Abstract
A group of Gram-negative bacteria, including the problematic pathogen Pseudomonas aeruginosa, has linked the steps in cell-wall recycling with the ability to manifest resistance to β-lactam antibiotics. A key step at the crossroads of the two events is performed by the protease AmpD, which hydrolyzes the peptide in the metabolite that influences these events. In contrast to other organisms that harbor this elaborate system, the genomic sequences of P. aeruginosa reveal it to have three paralogous genes for this protease, designated as ampD, ampDh2 and ampDh3. The recombinant gene products were purified to homogeneity and their functions were assessed by the use of synthetic samples of three bacterial metabolites in cell-wall recycling and of three surrogates of cell-wall peptidoglycan. The results unequivocally identify AmpD as the bona fide recycling enzyme and AmpDh2 and AmpDh3 as enzymes involved in turnover of the bacterial cell wall itself. These findings define for the first time the events mediated by these three enzymes that lead to turnover of a key cell-wall recycling metabolite as well as the cell wall itself in its maturation.
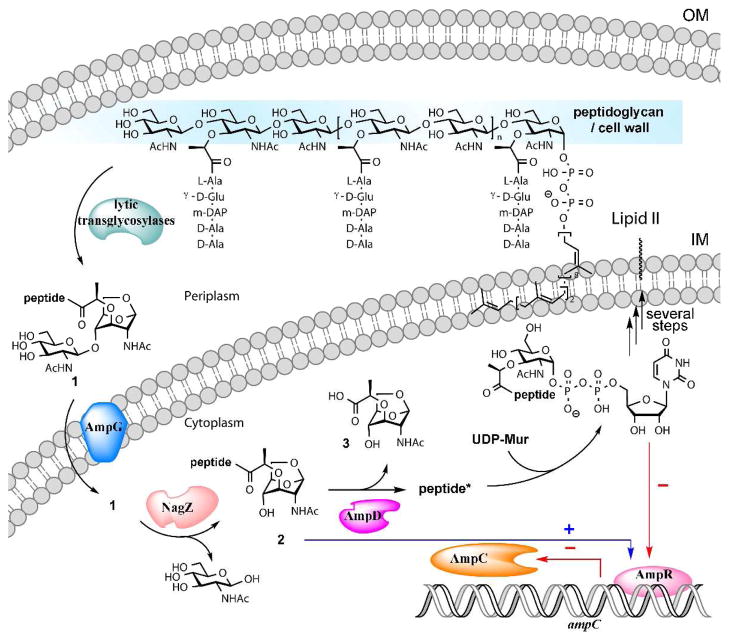
Members of Enterobacteriaceae and Pseudomonas aeruginosa have evolved an elaborate sequence of events that recycles more than 50% of the bacterial cell wall during normal growth for reasons that are not fully understood.1–5 The recycling also takes place when damage to the cell wall occurs. As damage to cell wall is also inflicted by exposure of bacteria to β-lactam antibiotics, these organisms have evolved a link between the biochemical steps of recycling and repair to unleashing of an inducible antibiotic-resistance mechanism involving the AmpC β-lactamase (Fig. 1).1,5–8
Figure 1.
The early events of bacterial cell-wall recycling and their link to antibiotic resistance. Lytic transglycosylases degrade the cell wall in the periplasm, located between the outer membrane (OM) and the inner membrane (IM).
