Abstract
Secondary B cell repertoire diversification occurs by somatic hypermutation (SHM) in germinal centers following antigen stimulation. In SHM, activation-induced cytidine deaminase (AID) mutates the variable region of the immunoglobulin (Ig) genes to increase the affinity of antibodies. Although somatic hypermutation (SHM) acts primarily at Ig loci, low levels of off-target mutation can result in oncogenic DNA damage, illustrating the importance of understanding SHM targeting mechanisms. A candidate targeting motif is the E box, a short DNA sequence (CANNTG) found abundantly in the genome and in many SHM target genes. Using a reporter assay in chicken DT40 B cells, we previously identified a 1928-bp portion of the chicken IgL locus capable of supporting robust SHM. Here, we demonstrate that mutation of all 20 E boxes in this fragment reduces SHM targeting activity by 90%, and that mutation of subsets of E boxes reveals a functional hierarchy in which E boxes within "core" targeting regions are of greatest importance. Strikingly, when the sequence and spacing of the 20 E boxes is preserved but surrounding sequences are altered, SHM targeting activity is eliminated. Hence, while E boxes are vital SHM targeting elements, their function is completely dependent on their surrounding sequence context. These results suggest an intimate cooperation between E boxes and other sequence motifs in SHM targeting to Ig loci and perhaps also in restricting mistargeting to certain non-Ig loci.
Introduction
A mutator enzyme, activation-induced cytidine deaminase (AID), is responsible for fine-tuning the antibody response to a specific antigen. Expressed in germinal center B cells, AID deaminates cytosines in the variable regions of the immunoglobulin (Ig) heavy and light (L) chain genes to initiate a process known as somatic hypermutation (SHM) (1). Following the conversion of cytosine to uracil, the base pair mismatch is either replicated over to produce a transition mutation, or processed in the base excision (BER) or mismatch (MMR) repair pathways to yield transitions and transversions at the position deaminated by AID or at nearby A:T pairs. Changes in the antibody binding site that increase affinity for antigen can then be selected for during affinity maturation.
AID also initiates double-stranded breaks in switch regions of Ig genes to change antibody isotype as part of class switch recombination (2), and in some species diversifies the Ig variable region through gene conversion (GCV) (3, 4). All of these processes require transcription (5), which has led to the idea that AID acts directly on exposed single-stranded DNA in transcription bubbles (6), possibly most frequently when RNA polymerase II has stalled (7–9).
The transcriptional requirement for SHM has helped define several aspects of how AID-mediated mutation is targeted on a local level, but given the vast number of transcribed genes in B cells (10), it fails to explain why Ig genes receive the brunt of mutation. An appealing hypothesis is that Ig loci contain unique cis-acting elements that provide for preferential recruitment of the SHM machinery. However, neither the variable region (11) nor the Ig promoters (12) are required for high levels of AID-dependent mutation. The Ig enhancers have been difficult to evaluate for SHM targeting activity, as deletion of these elements in different experimental contexts has resulted in contradictory effects on mutation frequency, and often is accompanied by a transcriptional defect (13).
It is now clear that AID is not active solely on its physiological targets in the Ig loci. AID is responsible for mutations in many non-Ig genes (14), including the proto-oncogene Bcl6 (15, 16), which can lead to lymphoma (17). Numerous translocations, including break points in both Ig and non-Ig genes, are also AID-dependent (18–20). Even though a majority of deamination events are abrogated by faithful DNA repair mechanisms (21, 22), AID’s promiscuity has reinvigorated the search for cis-elements that target—and perhaps mistarget—SHM.
Recent efforts to identify SHM/GCV regulatory sequences have turned to the chicken B cell line DT40, whose compact Ig light chain (IgL) locus undergoes a high rate of constitutive SHM/GCV (9, 23–28). Multiple groups have focused on the DNA sequences downstream of the IgL constant region, which can trigger AID-dependent mutation in reporter cassettes even when placed outside of the Ig loci (24, 26). Importantly, this DIVAC (diversification activator) region includes, but is not limited to, the defined IgL enhancer, which was previously shown not to be required for SHM in the IgL locus (9, 29). The identity of the functionally critical elements, however, is in dispute (9, 25), and thus far, no specific collection of DNA binding sites has been identified that can explain DIVAC function (24, 27).
Intriguingly, all of the studies of the DT40 IgL locus have narrowed in on regions containing E boxes (9, 25, 28). The E box motif is defined by the consensus sequence CANNTG, and serves as a binding site for class I helix-loop-helix DNA binding proteins, or E proteins, which have well characterized roles in B and T lymphocyte development (30). Although many other motifs are also present, the E box is of note because of its conspicuous presence in both Ig enhancers and numerous off-target genes (21, 31, 32).
In addition, the E box has previously been shown to stimulate SHM, either as part of an artificial insert in a murine Vκ transgene (33), or as part of the murine intronic and 3' Ig-κ enhancers when assayed in DT40 cells (34). These studies suggested that E boxes operate as potent SHM targeting, and perhaps mistargeting, elements, possibly functioning independently of other sequences. Other experiments have specifically implicated the E proteins encoded by E2a (E12 and E47), in SHM and GCV (35–37), but it remains unclear if E12, E47, or any other E protein actually binds E boxes within an endogenous Ig locus to promote SHM/GCV. Moreover, the suggestion that E boxes are sufficient for mutation recruitment/targeting is problematic, since the short E box motif is very abundant in the genome and therefore cannot by itself adequately explain how high-level SHM activity occurs only at Ig loci.
We decided to directly address the question of whether E boxes are involved in SHM targeting, and took advantage of our recent identification of a 1928-bp composite element with strong SHM targeting activity, which was obtained from the original 9.8-kb DIVAC, or ‘W,’ fragment (9, 26). The smaller size of this cis-element offered the unprecedented opportunity to explore the importance of the E box motif to DIVAC function within the natural sequence environment of a highly active DIVAC element. Point mutation of all 20 E boxes in this fragment resulted in a 10-fold loss of activity, demonstrating a large role for E protein binding sites in DIVAC function. Strikingly, however, when these E box motifs were spaced precisely as they are in the 1928-bp fragment but were embedded in scrambled sequence, they were unable to support substantial SHM. The dependence of E box function on the surrounding IgL sequence argues that there are strict contextual requirements that limit the ability of this frequently occurring motif to stimulate SHM.
Materials and Methods
Cell culture
DT40 cells were grown at 41°C with 5% CO2 in RPMI-1640 (Lonza) with the additional supplements: 10% fetal bovine serum (Lonza), 1% chicken serum (Sigma), 2 mM L-glutamine, penicillin/streptomycin, and 0.1 mM β-mercaptoethanol. All subcloned or transfected cells in 96-well plates were initially grown in medium with 20% FBS. The ψV−IgL−puroR DT40 cells used for transfection, the IgL(+)GFP2 AID−/− (ψV−IgLGFP2) and IgL(−)GFP2 (ψV−IgL−,GFP2) control cell lines, and cell lines with the W, 1928 and 751 fragments were previously described (9, 26).
Transfection
Stable cell lines expressing the GFP cassettes (with or without flanking sequences) were generated by electroporating 107 ψV−IgL−puroR cells with 40 µg NotI-linearized plasmid DNA at 25 µF and 700 V (Bio-Rad Gene Pulser). Transfectants were first selected with 10–15 µg/ml blasticidin (Invitrogen) and then screened for puromycin sensitivity (1 µg/ml puromycin, Sigma) with duplicate plating. Previously described PCR reactions confirmed gene targeting to the rearranged allele (26). At least two successful primary integrants for each targeting construct were subsequently subcloned by limiting dilution, and cultured for a total of 14 days before being assayed for GFP loss by flow cytometry.
Flow cytometry
At least 12 subclones for each primary integrant were evaluated for GFP expression by flow cytometry (FACSCalibur or FACScan, BD Biosciences). Each sample was first gated by forward side scatter (FSC) and side scatter (SSC) for live cells, followed by a GFP gate drawn one log below the primary GFP+ population (FlowJo software). GFP loss values above 50% were excluded from the fluctuation analysis to prevent inclusion of any cells that were GFP(−) at the time of subcloning.
