Abstract
Since epigenetic mechanisms convey information above and beyond the sequence of DNA, they are predicted to be critical in the complex regulation of brain development and explain the long-lived effects of environmental cues on pre- and early post-natal brain development. Neurons have a complex epigenetic landscape, which changes dynamically with transcriptional activity in early life. Here we summarize progress on understanding the discrete layers of the dynamic methylome, chromatin proteome, noncoding RNAs, chromatin loops, and long-range interactions in neuronal development and maturation. Many neurodevelopmental disorders have genetic alterations in these epigenetic modifications or regulators, and these human genetics lessons have demonstrated the importance of these epigenetic players and the epigenetic layers that transcriptional events lay down in the early brain.
Introduction
Epigenetics has historically referred to heritable modifications resulting in phenotypes that were not directly dependent on the DNA sequence inherited. However, in recent years the rapid expansion of epigenetic research into DNA methylation, histone modifications, and chromatin structure has broadened the definition of epigenetic heritability to include long-lived but reversible modifications to nucleotides or chromosomes. With this larger umbrella of epigenetic mechanisms has come greater complexity in defining the set of underlining epigenetic rules. Box 1 provides a current definition of epigenetics, lists the major categories of epigenetic layers and players, gives examples of what is and is not epigenetics, and dispels some epigenetic “urban myths”.
Box 1. What is epigenetics?
Epigenetics: Long-lived and reversible modifications to nucleotides or chromosomes that do not change the sequence but can alter gene expression and phenotype.
Epigenetic layers: DNA methylation, stably maintained histone modifications (e.g., H3K27me3, H3K9me3), chromatin loops, chromosomal organization and location within the nucleus, noncoding RNAs (bound to DNA)
Epigenetic regulators or “players”: chromatin-binding factors, chromatin remodeling complex proteins, noncoding RNAs, DNA methyltransferases, histone modifying enzymes
What is epigenetic? allelic differences in transcription not determined by sequence, cell lineage inheritance, long-lived modifications from past environmental exposures
What is NOT epigenetic? regulation of transcription without accompanying long-lived modifications, transcription factors (although they may direct epigenetic states)
Metastable: a physics term describing a state of precarious stability, which is also a useful term for labeling the long-lived but reversible characteristics of epigenetics [5]
Common epigenetic myths:
-
DNA sequence does not matter for epigenetic modifications.
While this statement may be true for allelic states such as imprinting and X chromosome inactivation, sequence features such as promoter CpG content or other nucleotide bias (such as G-skew on one DNA strand) can be very important in determining deposition of the epigenetic modification DNA methylation [6–8] and methylation patterns are heritable with genotype [9].
-
Epigenetic marks determine transcription (but not vice-versa).
While this was the historical thinking in the field, many recent examples demonstrate that transcription is required for the establishment of some epigenetic modifications. Epigenetic modifications may actually be historical marks of past transcriptional events that influence later transcriptional responsiveness.
-
Transcriptional differences between genetically identical individuals must be epigenetic and environmentally driven.
While an attractive idea, not all transcriptional differences are epigenetic and many epigenetic differences are stochastic, rather than environmentally determined.
-
DNA methylation patterns, once established in early development, are stably maintained.
While overall, methylation patterns appear to be much less variable between individuals or cell types than transcriptional patterns, there are some concrete examples of changes to methylation levels in neurons, following activity-dependent responses [1, 2, 10].
-
DNA methylation is always associated with transcriptional silencing.
While this is true for CpG-rich promoters that are subject to imprinting, X chromosome inactivation, or cell fate determination (Table 1), outside of these situations the rules change, and higher DNA methylation levels is generally associated with higher expression [11, 12].
Neuroscientists have understandably been drawn to the field of epigenetics because it can help explain complex transcriptional regulation that is not fully explained by inducible and tissue-specific transcription factors alone. Epigenetics is appealing as well because it offers a potential explanation for variable phenotypic outcomes from similar genetic backgrounds. Specifically, how do early life experiences during critical periods for learning and memory or establishment of the hypothalamic-pituitary-adrenal (HPA) axis have long lived effects on transcription lasting into adulthood? How are transcriptional patterns that define neuronal networks maintained and further refined with activity? How does drug abuse in adolescence set up a behavioral pattern of addiction? Since epigenetic modifications appear to be especially important for genes encoding proteins acting at the synapse [1–3], epigenomic investigations may point to potential “druggable” protein targets using existing medications in the treatment of neurodevelopmental and psychiatric disorders. In addition, since epigenetic mechanisms exist in a “metastable” state, an attractive possibility is the design of therapies to reverse epigenetic states in the brain without disrupting the underlying genetic state.
But caution should be exerted in not putting the cart before the horse in this line of research. Epigenetics is still a young field and the rules of how epigenetic modifications affect gene expression and may be involved in specific disease states have yet to be fully understood. With the advent of genome-wide technologies, the view of the epigenome has broadened considerably, but this new information has often challenged many of the past assumptions about epigenetic control of gene expression [4]. Here, we review the current understanding about the individual layers of epigenetic information and how they are important to neurons, primarily during early life. We define “layers” as structural components of DNA and chromatin, in contrast to “players” that are active complexes and enzymes required for epigenetic layers.
The neuronal methylome
The addition of a methyl group to the 5th carbon of cytosine (5meC) is primarily observed at CpG sites in the mammalian genome, although non-CpG methylation is also observed in plants as well as embryonic stem (ES) cells and neurons in mammals [13]. Brain DNA has one of the highest levels of 5meC in the human body. 5meC can also be converted to 5-hydroxymethylcytosine (5hmC) in the presence of TET1 [14, 15], and 5hmC is implicated in demethylation of the paternal genome in early mouse (M. Musculus) embryos and the self-renewal capacity of mouse embryonic stem cells, perhaps through demethylation of the Nanog promoter [16]. 5hmC levels are particularly high in brain compared to other tissues and Purkinje compared to granular neurons of the cerebellum [14]. Although 5hmC appears to be an important transition state in the dynamic methylation patterns of neurons, it is far less abundant than 5meC even in Purkinje neurons. Furthermore, most methods used to investigate DNA methylation that involve either bisulfite conversion or methyl-sensitive restriction enzymes cannot distinguish between 5meC and 5hmC. Methods for discriminating 5hmC are beginning to reveal differences in tissue and location from 5mC [17, 18], but the functional relevance of 5hmC is still unknown. Therefore, what we refer to in this review as “DNA methylation” and the “neuronal methylome” will actually be a collection of 5meC, 5hmC, and perhaps other less characterized modifications of cytosines [19].