Bacterial cell-wall recycling commences by degradation of the peptidoglycan, the major constituent of the cell wall, by the family of lytic transglycosylases.3,4,9,10 Whereas these enzymes turn over their polymeric substrates with some variations on the non-hydrolytic reaction that generates the fragmentation products, the major end product is N-acetyl-β-D-glucosamine-(1→4)-1,6-anhydro-N-acetyl-β-D-muramyl-peptide (1), with the full-length peptide being L-Ala-D-γ-Glu-meso-DAP-D-Ala-D-Ala (DAP stands for diaminopimelate).9
Metabolite 1 is internalized by the permease AmpG. Once in the cytoplasm, the glycosidase NagZ converts compound 1 to 2, which in turn serves as the substrate for the protease AmpD, which removes the peptide stem from the saccharide. The product of this reaction, compound 3, enters a sequence of biochemical events that synthesizes the building unit Lipid II, which is transported to the periplasmic side of the inner membrane to be used in assembly of the nascent peptidoglycan (Fig. 1). As a branching point in these events, metabolite 2 stimulates gene transcription to result in the production of an antibiotic-resistance enzyme, the AmpC β-lactamase (Fig. 1). Hence, the action of AmpD, at the crossroads of these events, shuts down the production of the AmpC β-lactamase and commits to recycling of the cell wall.3,5,11
In contrast to other organisms that possess this system, analyses of the genomic sequences of P. aeruginosa have led to the annotation of three paralogous genes for the protease AmpD. These are designated as AmpD, AmpDh2 and AmpDh3.12 AmpDh2 has a signal peptide that targets it to the periplasm and an anchor that is believed to insert it into the outer membrane.13 On the other hand, AmpD is believed to be cytoplasmic and the cellular location of AmpDh3 is currently unknown.13 Abrogation of the respective activities for these enzymes by mutational inactivation or by gene ablation led to a stepwise upregulation of antibiotic resistance, with the full effect achieved only by the loss of all three enzyme activities at the whole-organismal level.12–14 Interestingly, inactivation of ampD is not sufficient to affect fitness or virulence of P. aeruginosa in murine models of infection, but double or triple mutants with losses of additional activities of the other two enzymes (AmpDh2 and AmpDh3) would appear to be defective in both.13
Notwithstanding the careful analyses of the mutational effects at the whole-organismal level, the activities of these three AmpD enzymes of P. aeruginosa have not been studied with suitable substrates by enzymological analysis. This is largely due to the lack of availability of these substrates. We report herein our cloning of the genes ampD, ampDh2 and ampDh3 from the strain P. aeruginosa PA01. The respective proteins (AmpD, AmpDh2, and AmpDh3) were expressed and purified to homogeneity. We synthesized compounds 4–9 (Chart 1) by the methodology that we have reported earlier.15–17 Compounds 5 and 8 had not been made previously and were prepared for the first time for this study (described below), along with the other four known compounds.16,17 As indicated in Fig. 1, 1,6-anhydromuramyl derivatives 2 are bona fide substrates for AmpD. The variation among compounds 2 is in the length of the peptide stem, which is traced back to what is found in the cell wall. This is a uniquely bacterial peptide, whose full-length sequence is L-Ala-D-γ-Glu-meso-DAP-D-Ala-D-Ala. Maturation of cell wall generates variants of the peptide stem with loss of amino acids from the C-terminal end.3,18,19 As such, the tripeptide and tetrapeptide stems are more common, and the pentapeptide, which is biosynthetically introduced into the nascent peptidoglycan, is less so.3,18 We have synthesized all three versions (4–6). We have used compounds 7–9 as mimetics of the standard N-acetylglucosamine-based muropeptides from the cell wall (Chart 1). These compounds would allow detection of turnover of standard cell-wall peptidoglycan (which exists only in the periplasm), as opposed to that of the 1,6-anhydromuramyl derivatives that are formed by the action of lytic transglycosylases.
Chart 1.
