Abstract
Detection of pathogen-derived nucleic acids by pattern recognition receptors (PRRs) is essential for the host to mount an appropriate immune response, which for viruses involves the induction of type I interferons (IFNs). On the other hand, inappropriate activation of PRRs by self nucleic acids can lead to autoimmunity. Recent developments in PRR research have uncovered important new molecular details as to how Toll-like receptors and RIG-I-like receptors distinguish pathogen- from self-RNA, while the discovery of cytosolic DNA sensing pathways for IFN induction has revealed completely new innate signaling mechanisms, and also questions how innate immunity discriminates between self- and non-self DNA, if at all.
Keywords: Toll-like receptors, RIG-I, antiviral immunity, cytosolic DNA sensing, STING
Innate recognition of pathogens by nucleic acid sensing
The innate immune system is the most ancient form of host defence and is evolutionarily conserved throughout the animal kingdom. Once a microbe has overcome the physical and chemical barriers of epithelia, it will immediately be recognised by cells of the innate immune system, such as tissue-residing macrophages and dendritic cells (DCs), via cell-associated pattern recognition receptors (PRRs). PRRs recognise distinct, conserved microbial structures such as proteins, lipids, lipoproteins, glycans and nucleic acids of bacteria, viruses, parasites and fungi, referred to as pathogen-associated molecular patterns (PAMPs) [1]. Upon PAMP recognition, most PRRs activate a signaling cascade leading to upregulation of various immune genes, including inflammatory cytokines, chemokines and type I interferons (IFNs), in order to establish an appropriate immune response. As PAMPs usually differ from host molecules, they are thought to be well-suited for the initial discrimination of infectious non-self (pathogens) from non-infectious self. However, this paradigm of selective pathogen recognition is constantly being revisited, since PRRs also sense commensal microbes and endogenous host molecules released from damaged tissue, known as danger- (or damage-) associated molecular patterns (DAMPs). During infection, the simultaneous recognition of PAMPs and DAMPs most likely alerts the host immune system to the invasion of viable, harmful pathogens to trigger an even more robust immune response [2].
Extensive work over the past 15 years has led to the discovery and characterisation of various PRR families present in distinct cellular compartments. However very recently there has been an acceleration of research and discoveries relating to how, and indeed why, PRRs sense pathogen-derived nucleic acids. The innate immune system’s ability to detect RNA and DNA via endosomal and cytosolic PRRs (Figure 1, Table 1) is an essential mechanism to mount protective immune responses especially to viruses, as they do not have many other features suitable for detection. However, this bears the risk of self-recognition, as nucleic acids, unlike most other PAMPs, are highly abundant in the host. The new field of cytosolic DNA sensing in particular has seen a rapid development in the past few years, while new insights into how established PRRs, such as the Toll-like receptors (TLRs) and retinoic-acid-inducible gene I (RIG-I) like receptors (RLRs), engage with and discriminate microbial RNA have recently emerged and are described below.
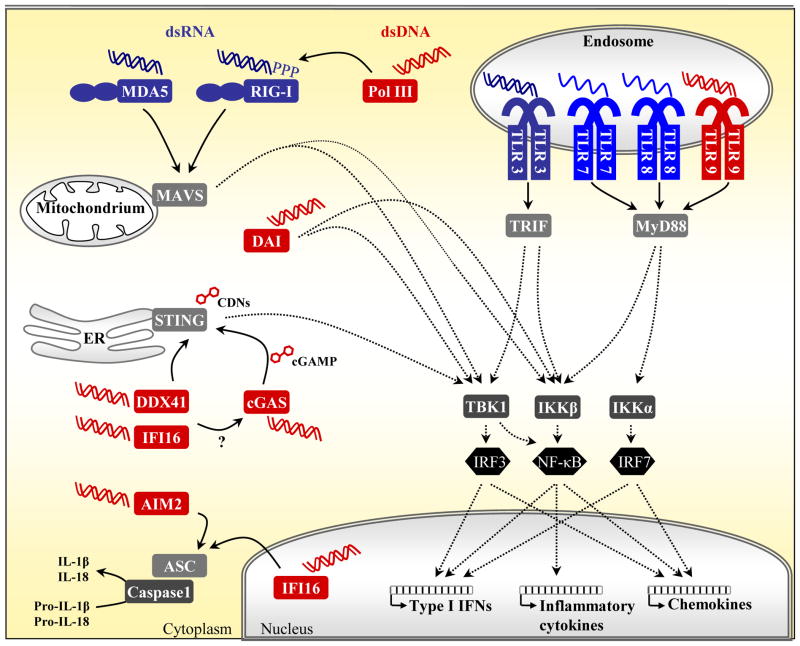
Figure 1. Nucleic acid sensing by PRRs in distinct cellular compartments.
