murine FLASH is covalently modified by SUMO at Lys residue 1792. Interestingly, ectopic expression of SUMO results in proteasome-dependent degradation of FLASH. A point mutant of FLASH with a mutated SUMO acceptor lysine residue, FLASHK1792R, is resistant to SUMO-induced degradation. Finally, we show that arsenic trioxide, a drug known to potentiate SUMO modification and degradation of PML, triggers recruitment of FLASH to PML bodies and concomitant loss of FLASH protein. Our data suggest that SUMO targets FLASH for proteasome-dependent degradation, which is associated with recruitment of FLASH to PML bodies.
Keywords: FLASH/Casp8AP2, SUMO, proteasome-dependent degradation
Introduction
FLASH (FLICE-associated huge protein)/Casp8AP2 (Caspase-8 associated protein 2) is a huge 1,962 amino acids long protein that has been implicated in numerous cellular processes, including apoptosis regulation, cell cycle, mRNA processing and transcriptional control.1-10 Knockout of the FLASH/Casp8AP2 gene in mice is lethal early in embryogenesis, indicating that FLASH is essential for life.11 Initially, FLASH was identified as an interacting molecule for Caspase-8 (FLICE) and was shown to potentiate death receptor CD95-induced Caspase-8 activation and apoptosis.1 In addition, FLASH has also been linked to TNF receptor-induced activation of the transcription factor NFκB through association with the signaling molecule TRAF2 and Caspase-8.12,13 Interestingly, a number of studies demonstrated that FLASH is mainly localized to the cell nucleus and at nuclear bodies (NBs). In this context, FLASH was shown to associate with Cajal bodies and to interact with histone gene promoters, thereby stimulating the expression of histone genes.2,3 Consequently, depletion of FLASH leads to accumulation of cells in S phase, presumably through histone depletion. A recent study has linked FLASH to histone gene pre-mRNA processing, documenting an additional role in the regulation of histone gene expression.5,6 FLASH does not localize exclusively to Cajal bodies, but a fraction of FLASH is associated with promyelocytic leukemia (PML) nuclear bodies.4 In response to CD95 receptor signaling, FLASH leaves the nuclear bodies and accumulates in the cytoplasm at the mitochondria, where it associates with Caspase-8 and mediates apoptosis.4,14 A recent study indicates that the E3 ubiquitin ligase Ro52 mediates nucleo-cytoplasmic translocation of FLASH,15 suggesting a potential mechanism for FLASH shuttling. Consistent with its role in apoptosis regulation, FLASH has been identified as a predictive marker for the therapy success of childhood acute lymphoblastic leukaemia and pediatric T-cell lymphoblastic lymphoma16,17
PML bodies are macromolecular nuclear domains present in almost all mammalian cells.18,19 Small ubiquitin-related modifier (SUMO) has been identified as a major regulator of PML body formation and homeostasis.20 On the one hand, PML itself is covalently modified with SUMO; on the other hand, PML contains a SUMO interacting motif (SIM) that mediates non-covalent interaction with SUMO-modified proteins.20,21 By means of such interactions PML builds up a protein meshwork, to which further SUMO-modified proteins can be recruited.20,21 Remarkably, it has been shown that SUMO modification of PML results in its proteasome-dependent degradation.22 Acute promyelocytic leukemia (APL) cells express an oncogenic PML-RAR fusion protein, which is causative for this disease. Treatment with the drug arsenic trioxide (ATO, As2O3) strongly potentiates SUMO-modification of PML-RAR and PML, which is associated with their recruitment to PML bodies and subsequent proteasome-dependent degradation mediated by the SUMO-dependent ubiquitin ligase RNF4.23-25 Interestingly, the chain-forming capacity of SUMO-2 and SUMO-3 appears to be critical for directing RNF4 to its substrate PML. These findings indicate that PML bodies can serve as sites of SUMO-regulated substrate ubiquitination and degradation.
