Abstract
The biochemical basis of circadian timekeeping is best characterized in cyanobacteria. The structures of its key molecular players, KaiA, KaiB, and KaiC are known and these proteins can reconstitute a remarkable circadian oscillation in a test tube. KaiC is rhythmically phosphorylated and its phospho-status is a marker of circadian phase that regulates ATPase activity and the oscillating assembly of a nanomachine. Analyses of the nanomachines have revealed how their timing circuit is ratcheted to be unidirectional and how they stay in synch to ensure a robust oscillator. These insights are likely to elucidate circadian timekeeping in higher organisms, including how transcription and translation could appear to be a core circadian timer when the true pacemaker is an embedded biochemical oscillator.
Introduction
The core mechanism of the circadian clock in eukaryotic cells is widely held to be based on a Transcription/Translation Feedback Loop (TTFL) [1,2], although there is recent evidence that this model may be incomplete or inaccurate [3,4]. In cyanobacteria, the initial evidence also supported a TTFL model [5]. However, our current understanding of the clock system in cyanobacteria is that a biochemical oscillator operates as the central “quartz crystal” of the clockwork [6], but this core pacemaker operates within (and regulates) a larger TTFL that controls outputs and replenishes the oscillator’s essential proteins [7,8]. The remarkable discovery that this core oscillator could be transplanted as a three-protein system to oscillate in vitro [6] has led some researchers to revisit the question of non-TTFL circadian clocks in eukaryotes – a search that has recently culminated in the discovery of circadian metabolic/redox oscillations that can operate in eukaryotes in the absence of transcription [9], and which resurrects an old literature on circadian clocks in enucleated algal cells [4]. Because we know the 3D-structures of the major protein players and the oscillator can be reconstituted in vitro, the cyanobacterial system constitutes a unique preparation to study the biochemistry, biophysics, and structural biology of post-translational circadian timekeeping.
To set the stage for the information that we summarize below, KaiC hydrolyzes ATP and this reaction may be an intrinsically constant-rate timer [10] that drives KaiC phosphorylation. The status of KaiC phosphorylation feeds back to regulate (i) the ATP hydrolytic rate (so that its intrinsic constant rate becomes rhythmic) and (ii) switching of the KaiABC nanocomplex between autokinase and autophosphatase states (Figure 1). An overly simplified analogy would be to consider KaiC ATPase activity as an “hourglass timer” which is restarted each cycle (the “hourglass is turned over”) by KaiC’s phosphorylation status, thereby converting the hourglass timer into an oscillation. Therefore, the extent of KaiC phosphorylation is both a marker of circadian phase [11,12] and a regulator of KaiC’s myriad activities (ATPase, autokinase, autophosphatase, phosphotransferase, and nanocomplex formation).
Figure 1.
The circadian in vitro oscillator that acts as a post-translational oscillator (PTO) in vivo. The reaction is composed of ATP hydrolytic activity and phosphorylation within KaiC, and rhythmic associations of KaiC with KaiA and KaiB to form a nanocomplex that regulates ATPase and phosphorylation activities. Cartoons depict associations among the KaiC hexamer (light blue), the KaiA dimer (pink), the KaiB tetamer (green) and phosphorylation on KaiC subunits (red dots). Rectangles show ATPase activity over time. Starting from the upper left corner, KaiA associates with unphosphorylated KaiC and stimulates autophosphorylation, which accelerates the rate of ATP hydrolysis. The stages of the reaction proceed as described in the text [41] in a counter-clockwise direction.
Architecture and function of KaiC, the central player
The N- and C-terminal domains (termed CI and CII, respectively) of KaiC exhibit sequence similarity with ATPases, such as the DnaB helicase and RecA recombinase. The crystal structure of S. elongatus KaiC resembles a double doughnut with CI and CII rings of similar height and width and a constricted waist region that is spanned by 15-amino acid linkers (Figure 2A) [13]. The conformation of the C-terminal region in CII differs considerably from the one in CI that constitutes the linker and winds along the outer surface of the CI/CII interface. Thus, CII C-terminal peptides form a crown-like arrangement of S-shaped loops ("tentacles"), comprised of residues E487 to I497, that rim the central KaiC channel and then protrude from the outer dome-shaped surface of CII.