Immune cells detect pathogen-derived nucleic acids by various PRR family members present in distinct cellular compartments. The four TLR family members TLR3, TLR7/8 and TLR9 are located in the endosome, where they detect dsRNA, ssRNA or unmethylated CpG DNA, respectively, leading to activation of TRIF- or MyD88-dependent pathways and the upregulation of type I IFNs, inflammatory cytokines and chemokines via IRF and NF-κB transcription factors. Cytosolic RNA detection is mediated by RLRs, i.e. RIG-I and MDA5, that subsequently signal via the mitochondria-localized signaling adaptor MAVS. Uncapped 5′-triphosphate (ppp) RNA is a potent RIG-I ligand, also generated by Pol III following binding to AT-rich dsDNA. However, other dsDNA-induced responses require the ER protein STING to activate the TBK1–IRF3 axis. Upstream candidate DNA receptors include DDX41 and the ALR family member IFI16. cGAS, on the other hand, was identified to synthesize the cyclic dinucleotide (CDN) cGAMP upon DNA stimulation, which then acts as a direct ligand for STING, similarly to bacterial CDNs. The cell type-specific DNA sensor DAI might transduce downstream signaling independently of STING. Furthermore, the ALRs AIM2 and IFI16 trigger the formation of an inflammasome following detection of cytosolic or nuclear DNA, respectively, by recruiting ASC and caspase-1 to induce processing of the inflammatory cytokines IL-1β and IL-18.
Table 1.
PRRs implicated in sensing microbial nucleic acidsa
| PRR | Location | Natural ligand | Detected pathogens | Adaptor | Ref. | |
|---|---|---|---|---|---|---|
| TLRs | TLR3 | Endosome | dsRNA | dsRNA virus (reovirus), ssRNA virus (RSV, EMCV, WNV), DNA virus (MCMV, HSV-1) | TRIF | [4] |
| TLR7/8 | Endosome | GU-rich ssRNA | ssRNA virus (VSV, IAV, HIV-1), endogenous retrovirus, Group B Streptococcus | MyD88 | ||
| TLR9 | Endosome | Unmethylated CpG DNA | MCMV, HSV1/2, ADV, bacteria, Plasmodium falciparum-generated hemozoin | MyD88 | ||
| mTLR13 | Endosome | Bacterial 23S rRNA | Bacteria | MyD88 | [12–14] | |
| RLRs | RIG-I | Cytoplasm | 5′ppp RNA, short dsRNA | ssRNA virus (e.g. Paramyxo-, Rhabdo-, Orthomyxo-, Flaviviridae), reovirus, DNA virus (EBV) | MAVS | [15] |
| MDA5 | Cytoplasm | Long dsRNA | ssRNA virus (Picorna-, Flaviviridae), reovirus, DNA virus (VACV) | MAVS | ||
| Pol III | Cytoplasm | AT-rich dsDNA | HSV-1, ADV, EBV, Legionella pneumophila | RIG-I | [45, 46] | |
| STING | Cytoplasm (ER) | CDNs | Listeria monocytogenes | STING | [63] | |
| cGAS | Cytoplasm | dsDNA | HSV-1, VACV | STING | [72] | |
| ALRs | IFI16 | Cytoplasm/nucleus | dsDNA | HSV-1, KSHV | STING | [50, 58] |
| AIM2 | Cytoplasm | dsDNA | VACV, L. monocytogenes, Francisella tularensis, Mycobacterium tuberculosis | ASC | [52–55, 83, 84] | |
| DAI | Cytoplasm | dsDNA | HSV-1 | ? | [43, 44] | |
| Ku70 | Cytoplasm | ds/ssDNA | HSV-2G | STING | [85] | |
| DDXs/DH Xs | DDX41 | Cytoplasm | dsDNA | HSV-1, VACV, ADV | STING | [49] |
| DHX9 | Cytoplasm | CpG-B | HSV | MyD88 | [48] | |
| DHX36 | Cytoplasm | CpG-A | HSV | MyD88 | [48] | |
| DDX9 | Cytoplasm | dsRNA | Reovirus, IAV | MAVS | [29] | |
| DDX1/ DDX21/ DHX36 | Cytoplasm | dsRNA | Reovirus, IAV | TRIF | [30] | |
| DDX3 | Cytoplasm | dsRNA | VSV | RLRs, MAVS | [27] | |
| DDX60 | Cytoplasm | ss/dsRNA, dsDNA | VSV, PV, SeV, HSV-1 | RLRs | [28] | |
Candidate RNA and DNA sensors are highlighted in light and dark grey, respectively. Abbreviations: RSV, respiratory syncytial virus; EMCV, encephalomyocarditis virus; WNV, West Nile virus; MCMV, murine cytomegalovirus; VSV, vesicular stomatitis virus; IAV, influenza A virus; HIV, human immunodeficiency virus; ADV, adenovirus; EBV, Epstein-Barr virus; PV, poliovirus; SeV, Sendai virus; VACV, vaccinia virus.