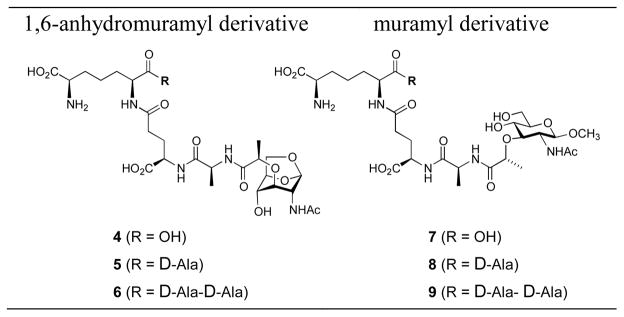
Chemical structures of substrates for AmpD enzymes used in this study
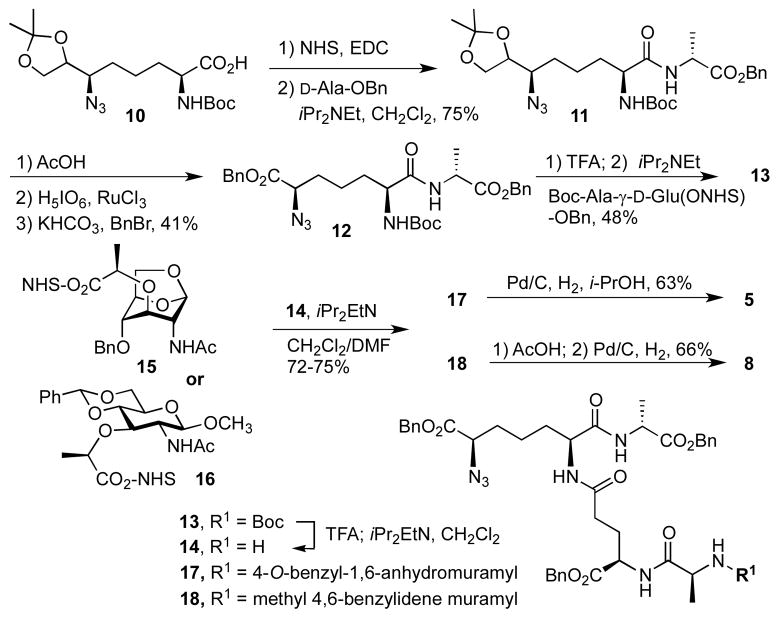
Syntheses of the tetrapeptide-containing derivatives (5 and 8) are shown in Scheme 1. The key tetrapeptide intermediate 13 was prepared by the same methodology developed for the synthesis of pentapeptide by our laboratory.16 The synthesis started with the acetonide 10, which was prepared in 13 steps according to the method of Hernández and Martín.20 The carboxylate moiety in compound 10 was activated by NHS/EDCI and the resulting NHS ester was treated with D-alanyl benzyl ester to give compound 11. The three-step transformation of the acetonide functionality to benzyl ester went smoothly using these conditions, which were used initially for the synthesis of pentapeptide.16
Scheme 1.
The Boc group in 12, was removed in the presence of trifluoroacetic acid and the resultant product was readily coupled to Boc-Ala-D-Glu(ONHS)-OBn to give the key tetrapeptide 13. The Boc group in 13 was removed and the resultant amine 14 was allowed to react with either 1,6-anhydromuramic NHS-ester 1517 or muramic NHS-ester 16.15,16 Global deprotection of 17 or 18 by catalytic hydrogenation (18 required acid treatment prior to hydrogenation) gave the final anhydromuramyl tetrapeptide 5 or muramyl tetrapeptide 8, respectively.
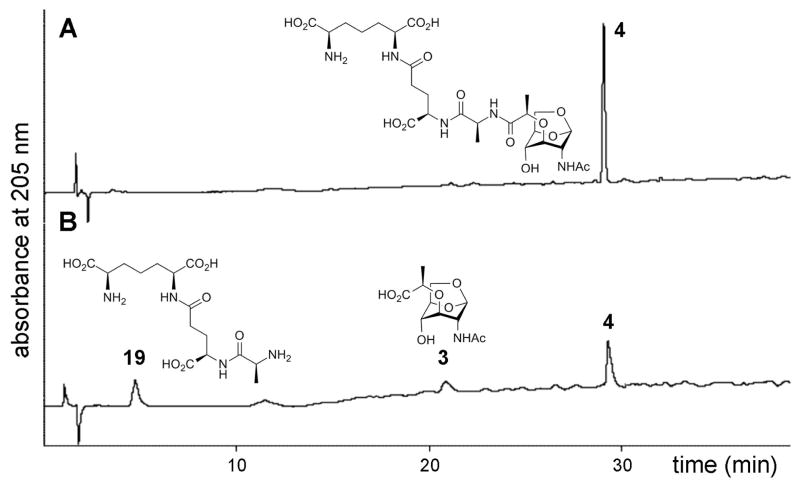
With the availability of all three enzymes and the six compounds as potential substrates for these enzymes, we performed kinetics. The kinetics were HPLC-based and we monitored quantitatively the consumption of the substrate and the formation of products. The products were individually analyzed by mass spectrometry as to delineate the nature of the individual reactions. This type of analysis is exemplified by turnover of compound 4 by the protease AmpD, as documented in Figure 2. The peak corresponding to 4 diminished as the reaction proceeded, with two additional peaks appearing, for which the chemical structures were assigned by mass spectrometry as compounds 3 and 19. Hence, the nature of the enzymatic reaction of AmpD with this compound is hydrolysis of the amide bond to the lactyl moiety. Quantification of the turnover with the use of an internal standard in each case was performed and the kinetic parameters were determined (Table 1). The very same type of analysis was performed for the reactions of each substrate with each enzyme, revealing that we were monitoring hydrolysis of the amide bond at the lactyl moiety in every case. The results are given in Table 1.