The relationship of DNA methylation to gene expression does not abide by one universal rule, but rather is heavily dependent on underlying sequence, transcription state, and location of the genomic DNA (Figure 1). The canonical view of DNA methylation as a repressive mark is observed for CpG island promoters of genes on the active versus inactive X chromosome (XCI) and imprinting control regions which are clear examples of allele-specific methylation [20, 21]. However, recent genome-wide DNA methylation studies have shown that high CpG methylation is common in gene bodies (genomic loci spanning exons and introns) where it positively correlates with transcription [22, 23]. Furthermore, gene bodies contained in partially methylated domains (PMDs, continuous domains of <70% methylated CpGs) [12] showed decreased expression compared to those contained in highly methylated domains (HMDs). Interestingly, tissue-specific differences in the presence of PMDs have revealed a subset of developmentally regulated genes that act at the synapse are highly methylated and expressed in human immature neurons (SH-SY5Y) cells compared to fetal fibroblasts [3] or placenta [24].
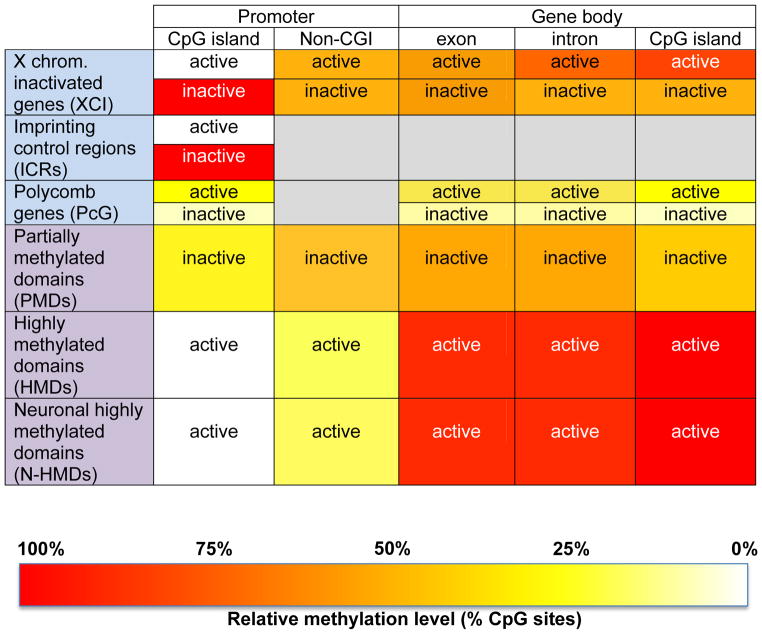
Figure 1. The context of sequence, transcription, and location in DNA methylation.
For the purpose of demonstrating the importance of genetic context in interpreting DNA methylation levels, genes are broken down into identifiable parts, with promoters separated as either CpG islands or non-CpG islands (non-CGI), and parts of the “gene body” (defined as the genomic locus from transcription start to end sites) separated as exons, introns, or non-promoter CpG islands. The methylation levels for each of these genomic locations are color-coded according to the heat map shown at the bottom, with red representing the highest methylation levels and white representing the lowest. Grey shading represents unrepresented or unknown categories. Rows with headings labeled in blue represent different subcategories of genes or controlling regions with distinct methylation patterns from other genes in the genome. Rows with headings labeled in purple represent the different whole genome landscape categories of partially methylated domains (PMDs, <70% methylated CpGs) [12], highly methylated domains (HMDs, >70% methylated CpG at non-CGIs) [3], or neuronal HMDs (N-HMDs, HMD in neuronal SH-SY5Y cells, but PMD in IMR90 fibroblasts and placenta) [24].
Neurons appear to have a unique methylomic landscape compared to other cell types. The contrast between very low levels of methylation in CpG islands and very high methylation in gene bodies is sharper in human brain and neurons compared to non-neuronal cells [3]. DNA methylation is highly dynamic in mammalian postnatal neurons and these cells express the corresponding DNA methyltransferases (DNMT) at high levels [25]. Importantly, mice deficient for both DNMT1 and DNMT3A in forebrain excitatory neurons showed deficits in learning and memory [26], similar to deficits in fear memory observed with DNMT1 chemical inhibition [27]. Likewise, DNMT3A regulates emotional behavior and spine plasticity [28] and ensures the expression of key neurogenic genes by targeting de novo non-promoter DNA methylation around active genes [29]. DNA methylation changes were observed at the Bdnf promoter in neuronal cultures following activity [30] and an in vivo paradigm of learning and memory resulted in significant changes to methylation patterns [31].
Dynamic changes in DNA patterns have been observed genome-wide in adult mouse dentate granule neurons after neuronal activation that remained stable for at least 24 h [1]. Interestingly, these activity-modified CpGs were specifically enriched in low-CpG density regions [1] that are more characteristic of PMDs [3] than the high CpG density regions typically examined for methylation differences. Furthermore, not all of the observed changes in DNA methylation correlated with an immediate transcriptional change, suggesting that altered methylomes may have subtle, long-lasting effects [1]. Interestingly, active demethylation in the adult brain can be potentially explained by the action of TET1, which hydroxylates 5mC to 5hmC, leading to demethylation [32]. Therefore, while once considered a stable mark in somatic tissues, DNA methylation is emerging as an important metastable epigenetic layer in postnatal neurons.