While pathogen-specific motifs have been identified for RNA detection, the ability of cytosolic DNA sensing mechanisms to discriminate pathogen from self nucleic acid is less clear. Understanding the mechanisms of nucleic acid sensing and signaling is of vast interest since it provides a molecular basis to explain both pathogen detection and autoimmunity. Although the picture is far from being complete, this review aims to capture the main recent developments in the fast-growing area of nucleic acid sensing and signaling.
Endosomal nucleic acid sensing by TLRs
The first identified and best characterized PRR family is the membrane-bound TLRs. With their recognition domain facing towards the extracellular space or the endosomal lumen, TLRs, respectively, detect either microbial surface molecules or pathogen-derived nucleic acids in the endocytic compartment. The endosomal TLRs, namely TLR3, 7, 8 and 9, sample the lumen for nucleic acids derived from phagocytosed microbes or infected apoptotic cells. Ligand-binding initiates the formation of a signaling platform at the cytosolic face of the TLR dimer, which subsequently leads to homotypic interaction with distinct TIR domain-containing adaptor proteins to selectively induce downstream signaling (Figure 1) [3]. Endosomal TLR3 signals solely via the adaptor TIR domain-containing adaptor inducing IFN-β (TRIF), while TLR7, 8 and 9 depend on myeloid differentiation factor-88 (MyD88). Both pathways subsequently activate the IκB kinase (IKK) complex leading to nuclear translocation of the transcription factor nuclear factor (NF)-κB to upregulate the expression of inflammatory cytokines and chemokines. IFN regulatory factor (IRF) transcription factors crucial for the induction of type I IFNs (IFN-α and IFN-β) are also activated by endosomal TLRs: for TLR3 the TRIF pathway signals through the noncanonical IKKs, TANK-binding kinase-1 (TBK1) and IKKε, to activate IRF3 and IFN-β expression, while for TLR7, 8 and 9 MyD88 activates IRF7 via IKKα leading to induction of IFN-α. The latter pathway is particularly important in plasmacytoid DCs (pDCs) that induce strong antiviral immune responses [4]. Although the main ligands for endosomal TLRs and their signaling mechanisms are now well established, new insights into the context and consequences of nucleic acid sensing by TLRs continue to emerge.
Potential clinical relevance of RNA sensing by TLRs
It has been known for some time that double-stranded RNA (dsRNA) is a key signature of viral infection, and that treatment of cells with the synthetic dsRNA analog polyinosinic-polycytidylic acid (Poly(I:C)) is an effective mimic of viral infection. TLR3 specifically recognizes dsRNA, such as the genome of reoviruses or dsRNA generated during replication of single-stranded RNA (ssRNA) viruses. Importantly, dsRNA is also produced by DNA viruses such as herpesviruses, likely due to overlapping converging transcription of viral genes [5]. Of note, TLR3 is one of the few PRRs linked to human disease, and in that context children with a defective TLR3 are known to be more susceptible to herpes simplex virus 1 (HSV-1) encephalitis [6]. In mice TLR3 deficiency has recently been shown to render astrocytes permissive to HSV and facilitates the establishment of a central nervous system infection [7]. Lafaille et al. then showed, by deriving induced pluripotent stem cells (iPSC) from TLR3-deficient patients, that TLR3-deficient neurons were highly susceptible to HSV-1 infection, providing a cellular basis for the increased susceptibility to HSV-1 encephalitis [8]. These studies demonstrate the physiological importance of nucleic acid detection by PRRs in infectious disease. Furthermore, in a separate study that elegantly demonstrated the potential clinical relevance of TLR3 detection of dsRNA, it was discovered that TLR3 sensing of retroviral expression vectors is essential for efficient nuclear reprogramming of iPSCs [9]. This provided an explanation as to why retrovirally delivered reprogramming factors generate iPSCs, whereas cell permeant proteins do not, since the retroviral RNA stimulates TLR3-dependent chromatin remodeling necessary for proper nuclear reprogramming.
TLR7 and TLR8 are known to sense guanosine/uridine (GU)-rich ssRNA of RNA viruses. Similar to TLR3, TLR7 has also been shown to have a role in responding to retroviruses. In this case TLR7 on B cells and/or DCs was shown by Yu et al. to be activated in response to endogenous retroviruses, leading to the production of protective anti-retroviral antibodies and suppression of viraemia in a mouse model [10]. It is conceivable thus that TLR7, and the highly related TLR8, may have a role in preventing endogenous retroviruses from causing human disease [11].