Here we demonstrate that murine FLASH is modified by SUMO at lysine residue 1792. In addition, we provide evidence that expression of SUMO results in reduced FLASH protein levels due to induction of FLASH degradation by the proteasome. A FLASH mutant with a mutated SUMO acceptor site, FLASHK1792R, is resistant to SUMO-mediated degradation. Finally, we show that treatment with ATO results in recruitment of FLASH to PML bodies and subsequent loss of FLASH expression. Taken together, our results suggest that FLASH is targeted for proteasome-dependent degradation by SUMO. Furthermore, FLASH degradation is associated with its recruitment to PML bodies, suggesting that PML bodies regulate FLASH degradation
Results
SUMO modification of murine FLASH at Lys1792
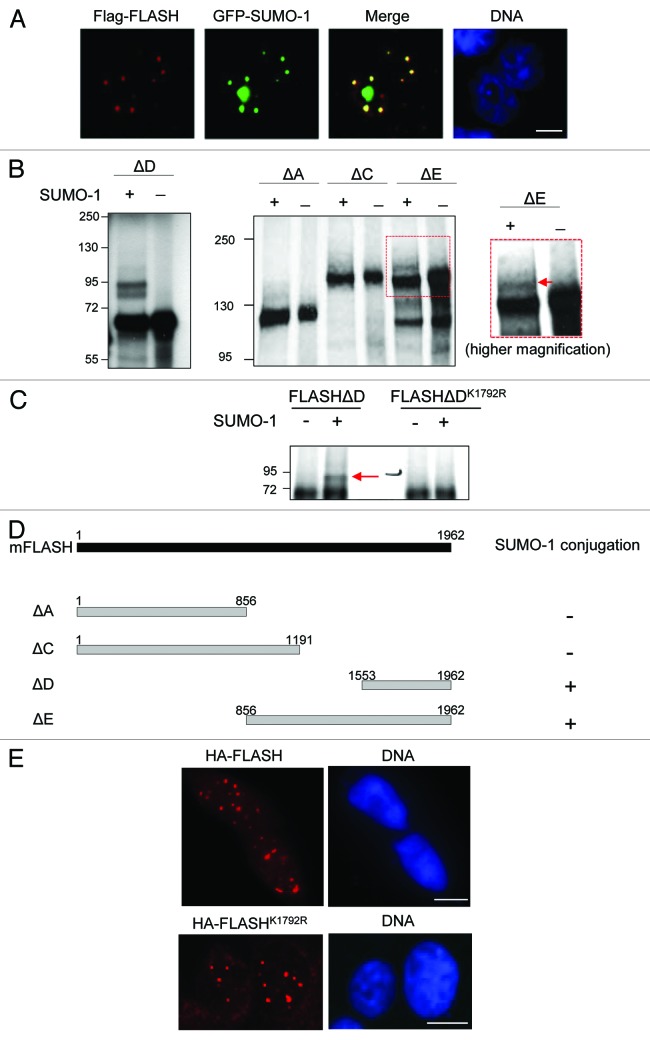
We and others have shown that FLASH localizes to nuclear bodies, including Cajal bodies and PML bodies.2-4 Since numerous SUMO-modified proteins have been found to localize to PML bodies,19 we performed confocal microscopy to analyze whether SUMO-1 co-localizes with FLASH. Indeed, Flag-FLASH and GFP-SUMO-1 partially co-localize in the cell nucleus in nuclear bodies (Fig. 1A).
Figure 1. FLASH is modified by SUMO-1 at lysine residue 1792 in vitro. (A) FLASH and SUMO-1 co-localize in nuclear bodies. HT1080 cells transiently expressing HA-FLASH and GFP-SUMO-1 were analyzed by immunofluorescence staining and confocal microscopy. (B) In vitro SUMO conjugation of FLASH. Different murine FLASH proteins (a schematic drawing is shown in d,) were in vitro translated and 35S-labeled. Subsequently in vitro SUMO conjugation assays were performed in absence and presence of SUMO-1. A representative autoradiogram is shown. (C) Lysine residue 1792 of FLASH is required for its SUMO modification. In vitro SUMO conjugation was performed and analyzed as described in (B). (D) Overview of the FLASH deletions used and the results of the in vitro SUMO conjugation assays. (E) Confocal image showing the subcellular localization of HA-FLASH and HA-FLASHK1792R in HT1080 cells. (Aand E). Scale bars: 20 µm.