Figure 2.
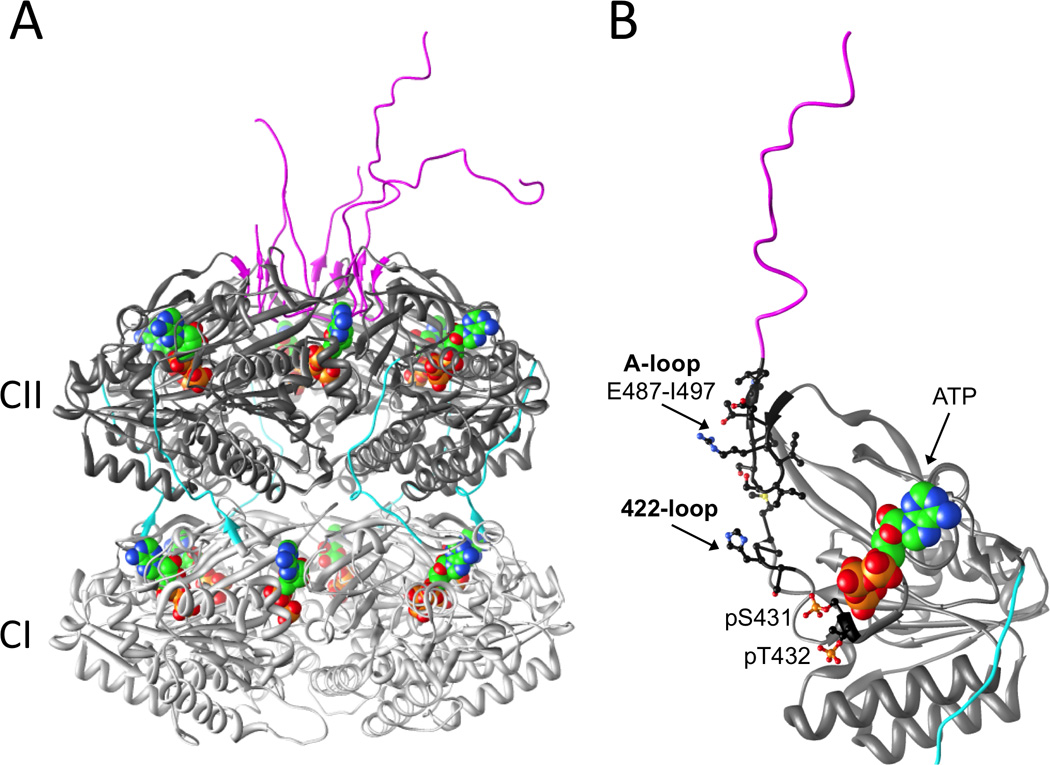
(A) Structure of the KaiC homo-hexamer (PDB ID code 3DVL) [13]. N-terminal CI and C-terminal CII domains from individual subunits are colored in light gray and dark gray, respectively, CI-CII linkers are cyan and C-terminal peptide tails are magenta. ATP molecules are shown in space filling mode with carbon, oxygen, nitrogen and phosphorus atoms colored in green, red, blue and orange, respectively. (B) An isolated KaiCll domain with A-loop and 422-loop as well as phosphorylation sites T432 and S431 highlighted.
ATP molecules are wedged between subunits in the CI and CII rings and bound to P-loop motifs (Figure 2) [13]. The two rings possess different functions; the CI ring harbors ATPase activity [10] and the CII ring catalyzes rhythmic phosphorylation and dephosphorylation reactions. The kinase activity [14,15] results in phosphorylation of Thr432 and Ser431 across CII subunit interfaces [13,16]. A third residue, T426, influences phosphorylation and dephosphorylation of T432/S431, possibly by acting as a labile phosphorylation site whereby a phosphate is shuttled between T426 and S431 [16,17]. The T432 residue is phosphorylated first in the cycle, followed by phosphorylation at S431 [11,12], and this strict order of phosphorylation can be explained with a kinetically controlled kinase activity, where the ATP γ-phosphate is closest to the hydroxyl group of T432 that is therefore phosphorylated before S431 [13,16]. The dephosphorylation reaction then proceeds in the same order (pT432 is followed by pS431), such that the overall process from the hypo- to the hyper- and back to the hypo-phosphorylated state of KaiC involves the following stages: TS → pTS → pTpS → TpS → TS [11,12]. The phosphorylation status of KaiC is therefore a marker for the phase of the in vitro oscillator [12].