Detection of bacterial ribosomal RNA by TLR13
The recent identification of bacterial 23S ribosomal RNA (rRNA) as a ligand for mouse TLR13 identifies bacterial rRNA as yet another pathogen nucleic acid PAMP. It had been appreciated that bacterial RNA could induce cytokines and IFNs in a MyD88-dependent manner, but the sensing mechanism or TLR involved was not known. Three recent studies pinpointed TLR13 as the PRR. Similar to TLR3 and 7, TLR13 resides in the endosome and Hidmark et al. showed that small interfering RNA (siRNA) targeting TLR13 inhibited the ability of Gram-positive bacterial RNA to induce cytokine production from DCs [12]. Two other studies demonstrated sequence-specific sensing by TLR13 of 23S rRNA [13, 14]. This establishes 23S rRNA as a bona fide PAMP and provides a rare example (to date) of PRR sensing of bacterial, as opposed to viral, RNA. Very interestingly, the RNA sequence recognized by TLR13 is within a region of RNA recognized by certain antibiotics, and clinical isolates of Staphylococcus aureus that were resistant to such antibiotics also lacked the ability to stimulate mouse TLR13 [13]. Thus the authors suggest that ancient antibiotic resistance has subverted TLR13-driven antibacterial immune responses, which may explain why TLR13 expression has been abandoned in certain mammals, including humans [13].
Cytosolic RNA sensing by helicases
Apart from endosomal sensing by TLRs, viral RNA also is detected in the cytosol by RLRs, which are DExD/H-box helicases. These PRRs are mobilized to detect viral RNA species during intracellular viral invasion and replication, and are potent inducers of type I IFNs in most cell types (in contrast to the more cell-restricted expression pattern of TLR3 and TLR7) leading to the establishment of the antiviral immune response. The RLR family consists of three members: retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) [15]. RIG-I and MDA5 share a similar domain structure, composed of two N-terminal caspase activation and recruitment domains (CARDs) required for downstream signaling, a central DExD/H-box RNA helicase domain with the capacity to hydrolyse ATP and a C-terminal domain (CTD). LGP2, however, lacks the CARD domains and therefore the signaling function. Unlike nucleic acid-sensing TLRs, which are sequestered in the endosomal compartment, the cytosolic RLRs are surrounded by various host RNAs, which raises the question of self versus non-self discrimination.
Activated RIG-I and MDA5 are subsequently recruited to the mitochondrial antiviral signaling (MAVS, also known as IPS-1, VISA, Cardif) adaptor, followed by homotypic CARD–CARD interaction with the adaptor and initiation of downstream signaling via MAVS oligomerisation (Figure 1). The mitochondrial location provides a platform for the recruitment of distinct signaling molecules to MAVS leading to activation of NF-κB and IRFs for gene induction.
Recent studies revealed that RIG-I indeed recognizes pathogen-specific molecular features of viral RNA that are absent in host RNA. The receptor is potently activated by short, uncapped 5′-triphosphate (5′ppp) ssRNA juxtaposed to a short region of dsRNA [16, 17]. This is a nucleic acid motif known to be present in some RNA virus genomes, very distinct from endogenous RNA transcripts that are ‘capped’ by a 7-methyl-guanosine group at the 5′ end. Both RIG-I and MDA5 also respond to dsRNA, but with different dependency on length. It was demonstrated that synthetic Poly(I:C), which forms dsRNA molecules longer than 1 kilo base pairs (kbp), was preferentially recognized by MDA5, but could be converted to a RIG-I ligand by shortening of the dsRNA length [18]. RIG-I was shown to respond to dsRNA fragments as short as 23bp with a preference for blunt-ended RNA [19]. The distinct ligand specificity of both receptors becomes apparent in vivo in the selective detection of RNA viruses, however, a subset of viruses are sensed by both (Table 1). The third member, LGP2, is also able to bind RNA, but the functional consequences are still not fully understood. While LGP2 was found to negatively regulate RIG-I signaling in vitro [20], studies of Lgp2-deficient mice also suggested positive roles of LGP2 in RIG-I and MDA5-mediated responses to RNA viruses [21, 22].
Structural basis of self- versus non-self RNA discrimination by RLRs
It was unclear for some time how RLRs bind their ligands, as none of the helicases contain a classical RNA-binding motif. However recent studies solving the crystal structure of both RIG-I and MDA5 in complex with RNA ligands have revealed the molecular mechanism of viral RNA sensing by RLRs. The CTD was shown to be involved in RNA recognition, and in the case of RIG-I a large, positively charged cleft in the CTD was required for 5′ppp dsRNA-binding. The structural analysis provided detailed insights into the ligand-binding and activation mechanism of RIG-I [23, 24]. RIG-I is held in an auto-repressed conformation, with the CARDs sequestered by the helicase subdomains and not available for signaling. Initial binding of the 5′ppp RNA motif to the CTD is followed by cooperative tight binding of ATP and RNA to the helicase domain, which leads to a conformational change that pushes the CARDs out of the complex.