The neuronal chromatin proteome
The neuronal methylome is intricately intertwined with the numerous proteins that bind to DNA in neuronal nuclei. The most abundant and best characterized factors in neurons are the histone core proteins H2, H3, and H4, linker histone H1, methyl CpG binding protein 2 (MeCP2), and chromatin insulator CTCF. The neuronal chromatin proteome is an important epigenetic layer because it both “reads” the methylome and influences the degree of compaction and accessibility of the DNA to transcription factors and the transcriptional machinery [33–35]. Like DNA methylation, the marks of the neuronal chromatin proteome can be both long-lived and dynamically altered in response to transcription or activity, resulting in metastable “chromatin states” defined by combinations of genome-wide analyses of histone modifications and transcription in the recent human ENCODE (Encyclopedia of DNA elements) project [36] (Figure 2).
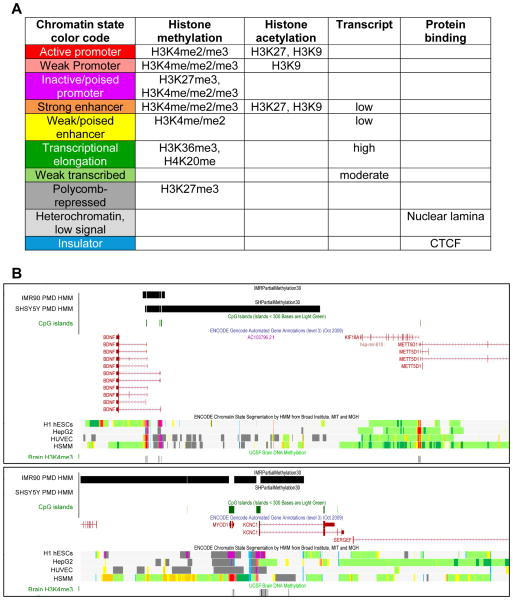
Figure 2. Chromatin states defined by histone modifications and protein binding.
A. A color-coded guide to the chromatin state maps derived from hidden Markov model (HMM) segmentation of histone modifications and CTCF binding sites in non-neuronal human cell lines from the human ENCODE project [36]. Histone methylation and acetylation was analyzed by ChIP-seq using antibodies to the specific modifications and sites listed. Transcript analysis was determined by RNA-seq on the same cell lines. Genes involved in developmental and tissue-specific functions exhibit H3K27 trimethylation (H3K27me3) blocks characteristic of polycomb-repressed genes (dark grey) and have inactive but “poised” bivalent promoter states (purple) of both silent (H3K27me3) and active (H3K4me3) histone modifications in human embryonic stem cells (hESCs).
B. Two examples of chromatin state maps combined with PMD state maps in the UCSC Genome Browser for neurologically relevant genes. PMDs mapped from HMM analysis of MethylC-seq data in fibroblast (IMR90) or neuronal (SH-SY5Y) cell lines [3] are shown as black bars, with gaps at CpG islands (green) because they were removed from the PMD analysis. ENCODE tracks (middle) include annotated genes and chromatin state segmentation by HMM color coded as in A. While neuronal tissue was not included in the preliminary ENCODE analysis of chromatin states, a track of “Brain H3K4me3” is shown at the bottom that identifies active genes in human adult brain. Notice the difference in chromatin states between regions covered by tissue-specific PMDs (BDNF, MYOD1, KCNC1, inactive poised promoter, polycomb-repressed, and heterochromatin) compared to genes with HMD in all tissues (METT5D1, KF18A, active promoter and transcribed). Also, notice the difference in chromatin states at the MYOD1 locus in human skeletal muscle myotubule (HSMM) cells compared to embryonic stem cells (H1 hESCs, inactive/poised promoter) and liver (HepG2) or umbilical vein epithelial cells (HUVEC) which have the H3K27me3 marks of polycomb repression. Both MYOD1 and KCNC1 are transcriptionally active in brain (H3K4me3 promoter peaks) and are within a neuronal highly methylated domain (N-HMD, defined as PMD in IMR90 but HMD in SH-SY5Y cells). In contrast, BDNF is inactive but poised for transcriptional activity differentially at different promoters and in cell lines and the alternative BDNF promoters are within a PMD in both SH-SY5Y and IMR90 cells. The nuclear lamina, a heterochromatic protein matrix at the nuclear periphery made of lamin and scaffold proteins, overlaps with both PMDs and light grey “off the map” locations in the chromatin state maps. CTCF (blue) shows multiple distinct sites genome-wide that are both intergenic and close to promoters and ubiquitous as well as tissue-specific.
While the specific chromatin states are less well defined for neurons as for the ENCODE cell lines, the importance of dynamic changes to chromatin states is acutely evident for neuronal development and activity. Activity and energy in the form of ATP is required for changes to chromatin states during neuronal lineage commitment in the epigenetic process of “chromatin remodeling” [37]. Interestingly, mutations in several chromatin remodeling factors, including ATRX and ARID1B are associated with human neurodevelopmental disorders [38–40]. Furthermore, neurons have a unique chromatin remodeling complex, defined by the neuron-specific subunit BAF53b, expressed in postmototic neurons around embryonic day 12.5 [41]. Deficiency of BAF53b in mouse results in significant deficits in long-term memory and transcriptional changes to the noncoding RNA miR132 implicated in synaptic plasticity [42].
Histone core proteins reflect and/or influence accessibility of the DNA through multiple post-translational modifications. Different combinations of histone core protein modifications make up what has been called the “histone code” which has been reviewed extensively [43–45] but debated as to whether most histone modifications fit the definition of being long-lived epigenetic states [46]. Perhaps the removal of histone methylation marks is particularly critical for neurons, as mutations in genes encoding histone demethylases have been found in human X-linked autism-spectrum disorders, including JMJD1C/TRP8 that demethylates H3K9 and KDM5C/JARD1C that demethylates H3K4 [47, 48].
Beyond the core nucleosome particle and associated modifications, other protein factors are important in defining chromatin states. The CCCTC-binding factor (CTCF) is a sequence-specific chromatin organizer of chromosomal domains [49]. Interestingly, CTCF has been implicated as a master regulator of alternative splicing of the highly alternatively spliced protocadherin gene clusters in neuronal connectivity [50], and protocadherin genes are also highly enriched within N-HMDs of the neuronal methylome [3, 24]. DNA methylation within gene bodies can influence alternative splicing by blocking CTCF binding and pausing of RNA Pol II near weak exons [51].