Since the murine FLASH protein harbors potential SUMO modification motifs in its primary amino acid sequence, we performed in vitro SUMO modification assays to find out whether FLASH is a direct SUMO-1 target protein. To this end, we in vitro translated different FLASH deletion mutants, which, together, span the entire FLASH molecule (for schematic overview see Fig. 1D) and analyzed their SUMO modification. Using this approach, we were able to clearly identify SUMO-1 conjugation of two overlapping FLASH polypeptides, FLASHΔD and FLASHΔE (Fig. 1B). The other FLASH polypeptides showed no SUMO-1 conjugation (Fig. 1B and D). Interestingly, FLASHΔE harbors one consensus SUMO modification site with Lys1792 as bona fide acceptor residue. We used site-directed mutagenesis and exchanged the potential acceptor Lys1792 to Arg (in the context of FLASHΔD). When we compared this mutant FLASH protein to the wild-type protein in the in vitro SUMO modification assay, no SUMOylation of the mutant protein was detected, whereas the wild-type form was SUMO-1 modified (Fig. 1C). Taken together, these data identify Lys1792 of FLASH as main acceptor residue for SUMO modification in vitro.
To determine whether SUMO modification might regulate FLASH localization, we compared the subcellular distribution of the full-length wild-type FLASH and FLASHK1792R proteins. Confocal microscopy revealed no obvious difference in the subcellular localization of wild-type and mutant FLASHK1792R, and both proteins showed the characteristic NB-associated subcellular distribution (Fig. 1E). Thus, we conclude that SUMO modification at Lys1792 is dispensable for the subcellular localization of FLASH.
SUMO expression results in reduced FLASH protein levels
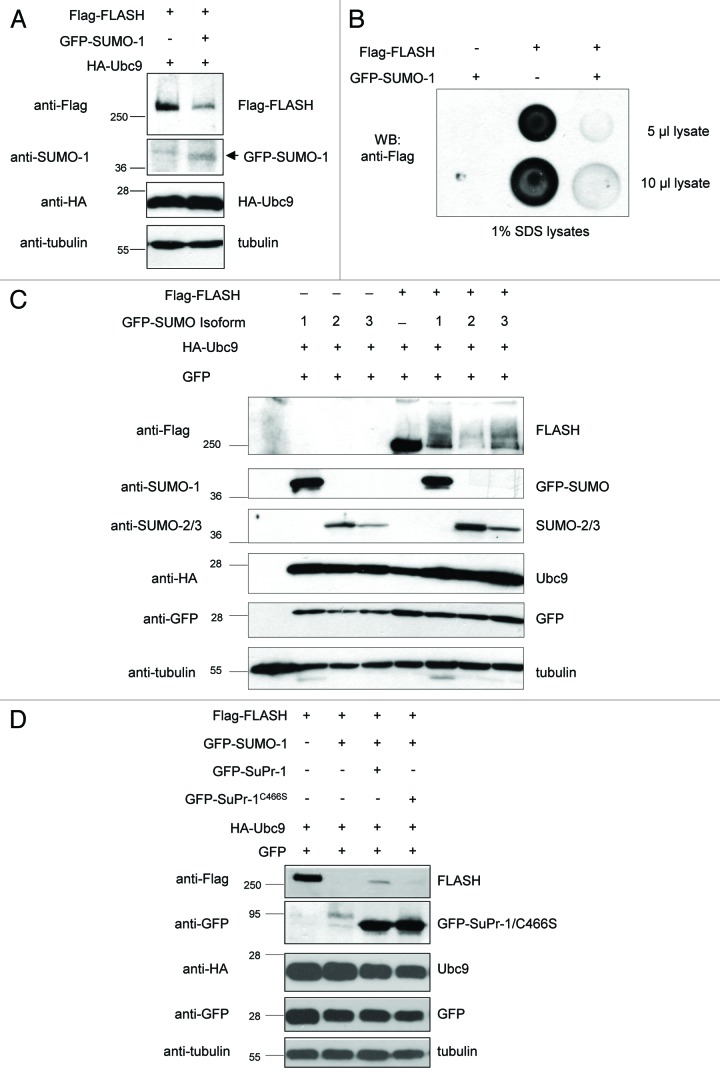
Interestingly, when we coexpressed SUMO-1 along with FLASH in 293T cell, we noticed a severe drop of FLASH protein levels in western blot analysis (Fig. 2A). To exclude the possibility that loss of FLASH is due to high molecular mass of SUMO-1-modified FLASH isoforms, which do not properly enter the SDS-PA gel, we performed dot blot analysis and directly spotted the cell lysates on the membranes to perform immunodetection with FLASH antibodies. Consistent with our results shown in Figure 1A, dot blot analysis verified a strong drop of FLASH protein levels upon expression of SUMO-1 (Fig. 2B). These results show that SUMO-1 expression leads to decreased FLASH protein levels.