GFP2 and GFP-d targeting constructs
GFP2 targeting constructs were created by modifying pIgL(−)GFP2 (26). pIgL(−)GFP-d was created by replacing the RSV promoter with a 418-bp fragment of the IgL V promoter. Modified cassettes were TOPO-TA cloned as intact BamHI fragments (Invitrogen) that could replace the GFP2 cassette (to contain homology arms) in pIgL(−)GFP2 following digestion with BamHI. First, the endogenous promoter was PCR amplified from DT40 genomic DNA (see Table S1 A). For the GFP-d cassette, an SV40 enhancer was amplified from pGL2-control and inserted downstream of the SV40 polyadenylation signal. For both GFP2 and GFP-d constructs, test fragments were inserted using the unique NheI/SpeI restriction sites. Cloning was performed either with traditional ligation with T4 DNA ligase (New England Biolabs) or the In-Fusion HD Cloning Kit (Clontech). The precise method used for each fragment can be discerned by the primer sequence, with the latter having long overlapping overhangs (see Tables S1 B and S1 D and E). For all PCR steps, the high-fidelity Phusion polymerase (New England Biolabs) was used to prevent inadvertent mutations, and each test fragment sequence and orientation was confirmed by DNA sequencing. Many test fragments involved PCR assembly of fragments amplified from different templates using primers with long overhangs (performed both traditionally or optimized as part of the In-Fusion kit). The QuikChange site-directed mutagenesis kit (Agilent Technologies) was used for constructs requiring a single base pair change, such as the core vs. non-core analysis of E boxes in the 1928 fragment (see Table S1 C). The tandem fragment targeting construct was created in an In-Fusion cloning reaction between NheI-digested pIgL(−)GFP2–1928m and a 1928 scram fragment PCR-amplified from pIgL(−)GFP2–1928 scram. The entire W fragment sequence is available under GenBank accession number FJ482234 (http://www.ncbi.nlm.nih.gov/nuccore/FJ482234).
Fragment synthesis
The following DNA sequences were synthesized by Blue Heron Biotechnology (Bothell, WA) and delivered in the Blue Heron pUC vector: 1928m, 1928 scram, 1928 scram-m, 751-Rag, 751-Ragm, 751mT, 751 E2A(+)m, and 751 E2A(−)m. Each was digested with NheI/SpeI and cloned into the GPF2 or GFP-d vectors. The 751 versions of 1928m, 1928 scram, 1928 scram-m were PCR-amplified from the delivered templates (see Table S1 B).
1928 fragment sequence scrambling
A customized Python script (T. Luong) was used to randomize the intervening sequences between each of the E boxes. The final assembled sequence was checked for the inadvertent creation of CpG islands using EMBOSS CpG plot/report and for E boxes and matched as closely as possible for overall CpG content.
RT-PCR analysis of GFP expression
Total RNA was isolated from each DT40 clone with the RNeasy Mini Kit (Qiagen), with 1 µg treated with DNase I (Invitrogen) and reverse transcribed by Superscript II (Qiagen) using random primers (Invitrogen). Duplicate Taqman qPCR reactions were performed with HotStarTaq (Qiagen) using company-specified cycling parameters (primers and probes, Table S1 F). cDNA values were normalized by respective amounts of 18S rRNA.
Results
SHM assay
To explore the role of E boxes in SHM, we used a previously described reporter system in the DT40 chicken B cell line (9, 26). The system relies on cells that have had their endogenous IgL locus replaced with a puromycin resistance gene (Fig. 1 A), allowing for efficient targeting of a GFP cassette, flanked by a test DNA fragment, to the region. Mutation of GFP, which produces a loss of fluorescence that is easily detected by flow cytometry, is both AID and DIVAC-dependent and thus serves as a proxy for SHM events (9, 26).
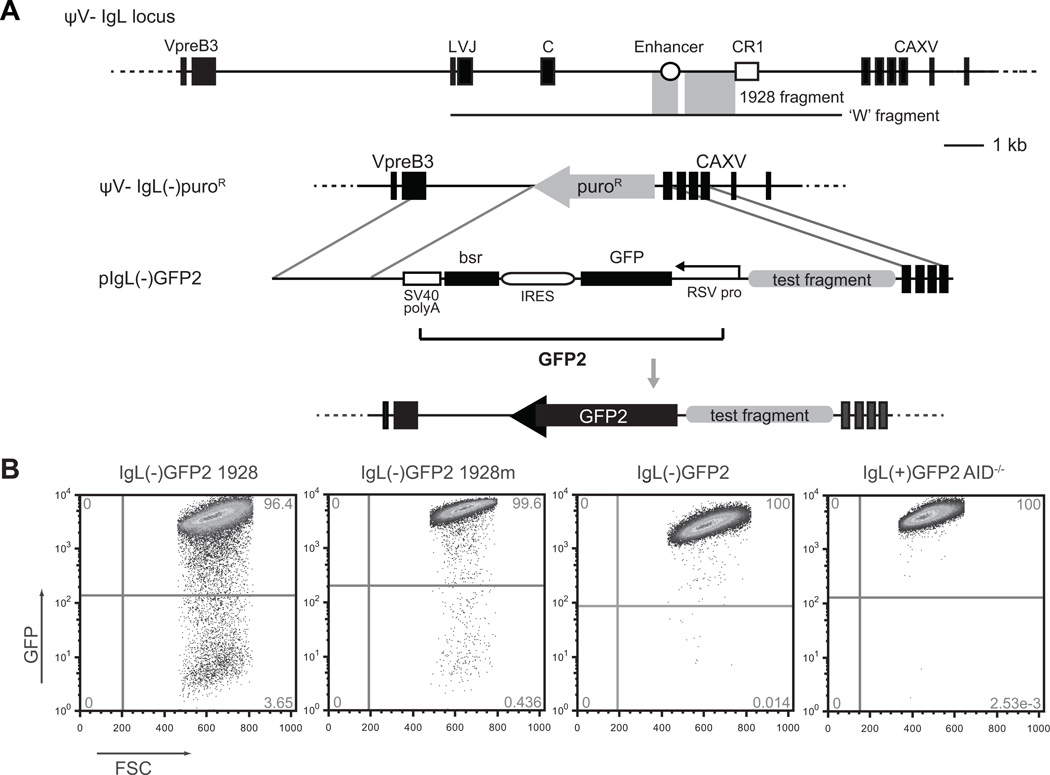
Figure 1. The DT40 SHM targeting assay.
(A) Schematic diagram of the DT40 ψVIgL locus and the experimental system used for assaying cis-elements for SHM targeting activity. The IgL locus, bounded by the VpreB and CAXV (carbonic anhydrase XV; predicted) genes, contains leader (L), VJ, and constant (C) exons (black boxes) along with an enhancer (open circle) and chicken repeat region 1 (CR1) (open square). The previously identified ‘W’ fragment (26) is illustrated by a black line, with gray shading highlighting the regions that comprise the 1928 fragment (9). Transfection of the ψV−IgL−puroR cell line with a plasmid containing a GFP2 reporter cassette, a given test fragment, and the appropriate homology arms, results in a new cell line that can be screened for targeted integration and assayed for SHM targeting activity through GFP loss. (B) Sample flow cytometry plots showing GFP expression/loss for a single subclone of the indicated cell lines. Each is a representative example of GFP loss observed after 14 days in culture.
The ‘GFP2’ cassette includes a Rous sarcoma virus (RSV) promoter to drive GFP expression, an internal ribosome entry site (IRES) to link expression to a blasticidin resistance gene, and finally, an SV40 virus polyadenylation signal. The strong RSV promoter in the cassette is unaffected by the presence or absence of enhancer elements, removing the problem of variable transcription that can complicate the interpretation of SHM activity (9, 26).
To test a DNA fragment for SHM targeting activity, the sequence is inserted next to the GFP2 cassette in the targeting vector (pIgL(−)GFP2), which has homology arms to promote replacement of the puromycin gene through homologous recombination. Following transfection into ψV−IgL(−)puroR cells, successful transfectants are selected for in blasticidin and screened for targeted integration by PCR. Multiple independent clones containing each test fragment are single-cell seeded and cultured for 2 weeks before being assayed by flow cytometry for loss of GFP expression (Fig. 1 B). Controls included in most experiments are the GFP2 cassette without any flanking DIVAC element (IgL(−)GFP2) and the GFP2 cassette flanked by the entire W fragment but in AID-deficient cells (IgL(+)GFP2 AID−/−). GFP loss is routinely less than 0.06% and 0.005%, respectively, for these two controls (see below) (9).
We confirmed that the previously identified 1928-bp fragment, which was derived by combining the most highly active sub-elements of the W fragment, supports robust SHM (3.6–6.07% GFP loss, Figs. 2 and 3). While the activity of the 1928 fragment is lower than the entirety of the W fragment [7.4–9.4% GFP loss (9, 26)] it retains a majority of activity despite being one-fifth its size. An even smaller 751-bp element, composed of the functionally most important 'core' portions of the 1928 fragment (9), also supports substantial SHM targeting activity (1.94–3.52% GFP loss, Fig. 2).