In contrast to CTCF which is ubiquitously expressed, has distinct binding sites, and a higher affinity for unmethylated DNA [49, 52], methyl CpG binding protein 2 (MeCP2) is highly expressed specifically in the central nervous system, binds widely throughout the genome, and has increased affinity for methylated DNA [53–55]. Mutations in MECP2 cause the X-linked autism-spectrum disorder Rett syndrome [56]. MeCP2 levels increase with neuronal maturity, becoming as abundant as histone proteins in mature neurons and displacing of portion of H1 linker histones [35]. Similar to histones, MeCP2 is extensively post-translationally modified by phosphorylation, acetylation, and ubiquitinylation [57]. Although the functional relevance of the “MeCP2 code” is much less clear than it is for the histone code, phosphorylation of S421 is implicated in psychostimulant responsiveness [58] and synaptic scaling [59], and differential phosphorylation of S80 and S229 of MeCP2 discriminate cofactor associations [57].
Long, non-coding RNAs as epigenetic players and layers in neurodevelopment
Recent lines of evidence have shown that long, non-coding RNAs (lncRNAs) are critical regulators of epigenetic mechanisms [60, 61] (Figure 3). That RNA is a regulatory keystone is not surprising given the hypothesis that the first living, biological molecules were RNA [62]. Furthermore, most cellular processes are governed by self-regulating feedback loops so the presence of RNA as a regulator of transcription fits with general biological principles. The advent of transcriptomic analyses revealed that many long and short non-coding transcripts exist [63] and that many exhibit tissue-specific patterns of regulation [64–66]. Many different forms of RNA that regulate transcription have been described, including microRNAs (miRNAs), long noncoding RNAs (lncRNAs) [67], enhancer RNAs (eRNAs) [68], and circular (circRNAs) [69]. While miRNAs and circRNAs play a critical role in neurodevelopment, they are not epigenetic layers per se, but can regulate other epigenetic layers and players as well as be epigenetically regulated themselves. For instance, the miRNAs miR-483-5p and miR-132 regulate MeCP2 levels [70, 71], and miR-132 itself is regulated by the neuronal chromatin remodeling complex containing BAF53b [42].
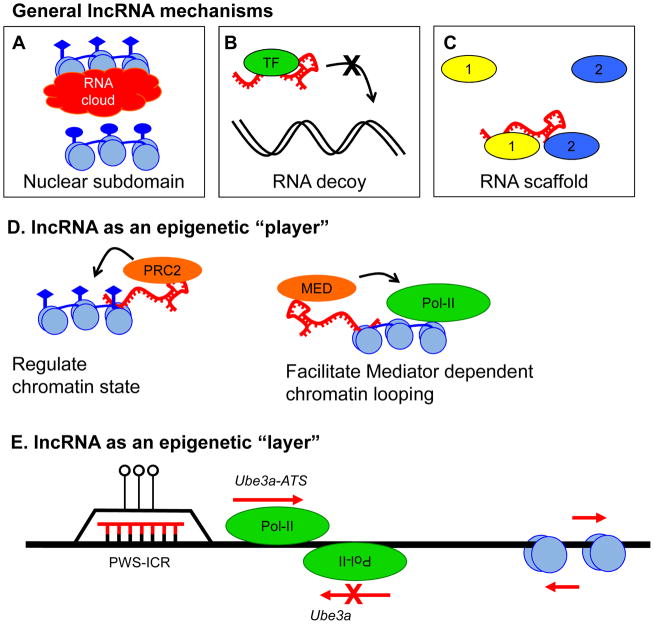
Figure 3. Noncoding RNA as an epigenetic layer and player.
A. LncRNAs can function to demarcate a nuclear subdomain by forming and “RNA cloud” (red), as seen for Xist and Kcnq1ot1, in order to coat chromosomes or genomic regions (blue) and regulate transcription [60]. Loci coated by the RNA cloud are most often silenced via recruitment of repressive chromatin complexes, but RNA clouds may also upregulate transcription [80]. B. LncRNAs (red) also act as molecular decoys either for DNA binding transcription factors (Gas5) or miRNAs (PTENP1). C. LncRNAs can have multiple protein interacting partners and thereby act as a molecular scaffold for larger complexes. D. In recent years a novel role for lncRNAs as epigenetic “players” has been described. Many lncRNAs were found to interact with the repressive chromatin modifier PRC2 [64, 79], and thereby regulate the chromatin structure of large domains, or even of whole chromosomes. Recently, activating lncRNAs were also described that bind to the Mediator complex and facilitate enhancer-promoter loops [3]. This function may potentially overlap with enhancer RNAs (eRNAs) which are transcribed bidirectionally from enhancers [68]. E. RNA also functions as an epigenetic “layer” by modifying the structure of the DNA or regulating sense transcription via antisense transcription. Such a role has been described at the neurodevelopmentally critical PWS/AS locus on chromosome 15q11-q13, where RNA:DNA hybrid (R-loop) formation at the Prader-Willi imprinting control region (PWS-ICR) protects against DNA methylation [8], and processive transcription to produce UBE3A-ATS silences paternal UBE3A in neurons [89]. Similarly, eRNAs may act as molecular signals to mark active enhancers in response to stimulation [68].
Here, we will focus on the role of lncRNAs since they are emerging as a key-regulator of epigenetic processes by targeting chromatin remodeling complexes (player) or by being a part of the chromatin structure (layer). LncRNAs certainly also have functions beyond directing epigenetic mechanisms, since they can regulate alternate splicing [72], miRNA abundance [69, 73] protein-protein interactions, and molecular signals [61]. Since many lncRNAs exhibit brain region specific patterns of expression [74], they are predicted to regulate the diversity of neuronal subtypes and populations in the brain. Comprehensive studies of lncRNA function during neurodevelopment have focused on control of neurogenesis from ESC to differentiated neurons in cell culture systems [75, 76], leaving unresolved their potential roles in the postnatal stages of neuronal maturation, and in mediating activity-dependent changes in chromatin structure.