Figure 2. SUMO expression results in reduced FLASH protein levels. (A) SUMO-1 expression results in reduced FLASH protein levels. The indicated constructs were transiently expressed in 293T cells and cell lysate were analyzed by immunoblotting as indicated. (B) Dot blot analysis of 293T cell lysates expressing the indicated expression constructs also shows reduction of FLASH protein levels upon SUMO-1 expression. (C) SUMO isoforms SUMO-1, SUMO-2 and SUMO-3 induce similar reduction in FLASH protein levels. The indicated constructs were transiently expressed in 293T cells, and cell lysate were analyzed by immunoblotting as indicated. (D) Expression of the SUMO protease SuPr-1 counteracts SUMO-mediated FLASH degradation. The indicated constructs were transiently expressed in 293T cells, and cell lysate were analyzed by immunoblotting as indicated.
We next aimed to test whether this effect is specific for SUMO-1 or can also be induced by the expression of SUMO-2 and SUMO-3 molecules. Although expression of each SUMO isoform resulted in profound reduction of FLASH protein levels, SUMO-2 appeared to be most effective in triggering FLASH degradation (Fig. 2C). These results suggest that FLASH degradation is not exclusively triggered by the chain-forming SUMO-2 and SUMO-3 isoforms and that—at least under the overexpression conditions used here—can be also mediated by SUMO-1.
SUMO modification has been shown to be reversible and can be removed by SUMO-specific proteases.26-28 SuPr-1(the mouse othologue of human SENP2) is a SUMO protease which has been previously shown to associate with PML bodies.27 Thus, we tested the effect of SuPr-1 on SUMO-dependent FLASH loss. Overexpression of SuPr-1 partially rescued SUMO-dependent degradation of FLASH (Fig. 2D). This effect was dependent on its peptidase activity, since a SuPr-1 mutant lacking peptidase activity, SuPr-1C466S, failed to interfere with FLASH degradation (Fig. 2D). Taken together, our results suggest that FLASH protein levels are regulated by its modification with SUMO.
SUMO mediates FLASH degradation
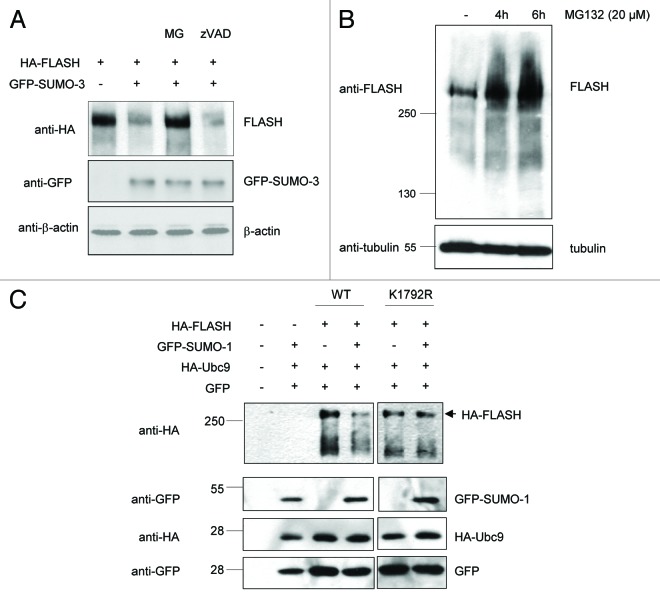
SUMO modification has been demonstrated to trigger increased PML body formation and PML degradation through the proteasome.22-25 To address whether SUMO might also induce proteasome-dependent degradation of FLASH, we inhibited the proteasome by using the inhibitor MG-132. Strikingly, proteasome-inhibition efficiently rescued FLASH depletion upon SUMO-3 expression, indicating that SUMO indeed triggers proteasome-dependent FLASH degradation (Fig. 3A). To test whether the endogenous FLASH protein is also subject to protein turnover, we inhibited the proteasome and analyzed the FLASH amounts by immunoblotting. FLASH protein amounts were clearly increased upon proteasome inhibition in a time-dependent fashion and slower migrating isoforms of FLASH accumulated upon proteasome inhibition (Fig. 3B). These data suggest that endogenous FLASH protein levels are balanced by proteasome-dependent breakdown, and proteasome inhibition results in accumulation of high-molecular FLASH isoforms.