KaiA stimulates KaiC phosphorylation that "ratchets" the clockwork
KaiC can auto-phosphorylate, especially at low temperatures (e.g., 4°C). At physiological temperatures, however, KaiA greatly stimulates the autokinase activity of KaiC [15,18]. Specifically, binding of KaiA homo-dimer to a KaiC C-terminal tail or "tentacle" activates the kinase in the CII ring [19,20], thus triggering phosphorylation of T432 and S431 residues (Figures 2, 3). Phosphorylation of T432 and S431/T426 leads to increased molecular interactions, salt bridges, and formation of additional hydrogen bonds [21–23]. These enhanced interactions render reverse reactions unfavorable and provide a "ratcheting" mechanism that can explain why time in the phosphorylation phase of the in vitro KaiABC reaction is unidirectional [21,24]. Therefore, "time doesn’t run backwards." Despite the presence of six C-terminal tentacles on KaiC, a single KaiA dimer is sufficient to convert KaiC from the hypo- to the hyper-phosphorylated form [25], thus implying an allosteric mechanism for KaiC phosphorylation [26]. On the basis of studies in which the C-terminal tentacles were deleted to various lengths, LiWang and coworkers concluded that the intramolecular action of KaiA binding to KaiC is to "pull" on KaiC’s buried A-loop, which straightens and exposes it (Figures 2B, 3) [27]. This exposure of the A-loop was hypothesized to move the ATP (bound to the P-loop) closer to T432/S431, possibly mediated by a β-strand segment that lies between the A-loop and P-loop (Figure 2B) [27].
Figure 3.

KaiA dimer (only C-terminal domains are depicted) interacting with a KaiCll C-terminal tentacle by pulling out the A-loop (highlighted in magenta) and unraveling it. The model is a fusion of the KaiC crystal structure [13] and the NMR structure of the KaiA-KaiC peptide complex [19]. See Figure 2 for KaiC color code.
Further support for this allosteric model and the general importance of the A-loop region for phosphorylation of KaiC comes from an analysis of the structure and dynamics of the KaiC A422V mutant. An alanine for valine exchange is a very conservative change, and yet this mutation has rather drastic consequences for function that include abnormal phase-resetting responses to dark pulses, abolished rhythms of KaiC accumulation, and a reduced amplitude of the KaiC phosphorylation rhythm [28]. Close inspection of the KaiC crystal structure reveals that the 422-loop is buried underneath the A-loop and that apical residues of the two loops are in direct contact. Therefore removal of the A-loop alters the constraints of the 422-loop and the region connecting A/V422 to the T432 and S431 phosphorylation sites (Figure 2B) [29]. Molecular dynamics (MD) simulations support the notion that the KaiA-mediated pulling of an A-loop alters the dynamics of KaiCll, triggering increased mobility and facilitating phosphorylation [29]. The consensus that emerges is that KaiA stimulates the KaiC kinase via a concerted allosteric mechanism [27,29] and that no large-scale conformational changes are required in either the CI or CII halves (as evidenced by the MD work) to move from the ST to the pTS and ultimately the pTpS state, using the aforementioned ratcheting that favors the forward reaction [21].
KaiC dephosphorylates by regenerating ATP
KaiC dephosphorylation has been generally assumed to involve a phosphatase activity that might require a considerable conformational adjustment to move pT432 and pS431 away from the kinase active site and reposition them at a putative phosphatase active site [21]. Those assumptions are probably incorrect. First, a number of studies using multiple techniques indicate that a large conformational change of KaiC is unlikely [13,16,20,22,30–32]. Second, signature motifs of common types of phosphatases [23] are missing in the KaiC sequence. Finally, two independent studies have confirmed that dephosphorylation of T432 and S431 proceeds – at least partially – via a phosphotransferase reaction that results in the regeneration of ATP from the phosphates at T432/S431 and the ADP bound at the P-loop (Figure 2) [23,33]. This phosphotransfer activity complicates accounting for the number of ATP molecules hydrolyzed over the daily clock cycle. In particular, the reported amount of 15 ATPs consumed per day per KaiC monomer [10] has to be considered a net balance because the absolute numbers of ATP molecules hydrolyzed and then regenerated in a 24-hour interval is not known. What is the basis of dephosphorylating by regenerating ATP? Possibly this is a mechanism to conserve ATP when its cellular concentration declines as a result of cells not being able to photosynthesize [34], thereby allowing the clock to keep running under extended dark exposure [35,36].