A neurodevelopmental role for lncRNAs has been shown in differentiation of neurons from human embryonic stem cells [75, 76] and in specifying subpopulations in the retina, where loss of function of two lncRNAs RNCR2 and Six3OS led to changes in neuronal populations [77, 78]. The loss of Six3OS may cause changes in chromatin states because of its interaction with the PRC2 complex member Ezh2, which mediates H3K27 methylation and interacts with thousands of additional lncRNAs [64, 79]. By binding PRC2, these lncRNAs act to repress target polycomb genes and provide specificity to a repressive complex that lacks a protein-DNA targeting factor. Although lncRNAs are also predicted to target activating complexes in a similar manner by binding Trithorax group proteins [80], a systematic identification of RNAs bound to activating complexes has not been performed.
While some lncRNAs appear to regulate only one or two genes [81], many lncRNAs apparently regulate multiple genes by forming a nuclear subdomain [64, 82–85]. Studies of the lncRNAs Xist and Kcnq1ot1 demonstrate how a lncRNA recruits repressive complexes to a whole chromosome (Xist) or an imprinted domain (Kcnq1ot1) [60]. Since neurons are postmitotic, formation of nuclear subdomains by lncRNAs may be one mechanism by which long-term epigenetic regulation occurs, although this has not yet been demonstrated. One nuclear lncRNA,116HG, is implicated in the pathogenesis of the imprinted neurodevelopmental disease Prader-Willi syndrome (PWS) [86] and expression of 116HG may regulate a large-scale chromatin decondensation [87], although the precise mechanism is unknown.
While emerging evidence points to lncRNAs as epigenetic “players” in neurodevelopment by directing epigenetic mechanisms, noncoding transcripts can also act as an epigenetic “layer” on chromosomes. As an example, transcription of Ube3a-ATS leads to paternal silencing of Ube3a in neurons. The precise mechanism of silencing is not known, but a strong similarity to the imprinted Airn/Igf2r locus suggests that the most likely mechanism is the act of transcription in the opposite orientation through the Ube3a gene and promoter rather than a direct action of the Ube3a-ATS ncRNA itself [88, 89]. Such a model needs to be confirmed, but strand-specific RNA-sequencing experiments in mice demonstrate that the paternal Ube3a promoter initiates transcription but terminates prematurely, supporting a polymerase clash model [90]. In neurons, expression of Ube3a-ATS coincides with a large-scale chromatin decondensation of the paternal allele in the presence of transcription [87], raising the possibility that transcription drives the change in chromatin structure, although it is not known whether this transcription acts as a “layer” of the chromatin or a “player” to recruit chromatin modifying complexes.
Noncoding RNA also appears to act as an epigenetic layer at enhancers. Activation of neurons with KCl treatment results in productive transcription at thousands of enhancers to form enhancer RNAs (eRNAs) [68]. Although the function of these eRNAs is unknown, they could be acting as signaling molecules to mark active enhancers, or as a molecular tag within the chromatin. Another possibility is that eRNAs could be part of the enhancer activity by binding Mediator in order to facilitate chromatin looping and neighboring gene activation [91].
An open question has been how CGI promoters are protected from de novo methylation during development. Recently, the formation of RNA:DNA hybrids called “R-loops” at CGI promoters was demonstrated as a protective mechanism from DNA methylation [92]. As a result, the presence of RNA as an RNA:DNA hybrid acts as an epigenetic layer that regulates the epigenetic layer of DNA methylation.
Chromatin loops and long-range interactions in neurodevelopment
Genomic chromatin loop organization is an important additional layer of epigenetic regulation containing several sub-layers at different scales (Figure 4). For example, the packaging of approximately two meters of genomic DNA into a series of “fractal globules” compartmentalizes DNA into open and closed chromatin loop regions in mammalian nuclei [93]. However, starting at the level of individual genes, chromosome conformation capture studies combinined with massively parallel sequencing have revealed complex looping interactions between enhancers and promoters. The CREB binding protein (CBP, encoded by CREBBP) is responsive to neuronal activity by recruiting RNA Pol II to enhancers such as Arc [68] (Figure 4A). As mentioned above, the chromatin factors CTCF and cohesin (SMCA1) mediate chromatin looping interactions between distal enhancers and alternate promoters of the protocadherin alpha gene locus [50]. Furthermore, in perhaps the most comprehensive study to date in non-neuronal cells, complex looping interactions were observed between transcription start sites in promoters and distal elements up to 120 kb away, with positive effects on gene expression [94].
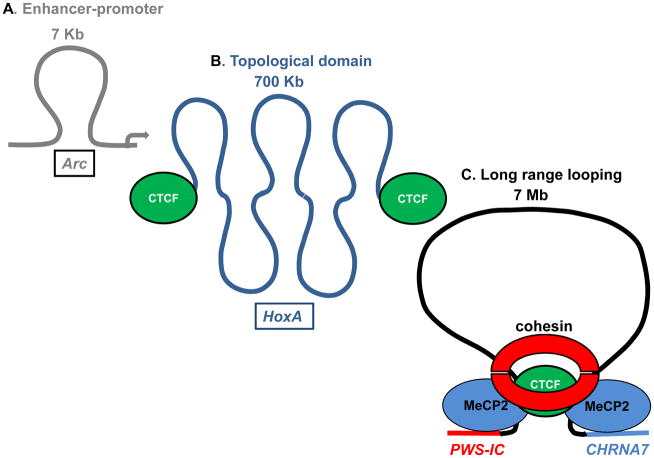
Figure 4. The epigenetic layer of chromatin looping is composed of at least three sublayers.
A. At the sub-layer of specific transcription units, enhancers and other distal units make contact with gene promoters such as Arc, a neuronal factor required for the regulation of AMPA receptors, via kb scale chromatin loops. Pol II (red) is recruited by enhancers where transcription initiates before transfer by looping to the Arc promoter [68]. B. Chromatin domains up to 1 Mb organize chromosomes into a series of functional units in mammalian cells. The insulating factor CTCF (green) regulates formation and maintenance of this intermediate layer in the HoxA locus and other domains genome wide [96]. C. Long range, multi-megabase chromatin loop interactions regulate gene expression between distant loci above the domain layer. At this layer, the PWS-IC (red) contacts the locus encoding CHRNA7 (blue) via a 7 megabase looping interaction via MeCP2 and potentially CTCF (green) and cohesion (red) [98, 99]. This long-range interaction modulates expression of CHRNA7 and other neurologic genes in 15q11–13 [98].