Figure 2.
The AmpD reaction analysis by HPLC. (A) Reaction mixture containing 100 nM purified AmpD and 3 mM of compound 4 was incubated at 25 °C. (B) Partial transformation identifies compounds 19 and 3 as products, with m/z values of 391 and 276, respectively.
Table 1.
Steady-state kinetic data for turnover of compounds by AmpD, AmpDh2 and AmpDh3.a
| Enzyme/Compds | Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) | |
|---|---|---|---|---|
| AmpD | 4 | 2.0 ± 0.1 | 24.2 ± 3.5 | 12100 ± 1800 |
| 5 | 1.2 ± 0.3 | 39.8 ± 3.1 | 33200 ± 7500 | |
| 6 | 1.9 ± 0.2 | 47.5 ± 5.5 | 25000 ± 4600 | |
| 7, 8, 9b | - | - | - | |
|
| ||||
| AmpDh2 | 4 | 1.5 ± 0.2 | 0.20 ± 0.02 | 130 ± 17 |
| 5 | 4.6 ± 0.5 | 20 ± 1.9 | 4300 ± 790 | |
| 6 | 1.9 ± 0.2 | 0.18 ± 0.02 | 95 ± 14 | |
| 7 | 1.5 ± 0.1 | 51.8 ± 8.9 | 34500 ± 4300 | |
| 8 | 1.8 ± 0.3 | 73.6 ± 5.3 | 40900 ± 6800 | |
| 9 | 1.4 ± 0.2 | 1.6 ± 0.2 | 1100 ± 150 | |
|
| ||||
| AmpDh3 | 4 | 3.2 ± 0.5 | 17.9 ± 1.3 | 5590 ± 610 |
| 5 | 4.5 ± 0.6 | 70.6 ± 6.6 | 15700 ± 1900 | |
| 6 | 1.4 ± 0.1 | 20.6 ± 1.9 | 14700 ± 1600 | |
| 7 | 2.2 ± 0.3 | 195 ± 24 | 88600 ± 13600 | |
| 8 | 1.4 ± 0.2 | 212 ± 19.3 | 151000 ± 20500 | |
| 9 | 2.4 ± 0.3 | 85.3 ± 10.2 | 35500 ± 4800 | |
The activities of all three enzymes decreased in the presence of EDTA, indicating that they are zinc proteases. Kinetic measurements were performed in the presence of 100 μM ZnCl2 as a supplement for AmpDh2 and AmpDh3.
No measurable activity was detected for AmpD with these compounds.