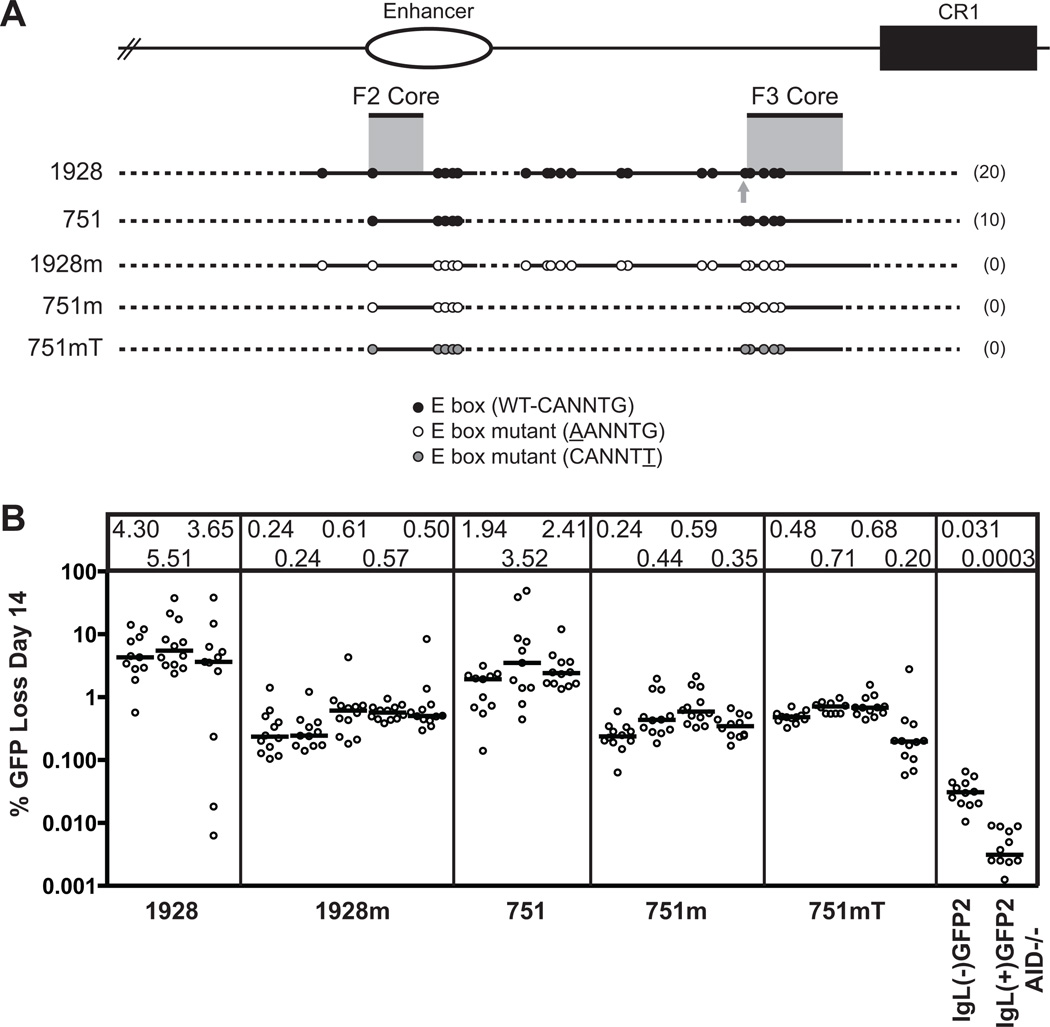
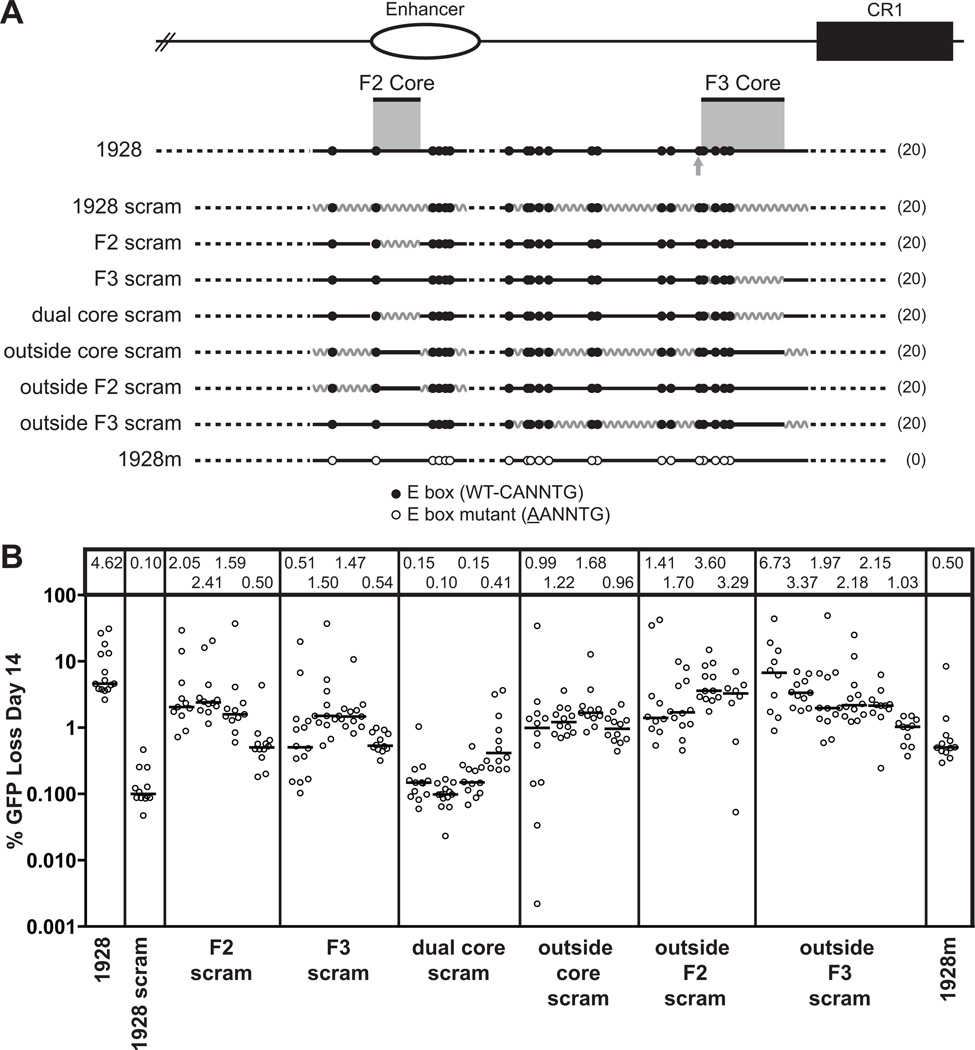
Figure 2. The E box motif is required for strong DIVAC activity.
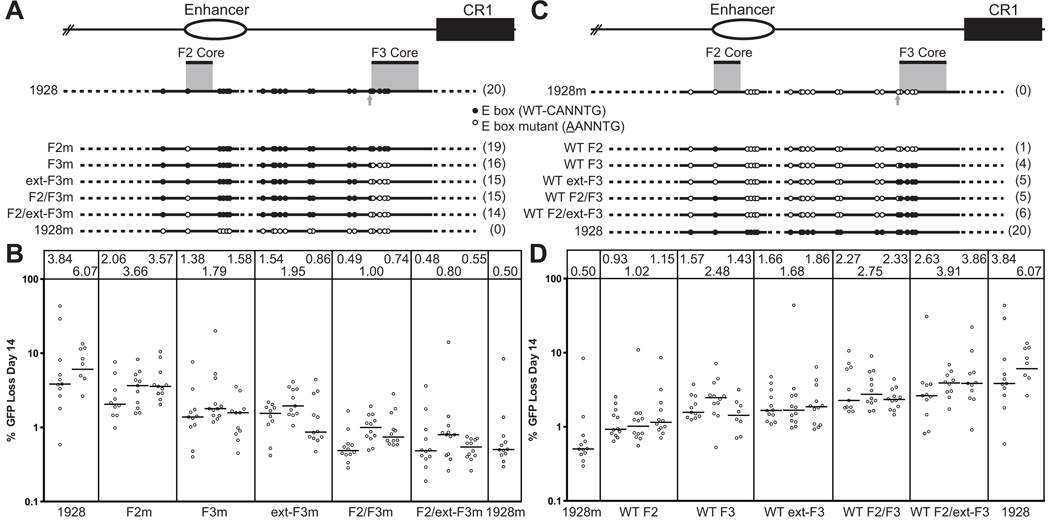
(A) Schematic diagram of the composite 1928 and 751 fragments and their position relative to the endogenous locus and to each of the previously identified “core” regions: the 200-bp F2 core and the 350-bp F3 core (gray shading). The gray arrow indicates the additional E box included in the extended F3 core. Top, The endogenous locus, including the enhancer (open oval) and CR1 element (black box), for orientation. Dotted lines denote missing sequence for each fragment tested, with the total number of wild-type E boxes for each fragment indicated in parentheses on the right; the central legend describes the two types of E box mutations. This and all subsequent schematic diagrams are drawn approximately to scale, with E box motifs enlarged for clarity. (B) Fluctuation analysis of GFP loss for cells lines expressing each of the listed fragments. Following transfection of ψV−IgL−puroR cells with the appropriate targeting construct, targeted integrants were identified and subcloned by limiting dilution. After a total of 14 days in culture, each subclone was assayed for GFP loss by flow cytometry. Each open circle denotes the percentage of GFP(−) cells in each cultured subclone, plotted on a logarithmic scale, and grouped together by primary clone. The median value of % GFP loss for each subclone set is represented by a bar and listed numerically in the box at the top of the graph.