While RIG-I primarily recognizes the terminal region of dsRNA (with a 5′ppp motif), MDA5 was shown to recognize the internal duplex structure of dsRNA [25]. This study also provides the rationale as to why MDA5 signals in response to long dsRNA, because MDA5 uses a molecule of long RNA as a signaling platform to oligomerize into a filament structure capable of activating MAVS. Thus remarkably, although RIG-I and MDA5 share a similar domain structure, their mechanism of viral dsRNA recognition are very distinct, yet correlate well with the nature of the RNA ligand being recognized. Altogether, these studies provide an exciting molecular insight into how RLRs respond to two key viral nucleic acid PAMPs that are distinct from the normal RNA content of a mammalian cell, namely 5′ppp RNA and long dsRNA.
Non-RLR helicases implicated in cytosolic RNA sensing
Apart from the pivotal role of RLR helicases in cytosolic RNA detection, other members of the DExD/H-box helicase superfamily were recently proposed to function as cytosolic RNA sensors (Table 1). DEAD box protein 3 (DDX3), already known to be an essential component of the virally activated TBK1 complex that phosphorylates IRF3 [26], was shown to bind viral RNA and proposed to subsequently signal to MAVS for IFN induction [27]. Similarly, DDX60 was demonstrated to interact with RLRs to promote their signaling following RNA virus infection [28]. In myeloid DCs (mDCs), DHX9 was implicated in Poly(I:C) and RNA virus-induced responses following binding to dsRNA and signaling via the adaptor MAVS [29]. Furthermore, mDCs expressed a complex composed of DDX1, DDX21 and DHX36 that mediated dsRNA-dependent immune responses via the adaptor TRIF, normally used by endosomal TLR3 [30]. It was suggested that the constitutively expressed DDX proteins might mediate the initial IFN production, before the IFN-induced RLRs come into play. However further more rigorous analysis of the role of these RNA helicases in directly sensing RNA will be required in order to exactly define their roles in comparison to RLRs, especially given that RNA helicases are implicated in almost all processes of RNA biogenesis and gene induction regardless of a PRR role. Furthermore, unlike the RLRs, which contain a defined signaling domain (CARD), it is currently unclear how RNA detection by the other RNA helicases would engage downstream signaling pathways.
Cytosolic DNA sensing and the role of STING
Endosomal TLR9 is the only DNA sensing TLR and specifically binds unmethylated cytosine-guanosine (CpG)-containing DNA motifs present in bacteria and DNA viruses, but which are rare in the mammalian genome [31]. However in 2005, several groups reported TLR9-independent immune activation by DNA. A study in DNase II-deficient mice demonstrated that the lethality caused by accumulating endogenous DNA and excessive IFN-β production was not reversed in mice lacking TLR9 or MyD88 [32]. Also the cytosolic delivery of mammalian or bacterial dsDNA into macrophages and DCs triggered TLR9-independent cytokine responses [33, 34]. Before any potential DNA sensor was identified, it became clear that the TBK1–IRF3 signaling axis was central for cytosolic DNA responses leading to the induction of type I IFNs and also for the adjuvant effect of DNA-based vaccines in vivo [35–37]. Later a transmembrane protein found in the endoplasmic reticulum (ER) termed stimulator of IFN genes (STING, also known as MPYS, MITA and ERIS) was identified and shown to function as a crucial adaptor upstream of TBK1 in the cytosolic DNA responsive pathway [38–41]. Studies of STING-knockout mice clearly demonstrated its essential role in the cytosolic DNA pathway [41]. STING-deficient immune cells, such as macrophages and DCs, were defective in type I IFN production following dsDNA and HSV-1 infection. STING was also required for protection against HSV-1 infection in vivo. Further studies demonstrated the mechanism whereby STING directs TBK1 to phosphorylate IRF3 by induced proximity as STING could recruit both TBK1 and IRF3 to its carboxyl terminal tail upon stimulation of cells with cytosolic DNA [42].