Figure 3. SUMO mediates proteasome-dependent degradation of FLASH. (A) Proteasome inhibition rescues SUMO-mediated FLASH degradation. The indicated constructs were transiently expressed in 293T cells, treated with the proteasome inhibitor MG-132 or the pan-caspase inhibitor zVAD as indicated, and cell lysates were analyzed by immunoblotting. (B) Proteasome inhibition leads to accumulation of endogenous FLASH protein levels in HT1080 cells. Cell lysates were analyzed by immunoblotting as indicated. (C) FLASHK1792R is refractory to SUMO-mediated degradation. The indicated constructs were transiently expressed in 293T cells, and cell lysates were analyzed by immunoblotting as indicated.
Next, we analyzed the role of the SUMO acceptor residue Lys1792 in SUMO-mediated FLASH degradation. Strikingly, FLASHK1792R was refractory to SUMO-mediated degradation, whereas the wild-type protein was readily degraded (Fig. 3C). These data suggest that Lys residue 1792 is critical for SUMO-dependent FLASH degradation.
Arsenic trioxide recruits FLASH to PML bodies and triggers FLASH degradation
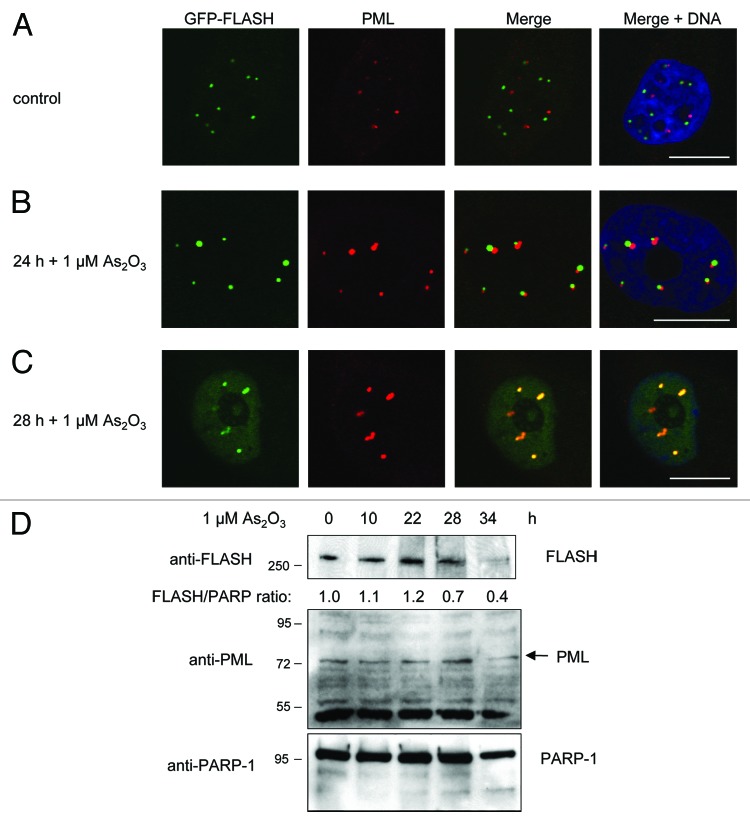
Arsenic trioxide (ATO) is an effective therapeutic drug, which is widely used to treat APL patients. Previous studies have established that ATO treatment results in increased SUMO modification of PML, potentiated PML body formation and subsequent degradation of PML through the proteasome.22-25 This prompted us to investigate whether ATO treatment might trigger the recruitment of FLASH to PML bodies by using confocal microscopy. Strikingly, whereas in untreated cells, no co-localization and only sporadic association of FLASH and PML was detectable (Fig. 4A), ATO treatment resulted in a time-dependent, gradual association and co-localization of FLASH with PML (Fig. 4B). Finally, complete co-localization of FLASH and PML at PML bodies was observed 28 h post ATO treatment (Fig. 4C). Of note, the size of PML bodies increased after ATO treatment, which is in line with previous reports.22-25
Figure 4. Arsenic trioxide treatment triggers recruitment of FLASH to PML bodies resulting in subsequent FLASH degradation. (A–C) HT1080 cell stably expressing GFP-FLASH were treated with arsenic trioxide (As2O3) and subsequently analyzed by immunofluorescence staining and confocal microscopy. Scale bars: 20 µm. (D) Immunoblot analysis of lysates of HT1080 cells stably expressing GFP-FLASH. The cells were treated with as indicated and subsequently harvested for immunoblot analysis.