Figure 3. Analysis of E boxes within the 1928 fragment.
(A and C) Schematic diagrams of the 1928 wild-type and mutant E box fragments, with elements as described in Fig. 2 A. (B and D) Fluctuation analysis of GFP loss at day 14, with median values indicated in the box above each subclone and represented as in Fig. 2 B. The same 1928 datasets are plotted in (B) and (D) for comparison. A representative subclone of the full E box mutant, 1928m (from Fig. 2 B), is also plotted to facilitate comparison.
E boxes are required for strong SHM activity
To determine conclusively whether E boxes contribute to DIVAC activity, we decided to entirely eliminate the motif from the relatively compact 1928 fragment. Of the residues in the E box consensus site, CANNTG, the cytosine has been previously replaced with an adenosine to prevent E protein binding (33, 34, 38). Thus, for each of the E boxes in 1928, we performed a C to A mutation, resulting in fragment 1928m (Fig. 2 A). These 20 point mutations reduced activity approximately 10-fold, to an average of 0.43% median GFP loss (Fig. 2 B). The remarkable loss of activity strongly suggests that the E box is a critical element for SHM. We verified this result in the smaller, but still highly active 751-bp fragment, which contains 10 E boxes (Fig. 2 A) (9). C to A mutation of each of these E boxes (fragment 751m) reduced activity of the 751 fragment approximately 6.5-fold, from 2.62% to 0.40% average median GFP loss (Fig. 2 B).
To confirm that the dramatic decrease in SHM activity of the C to A mutation was specific to the E box motif, we performed an alternative mutation, replacing the 3' G of the consensus E box sequence with T in the 751 fragment to generate fragment 751mT (Fig. 2 A). Since the E box is palindromic this mutation is equivalent to a C to A mutation on the other DNA strand. The G to T mutation resulted in a drop of activity commensurate with the C to A mutation (Fig. 2 B), providing further evidence that the E box motif, and not another DNA binding sequence, is required for SHM.
E boxes in core regions are especially important for SHM activity
We next wanted to determine whether particular E boxes are more important for DIVAC function than others. Given our previous finding that activity of the 1928 fragment is strongly dependent on a 200-bp portion of the IgL enhancer (F2 core) and a downstream stretch of 350 bp (F3 core) (9), we hypothesized that E boxes in these regions would be especially significant. There is one E box located in the first 22 bp of the 200-bp F2 core, and four E boxes in the 350-bp F3 core. A sixth E box is located immediately upstream of the F3 core. Since a previous deletion encompassing this box diminished activity (9), we included it as part of an extended F3 core, termed "ext-F3 core" (Fig. 3 A, C).
We tested the importance of these specific E boxes by making C to A point mutations, as before. Mutation of the single E box in the F2 core (F2m) resulted in a small but discernable drop in activity (Fig. 3 B). Mutation of the 4 or 5 E boxes in the F3 and ext-F3 cores (F3m and ext-F3m), respectively, produced a larger, approximately 4-fold, decrease in activity, and activity dropped even further when the 5 E boxes in the F2 and F3 cores (F2/F3m) were mutated. Strikingly, the combined mutation of the six E boxes in the F2 and ext-F3 cores (F2/ext-F3m) reduced activity nearly as much as mutation of all 20 E boxes in 1928 (0.48–0.80% vs. 0.23–0.61% median GFP loss, Fig. 3 B). The failure of the 14 remaining intact E boxes to support SHM demonstrates not only that non-core E boxes have little intrinsic activity, but also that they cannot substitute for E box motifs in the cores. DIVAC, then, is highly dependent on the core E boxes, whereas the non-core E boxes contribute far less.
To determine whether the core E boxes can support strong activity in the absence of the other E boxes, we performed a reciprocal series of experiments, reverting mutated E boxes to wild-type in the 1928m fragment (Fig. 3 C). Consistent with the results of Fig. 3 B, restoration of the single E box in the F2 core (WT F2) increased activity a small amount (about two fold), while restoration of the E boxes in the F3 (WT F3) or extended F3 (WT ext-F3) cores increased activity about four fold over that of 1928m (Fig. 3 D). Restoration of just the six E boxes in the F2 and ext-F3 cores raised GFP loss to about 75% of that of the intact 1928 fragment (2.63–3.91% vs. 3.65–6.07% median GFP loss, Fig. 3 D). Together, these data argue that the E boxes in both cores are required for full activity, and together can support high, but not full, activity. Therefore, the majority of E box function in DIVAC stems from a small subset of E boxes located in these previously identified regions. The results also indicate that the effects of mutating or restoring core E boxes is roughly additive, although we have not analyzed individual E boxes in the F3 core in this way.
E boxes by themselves cannot act to initiate SHM
The relatively small number of E boxes required for strong DIVAC activity raised the possibility that perhaps in certain configurations, like that of the IgL locus DIVAC region, E boxes are sufficient for SHM targeting. To investigate this, we created a new test fragment in which the 20 E box motifs are preserved but the sequence between each box is scrambled (Fig. 4 A, 1928 scram). Since the E boxes are embedded in the same base pair locations as the 1928 sequence, this design controls for potential spacing requirements of E boxes. In addition, each E box is flanked on each side by four base pairs of endogenous sequence (for a total conservation of 14 bp per box) to accommodate the possibility that E box function depends in part on the sequence of adjacent residues. Importantly, E47 occupancy observed in developing B cells from ChIP-seq experiments does not indicate that the protein has a preference for specific residues beyond these distances (38) (the sequence of 1928 scram, along with that of other key test fragments, is provided in Fig. S1).
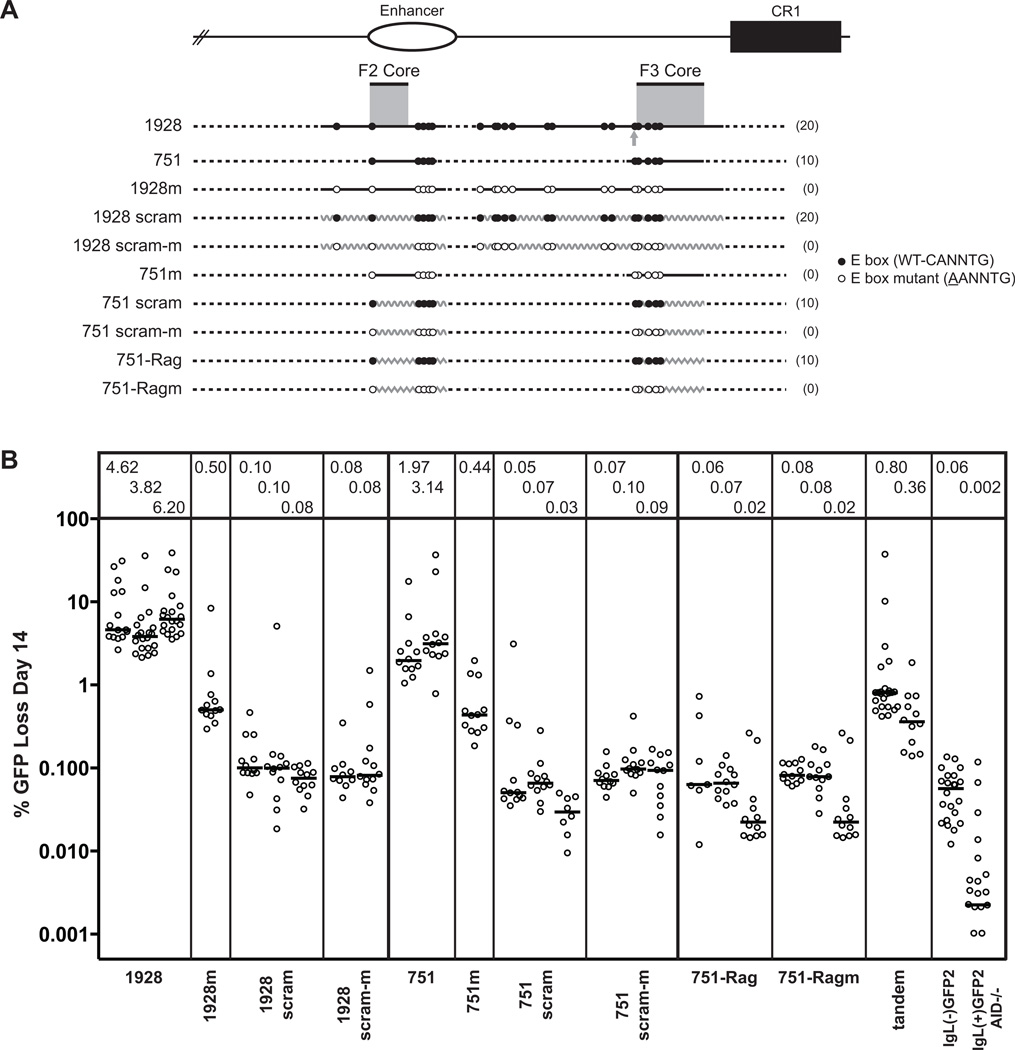
Figure 4. E boxes in DIVAC require immediately surrounding IgL sequence for SHM targeting.