Phosphorylation state mimics: a useful but potentially misleading tool
It is common practice to replace serine and threonine with aspartic or glutamic acid residues in proteins to mimic phospho-serine and phospho-threonine and many studies in regard to the structure and function of the KaiABC PTO have relied on this approach (e.g. the recent refs. [8,10,22,27,37–39]). However, it is important to keep in mind when using this mimicry that Asp and Glu are not perfect replacements of pSer or pThr. This is not just due to different sterics but also deviating negative charges (i.e. −1 for Asp and Glu and −2 for pSer and pThr at neutral pH). Crystal structures even at relatively low resolutions of around 3.5 Å can easily distinguish between unphosphorylated and phosphorylated Ser and Thr residues, and indeed crystallographic studies revealed important differences in the structure, phosphorylation states and electrostatic surface potentials (ESPs) of KaiC phosphorylation state mimics that do not seem to be generally considered when interpreting functional data employing KaiC mutants comprising E or D in place of the phosphorylation site residues T432 and/or S431. Moreover, functional studies uncovered a difference between the T432A/S431A mutant and authentic unphosphorylated KaiC [40]. X-ray crystallography revealed the following phosphorylation patterns in the structures of KaiC mutants [16,32]: T432E/S431A (pTS mimic) has T426 phosphorylated in four subunits, in S431D (TpS mimic) three out of six T432 residues carried a phosphate, and T432E/S431E (pTpS mimic) exhibited a new phosphorylation site at S320 in two subunits and its ESP on the CII dome surface matched neither that of hyper- nor that of hypophosphorylated KaiC.
Maintaining synchronous KaiC hexamers and robust rhythmicity
KaiB associates with KaiC during dephosphorylation and this association correlates with the phase of KaiC subunit exchange, suggesting a functional link between KaiB’s binding to KaiC and its monomer exchange (Figure 1) [41]. This subunit monomer exchange is important in terms of maintaining synchrony of the phosphorylation status among all the KaiC hexamers in the population of hexamers present in the in vitro reaction (and presumably in vivo as well) [41,42]. By maintaining synchrony among the KaiC hexamers, robust rhythmicity is achieved, which has been experimentally demonstrated to be a property of the in vitro reaction for at least 10 days [42]. An equally important factor for maintaining synchrony among KaiC hexamers is the sequestration and progressive inactivation of KaiA dimers during the phosphorylation phase [12,26,37]. Therefore, it appears that there are at least two mechanisms for maintaining KaiC phospho-synchrony and robust rhythmicity: KaiC monomer exchange/KaiB binding during the KaiC dephosphorylation phase, and KaiA sequestration.
The timing and impact of the binding of KaiA and KaiB to KaiC are critical for maintaining synchrony among KaiC hexamers, thereby promoting high-amplitude oscillations that persist for many cycles. But where do KaiA and KaiB bind to KaiC? As mentioned above, at the beginning of the phosphorylation phase, KaiA associates with the C-terminal tentacles of KaiC (Figures 2, 3); at the end of the phosphorylation phase, KaiA is sequestered by KaiC (or by a KaiB-KaiC complex, Figure 4) to an unknown site, but it might be near the waist/linker region of KaiC [32,37]. The identity of the binding site(s) of KaiB to KaiC has recently become contested. In 2008, we presented a three-dimensional model of the KaiBC complex based on negative stain and cryo EM that features two KaiB dimers bound to the dome-shaped surface of KaiCll (Figure 4) [30]. LiWang and coworkers recently used solution NMR and gel filtration chromatography to study the interactions between KaiB and separate CI and CII rings as well as monomeric CI and CII domains and arrived at the conclusion that KaiB binds to the CI ring [38]. However, EM studies of complexes between KaiB and KaiC S431D mutant hexamers carrying C-terminal His6 tags labeled with nickel(II) nitrilotriacetic acid gold nanoparticles (Ni-NTA-Nanogold) revealed that gold is located on the same side as KaiB, thus providing unequivocal evidence that B is bound to CII [43].