Next, on a broader scale sub-level of chromatin organization, chromatin looping can organize genomic regions into functional domains. While enhancer-promoter looping interactions are typically in the 1–100 kb range, the genome appears to be organized into a series of larger topologically grouped domains. Evidence for this comes from the chromatin structure assay called HiC, a derivative chromosome conformation capture (3C) assay, performed in non-neuronal cell lines. From this genome-wide analysis, a series of megabase (Mb) sized chromatin looped “topological” domains were observed bound by CTCF, transfer RNAs and retro-transposons [95]. Remarkably, these topological looped domains were conserved across cell types and between species. As a positive control, results from an earlier analysis of looping patterns in the HoxA cluster (Figure 4B) were confirmed that organize this locus into two functional domains [96]. The identification of megabase-sized looped chromatin domains is also supported by results using an independent ChIA-PET technique which mapped CTCF looped domains in ES cells, and identified a portion of these that contained repressed genes located at the nuclear lamina [97]. As with enhancer-promoter loop interactions, chromatin domains have been characterized primarily in non-neuronal cells, but the mapping of regulatory elements in multiple tissues supports the hypothesis that the neuronal genome is also subdivided into a series of co-regulated domains [97].
Although stable topological domains may be common in multiple cell types, the additional epigenetic mechanisms that specify gene expression in neurons may not occur at this layer of chromosomal organization. In the case of specific neuronal genes, regulation may occur at the level of long-range multi-megabase looping interactions. The first example of regulation by long-range chromatin looping was shown at the human 15q11–13 locus [98] (Figure 3C). In this study, chromosome conformation capture on microarray (4C) analysis in neurons revealed that the Prader-Willi imprinting control region forms a series of long-range interactions with loci throughout 15q11–13. One of these interactions was observed over 7 megabases between the Prader-Willi ICR and the CHRNA7 locus (encoding nicotinic acetylcholine receptor alpha 7 subunit). Furthermore, deficient MeCP2 expression in Rett and autistic brain cortices was correlated with reduced CHRNA7 expression [98] suggesting that MeCP2 and perhaps the CTCF/cohesin complex [99] are involved in this long range loop interaction (Figure 3C) that was previously identified at the Gtl2/Dlk1 locus [39]. MeCP2 and CTCF also were shown to form long-range complexes with both ATRX and cohesion [39], which are implicated in human neurodevelopmental disorders. A prime example of tissue-specific epigenetic regulation by chromatin looping was shown by MeCP2 ChIP-loop analysis of the striatum where MeCP2 anchoring and silent chromatin looping prevented the ectopic expression of Dlx5 [100]. In summary, multiple levels of chromatin looping constitute the epigenetic layer of chromatin organization that contribute to neuronal gene expression and phenotype.
Higher-order nuclear organization of neuronal nuclei
The last epigenetic layer is one that can be observed at the light microscopic level as the high degree of nuclear architechiture in neurons (recently reviewed in [101]). One form of nuclear compartmentalization is the existence of chromosome territories and the propensity of a genomic region to associate more readily with other regions from the same chromosome as compared to direct inter-chromosomal interactions [93, 102]. Interestingly, chromosome territories defined by DNA fluorescence in situ hybridization (FISH) also appear to exhibit higher-order organization with exons of some genes appearing to be present beyond the periphery of their chromosome territories [103]. Such higher-order organization could relate to the formation of transcription factories, which are thought to facilitate transcription of large domains or repeated transcription events on gene loops [104]. However, debate exists as to whether the formation and localization of transcription factories are a consequence or a determinant of transcription [105]. Furthermore, transcription factories are defined as distinct foci of RNA Pol II by immunofluorescence and active transcription, and investigation of transcription factories alone does not address the additional layer of chromosome-wide domains of association and transcription regulation.
Some chromosomal organizations appear to be non-random in neuronal nuclei, including the homologous pairing of maternal and paternal alleles of human 15q11–13 loci in differentiated neurons that is disrupted by loss of MeCP2 [106] or maternal duplication of chromosome 15q [99, 107]. In mouse ES cells and other tissues, homologous pairing is observed in a subset of both imprinted and nonimprinted gene loci and corresponds with transcriptional activity [108]. Therefore, it may not be the specific epigenetic marks at imprinted loci per se that are driving non-random interactions within neuronal nuclei, but the transcriptional events and locations in common between chromosomal loci containing complex epigenetic regulation at multiple layers.
Concluding remarks
In conclusion, an emerging theme in common to all the epigenetic layers is that they are more likely to be the consequences of past transcriptional events rather than simple signposts of current transcriptional activity. But by poising specific genes either positively or negatively for transcriptional responsiveness to later activity, epigenetic layers and players can have profound effects on phenotype, as evident from the multiple human diseases with specific deficiencies (Table 1). New technologies have emerged for visualizing and cataloguing the epigenetic layers at genome-wide resolution that will be important to utilize in neuronal systems and brain tissue (Table 2). Perhaps by viewing the epigenetic layers through the lens of cellular historians, we can more thoughtfully interpret the evidence laid down in multiple layers to find more long-lasting treasures relevant to human neurodevelopmental disorders (Box 2). Since by definition, an epigenetic “metastable” state means that the state is precarious and thereby changeable using the proper conditions, epigenetic layers are strong candidates for targeted therapeutic strategies for neurodevelopmental disorders such as autism, schizophrenia, and intellectual disability.
Table 1.