(A) Schematic diagram of the composite 1928 and 751 fragments, with elements as described in Fig. 2 A. The curved and jagged gray zigzag line represents scrambled and Rag 1 intronic sequence, respectively. The tandem fragment is comprised of the 1928 scram fragment placed 5’ to the 1928m fragment. (B) Fluctuation analysis of GFP loss at day 14, with median values indicated in the box above each subclone and represented as in Fig. 2 B. For reference, representative subclones of the 1928m and 751m fragments (from Fig. 2 B) are shown.
Despite the presence of 20 properly spaced E boxes, 1928 scram produced only 0.1% GFP loss after two weeks in culture (Fig. 4 B). This extremely low level of activity is barely above the background of the GFP2 cassette with no DIVAC element (IgL(−)GFP2), demonstrating that E boxes alone cannot support SHM. Indeed, mutation of the 20 E boxes in the scrambled fragment (1928 scram-m) did not further reduce activity, indicating that E boxes are incapable of stimulating even a small amount of SHM on their own (Fig. 4 B).
We also performed the scrambling experiment with the 751-bp fragment and its 10 E boxes (751 scram) and observed the same extremely low, background-level of activity (<0.07% median GFP loss, Fig. 4 B). To confirm that our result with the scrambled sequence was not due to particular features of its randomization (described in Materials and Methods), we also embedded the E boxes from the 751 fragment into a different sequence context—in this case, a portion of the chicken Rag1 intron, which has no DIVAC activity in the GFP2 assay (data not shown), and which itself is devoid of E boxes. As before, we included a four base pair buffer on both sides of each E box. This E box-containing fragment (751-Rag) also exhibited levels of activity on par with the background (<0.07% median GFP loss, Fig. 4 B), and mutation of the 10 E boxes in this context (751-Ragm) produced no additional drop in activity (Fig. 4 B). Collectively, these results reveal that despite the clear requirement for E boxes in DIVAC, the function of the motif is entirely dependent on an appropriate surrounding sequence such as that provided by the IgL locus.
E boxes in DIVAC require immediately surrounding IgL sequence
To begin to define the context requirements of E boxes, we created a "tandem" test fragment by attaching the 1928 sequence with mutant E boxes to the scrambled 1928 sequence with intact E boxes (1928m adjacent to 1928 scram). Although completely artificial, this tandem construct theoretically contains all of the required DNA elements for full activity in either its left or right half. The activity of the 3.9-kb tandem fragment was quite low (0.36–0.80% median GFP loss, Fig. 4 B), 8.5-fold below the wild-type 1928 fragment, and consistent with the additive activity of each of its two halves. This suggests that the scrambled sequence is not detracting from the low but detectable activity of the E box mutant half of the fragment. While the interpretation of this experiment is limited by its particular spacing arrangements, it is evident that non-E box DIVAC elements cannot cooperate with the intact E boxes located less than 2 kb away. The inability of the joined fragments to complement each other’s particular deficiencies suggests the need for the E box motif to be contiguous with cooperative, supportive sequence.
Exploring the role of non-E box sequences in DIVAC
With our finding that the function of E boxes is highly context-dependent, we were interested in learning which of the non-E box sequences in DIVAC are contributing to function. Previous attempts to define critical sequences, E box or not, have largely relied on deletions (9, 23, 25–27), but they have not been designed with the knowledge that the core E boxes are critical for DIVAC activity. As a consequence, previous analyses did not distinguish between important non-E box sequences and the critical E box motifs themselves.
To avoid this complication, we left all of the E boxes intact (together with four flanking residues on each side, as above), and scrambled different portions of the 1928 fragment. We attacked the problem broadly, scrambling each core individually or together, as well as the non-core portions from the two fragments comprising 1928, DIVAC 2 (650 bp) and DIVAC 3 (1.4 kb) (Fig. 5 A) (9).
Figure 5. The role of non-E box sequence in DIVAC function.
(A) Schematic diagram of the composite 1928 fragment, with elements as described in Fig. 2 A. The curved gray zigzag line indicates segments that were scrambled. (B) Fluctuation analysis of GFP loss at day 14, with median values indicated in the box above each subclone and represented as in Fig. 2 B. For comparison, representative subclones of the 1928, 1928 scram, and 1928m fragments (from Figs. 2 B and 4 B) are shown.
Consistent with previous findings (9), scrambling the F2 core (F2 scram) substantially reduced activity, and the effect was even stronger when the F3 core was scrambled (Fig. 5 B). The scrambling of both core regions (dual core scram) almost completely debilitated the 1928 fragment (0.1–0.41% median GFP loss), demonstrating that the most crucial non-E box sequences reside in the cores.
When all of the non-core sequences were scrambled, keeping all E boxes intact (outside core scram), activity dropped by four-fold relative to the wild-type 1928 fragment (Fig. 5 B). Notably, scrambling the non-core portions of DIVAC 3 (outside F3 scram) generally produced less of a defect than scrambling the non-core portions of DIVAC 2 (outside F2 scram), even though the F3 region is twice as large as the F2 region. The non-core portions of DIVAC 2 in particular appear to contain helpful sequences, although we cannot exclude the possibility of uneven negative effects from the scrambling in this, or indeed any, fragment. Together, these data indicate that non-E box sequences in the cores are particularly important, but they also make a significant contribution outside of the cores. Inside the cores, their role is likely closely entwined with the function of the E boxes, while outside, where E box function is minor, their interdependency with E boxes is less clear.
E boxes are also required for DIVAC function in a modified GFP cassette driven by the IgL promoter
Prior efforts to identify the cis-elements involved in SHM targeting have ruled out an explicit requirement for immunoglobulin promoters, but it has also been shown that not all heterologous promoters are as effective as an Ig promoter in promoting SHM, even if they drive equivalent or higher levels of transcription (29). This observation suggests that Ig promoters might be unique in some way, conceivably in the manner in which they interact with nearby cis-elements to support SHM. We therefore wanted to determine whether DIVAC is as strongly dependent on E boxes in the context of an Ig promoter as in the context of the RSV promoter.
To address this, we replaced the RSV promoter in the GFP2 cassette with the endogenous chicken IgL V region promoter (IgL-pro), which is strongly enhancer-dependent (Fig. 6 A). As expected, cells transfected with the IgL-pro driven cassette expressed significantly less GFP than the RSV-driven cassette, as indicated by mean GFP fluorescence levels (Fig. 6 B, middle and left panels). To boost transcription, we added a downstream SV40 enhancer to generate the GFP-d (downstream SV40 enhancer) reporter (Fig. 6 A). The SV40 enhancer has been used by others to rescue IgL-pro driven transcription, and is free of intrinsic SHM targeting activity (23). GFP expression was substantially increased upon addition of the SV40 enhancer, although it remained lower than with GFP2 (Fig. 6 B, right and left panels). This suggests that while the combination of IgL-pro and SV40 enhancer drives substantial levels of transcription, the levels remain lower than with the RSV promoter.