Figure 4.
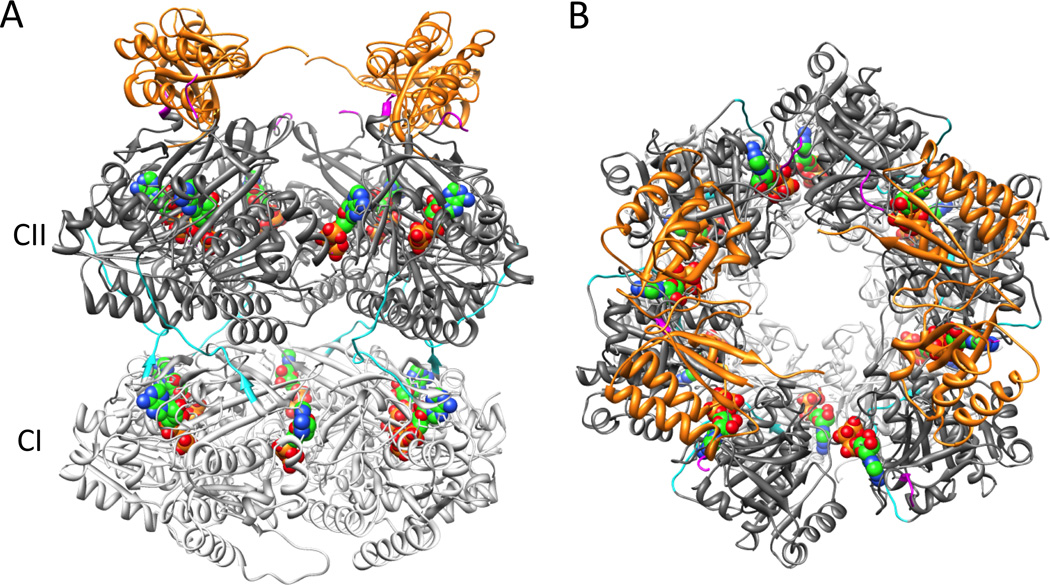
EM-based model of the KaiB-KaiC complex [30], viewed (A) from the side and (B) rotated by 90° around the horizontal and viewed along the central KaiC channel from the CII side. KaiB dimers are colored in orange and the coloring code for KaiC matches that in Figure 2.
Entraining the PTO pacemaker embedded within a TTFL slave oscillator
Thus, the underlying circadian system in cyanobacterial cells appears to comprise two subcomponents: a post-translational oscillator (PTO) and a transcriptional/translational feedback loop (TTFL) [7,8]. The TTFL drives global gene expression and the PTO is the core “quartz crystal” of the timekeeper. Modeling indicates a mechanism by which the TTFL can feed into the PTO such that new synthesis of clock proteins can phase-shift or entrain the core PTO pacemaker. This prediction was experimentally tested and confirmed by entraining the in vivo circadian system with cycles of new clock protein synthesis that modulate the phosphorylation status of the clock proteins in the PTO [8]. Consequently the TTFL can provide entraining input into the PTO.