Epigenetic layers and players and their human disease associations
| Full name and function | Human neuronal disease association | Refs | |
|---|---|---|---|
| Layers | Modifications of DNA or chromatin | ||
| 5hmeC | 5-hydroxymethylcytosine | [13] | |
| 5mC | 5-methylcytosine | [14, 15] | |
| PMD | Partially methylated domain | [12] | |
| HMD | Highly methylated domain | [3] | |
| N-HMD | Neuronal highly methylated | [3, 24] | |
| ICR | Imprinting control region | Prader-Willi, Angelman | [20, 21] |
| PcG | Polycomb group gene | [36] [24] | |
| H3K27me3 | Histone H3 trimethylation at lysine 27 | [36] | |
| H3K4me3 | Histone H3 trimethylation at lysine 4 | [36] | |
| H3K36me3 | Histone H3 trimethylation at lysine 36 | [36] | |
| H3K27ac | Histone H3 acetylation at lysine 27 | [36] | |
| H3K9ac | Histone H3 acetylation at lysine 9 | [36] | |
| CGI | CpG island, nonrandom cluster of CpG sites, association with gene promoters | [92] | |
| R-loop | RNA/DNA hybrid formed at transcribed regions with high G skew, protects active promoter CGIs from DNA methylation | [92] | |
| eRNA | Enhancer RNA, noncoding RNAs transcribed from active enhancers, implicated in long- range looping interactions | [68] | |
| antisense RNA | Noncoding RNA transcribed in the opposite strand orientation from a protein coding gene, inhibition of transcriptional elongation of sense transcript | [70] [91] | |
| Players | Enzymes or protein complexes that direct epigenetic layers | ||
| DNMT1 | DNA methyltransferase 1, maintenance DNA methylation | [25] [27] | |
| DNMT3A | DNA methyltransferase 3A, de novo DNA methylation | [26] [28] | |
| DNMT3B | DNA methyltransferase 3B, de novo DNA methylation | immunodeficiency, centromeric instability, facial dysmorphism (ICF) | [109] |
| PRC1, PRC2 | Polycomb repressive complex 1 and 2, regulation of polycomb group genes | [64, 79] | |
| EZH2 | Enhancer of Zeste, Drosophila homologue 2, methyltransferase for H3K27 | [64, 79] | |
| MeCP2 | Methyl CpG binding protein 2, abundant nuclear factor in mature neurons, activity- dependent transcriptional responses | Rett syndrome, MECP2 duplication syndrome | [56] [110] |
| ATRX | Nuclear factor with Zinc finger, ATPase, and helicase domains, similarities to SNF2H chromatin remodeling protein | Alpha-thalassemia/Mental retardation, X linked | [111] |
| BAF53b | Actin-like component of neuronal chromatin remodeling complex, implicated in synaptic plasticity | [41] [42] | |
| ARID1B | Component of neuronal BAF chromatin remodeling complex | Coffin-Siris syndrome | [40, 112] |
| JMJD1C/TRP8 | Histone demethylase for H3K9, hormone-dependent transcriptional activation | X-linked intellectual disability, autism | 45 [113] |
| KDM5C/JARD1C | Histone demethylase of H3K4, gene repression | X-linked intellectual disability, autism | [47, 114] |
| SMCA1/cohesin | Cohesion, structural maintenance of chromosomes 1A, chromatin looping | Cornelia de Lange | [115] |
| Mediator | Multiprotein complex and general regulator of transcription, transcriptional coactivator, chromatin looping, activating-RNA binding | [91] | |
| CTCF | CCCTC-binding factor, chromatin insulator, chromatin looping | [51] [50] | |
| CREBBP (CBP) | CREB binding protein, H3K56 acetyltransferase, activity- dependent transcriptional responses | Rubinstein-Taybi syndrome | [116] [117] |
| TET1 | TET oncogene, family member 1, methylcytosine dioxygenase, converts 5mC to 5hmC resulting in demethylation | [14, 15] | |
| MLL1 | Mixed lineage leukemia 1, H3K4 methyltransferase | Wiedemann-Steiner syndrome | [118] |
| lncRNA | Long, non-coding RNA, >200nt with low-protein coding potential | [72] [73] | |
| Xist | X chromosome inactivation specific transcript | [60] |
Table 2.
Methods utilized for identifying epigenetic layers and players
| Method abbreviation | Method description | Refs |
|---|---|---|
| ChIP-seq | Chromatin immunoprecipitation with high throughput sequencing to identify DNA bound to target proteins | [36] |
| RNA-seq | RNA-sequencing to identify genome-wide transcriptome | [36] |
| 3C, 4C | Chromosome conformation capture (3C) on microarray (4C) | [98] [39] |
| HiC | 3C variant interrogating genome wide with high throughput sequencing | [95] [96] |
| HMM | Hidden Markov model, a computer learning tool used for defining and segmenting chromatin and methylation states based on sequencing data | [36] [3] |
| MethylC-seq | Methyl cytosine bisulfite conversion plus high throughput sequencing | [3, 12] |
| FISH | Fluorescent in situ hybridization to characterize the subcellular localization and expression of RNAs and homologous pairing of chromomsomes | [60] [86] [87] |
| ChIA-PET | Chromatin interaction analysis with paired end tag sequencing | [97] |
| ChIP-loop | Chromatin immunoprecipitation combined with loci specific PCR | [100] |
Box 2. Outstanding questions.
Why do partial methylated domains exist and how are they important for developing neurons?
How and why do neurons change methylation patterns throughout life?
How does a threshold of transcriptional activity in neurons set up a pattern of epigenetic memory and transcriptional responsiveness of a gene after the initial stimulus is gone?
What is the “MeCP2 code” of isoform differences and post-translational modifications?
Which noncoding RNAs provide specificity and/or long-lasting memory to epigenetic mechanisms in neurons?
How do lncRNAs function in neurons, and what role do they have in the pathogenesis of neurodevelopmental disorders?
How do genomic copy number variations affect epigenetic layers, such as DNA methylation and large-scale chromatin organization, in human neurodevelopmental disorders?
How are chromatin topological domains formed and maintained across cell type and species?
How can therapies shift epigenetic states?
How does the dynamic epigenetic state fit together with dynamic transcriptional loops?
Highlights.
Epigenetic modifications are long-lived layers of past transcriptional events.
The relationship of DNA methylation to transcription depends on genomic context.