However, how DNA actually activates the STING–TBK1–IRF3 signaling axis has been a subject of intense research, and the exact mechanisms are still unclear. Many cytosolic DNA sensors upstream of the STING pathway have been proposed, the idea being that direct engagement of cytosolic DNA by such a sensor somehow mobilizes STING to engage TBK1 and ultimately cause type I IFN induction. Such pathways would also be subject to negative regulation in order to avoid constant IFN induction by endogenous DNA (Box 1). The first candidate cytosolic DNA receptor, DNA-dependent activator of IRFs (DAI, also known as ZBP1), was identified in 2007, even before STING was implicated in DNA responses. DAI was shown to trigger IFN-β gene expression in murine L929 fibroblasts following binding to dsDNA, and interaction with TBK1 and IRF3 [43, 44]. However, Dai-knockout mice and knockout cells, such as mouse embryonic fibroblasts, macrophages and DCs, responded normally to DNA [37], while there is very little evidence to date that DAI engages the STING pathway. This suggests a restricted, and likely cell type-specific, role for DAI in the cytosolic DNA sensing pathway. Another DNA sensor, which responds to AT-rich DNA (a hallmark of some pathogen genomes), is RNA polymerase III (Pol III) (Figure 1). Pol III was shown to transcribe AT-rich dsDNA, such as Poly(dA:dT), into 5′ppp-containing dsRNA, which subsequently functions as a ligand for RIG-I [45, 46]. This explains why the adaptor MAVS was found to be involved in some DNA responses, as observed for Poly(dA:dT)-stimulated HEK293 cells that are generally unresponsive to DNA ligands [35, 47]. Thus the Pol III sensing pathway, whose physiological relevance is yet to be fully established, operates via MAVS and not STING. Other candidate DNA sensors are listed in Table 1. Below we highlight the DDX proteins and pyrin and HIN200 domain-containing (PYHIN) proteins as cytosolic DNA sensors since there have been multiple publications on their roles and mechanisms of action in DNA sensing in the past three years.
Box 1. Negative regulators of DNA sensing.
Compartmentalisation and specificity for microbial DNA generally prevents self DNA recognition by TLR9 in the endosome. However, in the case of cytosolic and potentially nuclear DNA sensors this mechanism of discrimination falls apart. Instead any mislocated, naked dsDNA of microbial or host origin is likely recognized to alarm the innate immune system of ‘danger’. This ensures host defense against many pathogens, especially intracellular bacteria and viruses, but can have detrimental consequences in cases where accumulated host DNA can lead to the development of severe autoimmune diseases. Thus deoxyribonucleases (DNases) are located at extra- and intracellular locations to immediately degrade endogenous DNA released from apoptotic and necrotic cells. DNase I is found in the extracellular space and serum and mutations of this enzyme have been linked with systemic lupus erythematosus (SLE)-like symptoms [73, 74]. Although TLR7 and TLR9 were shown to play roles in disease pathogenesis [4], cytosolic DNA sensors are likely to be involved. DNase II, on the other hand, is important for clearance of engulfed DNA in macrophage lysosomes and a defect of this enzyme leads to massive type I IFN production causing embryonic lethality in mice [75]. Cytosolic ER-associated DNase III, termed three prime repair exonuclease 1 (TREX1), digests reverse-transcribed ssDNA intermediates from endogenous retroelements. Mutations in TREX1 are associated with Aicardi-Goutières syndrome (AGS), chilblain lupus and SLE [76]; and STING has been implicated in the development of AGS in a mouse model [77].
During pathogen infection, negative regulators are required to prevent overwhelming immune responses and restore homoeostasis. For example, the IFN-inducible E3 ligase TRIM21 was reported to control cytosolic DNA responses by mediating DDX41 degradation [78]. Pathogens have also evolved strategies to block this cytosolic DNA detection system, as has been found for many other PRR pathways [79]. For example, DAI-mediated NF-κB activation is inhibited by M45 of murine cytomegalovirus [80]. Also IFI16 was shown to be targeted by human herpesvirus proteins, including the human cytomegalovirus protein pUL83 [81] and the HSV-1-encoded ICPO, which mediates IFI16 degradation [60]. The poxvirus protein M13L contains a pyrin domain and was shown to inhibit inflammasome activity by binding to the adaptor ASC [82]. It will be interesting to determine if this viral protein also directly interferes with DNA sensors containing a pyrin domain, namely ALRs.
Role of DEAD box proteins in DNA sensing
Similar to the case with cytosolic RNA sensing, DEAD box helicases have also been implicated as sensors of cytosolic DNA (Table 1). In pDCs, DHX9 and DHX36 were shown to interact with MyD88 and trigger gene expression in response to cytosolic CpG DNA [48]. While DHX9 primarily activated the NF-κB pathway to induce the proinflammatory cytokines TNF-α and IL-6, DHX36 signalled via IRF7 to upregulate IFN-α. How these signaling pathways are selectively regulated downstream of the common adaptor MyD88 remains to be determined, and a rationale is needed to explain how in some cell types DHX9 and DHX36 sense RNA leading to MAVS- and TRIF-dependent signaling (see above), while in other cell types these helicases sense DNA and engage with the MyD88 pathway. A further DDX protein, DDX41, is now strongly implicated in cytosolic DNA sensing, and was shown to interact with the STING–TBK1–IRF3 signaling axis [49]. A role for DDX41 in gene induction and activation of the transcription factors NF-κB and IRF3 was demonstrated in mDCs and human monocytes, using siRNA targeting DDX41. Furthermore DDX41 was shown to interact with dsDNA through its DEAD domain, and also to recruit STING (via the same domain) upon DNA transfection or DNA virus infection.