We next wanted to determine whether ATO treatment results in PML degradation and loss of FLASH protein. As expected, 36 h after ATO treatment, we detected a clear reduction in PML protein levels, which is indicative for PML degradation (Fig. 4B). Remarkably, also FLASH protein levels were reduced upon ATO treatment, arguing that ATO trigger degradation of the FLASH protein (Fig. 4D). Taken together, our results indicate that ATO treatment triggers recruitment of FLASH to PML bodies, which presumably leads to FLASH protein degradation.
Discussion
Previous work identified FLASH as a multifunctional protein and component of nuclear bodies, such as Cajal bodies and PML bodies. Whereas FLASH has been shown to co-localizes to high extent with NPAT,3,29 only a minor fraction of FLASH appears to co-localize with PML bodies.4,14 One explanation for this observation might be that association of FLASH with PML bodies is a dynamic process regulated by its SUMO modification, which finally may result in FLASH degradation at PML bodies. In accordance with this interpretation, we found that co-localization between FLASH and PML is profoundly increased in response to ATO treatment. ATO has been established to potentiate SUMO modification and subsequent proteasome-dependent degradation of PML through the SUMO-interacting E3 ubiquitin ligases RNF4 or Arkadia/RNF165.22,24,25,30 PML body recruitment of FLASH is followed by reduction of FLASH protein levels, suggesting that FLASH is degraded upon ATO treatment. Thus, our results suggest that FLASH, similar to PML, is degraded in a SUMO-dependent fashion in association with PML bodies. This interpretation is in line with our findings that ectopic expression of SUMO triggers proteasome-dependent degradation of FLASH. In addition, a FLASH mutant lacking Lys1792, the bona fide SUMO acceptor residue, is unresponsive to SUMO-mediated degradation. Although the detailed mechanism of FLASH degradation remains to be identified, it is tempting to speculate that FLASH degradation might use a similar SUMO-dependent poly-ubiquitination mechanism as recently uncovered for PML and the oncogenic PML-RARα fusion protein, which is mediated by the RNF4 ubiquitin ligase.24,25
The herein identified SUMO acceptor site Lys1792 of murine FLASH is an analogous site to the previously identified acceptor site Lys1813 in the human FLASH protein.31 Exchange of SUMO acceptor lysine residue 1823 to arginine did not change the localization pattern of human FLASH. However, it reduces the transactivating function of a FLASH-Gal4 fusion.31 These results, along with our study, suggest that SUMO modification of FLASH might have pleiotropic species- and cell type-specific functions.
ATO treatment results in cell cycle arrest and induction of apoptosis in APL blast through mediating degradation of the oncogenic PML-RARα fusion protein.22-25 FLASH/Casp8AP2 has been shown to play an essential role in regulation of the cell cycle, in particular in S phase progression.2 It is therefore tempting to speculate that proteasome-dependent degradation of FLASH/Casp8AP2 by ATO might contribute to cell cycle arrest in APL blasts. Future studies will be important to address this question.
Consistent with our findings that FLASH is targeted for degradation by the proteasome, it has been previously demonstrated that FLASH is an unstable protein that is degraded upon UV damage.32 It remains to be studied in the future whether UV-induced FLASH degradation may be linked to SUMO conjugation, or whether it follows a different mechanism of degradation. Furthermore, the functional consequences of SUMO-linked FLASH degradation need to be determined in the future. Interestingly, a recent publication identified a cluster of SUMO-interacting motifs (SIMs) in the FLASH protein.33 It will be interesting to see whether non-covalent interaction of FLASH with SUMO through the SIM’s plays a role in the SUMO-regulated FLASH degradation proposed in this study. In summary, our results presented here strongly suggest that SUMO mediates proteasome-dependent degradation of FLASH, presumably through potentiating the association of FLASH with PML bodies.
Materials and Methods
Cell lines, cell culture, transduction and transfection
293T and HT1080 (both obtained from ATCC) were maintained in DMEM (Gibco) supplemented with 10% FCS, 1% (w/v) penicillin/streptomycin, 2 mM L-Glutamine, 1mM Sodium Pyruvat and 20 mM Hepes buffer at 37°C at 5% CO2. Transient transfections were done using Lipofectamine 2000 (Invitrogen) or by standard calcium phosphate precipitation. HT1080 cells stably expressing GFP-FLASH have been generated by transient transfection and subsequent selection with Hygromycin. The resulting cell pool was enriched for GFP-FLASH expressing cells by FACS sorting and used for the experiments in absence of the selection drug.