Noncoding RNAs act as epigenetic layers and players influencing chromatin states.
Chromatin loops and long-range interactions are observed in neurodevelopment.
Acknowledgments
We thank the UC Davis Epigenome Club for helpful discussions and Dr. Diane Schroeder for critical reading of manuscript. Funding for research provided by the National Institutes of Health, Department of Defense Congressionally Mandated Directed Research Program, International Rett Syndrome Foundation, and the Prader-Willi Syndrome Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nature neuroscience. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson ED, et al. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder DI, et al. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Research. 2011;21:1583–1591. doi: 10.1101/gr.119131.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaSalle JM, Yasui DH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics. 2009;1:119–130. doi: 10.2217/epi.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakyan VK, et al. Metastable epialleles in mammals. Trends in genetics: TIG. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 6.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 7.Rollins RA, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginno PA, et al. R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gertz J, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS genetics. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JU, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroeder DI, et al. The human placenta methylome. Proceedings of the National Academy of Sciences of the United States of America; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009 doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie W, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruzov A, et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell research. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin SG, et al. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Research. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutcliffe JS, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 21.Dittrich B, et al. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nature Genetics. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- 22.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 23.Rauch TA, et al. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder DI, et al. The human placental methylome; Proc Natl Acad Sci U S A. in press. [Google Scholar]
- 25.Feng J, et al. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 26.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 28.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 31.Miller CA, et al. Cortical DNA methylation maintains remote memory. Nature neuroscience. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JU, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellen M, et al. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qureshi IA, et al. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell research. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picketts DJ, et al. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 39.Kernohan KD, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Developmental cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 40.Halgren C, et al. Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clinical genetics. 2012;82:248–255. doi: 10.1111/j.1399-0004.2011.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olave I, et al. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes & development. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel-Ciernia A, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nature neuroscience. 2013 doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Current Opinion in Genetics & Development. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaPlant Q, Nestler EJ. CRACKing the histone code: cocaine’s effects on chromatin structure and function. Hormones and behavior. 2011;59:321–330. doi: 10.1016/j.yhbeh.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margueron R, et al. The key to development: interpreting the histone code? Current Opinion in Genetics & Development. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Ptashne M. Epigenetics: Core misconcept. Proceedings of the National Academy of Sciences of the United States of America; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 48.Tahiliani M, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 49.Lee BK, Iyer VR. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. The Journal of biological chemistry. 2012;287:30906–30913. doi: 10.1074/jbc.R111.324962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golan-Mashiach M, et al. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Research. 2012;40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herold M, et al. CTCF: insights into insulator function during development. Development. 2012;139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 53.Gonzales ML, LaSalle JM. The role of MeCP2 in brain development and neurodevelopmental disorders. Current psychiatry reports. 2010;12:127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adkins NL, Georgel PT. MeCP2: structure and function. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2011;89:1–11. doi: 10.1139/O10-112. [DOI] [PubMed] [Google Scholar]
- 55.Na ES, Monteggia LM. The role of MeCP2 in CNS development and function. Hormones and behavior. 2011;59:364–368. doi: 10.1016/j.yhbeh.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 57.Gonzales ML, et al. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Molecular and cellular biology. 2012;32:2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng JV, et al. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nature neuroscience. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong X, et al. MeCP2 phosphorylation is required for modulating synaptic scaling through mGluR5. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:12841–12847. doi: 10.1523/JNEUROSCI.2784-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 61.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cech TR. The RNA worlds in context. Cold Spring Harb Perspect Biol. 2012;4:a006742. doi: 10.1101/cshperspect.a006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 64.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 70.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 71.Han K, et al. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes & development. 2013;27:485–490. doi: 10.1101/gad.207456.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mercer TR, et al. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng SY, et al. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mercer TR, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rapicavoli NA, et al. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rapicavoli NA, et al. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6:129–137. doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 81.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Redrup L, et al. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 84.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vitali P, et al. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J Cell Sci. 2010;123:70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- 87.Leung KN, et al. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum Mol Genet. 2009;18:4227–4238. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 89.Rougeulle C, et al. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 90.Numata K, et al. Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic Acids Res. 2011;39:2649–2657. doi: 10.1093/nar/gkq1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ginno PA, et al. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanyal A, et al. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 97.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yasui DH, et al. 15q11.2–13.3 chromatin analysis reveals epigenetic regulation of CHRNA7 with deficiencies in Rett and autism brain. Human Molecular Genetics. 2011;20:4311–4323. doi: 10.1093/hmg/ddr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meguro-Horike M, et al. Neuron-specific impairment of inter-chromosomal pairing and transcription in a novel model of human 15q-duplication syndrome. Human Molecular Genetics. 2011;20:3798–3810. doi: 10.1093/hmg/ddr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Horike S, et al. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 101.Takizawa T, Meshorer E. Chromatin and nuclear architecture in the nervous system. Trends in neurosciences. 2008;31:343–352. doi: 10.1016/j.tins.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 102.Cremer T, et al. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 103.Boyle S, et al. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011;19:901–909. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 105.Morey C, et al. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thatcher KN, et al. Homologous pairing of 15q11–13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Human Molecular Genetics. 2005;14:785–797. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- 107.Hogart A, et al. Chromosome 15q11–13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J Med Genet. 2009;46:86–93. doi: 10.1136/jmg.2008.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krueger C, et al. Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PloS one. 2012;7:e38983. doi: 10.1371/journal.pone.0038983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hansen RS, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Van Esch H, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. American Journal of Human Genetics. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gibbons RJ, et al. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 112.Nord AS, et al. Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. European journal of human genetics: EJHG. 2011;19:727–731. doi: 10.1038/ejhg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Castermans D, et al. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. European journal of human genetics: EJHG. 2007;15:422–431. doi: 10.1038/sj.ejhg.5201785. [DOI] [PubMed] [Google Scholar]
- 114.Jensen LR, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. American Journal of Human Genetics. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Deardorff MA, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. American Journal of Human Genetics. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Das C, et al. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 118.Jones WD, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. American Journal of Human Genetics. 2012;91:358–364. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]