But what are the metabolic input signals and how are they related to the daily light/dark cycle that entrains the clock? Four hour dark pulses readily reset the phase of cyanobacterial cells that are free-running in LL [28]. Insight into phase-shifting has recently been gained by way of two sources that were identified to feed into the KaiABC oscillator and thus allow cyanobacteria to keep track of the daily light/dark cycle. Rust, Golden and O’Shea demonstrated that KaiC phosphorylation is directly influenced by the cellular ATP/ADP ratio that is itself dependent on photophosphorylation [34]. Interestingly, the ATP/ADP ratio also affects KaiC phosphorylation in the in vitro KaiABC PTO and was demonstrated to be sufficient to reset the phase of phosphorylation. In addition to the ATP/ADP ratio, the phase of the KaiABC PTO is influenced by the oxidation state of the plastoquinone (PQ) pool [44]. PQ appears to interfere with KaiA-stimulated KaiC phosphorylation in at least two ways. Thus, in its oxidized but not in the reduced form, it causes KaiA aggregation by preferably binding to the N-terminal receiver domain, as was demonstrated with dibromothymoquinone, a water soluble PQ analog [44]. Using gel-based and crystallographic assays with the KaiA-dibromothymoquinone complex, we established that the PQ analog interferes with KaiA binding to the KaiC C-terminus, thereby hampering KaiC phosphorylation [45]. Both cues, ATP/ADP ratio and the pool of oxidized PQ, affect KaiC phosphorylation and operate in tandem [44]. The ATP/ADP ratio slowly declines during the night and thus serves as a measure for the duration of darkness, eventually inducing a phase shift. Conversely, diminishing light leads to the rapid oxidation of the PQ pool and signals impending darkness.
Coupling of PTO and TTFL to orchestrate global circadian regulation
The clock-controlled output pathway that governs transcriptional oscillation of clock-regulated genes involves the histine kinase SasA and the transcription factors RpaA and RpaB that may constitute a classic two-component system, whereby SasA senses the phosphorylation state of KaiC, autophosphorylates and subsequently effects phosphorylation of the transcription factors that bind to the kaiBC promoter [46–48]. As in the case of KaiB discussed above, two different models of the interaction between KaiC and SasA have been proposed, one which proposes SasA to bind to the Cll dome of KaiC [32] and the other to the CI domain [49,50]. The end result of this SasA-mediated coupling between PTO and TTFL is to globally regulate [51–53] via circadian changes in chromosomal topology [54] and/or the transcriptional factors RpaA/RpaB [47,48,55] the expression of output genes but also to replenish the central clock components KaiA, KaiB, and KaiC, thereby completing a transcription and translation loop [8].
Conclusion and implications for eukaryotic clocks
A system composed of a mass-action biochemical pacemaker embedded within a transcription/translation loop has important implications. Biochemical reactions that involve small numbers of molecules (e.g., transcription factors) are intrinsically noisy, being dominated by large concentration fluctuations. However, a PTO that is rooted in the phosphorylation status of thousands of molecules would be expected to be robust in the face of noise, as we modeled for the cyanobacterial system [8]. Not only is the circadian system resilient within the cyanobacterial cell, but it is robust among a population of cells [56]. Resilience of the daily timekeeper is particularly important for cells that must keep accurate track of time in the face of cell division, when a TTFL might become perturbed because the ratio of DNA to transcriptional factors can change during replication and when DNA can become less accessible during chromosomal condensation in preparation for division [4]. The cyanobacterial clock does an excellent job of maintaining a consistent circadian period while simultaneously controlling the timing of cell division [40,56,57].
Can the studies in cyanobacteria lead us to new insights into the eukaryotic clockwork, which is currently thought to have a TTFL as the core timing circuit? Early evidence for a TTFL as the core pacemaker in the cyanobacterial system came from numerous studies that showed the same phenomena which has been used to support a TTFL model in eukaryotes [5,8,35]. However, the data summarized here indicate that the central TTFL dogma has been replaced by a PTO/TTFL system in cyanobacteria [6–8,58]. Could self-sustained biochemical core oscillators underlie eukaryotic clocks? The introductory paragraph of this paper briefly mentioned some of the studies that are beginning a re-evaluation of the core-TTFL model for eukaryotic clocks. The advantages that accrue to the cyanobacterial system by having a post-translational mechanism at its core are also relevant to eukaryotic clocks [4,21]. For example, individual mammalian fibroblasts express cell-autonomous, self-sustained circadian oscillations of gene expression that are largely unperturbed by cell division [59,60] in a fashion reminiscent of cyanobacteria [4,56,57]. Our perspective (admittedly a biased viewpoint) is that the results with cyanobacteria have important implications for eukaryotic clock systems in that they can explain how a TTFL could appear to be a core circadian clockwork when in fact the true pacemaker is an embedded biochemical oscillator [4,8,21].
Highlights.