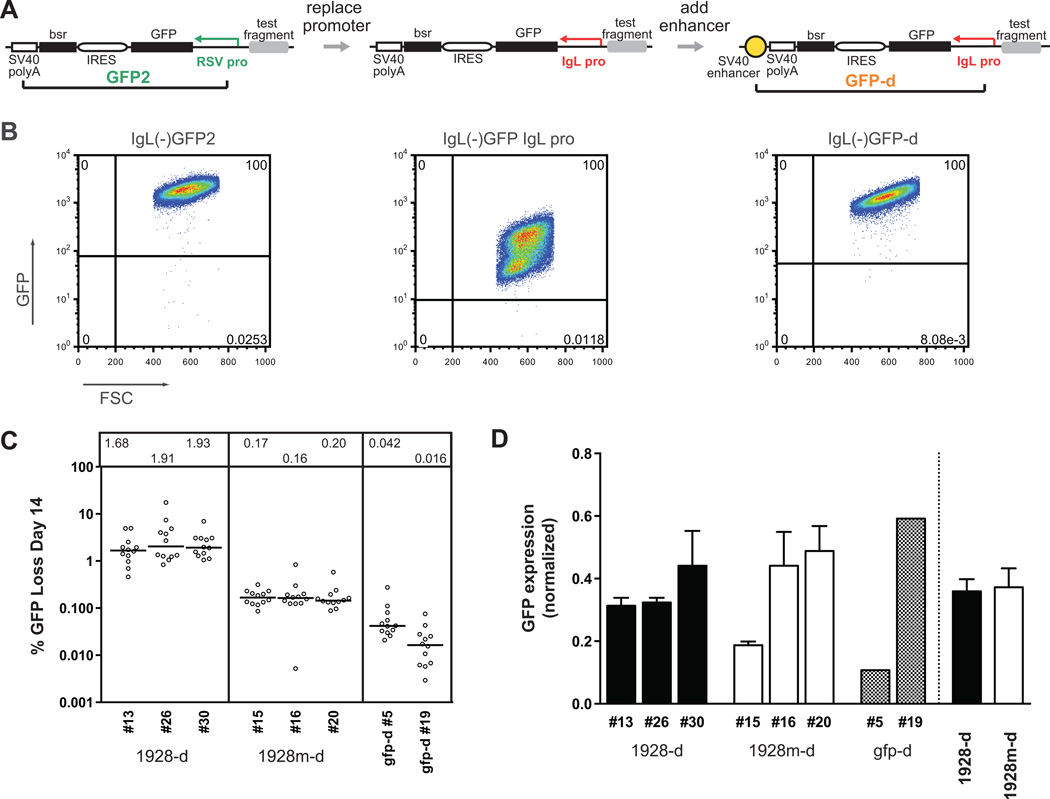
Figure 6. A modified GFP cassette to assay for SHM targeting using the IgL promoter.
(A) Schematic diagrams of all tested GFP cassettes, and development of the GFP-d assay. Left, the GFP2 reporter, which is driven by the strong heterologous RSV promoter (green). Middle, replacement of the RSV promoter with the chicken IgL promoter (red). Right, addition of a downstream SV40 enhancer element (yellow) to boost transcription, creating the GFP-d cassette. Cassette elements are drawn to scale. (B) Sample FACS plots for GFP expression/loss for each cassette in (A), without any flanking DIVAC sequence. Shown are primary transfectants for both IgL promoter cassettes, and a representative subclone of the standard GFP2 cassette, each 14 days after culture. Gates are drawn a log below the lower boundary of the GFP+ population. (C) Fluctuation analysis of GFP loss at day 14, with median values indicated in the box above each subclone and represented as in Fig. 2 B. (D) Transcript levels of the GFP coding region for each individual clone, as assayed by Taqman quantitative RT-PCR. For each, expression is normalized to the respective 18S rRNA signal. For 1928-d and 1928m–d, bar height indicates the average signal from three independently derived RNA samples for each subclone (left of dotted line), or the average of all data collected from clones of that type (right). Data for cell lines with mutant E box fragments are displayed with white bars; cassette-only clones are patterned. Error bars mark the standard error of the mean (SEM).
As expected, the GFP-d cassette lacking a DIVAC sequence produced very little GFP loss after two weeks in culture (<0.042% median GFP loss, Fig. 6 C). The addition of a flanking DIVAC sequence, such as the 1928 fragment (construct 1928-d), resulted in a substantial increase in GFP loss (mean median of 1.82%, Fig. 6 C), suggesting that the GFP-d system is also DIVAC-dependent. GFP-d does not appear to support mutation as efficiently as GFP2 (3.6–6.02% GFP loss with the 1928 fragment), perhaps due to lower levels of transcription from the former.
It was important to determine whether addition of the 1928 fragment (which contains an enhancer) to GFP-d increases GFP loss simply by increasing GFP transcription. Using quantitative RT-PCR, we observed that GFP-d transcript levels were somewhat variable with a variety of test fragments (Fig. 6 D and data not shown). For example, one clone containing GFP-d (#19) yielded GFP transcript levels comparable to that of 1928-d clones, but another (#5) did not (Fig. 6 D). Both GFP-d clones, however, yielded much lower levels of GFP loss than clones containing 1928-d (Fig. 6 C), supporting the conclusion that the GFP-d system is DIVAC dependent by a mechanism that does not simply involve increased transcription.
We then tested whether DIVAC remains dependent on E boxes with the IgL promoter. In the context of the GFP-d reporter, mutation of all 20 E boxes in the 1928 fragment (1928m-d) reduced GFP loss from 1.82% to just 0.14–0.17% (Fig. 6 C). To ensure that the dramatic 10-fold drop in GFP loss was not due to reduced transcription, we compared GFP transcript levels in 1928-d and 1928m-d clones and found no consistent differences (Fig. 6 D; average of 0.36 ± 0.12 for 1928-d vs. 0.37 ± 0.18 for 1928m–d). We conclude that the large difference in measured SHM activity between cassettes with intact or mutant E boxes is independent of transcription, and therefore that E boxes are also required for strong DIVAC function with the IgL promoter.
The role of E boxes predicted to bind E12/E47
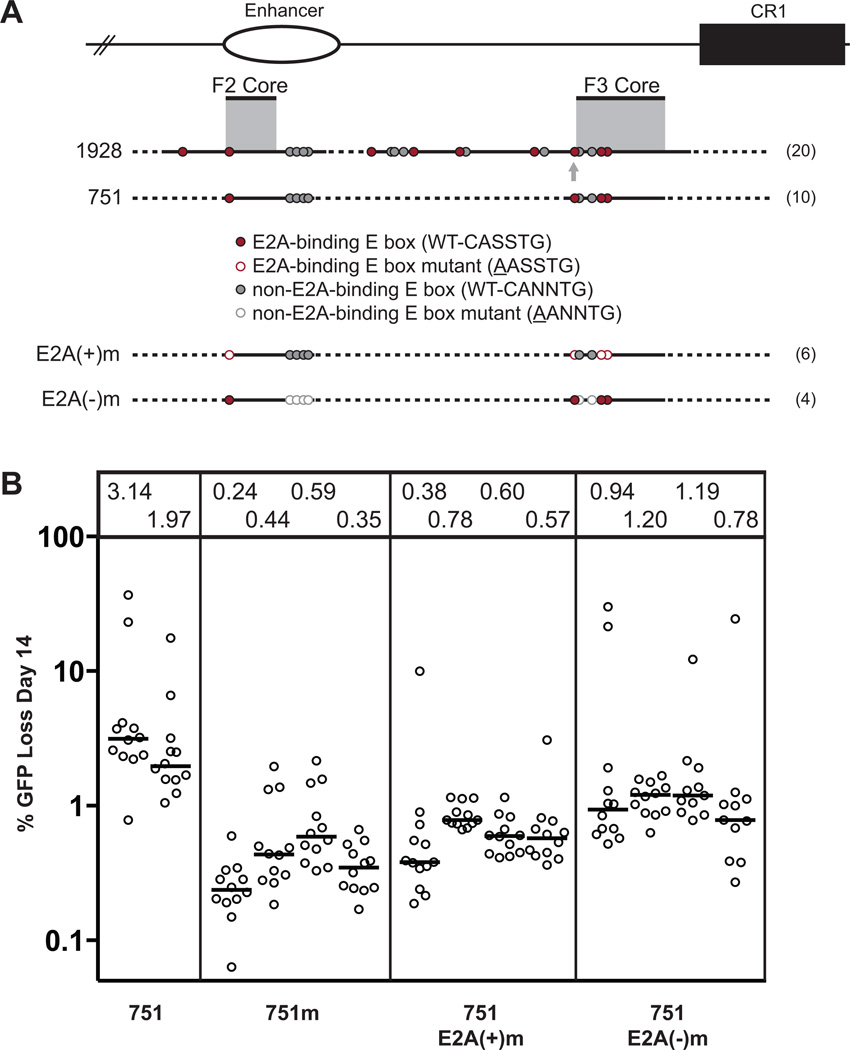
Since several studies have specifically linked the E2a-encoded proteins E12 and E47 (hereafter, E2A proteins) to SHM and GCV (35–37), one possibility is that E box functionality might be determined by whether the motif can be bound by these proteins. E2A proteins prefer the motif CAGCTG, and a recent genome wide analysis revealed a general binding motif of CASSTG (where S = G or C), where the C residue is not flanked on its 5' side by T on either strand (38). Classification of the 20 E boxes in the 1928 fragment according to this criterion reveals that 9 are predicted to bind E2A (E2A+), as are 4 out of 6 E boxes in the F2 core plus ext-F3 core (Fig. 7 A). To determine whether SHM targeting activity is primarily determined by the potential to bind E2A proteins, we mutated the four core E2A+ E boxes in the context of the 751 fragment (751 E2A(+)m). This resulted in a roughly 75% drop in activity (1.97–3.14% with the intact 751 fragment versus 0.38–0.78% GFP loss with 751 E2A(+)m; Fig. 7 B). The 751 fragment also contains six E2A(−) E boxes (two inside and four outside the core regions) (Fig. 7 A). Mutation of these six E boxes (751 E2A(−)m) resulted in a 60% drop in activity compared to the intact 751 fragment, while mutation of all 10 E boxes (751m) resulted in an 84% drop in activity (Fig. 7 B). Therefore, both E2A(+) and E2A(−) E boxes contribute substantially to DIVAC function in the 751 fragment.