Antibodies
The following antibodies were used: rabbit anti-PML (a kind gift from Peter Hemmerich, Jena), GFP (FL, Calbiochem), Flag (M2) from Sigma, actin (C4) from MP Biomedicals, HA (12CA5) from Roche and tubulin from Sigma. The affinity-purified rabbit FLASH antibodies have been previously described.4
Expression constructs
FLASH expression constructs have been previously described1,4 or generated by standard PCR techniques. FLASHK1792R constructs were generated by site-directed mutagenesis and cloned in the respective target vectors. All constructs were verified by DNA sequencing.
Immunofluorescence staining and confocal microscopy
Cells were seeded onto coverslips and transfected as indicated in the individual experiments. Cells were fixed in 4% paraformaldehyde/PBS solution for 10 min at RT. After washing once with PBS, cells were blocked in 10% goat serum/PBS for 1 h at RT. Cells were incubated either with anti-PML or anti-HA antibodies for 1 h at RT. After washing with PBS, cells were incubated with secondary antibodies Alexa Fluor 594 donkey anti-rabbit (Invitrogen) in PBS with Hoechst 1:1,000 w/v (Sigma) and mounted on glass slides with Mowiol (Sigma). Images were taken using a confocal laser scanning microscope (FluoView1000, Olympus) with a 60× oil objective using the sequential scanning mode. All images were collected and processed using the FluoView Software (Olympus) and Adobe Photoshop.
Immunoblot analysis
Immunoblotting was performed as published,34 and proteins were detected by enhanced chemiluminescence (SuperSignal West Dura and Femto, Pierce).
In vitro SUMO modification assays
FLASH proteins were in vitro translated and labeled with 35S-methionine as described previously.4 In vitro SUMO modification reactions were performed using the In vitro SUMO-1 Conjugation Kit (Boston Biochem) according to the instructions of the manufacturer. After incubation for 30 min at 30°C, reactions were stopped by adding 5× SDS loading buffer. After separation by SDS-PAGE, gels were fixed, dried and exposed to X-ray films.
Acknowledgments
We are grateful to Eva Krieghoff-Henning for generation of GFP-FLASH-expressing cells, to Kathrin Schultheiss for help with the experiments, to S. Yonehara for providing FLASH expression constructs, to Ron Hay for the Ubc9 and SUMO constructs, to Leonard Zon for the SuPr-1 constructs and to Peter Hemmerich for the rabbit PML antibody. We thank the members of the Hofmann lab for fruitful discussions. This work was supported by a grant from the Deutsche Krebshilfe to T.G.H.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
Conceived and designed the experiments: A.V., T.G.H. Performed the experiments: A.V. Analyzed the data: A.V., T.G.H. Wrote the paper: T.G.H.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24943
References
- 1.Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature. 1999;398:777–85. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- 2.Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, et al. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci USA. 2006;103:14808–12. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcaroli D, Dinsdale D, Neale MH, Bongiorno-Borbone L, Ranalli M, Munarriz E, et al. FLASH is an essential component of Cajal bodies. Proc Natl Acad Sci USA. 2006;103:14802–7. doi: 10.1073/pnas.0604225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milovic-Holm K, Krieghoff E, Jensen K, Will H, Hofmann TG. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007;26:391–401. doi: 10.1038/sj.emboj.7601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–78. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XC, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, et al. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol Cell Biol. 2011;31:1492–502. doi: 10.1128/MCB.00979-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kino T, Chrousos GP. Tumor necrosis factor alpha receptor- and Fas-associated FLASH inhibit transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J Biol Chem. 2003;278:3023–9. doi: 10.1074/jbc.M209234200. [DOI] [PubMed] [Google Scholar]
- 8.Alm-Kristiansen AH, Lorenzo PI, Molværsmyr AK, Matre V, Ledsaak M, Sæther T, et al. PIAS1 interacts with FLASH and enhances its co-activation of c-Myb. Mol Cancer. 2011;10:21. doi: 10.1186/1476-4598-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alm-Kristiansen AH, Saether T, Matre V, Gilfillan S, Dahle O, Gabrielsen OS. FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene. 2008;27:4644–56. doi: 10.1038/onc.2008.105. [DOI] [PubMed] [Google Scholar]