-
–
A biochemical circadian oscillator can be reconstituted with three proteins and ATP
-
–
The biochemical oscillator drives a transcription/translation feedback loop in vivo
-
–
Activities of the core protein include ATPase, kinase and phospho-transferase
-
–
Structural insights explain unidirectionality, synchrony, robustness, and entrainment
Acknowledgements
We are grateful to the National Institutes of Health for supporting clock research in our labs (R01 GM067152 and R01 GM088595 to CHJ and R01 GM073845 to ME).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been annotated and highlighted as:
• of special interest
•• of outstanding interest
- 1.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 2.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 3.Dibner C, Sage D, Unser M, Bauer C, d'Eysmond T, Naef F, Schibler U. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson CH. Circadian clocks and cell division: what’s the pacemaker? Cell Cycle. 2010;9:3864–3873. doi: 10.4161/cc.9.19.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 6. Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. •• Although this paper was not published within the past 2–3 years, it is already a classic demonstration (an in vitro circadian oscillator composed of only a few proteins) that has precipitated a major re-evaluation of everything that we think we know about circadian mechanisms in all organisms, including mammals.
- 7.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. •• This study experimentally and theoretically investigates the dynamics of a circadian clock in which a PTO is coupled to a TTFL; the results have important implications for all clock systems (including eukaryotic clocks) in that they can explain how a TTFL can appear to be a core circadian clockwork when in fact the true pacemaker is an embedded biochemical oscillator.
- 9.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. The ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rust MJ, Markson JS, Lane WS, Fisher DS, O'Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell. 2004;15:375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattanayek R, Mori T, Xu Y, Pattanayek S, Johnson CH, Egli M. Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS ONE. 2009;4:e7529. doi: 10.1371/journal.pone.0007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Mori T, Qin X, Yan H, Egli M, Johnson CH. Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLoS ONE. 2009;4:e7509. doi: 10.1371/journal.pone.0007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechocoscus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA. 2004;101:10925–10930. doi: 10.1073/pnas.0403037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M. Analysis of KaiA-KaiC protein interactions in the cyanobacterial circadian clock using hybrid structural methods. EMBO J. 2006;25:2017–2038. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CH, Egli M, Stewart PL. Structural insights into a circadian oscillator. Science. 2008;322:697–701. doi: 10.1126/science.1150451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murayama Y, Mukaiyama A, Imai K, Onoue Y, Tsunoda A, Nohara A, Ishida T, Maéda Y, Terauchi K, Kondo T, Akiyama S. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 2011;30:68–78. doi: 10.1038/emboj.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egli M, Mori T, Pattanayek R, Xu Y, Qin X, Johnson CH. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51:1547–1558. doi: 10.1021/bi201525n. • This paper and ref. 33 provide evidence that dephosphorylation of KaiC proceeds at least in part via a phospho-transferase mechanism that regenerates ATP.
- 24.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi F, Ito H, Fujita M, Iwase R, Uzumaki T, Ishiura M. Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem Biophys Res Comm. 2004;316:195–202. doi: 10.1016/j.bbrc.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 26.van Zon JS, Lubensky DK, Altena PRH, ten Wolde PR. An allosteric model of circadian KaiC phosphorylation. Proc Natl Acad Sci USA. 2007;104:7420–7425. doi: 10.1073/pnas.0608665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y-I, Dong G, Carruthers C, Golden SS, LiWang A. A day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105:12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J Bacteriol. 2005;187:2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egli M, Pattanayek R, Sheehan JH, Xu Y, Mori T, Smith JA, Johnson CH. Loop-loop interactions regulate KaiA-stimulated KaiC phosphorylation in the cyanobacterial KaiABC circadian clock. Biochemistry. 2013;52 doi: 10.1021/bi301691a. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattanayek R, Williams DR, Pattanayek S, Mori T, Johnson CH, Stewart PL, Egli M. Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J. 2008;27:1767–1778. doi: 10.1038/emboj.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akiyama S, Nohara A, Ito K, Maéda Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell. 2008;29:703–716. doi: 10.1016/j.molcel.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 32. Pattanayek R, Williams DR, Rossi G, Weigand S, Mori T, Johnson CH, Stewart PL, Egli M. Combined SAXS/EM based models of the S. elongatus posttranslational oscillator and its interactions with the output His-kinase SasA. PLoS ONE. 2011;6:e23697. doi: 10.1371/journal.pone.0023697. • Uses a hybrid structural biology approach including X-ray crystallography, electron microscopy and small angle X-ray scattering to analyze the three-dimensional structures of the binary KaiAC and KaiBC complexes and the ternary KaiABC complex.