Figure 7. Analysis of E boxes within the 751 fragment.
(A) Schematic diagram of the composite 1928 and 751 fragments, with elements as described in Fig. 2 A. Additionally, each E box is classified as to whether E2A proteins are predicted to bind to its particular sequence. E2A(+) E boxes, which conform to the CASSTG motif (where S = G or C), along with other constraints (see text for full description), are indicated in red. Intact and mutant E boxes of each type are represented by filled and open circles, respectively. (B) Fluctuation analysis of GFP loss at day 14, with median values indicated in the box above each subclone and represented as in Fig. 2 B. For comparison, all of the 751m fragment datasets from Fig. 2 B are shown.
Discussion
Using a reporter assay in conjunction with a previously identified 1928-bp DIVAC fragment derived from the chicken IgL locus, we found a 10-fold reduction in SHM targeting upon disruption of all E box sequences. This is the largest role yet observed for any single cis-element in the targeting of SHM, and represents the first demonstration of a crucial role for E boxes in a highly active SHM targeting region. Notably, this active region contains over twice as many E boxes as chance alone would predict for its size. At the same time, we found that E boxes cannot on their own target SHM, implicating other, as-yet unidentified, elements as cooperative partners necessary for E box-mediated targeting, and perhaps mistargeting, of SHM.
A central role for E boxes in SHM targeting
Our study builds on a number of previous studies implicating E boxes in the targeting of SHM (33–36). The most recent of these, by Tanaka et al. (34), found that the three E boxes in the murine Igκ intronic and 3' enhancers were required to detect SHM reliably in a GFP reversion assay in DT40 cells. The authors concluded that E boxes were required and sufficient for the targeting of SHM, at least in the context of the enhancer elements examined. Our results substantially advance our understanding of this phenomenon and call into question both of these conclusions. Using two different point mutations to inactivate all of the E boxes present in various DIVAC fragments, we observed substantial residual DIVAC function associated with the remaining sequences, (approximately 10-fold above background; Fig. 2). We conclude that although E boxes contribute to as much as 90% of SHM targeting activity, they are not prerequisites for it. Furthermore, our finding that various DIVAC fragments with intact E boxes but with other sequences altered have only background levels of activity (Fig. 4), strongly argues against the notion that E boxes are sufficient for SHM targeting. The distinct conclusions arising from the two studies are likely due to the weak signal provided by the GFP reversion assay used by Tanaka et al. (34) and their use of murine enhancers that we (9) and others (39) have shown have poor SHM targeting activity in DT40 cells. In contrast, our analysis rests on a robust SHM assay and DIVAC sequences that contain much or most of the known DIVAC function of the chicken IgL locus (26).
We have also confirmed that E boxes are critical for SHM activity when the IgL promoter drives transcription (GFP-d system). This is significant in light of our previous finding that the IgL promoter is specialized for SHM/GCV (29), and therefore might behave differently than the RSV promoter used in GFP2. However, we found that mutation of the 20 E boxes in the 1928 fragment in the GFP-d cassette reduced activity to the same degree as in the GFP2 system (over 10-fold, Fig. 6 C). Importantly, we did not observe a decrease in transcription upon E box mutation, even in the more sensitive GFP-d assay (Fig. 6 D), consistent with results obtained upon mutation of the E boxes in the murine Igκ enhancers (34, 40) and upon deletion of E2a in DT40 cells (35).
Although we have found naturally derived Ig cis-elements to be strongly dependent on E boxes for SHM targeting when driven by an Ig promoter, this result has yet to be extrapolated to the entirety of an endogenous Ig locus. For technical reasons, we performed the E box mutations in fragments 1928-bp and smaller, leaving open the possibility that the nearly 8 kb of remaining chicken IgL DIVAC sequence contains elements that can compensate for the loss of E boxes. Redundancy is common in biological systems where stability and precision are important (41), and represents a significant challenge to the study of SHM targeting sequences in chicken IgL. Redundancy is likely to pose an even bigger challenge in the larger and more elaborate Ig loci of mammals. By focusing on a smaller element derived from the most active sub-fragments of chicken IgL, we aimed to minimize complexity and reveal at least some of the underlying motifs driving SHM. As such, we view the crippling of activity observed in the 1928 E box mutant as evidence of a genuine, if not unique, mode of SHM targeting.
Our experiments provide the first direct test of whether E boxes are sufficient for SHM targeting, with the results indicating that they are unable to initiate SHM activity above background in either of two distinct sequence environments, and in DIVAC fragments of two different sizes (Fig. 4). There are, however, some caveats associated with these experiments. While we took precautions to minimize confounding effects of the new sequences (see Materials and Methods), we cannot rule out the possibility that negative regulatory elements were inadvertently created that masked E box function, although this would have to be true for both of the sequence contexts examined. For the most part, experiments with normal-scramble hybrid fragments did not suggest accidental negative elements, with the activity of hybrid fragments largely in agreement with the results of our earlier deletion analysis (9). We note, however, that the "outside core scram" fragment (Fig. 5) was less active than expected from the remaining normal sequence, raising the possibility of a negative effect of scrambled sequences outside of the cores, at least in this particular context. Most of these non-core sequences would not be present in the context of the 751 fragment, making it unlikely that this was relevant to the profound lack of activity when the E boxes in this fragment were embedded in different sequence contexts (Fig. 4). In general, there is no reason to suspect these sequence contexts are qualitatively different from random sequences in the genome.
Overall, our data strongly argue that E boxes are dependent on surrounding supportive sequences for SHM targeting activity. The results provide experimental evidence for a context–based restriction on E box DIVAC function, which helps explain how a ubiquitous motif can serve as a targeting element for Ig loci, and possibly in less potent form, for genes scattered across the genome.
Our data demonstrate that E boxes located in the two core regions are especially important. This is most clearly illustrated by our finding that activity was strongly reduced when the six core E boxes are mutated, but only slightly reduced when the 14 non-core E boxes were mutated (Fig. 3). These results, combined with our previous finding that deletion of the two cores from DIVAC 1928 or even larger DIVAC fragments eliminated activity (9), strongly argues that while non-core E boxes contribute to DIVAC function, they do so only in the presence of E box-competent core regions. This emphasizes the importance of sequence context in determining whether E boxes are able to stimulate SHM. Intriguingly, our previous deletion series suggested there was functional synergy between the F2 and F3 cores (9). The contribution of core E boxes to activity, however, appears additive, so synergy between the cores is likely due to cooperation requiring non-E box sequences.
What determines the E box hierarchy is unclear, although the cores seem to provide a particularly hospitable environment. Most core E boxes are predicted to be suitable binding sites for E2A proteins, yet there are five E2A+ boxes remaining outside the cores that are insufficient for activity (Fig. 3). The presence of a single, functionally important, E box in the F2 core suggests that high E box density is not required for the function of a DIVAC element, although it might contribute to the activity of the F3 core (which contains 5 E boxes). This issue, and the role of individual E boxes in the F3 core, would best be explored with a more sensitive DIVAC assay.