- 10.Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, et al. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–67. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- 11.De Cola A, Bongiorno-Borbone L, Bianchi E, Barcaroli D, Carletti E, Knight RA, et al. FLASH is essential during early embryogenesis and cooperates with p73 to regulate histone gene transcription. Oncogene. 2012;31:573–82. doi: 10.1038/onc.2011.274. [DOI] [PubMed] [Google Scholar]
- 12.Choi YH, Kim KB, Kim HH, Hong GS, Kwon YK, Chung CW, et al. FLASH coordinates NFkappa B activity via TRAF2. J Biol Chem. 2001;276:25073–7. doi: 10.1074/jbc.M102941200. [DOI] [PubMed] [Google Scholar]
- 13.Jun JI, Chung CW, Lee HJ, Pyo JO, Lee KN, Kim NS, et al. Role of FLASH in caspase-8-mediated activation of NFkappaB: dominant-negative function of FLASH mutant in NFkappaB signaling pathway. Oncogene. 2005;24:688–96. doi: 10.1038/sj.onc.1208186. [DOI] [PubMed] [Google Scholar]
- 14.Krieghoff E, Milovic-Holm K, Hofmann TG. FLASH meets nuclear bodies: CD95 receptor signals via a nuclear pathway. Cell Cycle. 2007;6:771–5. doi: 10.4161/cc.6.7.4046. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Kamitani T. Cytoplasmic relocation of Daxx induced by Ro52 and FLASH. Histochem Cell Biol. 2010;134:297–306. doi: 10.1007/s00418-010-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flotho C, Coustan-Smith E, Pei D, Iwamoto S, Song G, Cheng C, et al. Genes contributing to minimal residual disease in childhood acute lymphoblastic leukemia: prognostic significance of CASP8AP2. Blood. 2006;108:1050–7. doi: 10.1182/blood-2006-01-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callens C, Baleydier F, Lengline E, Ben Abdelali R, Petit A, Villarese P, et al. Clinical impact of NOTCH1 and/or FBXW7 mutations, FLASH deletion, and TCR status in pediatric T-cell lymphoblastic lymphoma. J Clin Oncol. 2012;30:1966–73. doi: 10.1200/JCO.2011.39.7661. [DOI] [PubMed] [Google Scholar]
- 18.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–70. doi: 10.1016/S0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–9. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- 20.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–9. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–54. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honoré N, Doubeikovsky A, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–71. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lallemand-Breitenbach V, Zhu J, Chen Z, de Thé H. Curing APL through PML/RARA degradation by As2O3. Trends Mol Med. 2012;18:36–42. doi: 10.1016/j.molmed.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–55. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 25.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–46. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 26.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10:831–42. doi: 10.1016/S1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 27.Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, et al. SUMO-1 protease-1 regulates gene transcription through PML. Mol Cell. 2002;10:843–55. doi: 10.1016/S1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann TG, Jaffray E, Stollberg N, Hay RT, Will H. Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J Biol Chem. 2005;280:29224–32. doi: 10.1074/jbc.M503921200. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Evans HG, Evans DR. FLASH knockdown sensitizes cells to Fas-mediated apoptosis via down-regulation of the anti-apoptotic proteins, MCL-1 and Cflip short. PLoS ONE. 2012;7:e32971. doi: 10.1371/journal.pone.0032971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erker Y, Neyret-Kahn H, Seeler JS, Dejean A, Atfi A, Levy L. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol. 2013 doi: 10.1128/MCB.01019-12. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alm-Kristiansen AH, Norman IL, Matre V, Gabrielsen OS. SUMO modification regulates the transcriptional activity of FLASH. Biochem Biophys Res Commun. 2009;387:494–9. doi: 10.1016/j.bbrc.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 32.Bongiorno-Borbone L, De Cola A, Barcaroli D, Knight RA, Di Ilio C, Melino G, et al. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene. 2010;29:802–10. doi: 10.1038/onc.2009.388. [DOI] [PubMed] [Google Scholar]
- 33.Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem. 2012;287:42071–83. doi: 10.1074/jbc.M112.410985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, et al. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol. 2008;10:812–24. doi: 10.1038/ncb1743. [DOI] [PubMed] [Google Scholar]