- 33.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287:18030–18035. doi: 10.1074/jbc.M112.350660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. • This study shows that metabolism can be linked to the central oscillator to accomplish entrainment.
- 35.Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomita J, Nakajima M, Kondo T, Iwasaki H. Circadian rhythm of KaiC phosphorylation without transcription-translation feedback. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 37.Qin X, Byrne M, Mori T, Zou P, Williams DR, Mchaourab H, Johnson CH. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA. 2010;107:14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang Y-G, Tseng R, Kuo N-W, LiWang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci USA. 2012;109:16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phong C, Markson JS, Wilhoite CM, Rust MJ. Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc Natl Acad Sci USA. 2013;110:1124–1129. doi: 10.1073/pnas.1212113110. • Demonstrates that KaiC consists of two timers, an N-terminal ATPase domain that ticks input-independently and a C-terminal kinase domain that integrates light-effected cues (cellular ATP/ADP ratio and oxidized plastoquinone) in a phase-dependent manner.
- 40. Dong G, Yang Q, Wang Q, Kim YI, Wood TL, Osteryoung KW, van Oudenaarden A, Golden SS. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140:529–539. doi: 10.1016/j.cell.2009.12.042. • Provides first molecular mechanism for the linkage between the central clock proteins and circadian gating of cell division.
- 41. Mori T, Williams DR, Byrn M, Qin X, Egli M, Mchaourab H, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. • First visualization (by EM) of clock proteins “doing their thing” as they rhythmically assemble and disassemble the circadian nanomachine complex.
- 42.Ito H, Kageyama H, Mutsuda M, Nakajima M, Oyama T, Kondo T. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat Struct Mol Biol. 2007;11:1084–1088. doi: 10.1038/nsmb1312. [DOI] [PubMed] [Google Scholar]
- 43.Pattanayek R, Yadagirib KK, Ohi MD, Egli M. Nature of KaiB-KaiC Binding in the Cyanobacterial Circadian Oscillator. Cell Cycle. 2013;12 doi: 10.4161/cc.23757. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y-I, Vinyard DJ, Ananyev G, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial oscillator. Proc Natl Acad Sci USA. 2012;109:17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pattanayek R, Sidiqi SK, Egli SK. Crystal structure of the redox-active cofactor DBMIB bound to circadian clock protein KaiA and structural basis for DBMIB’s ability to prevent stimulation of KaiC phosphorylation by KaiA. Biochemistry. 2012;51:8050–8052. doi: 10.1021/bi301222t. • Provides evidence that binding of oxidized plastoquinone to KaiA dimer causes aggregation of KaiA and weakly inhibits binding of the KaiC C-terminal to KaiA, thus preventing the latter from stimulating the KaiC auto-kinase activity.
- 46.Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 47.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanaoka M, Takai N, Hosokawa N, Fujiwara M, Akimoto Y, Kobori N, Iwasaki H, Kondo T, Tanaka K. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J Biol Chem. 2012;287:26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang YG, Kuo NW, Tseng R, LiWang A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2011;108:14431–14436. doi: 10.1073/pnas.1104221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valencia JS, Bitou K, Ishii K, Murakami R, Morishita M, Onai K, Furukawa Y, Imada K, Namba K, Ishiura M. Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells. 2012;17:398–419. doi: 10.1111/j.1365-2443.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 52.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woelfle MA, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci USA. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 57.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwicker D, Lubensky DK, ten Wolde PR. Robust circadian clocks from coupled protein modification and transcription–translation cycles. Proc Natl Acad Sci USA. 2010;107:22540–22545. doi: 10.1073/pnas.1007613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 60.Yeom M, Pendergast JS, Ohmiya Y, Yamazaki S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc Natl Acad Sci USA. 2010;107:9665–9670. doi: 10.1073/pnas.0914078107. [DOI] [PMC free article] [PubMed] [Google Scholar]