Role of PYHIN proteins in DNA sensing
Apart from DDX41, the PYHIN protein IFI16 (and its apparent functional ortholog in mice, p204) has been shown to be a DNA sensor operating upstream of STING–TBK1–IRF3 [50] (Figure 1). IFI16 was affinity purified from cytosolic extracts of human monocytes using transfected 70bp-dsDNA from vaccinia virus (VACV70mer) as bait. IFI16 recruited STING to activate the TBK1–IRF3 axis, and knockdown of murine p204 inhibited IRF3 and NF-κB nuclear translocation and gene induction in response to DNA and HSV-1 infection. IFI16 and p204 contain an N-terminal pyrin domain, and two DNA binding HIN200 domains, and IFI16 was confirmed to directly bind VACV70mer in vitro [50]. Besides inducing type I IFNs, cytosolic microbial and mammalian dsDNA also triggers inflammasome activation leading to production of the inflammatory cytokines IL-1β and IL-18 [51]. The PYHIN protein absent in melanoma 2 (AIM2), which has a pyrin domain and just one HIN200 domain, was identified by four independent groups as the DNA sensor that interacted with the adaptor protein apoptosis-associated speck-like protein containing a CARD (ASC) followed by caspase-1 recruitment to form the AIM2 inflammasome [52–55].
The structure of the PYHIN proteins is consistent with their proposed role as cytosolic DNA sensing PRRs, as they possess a defined signaling (pyrin) and DNA binding (HIN200) domain. Thus IFI16, p204 and AIM2 form a new family of PRRs termed AIM2-like receptors (ALRs) [56]. Furthermore the structure of both an IFI16 and AIM2 HIN domain in complex with dsDNA has recently been solved. This showed that the DNA–receptor complex is formed by electrostatic interactions between the positively charged HIN domain and the dsDNA sugar-phosphate backbone, in a DNA sequence-independent manner [57]. Hence this analysis of how PYHIN proteins engage with DNA provides a rationale as to why dsDNA of any sequence or species seems to be immune-stimulatory if introduced into the cytosol. This suggests that in contrast to cytosolic pathogen RNA, with its clearly defined non-self molecular signatures, cytosolic DNA should be viewed more as a DAMP than as a PAMP.
Studies into the role of PYHIN proteins as DNA sensors have also questioned the idea that it is DNA in the cytosol only, and not in the nucleus, that is immune stimulatory. Although this raises questions as to why host nuclear DNA does not cause an immune response, it is consistent with the fact that many DNA viruses replicate in the nucleus. As such Kerur et al. showed that IFI16, which is located in the nucleus in many cell types, sensed Kaposi’s sarcoma-associated herpes virus (KSHV) in the nucleus of endothelial cells leading to formation of an IFI16–ASC–caspase-1 inflammasome in the cytosol [58]. The ability of IFI16 to directly interact with ASC is still controversial, since previous work showed that the two proteins could not co-immunoprecipitate together [52], while ASC was not required for p204-dependent IFN induction by DNA [50]. Furthermore, IFI16 detected HSV-1 DNA in the nucleus of permissive cells, leading to type I IFN induction [59, 60]. How nuclear sensing of DNA by IFI16 engages the cytosolic STING–TBK1–IRF3 pathway is still an open question.
A novel host second messenger activates STING
Cyclic dinucleotides (CDNs), such as cyclic diguanylate or diadenylate monophosphate (c-di-GMP or c-di-AMP), are bacterial nucleic acid second messengers thought to be PAMPs, because when these are introduced into mammalian cells, similar to dsDNA, they cause STING-dependent type I IFN induction [61, 62]. In fact CDNs were shown to directly bind to and activate STING [63], which was recently confirmed by several crystal structures of a complex between the carboxyl region of STING and a CDN [64–68]. Although this appears to be a case of host recognition of yet another pathogen nucleic acid PAMP, the discovery that STING is activated by a novel host cell second messenger, of similar structure to CDNs (described below), suggests that bacterial CDNs may be mimicking this second messenger in order to stimulate STING-dependent type I IFN induction for the benefit of the bacterial pathogen [69]. In a related example, Mycobacterium tuberculosis has been shown to stimulate the p204–STING–TBK1–IRF3 pathway to enable long term infection in vivo [70].