The important observation from these data is that AmpD is the true protease for the recycling process in P. aeruginosa, as its function is exclusive for turnover of 1,6-anhydromuramyl variants 4–6. This enzyme does not turn over variants 7–9, which are mimetics of the standard peptidoglycan. The AmpD enzyme has evolved for turnover of 1,6-anhydromuramyl compounds, akin to the reaction that we have documented earlier for the enzyme from Citrobacter freundii.17
On the other hand, AmpDh2 and AmpDh3 exhibit activities with both types of substrates. However, the activities of AmpDh2 and AmpDh3 with the 1,6-anhydromuramyl compounds 4–6 are a mere 6% and 12%, respectively, of the total activity (as assessed by the kcat/Km values). For these two enzymes the overwhelming activity is documented for derivatives 7–9 as substrates. Hence, it is clear that AmpDh2 and AmpDh3 have evolved as enzymes for processing of the peptidoglycan components of the cell wall in the periplasmic space. The mere 6–12% hydrolytic activity with the 1,6-anhydromuramyl compounds 4–6 that we measure for these two enzymes are nonetheless real, a subject to which we return below. However, this marginal activity is likely to be adventitious. Since these enzymes clearly prefer the forms of the peptidoglycan found in the cell wall, they are likely counterparts to the cell-wall amidases such as AmiA, B, C and D, which have been known in Escherichia coli.3
The AmpD enzyme, the bona fide recycling protease, reaches saturation (Km) at low millimolar range with its substrates (Table 1), indicating that the intracellular concentrations of metabolites 4–6 during active cell-wall recycling must be in that range. The rather high concentration is not uncommon for other important bacterial metabolites as well.15,21,22 This observation regarding the Km of metabolites 4–6 holds true for the other two enzymes as well, although their true function, per our findings, is processing of cell wall in the periplasm. We also note that all three enzymes exhibit modest preferences for the tetrapeptide stems in the substrates (kcat/Km effect). Whereas the importance of this observation is not immediately apparent, we are aware that muropeptides with tetrapeptide stem are among the most abundant components in the peptidoglycan analyses of E. coli and P. aeruginosa.3,18 It is the reaction of DD-carboxypeptidases, which generates the tetrapeptide version.3,15,23,24
If AmpDh2 and AmpDh3 were to be periplasmic enzymes, as proposed in this report, then they will never come in contact with compounds 4–6, the cytoplasmic metabolites, notwithstanding their marginal ability to turn them over. This is a curiosity, as we have noted that inactivation of the genes for these two enzymes results in stepwise and incremental increase in antibiotic resistance.12,13 However, AmpDh2 and AmpDh3 do encounter compound 1 in the periplasm. Could the effect be manifested by AmpDh2 and AmpDh3 removing the peptide from metabolite 1, at least for a fraction of its total concentration? Indeed, this is the case. Compound 1, which we synthesized by a reported method,25 is a poor substrate for both AmpDh2 and AmpDh3: kcat = 0.45 ± 0.03 s−1, Km = 1.0 ± 0.1 mM and kcat/Km = 440 ± 53 M−1s−1 for AmpDh2; kcat = 2.2 ± 0.2 s−1, Km = 1.2 ± 0.3 mM and kcat/Km = 1800 ± 480 M−1s−1 for AmpDh3. Hence, the primary reaction of AmpDh2 and AmpDh3 is turnover of the normal cell-wall peptidoglycan. However, these two enzymes commence removal of the peptide from a fraction of 1 in the periplasm, prior to the completion of the process by the action of AmpD within the cytoplasm.
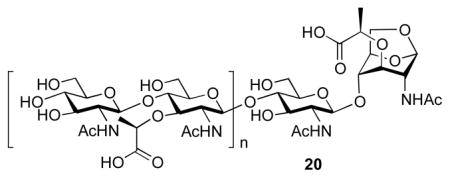
That the cell-wall surrogate compounds 7–9 were turned over by AmpDh2 and AmpDh3 was intriguing. However, do these enzymes actually turn over the cell wall itself? To address this issue, we prepared the P. aeruginosa sacculus according to a reported method.9,26 Overnight incubation of the polymeric sacculus with AmpDh2 produced products with oligomeric cell-wall sugars without the peptide stem. Mass spectrometric detection identified m/z values of 479+, 957+, 1435+, 9572+, 11962+, 14362+, 11173+, and 12763+, corresponding to the general formula, (NAG-NAM)n-NAG-anhMur (structure 20, n = 0–7). The reaction with AmpDh3 also produced products (20), but its quantity was three times less than that with AmpDh2.
In summary, we have provided the first enzymological analyses of reactions of AmpD, AmpDh2 and AmpDh3 from P. aeruginosa with unique synthetic substrates (three bacterial metabolites and three structural surrogates for the cell wall). This analysis identifies AmpD of P. aeruginosa as the cytoplasmic protease for commitment to the cell-wall recycling events and for the reversal of the activation of the antibiotic-resistance pathway. The enzymes AmpDh2 and AmpDh3 exhibited marginal activities with the 1,6-anhydromurmyl compounds 4–6 as substrates. Their function(s) is turnover of cell wall within the periplasmic space. Nonetheless, the marginal activity in turnover of metabolite 1 with AmpDh2 and AmpDh3 explains the stepwise effect observed in augmenting derepression of AmpC β-lactamase by eliminations of these enzymes one-by-one.12,13
Supplementary Material
Acknowledgments
The authors declare no competing financial interest. This work was supported by a grant from the NIH (GM61629). The Mass Spectrometry & Proteomics Facility of the University of Notre Dame is supported by grant CHE0741793 from the NSF.