Our data also indicate that E boxes located in non-enhancer regions of the chicken IgL locus contribute to SHM targeting. This is consistent with our previous analyses indicating a wide distribution of sequences with DIVAC function (9, 26), and might be a feature shared with cis-elements in other species.
The role of non-E box sequence
The results obtained with normal-scramble hybrid fragments with intact E boxes (Fig. 5) indicate that important non-E box sequences are scattered throughout DIVAC, but are particularly concentrated in the core regions. Although we have not yet identified specific motifs that could explain supportive function, our results are consistent with previous studies implicating NF-κB and PU.1/IRF4 sites, both of which are located in the F2 core (9, 24, 27). We note that deletions affecting the non-core regions of DIVAC 3 (9) are more detrimental to activity than scrambling of the non-core, non-E box portions of DIVAC 3 (Fig. 5). This argues that the E boxes in these regions are able to exert some activity in different contexts while the non-E box sequences are of little importance.
The localization of critical E box and non-E box sequences to just two core regions totaling less than 600 bp might appear at odds with the highly dispersed nature of DIVAC. The cores, however, still rely on non-core sequence to augment their activity, and themselves are discrete, non-contiguous units, providing further evidence of the complex redundancy in just a fraction of the chicken IgL locus. We speculate that other Ig loci (and perhaps the most highly targeted non-Ig genes) are also organized around multiple SHM cores, or ‘hubs,’ that are only fully active upon coordination with nearby sequences.
Candidate E box binding factors
It seems likely that E boxes contribute to DIVAC function by serving as binding sites for one or more E proteins, although this has yet to be proven. The most obvious E protein candidates are those encoded by E2a, including E47, which has been shown to bind the IgL locus in DT40 cells (37). Disruption of E2a in DT40 cells reduces both SHM and GCV, although there is disagreement concerning the mechanism of this (35, 36). Notably, two-thirds of the E boxes in the DIVAC cores conform to the subtype preferred by E2a-encoded proteins (Fig. 7). Mutation of just the E2A(−) E boxes, however, still reduced activity substantially (Fig. 7), suggesting either that chicken E12/E47 can bind these non-canonical sites or that other E proteins contribute to DIVAC function. We note that mutation of just 20 nucleotides in DIVAC 1928 reduced SHM of GFP 10-fold, twice as much as disruption of the E2a gene reduced IgL SHM (35). While it is unclear if the results of the two types of assays involved can be directly compared, existing data leave open the question of the identity of the trans factor(s) that bind E boxes to mediate DIVAC function.
In summary, our work has shown the E box motif to be a critical element for the targeting of SHM, and has further revealed that E boxes are critically dependent on surrounding DNA sequences for function. We hypothesize that such dependency reflects a mechanism in which several trans factors are required to collaborate with the proteins acting on E boxes to achieve high levels of AID-mediated mutation at Ig loci. Varying degrees of congruency between active DIVAC sequences and sequence combinations elsewhere in the genome might explain the wide range of mutation frequencies seen at non-Ig genes (14, 21). Some support for this idea comes from a recent analysis of the sequences surrounding the transcription start sites of non-Ig genes, which found that AID mediated deamination correlates with the tri-localization of the E box motif with the binding sites for Yy1 and C/EBPβ (31). Our findings suggest that it will be informative to analyze enhancer and other sequences located distal to the transcription start site in AID-target genes for motifs that collaborate in the targeting of SHM.
Supplementary Material
Acknowledgements
We thank Dr. Kristin M. Kohler for helpful advice and discussions. We also thank Dr. ThaiBinh Luong for writing the Python script used for scrambling the 1928 DIVAC sequence, which provided preliminary information on CpG content for each intervening sequence.
Abbreviations used in this paper
- AID
activation-induced cytidine deaminase
- CR1
chicken repeat 1
- DIVAC
diversification activator
- GCV
gene conversion
- SHM
somatic hypermutation
Footnotes
Funding for this work was provided by the Howard Hughes Medical Institute. J. McDonald was supported in part by Training Grant T32AI07019 from the National Institutes of Health. J–M. Buerstedde was supported by a Marie Curie International Fellowship from the European Union.
References
- 1.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 4.Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang SY, Fugmann SD, Gramlich HS, Schatz DG. Activation-induced cytidine deaminase-mediated sequence diversification is transiently targeted to newly integrated DNA substrates. J. Biol. Chem. 2007;282:25308–25313. doi: 10.1074/jbc.M704231200. [DOI] [PubMed] [Google Scholar]
- 6.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 7.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler KM, McDonald JJ, Duke JL, Arakawa H, Tan S, Kleinstein SH, Buerstedde JM, Schatz DG. Identification of core DNA elements that target somatic hypermutation. J. Immunol. 2012;189:5314–5326. doi: 10.4049/jimmunol.1202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr., Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez Fernandez A, Pannell R, Neuberger MS, Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 12.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 13.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, Dalla-Favera R. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd., Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 18.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, Casellas R, Nussenzweig MC. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 22.Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, Wilpan RY, He Y, King BL, Mills KD. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat. Immunol. 2010;11:820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothapalli N, Norton DD, Fugmann SD. Cutting Edge: A cis-acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J. Immunol. 2008;180:2019–2023. doi: 10.4049/jimmunol.180.4.2019. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Tian M. NF-kappaB family of transcription factor facilitates gene conversion in chicken B cells. Mol. Immunol. 2009;46:3283–3291. doi: 10.1016/j.molimm.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Kothapalli NR, Collura KM, Norton DD, Fugmann SD. Separation of mutational and transcriptional enhancers in Ig genes. J. Immunol. 2011;187:3247–3255. doi: 10.4049/jimmunol.1101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blagodatski A, Batrak V, Schmidl S, Schoetz U, Caldwell RB, Arakawa H, Buerstedde JM. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS Genet. 2009;5:e1000332. doi: 10.1371/journal.pgen.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H, Tian M. Transcription factors PU.1 and IRF4 regulate activation induced cytidine deaminase in chicken B cells. Mol. Immunol. 2010;47:1383–1395. doi: 10.1016/j.molimm.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y, Tian M. The recruitment of activation induced cytidine deaminase to the immunoglobulin locus by a regulatory element. Mol. Immunol. 2010;47:1860–1865. doi: 10.1016/j.molimm.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Yang SY, Fugmann SD, Schatz DG. Control of gene conversion and somatic hypermutation by immunoglobulin promoter and enhancer sequences. J. Exp. Med. 2006;203:2919–2928. doi: 10.1084/jem.20061835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 31.Duke JL, Liu M, Yaari G, Khalil AM, Tomayko MM, Shlomchik MJ, Schatz DG, Kleinstein SH. Multiple Transcription Factor Binding Sites Predict AID Targeting in Non-Ig Genes. J. Immunol. 2013;190:3878–3888. doi: 10.4049/jimmunol.1202547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E Box Motif CAGGTG Enhances Somatic Hypermutation without Enhancing Transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka A, Shen HM, Ratnam S, Kodgire P, Storb U. Attracting AID to targets of somatic hypermutation. J. Exp. Med. 2010;207:405–415. doi: 10.1084/jem.20090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoetz U, Cervelli M, Wang YD, Fiedler P, Buerstedde JM. E2A expression stimulates Ig hypermutation. J. Immunol. 2006;177:395–400. doi: 10.4049/jimmunol.177.1.395. [DOI] [PubMed] [Google Scholar]
- 36.Kitao H, Kimura M, Yamamoto K, Seo H, Namikoshi K, Agata Y, Ohta K, Takata M. Regulation of histone H4 acetylation by transcription factor E2A in Ig gene conversion. Int. Immunol. 2008;20:277–284. doi: 10.1093/intimm/dxm140. [DOI] [PubMed] [Google Scholar]
- 37.Yabuki M, Ordinario EC, Cummings WJ, Fujii MM, Maizels N. E2A acts in cis in G1 phase of cell cycle to promote Ig gene diversification. J. Immunol. 2009;182:408–415. doi: 10.4049/jimmunol.182.1.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kothapalli NR, Norton DD, Fugmann SD. Classical Mus musculus Igkappa enhancers support transcription but not high level somatic hypermutation from a V-lambda promoter in chicken DT40 cells. PloS ONE. 2011;6:e18955. doi: 10.1371/journal.pone.0018955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inlay MA, Tian H, Lin T, Xu Y. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating Vkappa Jkappa rearrangement. J. Exp. Med. 2004;200:1205–1211. doi: 10.1084/jem.20041135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.