At the beginning of 2013, a fascinating new signaling pathway activated by cytosolic dsDNA was discovered by the group of Zhijian J. Chen that provides a key missing link between ‘upstream’ DNA sensing and STING activation. They discovered a factor newly synthesized in mammalian cells following DNA stimulation that activated the STING-dependent pathway, and then isolated and identified this factor as cyclic-GMP-AMP (cGAMP) [71]. cGAMP directly bound to STING, and caused IRF3 activation. In an accompanying paper they also identified cGAMP synthetase (cGAS) as a cyclase that generated cGAMP from ATP and GTP following DNA stimulation [72]. This new signaling axis is similar to the classic cAMP second messenger system where adenylate cyclase generates cAMP from ATP downstream of G protein-coupled receptors. Although the authors showed that cGAS directly bound to DNA, it remains to be established whether cGAS is a bona fide cytosolic DNA sensor with broad relevance in different cell types and whether other DNA sensors, such as IFI16 and DDX41, engage the cGAS–cGAMP system in order to activate STING.
Concluding remarks
Rapid progress has been made in recent years in understanding the physiological relevance of PRR detection of both self and microbial nucleic acids. Furthermore, elegant mechanisms for discriminating self from non-self RNA have been revealed at the molecular level. A whole new field of PRR research has opened up with the appreciation that cytosolic dsDNA is immune stimulatory, leading to the identification of many new putative DNA sensing PRRs, and the discovery of new signaling molecules such as STING and cGAS. Studies into the role of the PYHIN family in DNA sensing have questioned the paradigm that it is only cytosolic, and not nuclear DNA that is immune stimulatory, and further work is needed to delineate the factors involved in nuclear DNA sensing and how potential immune responses to normal host DNA in the nucleus are suppressed. Given the observation that DNA sensing (at least by the PYHIN proteins) is sequence-independent and merely relies on a dsDNA backbone, it seems that although the innate immune system recognizes pathogen RNA as ‘stranger’, DNA is rather recognized as ‘danger’. The in vivo relevance of many of the proposed DNA sensors (Table 1) will also need to be established, which will help to determine which ones have cell type-specific roles and how much redundancy exists, which will help define a hierarchy of sensors with importance in autoimmunity and infectious diseases.
Highlights.
Endosomal TLRs have new functions in sensing RNA.
Mechanisms are known for how RIG-I and MDA5 discriminate between host and viral RNA.
Multiple DEAD box helicases have been proposed as nucleic acid sensors.
New signaling pathways have been found controlling cytosolic DNA responses.
Acknowledgments
Work in the authors’ laboratory is supported by grants from Science Foundation Ireland (11/PI/1056) and the NIH (AI093752).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- 3.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 5.Weber F, et al. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 7.Reinert LS, et al. TLR3 deficiency renders astrocytes permissive to herpes simplex virus infection and facilitates establishment of CNS infection in mice. J Clin Invest. 2012;122:1368–1376. doi: 10.1172/JCI60893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafaille FG, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491:769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu P, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Mankan AK, Hornung V. Retroviral danger from within: TLR7 is in control. Immunity. 2012;37:763–766. doi: 10.1016/j.immuni.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Hidmark A, et al. Cutting edge: TLR13 is a receptor for bacterial RNA. J Immunol. 2012;189:2717–2721. doi: 10.4049/jimmunol.1200898. [DOI] [PubMed] [Google Scholar]
- 13.Oldenburg M, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 14.Li XD, Chen ZJ. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife. 2012;1:e00102. doi: 10.7554/eLife.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 17.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 18.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques JT, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 20.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 21.Satoh T, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkataraman T, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 23.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 24.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 26.Schroder M. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem Pharmacol. 2010;79:297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 27.Oshiumi H, et al. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita M, et al. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, et al. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol. 2011;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 32.Okabe Y, et al. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda K, et al. Macrophage activation by a DNA/cationic liposome complex requires endosomal acidification and TLR9-dependent and -independent pathways. J Leukoc Biol. 2005;77:71–79. doi: 10.1189/jlb.0204089. [DOI] [PubMed] [Google Scholar]
- 34.Yasuda K, et al. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J Immunol. 2005;174:6129–6136. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 35.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 36.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 38.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa H, et al. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu YH, et al. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng G, et al. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci U S A. 2007;104:9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim T, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 52.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandes-Alnemri T, et al. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 55.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 56.Keating SE, et al. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li T, et al. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orzalli MH, et al. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McWhirter SM, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sauer JD, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin Q, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouyang S, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang YH, et al. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 67.Shang G, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 68.Shu C, et al. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monroe KM, et al. Induction of type I interferons by bacteria. Cell Microbiol. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manzanillo PS, et al. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun L, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Napirei M, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 74.Yasutomo K, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 75.Yoshida H, et al. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 76.Stetson DB, et al. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, et al. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2012;14:172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rebsamen M, et al. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cristea IM, et al. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnston JB, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 83.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saiga H, et al. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol. 2012;24:637–644. doi: 10.1093/intimm/dxs062. [DOI] [PubMed] [Google Scholar]
- 85.Zhang X, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]