Footnotes
Supporting Information. Experimental procedures for syntheses of the new compounds, cloning, enzyme purification, kinetics, MS analyses of products and copies of NMR spectra of the new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Park JT, Uehara T. Microbiol Mol Biol Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reith J, Mayer C. Appl Microbiol Biotechnol. 2011;92:1–11. doi: 10.1007/s00253-011-3486-x. [DOI] [PubMed] [Google Scholar]
- 3.van Heijenoort J. Microbiol Mol Biol Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vollmer W, Joris B, Charlier P, Foster S. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JW, Fisher JF, Mobashery S. Ann NY Acad Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher JF, Mobashery S. Enzymology of Antibacterial Resistance. In: Mander L, Liu HW, editors. Comprehensive Natural Products Chemistry II, Chemistry and Biology. Elsevier; Oxford: 2010. pp. 443–487. [Google Scholar]
- 7.Jacobs C, Frere JM, Normark S. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 8.Normark S. Microb Drug Resist. 1995;1:111–114. doi: 10.1089/mdr.1995.1.111. [DOI] [PubMed] [Google Scholar]
- 9.Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, Mobashery S. J Am Chem Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheurwater E, Reid CW, Clarke AJ. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Boudreau MA, Fisher JF, Mobashery S. Biochemistry. 2012;51:2974–2990. doi: 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan C, Moya B, Perez JL, Oliver A. Antimicrob Agents Chemother. 2006;50:1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moya B, Juan C, Alberti S, Perez JL, Oliver A. Antimicrob Agents Chemother. 2008;52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langaee TY, Gagnon L, Huletsky A. Antimicrob Agents Chemother. 2000;44:583–589. doi: 10.1128/aac.44.3.583-589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hesek D, Suvorov M, Morio K, Lee M, Brown S, Vakulenko SB, Mobashery S. J Org Chem. 2004;69:778–784. doi: 10.1021/jo035397e. [DOI] [PubMed] [Google Scholar]
- 16.Lee M, Hesek D, Shah IM, Oliver AG, Dworkin J, Mobashery S. Chembiochem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MJ, Zhang WL, Hesek D, Noll BC, Boggess B, Mobashery S. J Am Chem Soc. 2009;131:8742–8743. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cigana C, Curcurù L, Leone MR, Ieranò T, Lorè NI, Bianconi I, Silipo A, Cozzolino F, Lanzetta R, Molinaro A, Bernardini ML, Bragonzi A. PLoS ONE. 2009;4:e8439. doi: 10.1371/journal.pone.0008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suvorov M, Fisher JF, Mobashery S. Bacterial Cell Wall: Morphology and Biochemistry. In: Goldman E, Green LH, editors. Practical Handbook of Microbiology. 2. CRC Press; 2009. [Google Scholar]
- 20.Hernández N, Martín VS. J Org Chem. 2001;66:4934–4938. doi: 10.1021/jo0155714. [DOI] [PubMed] [Google Scholar]
- 21.Toth M, Frase H, Antunes NT, Smith CA, Vakulenko SB. Protein Sci. 2010;19:1565–1576. doi: 10.1002/pro.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth M, Zajicek J, Kim C, Chow JW, Smith C, Mobashery S, Vakulenko S. Biochemistry. 2007;46:5570–5578. doi: 10.1021/bi6024512. [DOI] [PubMed] [Google Scholar]
- 23.Sauvage E, Powell AJ, Heilemann J, Josephine HR, Charlier P, Davies C, Pratt RF. J Mol Biol. 2008;381:383–393. doi: 10.1016/j.jmb.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Q, Meroueh SO, Fisher JF, Mobashery S. J Am Chem Soc. 2008;130:9293–9303. doi: 10.1021/ja801727k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesek D, Lee M, Zhang W, Noll BC, Mobashery S. J Am Chem Soc. 2009;131:5187–5193. doi: 10.1021/ja808498m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suvorov M, Lee M, Hesek D, Boggess B, Mobashery S. J Am Chem Soc. 2008;130:11878–11879. doi: 10.1021/ja805482b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.